Abstract
Cell death is a prevalent, well-controlled and fundamental aspect of development, particularly in the nervous system. In Drosophila, specific neural stem cells are eliminated by apoptosis during embryogenesis. In the absence of apoptosis, these stem cells continue to divide, resulting in a dramatically hyperplastic central nervous system and adult lethality. Although core cell death pathways have been well described, the spatial, temporal and cell identity cues that activate the cell death machinery in specific cells are largely unknown. We identified a cis-regulatory region that controls the transcription of the cell death activators reaper, grim and sickle exclusively in neural stem cells. Using a reporter generated from this regulatory region, we found that Notch activity is required for neural stem cell death. Notch regulates the expression of the abdominalA homeobox protein, which provides important spatial cues for death. Importantly, we show that pro-apoptotic Notch signaling is activated by the Delta ligand expressed on the neighboring progeny of the stem cell. Thus we identify a previously undescribed role for progeny in regulating the proper developmental death of their parental stem cells.
Programmed cell death is critical for the normal development of multi-cellular organisms, removing unneeded cells and controlling cell number.1 When cell death is inhibited, tissues can become enlarged and disorganized, resulting in decreased organismal viability.2 The developing nervous system is particularly susceptible to altered levels of programmed cell death, which may be important for both robustness and flexibility in the nervous system development.3, 4 Understanding the pathways that regulate developmental death will provide insight not only into the developmental program but also into the pathophysiology of nervous system diseases such as neurodegeneration. In addition, regulators of developmental death may be disrupted in cancer, making them potential new targets for chemotherapeutics.
Elimination of stem cell populations is particularly important for normal development and homeostasis, as inappropriately surviving stem cells can continue to produce progeny and become tumorigenic.5, 6 Drosophila neural stem cells, or neuroblasts (NBs), of the central nervous system (CNS) provide a well-characterized and genetically accessible model to dissect the developmental regulation of stem cell death. In embryogenesis, NBs divide asymmetrically giving rise to the neurons and glia of the CNS.7, 8 At the end of embryogenesis, the majority of NBs in the ventral nerve cord (VNC) undergo apoptosis (Figure 1a).9, 10, 11, 12 This death is limited to NBs in the abdominal segments of the VNC. When developmental apoptosis of these stem cells is blocked, they continue to proliferate, resulting in a massively enlarged CNS and poor organismal survival.6, 13
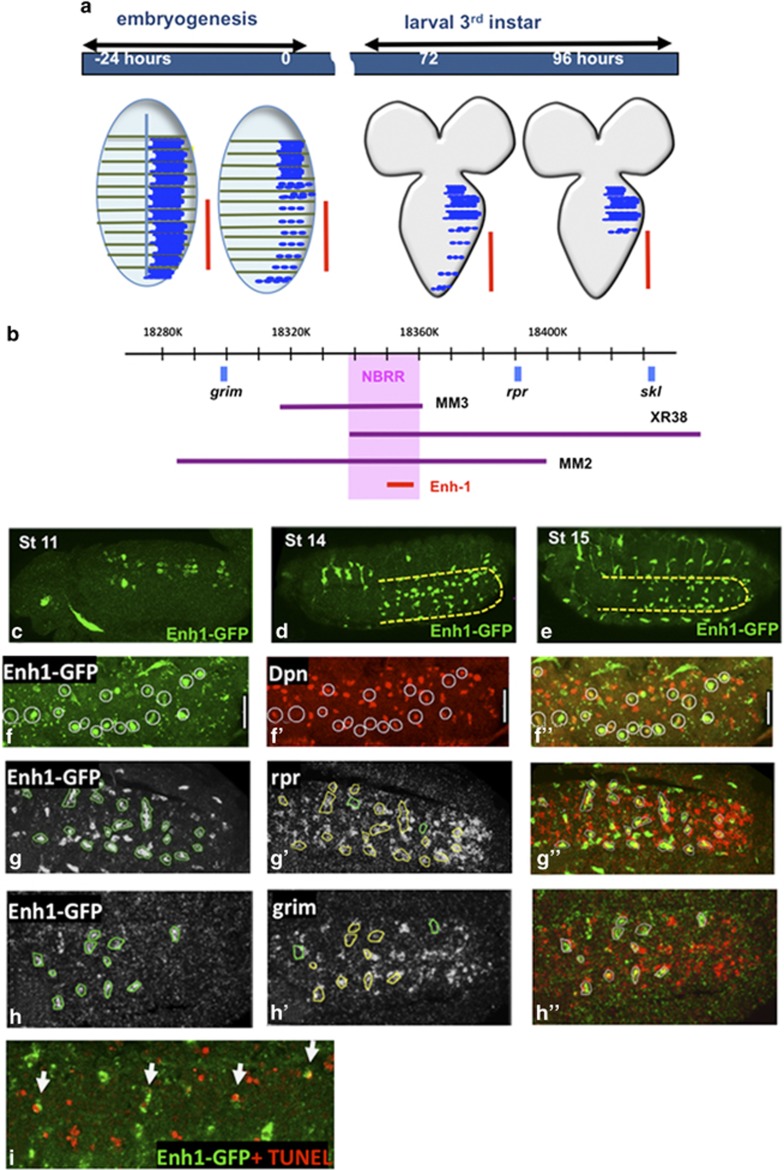
Figure 1.
Enhancer1-GFP is expressed in doomed NBs. (a) Apoptosis eliminates NBs in the ventral nerve cord at two times during development. In late embryogenesis, 27 of the 30 NBs in each hemisegment of the central abdominal region (marked with red line) undergo apoptosis. The remaining three post-embryonic NBs (pNBs) in each abdominal hemisegment die in the mid-third instar.11 (b) The genetically defined NBRR region (pink) is required for the expression of rpr, grim and skl to eliminate abdominal NBs in the embryo and the remaining pNBs in larvae.13 The Enhancer-1-GFP reporter contains 5 kb from the proximal end of the NBRR. (c–e) Enh1-GFP expression in embryos, as detected by GFP RNA in situ (green). In stage 11 embryos (c) (dorsal view), Enh1-GFP is expressed in two pairs of NBs in each abdominal hemisegment. In stage 14 (d) and stage 15 (e) embryos (ventro-lateral view), Enh1-GFP expression is restricted to a subset of abdominal NBs (yellow outline). Tracheal expression is an artifact of the in situ protocol. (f–h) Single confocal sections to show Enh1 co-labeling. (f–f′′) Enh1-GFP is restricted to NBs, as demonstrated by co-labeling with anti-Deadpan (Dpn, red) in wor>p35 embryos. In wild-type embryos, many Enh1-GFP expressing cells also express rpr (red) (g–g′′) and/or grim (red) mRNA (h–h′′). Yellow circles mark co-expressing NBs, green circles mark Enh1-GFP expressing cells that are not expressing rpr or grim. Other cells in the VNC also express rpr or grim. These are likely neurons that die during embryonic development.9, 58 For all embryos, anterior is left (i) Some Enh1-GFP expressing NBs are apoptotic at stage 16, as detected by TUNEL (red). Note that TUNEL positive neurons are present and do not express Enh1-GFP
Most developmental apoptosis in the fly is initiated by the activation of the RHG genes: reaper (rpr), head involution defective (hid), grim and sickle (skl).9 RHG proteins initiate apoptosis by binding to the caspase inhibitor DIAP1, releasing activated caspases, and by altering mitochondrial dynamics.14 RHG activity is controlled mainly at the level of transcription.15 In the doomed cells, the transcription of multiple RHG genes is initiated 1–2 h before the cells begin to show signs of cell death.16, 17, 18, 19 Importantly, the RHG genes are clustered together in the genome, and there is evidence that shared cis-regulatory regions control the transcription of more than one RHG gene.13, 20, 21
We previously identified a 20 kb genomic region, the neuroblast regulatory region (NBRR), between rpr and grim, which regulates NB death by controlling the expression of grim, rpr and skl specifically in the doomed NBs in the embryo (Figure 1b).13 Deletion of the NBRR (Df[3L]MM3) results in the suppression of grim, rpr and skl expression specifically in NBs. Here we identify a cis-regulatory element within the NBRR that drives expression specifically in the abdominal NBs before they begin to die. We show that Notch (N) signaling influences the activity of this regulatory element. N is involved in many binary cell fate decisions, and is required for stem cell viability in mammals.22, 23 Here we show that N is required for stem cell elimination. We find that N activates NB death by promoting the expression of the homeobox gene abdominalA (abdA) in the doomed NBs. abdA controls the spatial specificity of NB death, and is necessary and sufficient for the activation of the NB enhancer. These data show how N, a gene that can be both an oncogene and a tumor suppressor, regulates the death of stem cells in a context-dependent manner.
Importantly, we show that N-dependent activation of NB death requires the expression of the N ligand Delta (Dl) on the glial progeny of the NB. This highlights a non-autonomous role of the stem cell progeny in activating the death of their parental stem cells. Similar principals may underlie the regulation of developmental cell death in additional cell types and other organisms.
Results
NBRR enhancer 1 drives expression in neuroblasts fated to die
To dissect the NBRR, we generated reporters containing fragments of the genomic region. These fragments were chosen based on evolutionary conservation and transcription factor high-occupancy target (HOT) spot analysis.24 A GFP reporter (Enhancer1-GFP, Enh1-GFP) containing 5 kb from the proximal end of the NBRR is expressed specifically in the abdominal NBs starting at embryonic stage 11, peaking at stage 15, 1 or 2 h before the time of highest NB death (Figures 1c–e). Co-labeling with the NB marker Deadpan (Dpn) shows Enh1-GFP is specifically expressed in NBs (Figure 1f), and not in the neurons that also die at this stage. Enh1-GFP expression co-localizes in NBs with rpr and grim mRNA and with apoptotic markers such as TUNEL (Figures 1g–i). Taken together with our genetic data indicating that this genomic region is required for rpr, grim and skl expression and NB apoptosis, we conclude that Enh1-GFP is an accurate reporter of the regulatory sequences important for rpr, grim and skl expression in abdominal NBs, providing us with a tool to examine the upstream regulation of NB death.
Notch signaling is necessary for Enh1 activity
To identify novel regulators of embryonic NB death, we conducted an RNAi screen for transcription factors and DNA binding proteins required in the NB or its progeny for NB death (R Arya, T Sarkissian, F Rodriguez, Y Tan and K White, unpublished). Genes were knocked down in embryonic NBs and their progeny with worniu-gal4 (wor-gal4, Supplementary Figure S1),25 driving UAS-RNAi lines.26 NB survival was assayed in third instar larvae with PCNA-GFP, which marks the dividing NBs (data not shown). Knockdown lines that resulted in persistent dividing NBs in the abdominal segments of the late larval VNC were considered candidates.
One of the strongest hits from our screen was Notch (N). Notch is involved in many binary cell fate decisions and the outcome of N signaling is often context dependent. In mammalian neural stem cells, N is important to preserve viability and ‘stemness', but N can also promote differentiation and act as a tumor suppressor.22, 23 N regulates the life or death decisions of post-embryonic neurons in Drosophila.27, 28, 29 A role for N in promoting the death of NBs has not been previously described.
Knockdown of N in NBs and their progeny with eight different RNAi lines (wor>Ni) resulted in increased NB survival in the abdominal region of late third instar larvae (Figures 2a and b). Knockdown of mastermind (mam), a transcriptional co-activator in the N pathway,30 also results in ectopic NB survival (Figure 2c). Knockdown of N with elav-gal4, which is expressed in neurons and weakly in some NBs31 (Supplementary Figure S1) also resulted in increased NB survival (Figure 2d), while knockdown in glia, with repo-gal4 had no effect (Figure 2e). N signaling is seen in embryonic NBs just before the initiation of NB death.32 Taken together, these data support the conclusion that N signaling is required in the abdominal NBs for NB death in the embryo.
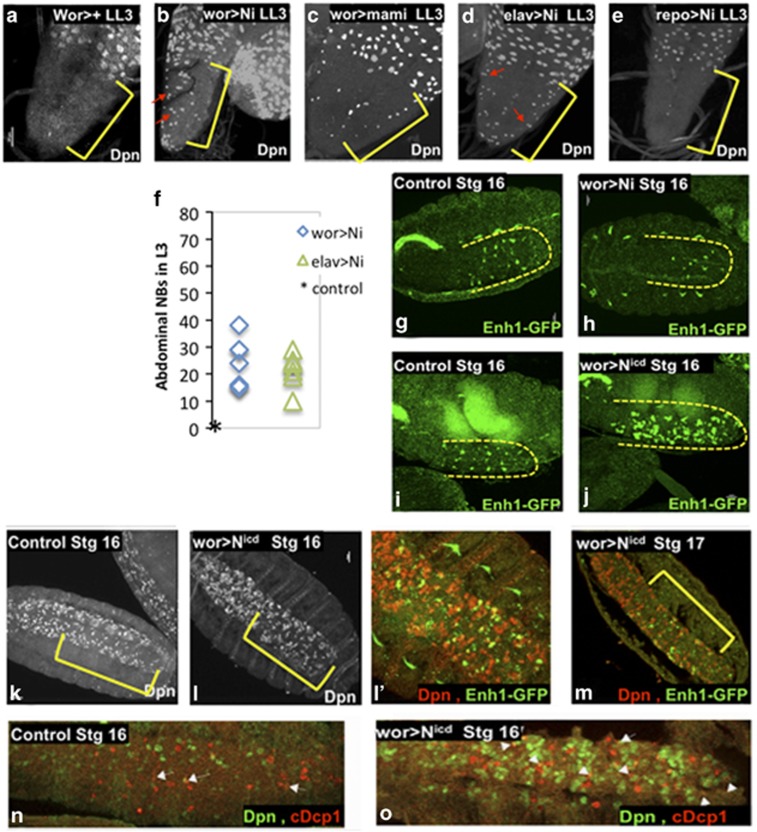
Figure 2.
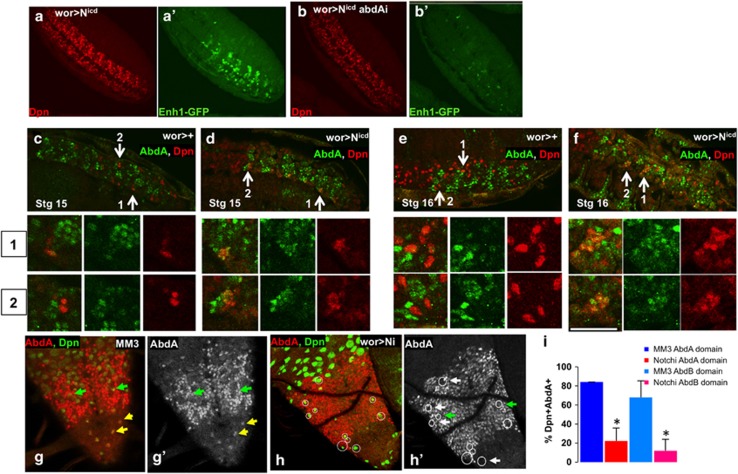
N is necessary for NB death and regulates Enh1. In control late third instar larvae there are no NBs in the abdominal VNC ((a), wor-gal4>+). Knockdown of N in NBs and progeny results in ectopic abdominal NB survival in late third instar VNCs, as detected by Dpn ((b) wor>Ni), as does knockdown of mastermind ((c) wor>mami). We can detect surviving NBs in ectopic locations (red arrows). Yellow brackets mark the central abdominal VNC, as marked by abdA expression (not shown). Knockdown of N with elav-gal4 gives a similar phenotype (d), while knockdown of N in glia has no effect ((e) repo>Ni) (f) Dpn positive NBs in the abdA domain were counted in late third instar larvae. No Dpn positive NBs are seen in controls at this stage (*). N is required for Enh1 expression, as N knockdown in NBs and their progeny results in decreased Enh1-GFP in the embryonic VNC ((g) control, (h) wor>Ni). Conversely, expression of activated N leads to a substantial increase in Enh1-GFP levels ((i) control, (j) wor>Nicd), although neither the timing of expression nor the anterior/posterior distribution of Enh1-GFP is significantly altered. Expression of activated N has additional effects on the CNS. The number of Dpn positive cells is dramatically increased (Dpn, (k) control, (l) wor>Nicd). Enh1-GFP is localized in clusters of Dpn expressing cells ((l′) Dpn-red and Enh1-GFP green). Most of the abdominal NBs are eliminated by the end of embryogenesis in wor>Nicd (m wor>Nicd Dpn-red and Enh1-GFP green, stage 17). Yellow brackets mark the central abdominal region. Increased NB death can be detected in wor>Nicd (o) with cleaved Dcp1 in NBs (cDcp1 red, Dpn green, white arrows show examples of co-localization). In control embryos, most of the NB death has already occurred at this stage although neuronal death continues (n)
Knockdown of N in NBs with wor>Ni profoundly reduced Enh1-GFP expression in the embryo (Figure 2g and h), supporting a role for N in embryonic NB death. Conversely, expression of an activated form of N (Nicd)33 resulted in very-high levels of Enh1-GFP in abdominal NBs (Figures 2i, j and l′). Neither the timing, nor the spatial distribution of Enh1-GFP expression is influenced by N mis-activation, suggesting that positional and temporal cues regulate the competence of NBs to express Enh1 in response to N activation.
Nicd expression results in increased Dpn expressing NBs cells32 (Figures 2k and l). However, the majority of these NBs are eliminated in late embryogenesis (Figures 2m). Apoptosis of abdominal NBs is increased on Nicd expression (Figures 2n and o). Thus, activation of N is sufficient to drive increased Enh1 activity and to promote the death of many NBs, but this activity is context dependent.
abdA is necessary and sufficient for Enh1-GFP expression and NB death
We hypothesized that the spatial constraint on Enh1 activation by N could be provided by the abdominalA (abdA) Hox gene. Hox genes are critical for the anterior/posterior patterning of the organism.34, 35 In the embryo, abdA has been shown to have a role in abdominal NB death; however, it was not clear whether this was due to the general effects on embryonic patterning.35 In the larvae, a pulse of abdA in the post-embryonic NBs (pNBs) initiates the death of the three pNBs that normally survive in each abdominal hemisegment.11 If abdA regulates NB death through Enh1, it should be necessary and sufficient for Enh1-GFP expression.
We knocked down abdA in embryonic NBs (wor>abdAi) to test whether abdA is required for Enh1 activity and NB death. abdA knockdown dramatically increased the number of NBs surviving into larval life in the abdominal neuromeres (Figures 3a, b and d). We examined Enh1-GFP expression in wor>abdAi embryos, and found it was dramatically reduced (Figures 3e and f). Abdominal VNC expression of both rpr and grim was also reduced in wor>abdAi embryos at stage 15/16, during the period of NB death (Figures 3g–j). These data indicate that abdA is required in the NBs or their progeny for Enh1 activity and NB death in the embryo.
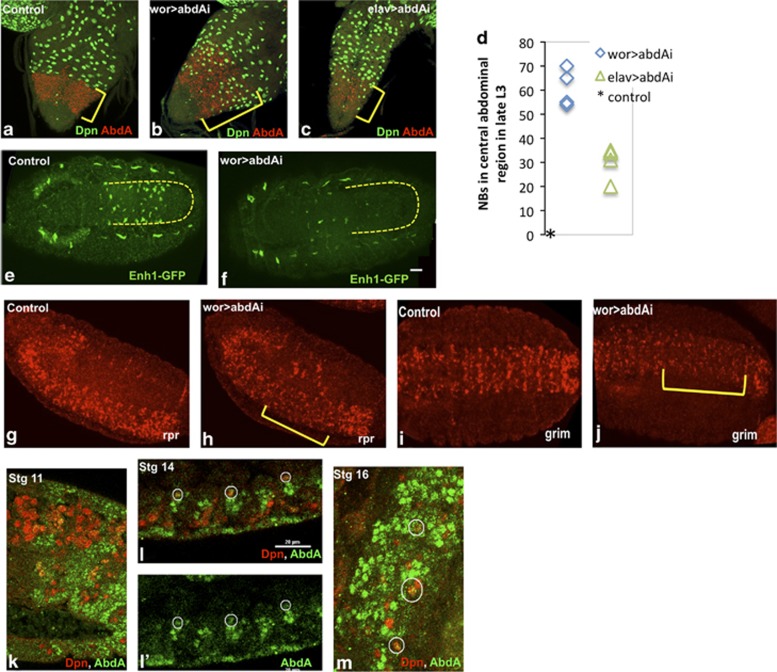
Figure 3.
abdA is required for Enh1 activity and NB death. Knockdown of abdA in NBs results in ectopic neural stem cell survival in late L3 larvae, detected with anti-Dpn staining ((a) control, (b) wor>abdAi) (c) Knockdown of abdA with elav-gal4 results in some ectopic survivors in the central abdominal region, but the phenotype is much weaker than in wor-gal4 mediated knockdown. Yellow brackets mark the central abdominal segments, as detected by abdA staining (red) (d) Dpn positive NBs in the abdA expression domain were counted in late third instar larvae. No Dpn positive NBs are seen in controls at this stage (*). abdA knockdown in NBs doubles the number of NBs that survive the embryonic phase of death. Enh1-GFP levels, as detected by in situ, are greatly reduced in the absence of abdA. wor>abdAi (f) embryos show reduced GFP expression in the VNC (marked by yellow outline) as compared with control. (e) Background staining in the tracheal system remains. In situ for rpr ((g) control, (h) wor>abdAi) and grim mRNA ((i) control, (j) wor>abdAi) show reduced levels of expression in the abdominal neuromeres when abdA is knocked down. A pulse of abdA expression detects NB fated to die. (k) In stage 11 embryos, abdA expression (green) is detected in many NBs (Dpn-red). Later in embryogenesis expression is detected in some abdominal NBs (white circles) at stage 14 (l–l′) and at stage 16 (m) Many of the NB progeny, which are adjacent to the NBs, express abdA at these stages
To distinguish whether abdA is required autonomously in NBs for NB death, we knocked down abdA with elav-gal4, which drives expression in neurons and weakly in some NBs (Supplementary Figure S1). A small number of ectopic NBs were detected in elav>abdAi larvae, but the phenotype was much weaker than that seen in wor>abdAi (Figures 3b–d). Knockdown of abdA in glia, with repo-gal4 did not result in ectopic NB survival (data not shown). These results indicate that abdA functions autonomously in embryonic NBs to activate Enh1 and initiate NB death.
Later in larval life, a pulse of abdA expression in the abdominal pNBs precedes their death.11 In embryos, expression of abdA is seen in some abdominal NBs early in neurogenesis, as well as in many other cells (Figure 3k).35 We detected weak expression of abdA transiently in some abdominal NBs later in embryogenesis (Figures 3l and m). Thus in both the embryo and the larvae, a pulse of abdA expression precedes death, and serves as a trigger to turn-on the RHG genes by activating Enh1.
Ectopic abdA expression is sufficient to cause increased and ectopic Enh1-GFP expression and NB death. We drove expression of UAS-abdA with wor-gal4 (wor>abdA). Enh1-GFP expression is increased in abdominal NBs, and is misexpressed in thoracic NBs in wor>abdA embryos (Figures 4a and b). NB death is increased in response to abdA overexpression in NBs35 (Figures 4d and e). Interestingly, although the wor-gal4 driver is active from stage 11 (Supplementary Figure S1), we do not see increased expression of Enh1-GFP before stage 13, suggesting that NBs only become competent to respond to abdA in later embryonic stages.
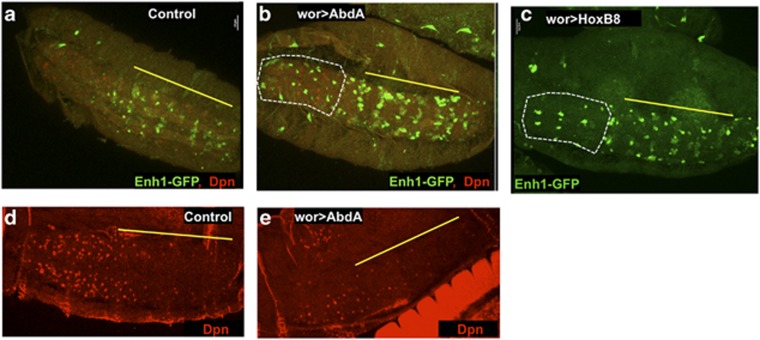
Figure 4.
abdA is sufficient to drive increased and ectopic Enh1-GFP expression and NB death. Misexpression of abdA in the embryo increases Enh1 expression in the abdominal domain (yellow line) and expands Enh1-GFP expression into the thoracic neuromeres (white outline) ((a) enh1-GFP/+, (b) wor-gal4 UAS-abdA, anti-GFP green, anti-Dpn-red). Expression of mouse HoxB8 also results in ectopic Enh1 expression in thoracic lineages (white outline) ((c) wor>HoxB8). NB death is increased on abdA mis-expression, as seen in stage 17 embryos (anti-Dpn, (e) wor>abdA versus control in (d))
Misexpression of other Hox genes has been shown to increase pNB death in larvae.11 We found that both AbdB and Ubx were sufficient to drive ectopic expression of Enh1-GFP, and to cause extensive cell death (data not shown). Interestingly, expression of the mammalian Hox genes HoxA9 and HoxB8 also result in ectopic Enh1-GFP expression (Figure 4c). Taken together these data indicate that Hox genes are broadly able to activate Enh1-GFP in specific NBs. This may reflect promiscuity of Hox binding to sites within Enh1, or another target.
Notch acts upstream of abdA to activate NB death
Both N and abdA regulate the activity of the Enh1-GFP reporter. We tested whether N acts upstream or downstream of abdA to regulate Enh1 activity and NB death. Expression of activated N with wor-gal4 resulted in increased NB number in the embryo and high levels of Enh1-GFP (Figures 5a and a′). If abdA is knocked down in Nicd expressing NBs, the number of NBs is still elevated, but Enh1-GFP levels are greatly reduced (Figures 5b and b′), similar to what is seen with abdA knockdown alone (Figures 3e and f). This shows that the effect of Nicd on Enh1 activity is dependent on abdA. abdA overexpression results in increased Enh1 expression (Figure 4b). Concurrent N knockdown does not decrease Enh1 expression (data not shown), supporting a role for N upstream of abdA in regulating Enh1 activity.
Figure 5.
Notch acts upstream of abdA to promote abdominal NB death. Expression of activated N (a wor>Nicd, Dpn-red, Enh1-GFP anti-GFP green) results in increased NB number (a) and increased Enh1-GFP activity (a′). Simultaneous knockdown of abdA (b wor>Nicd, abdAi) does not decrease NB numbers (b), but profoundly influences Enh1-GFP expression (b′). This indicates that N activation of Enh1 depends on the activity of abdA. (c–f) Activated N drives greatly increased abdA expression (green) in stage 15 (c control, d wor>Nicd ) and stage 16 (e control, f wor>Nicd) embryonic NBs (Dpn-red, arrows mark co-labeled cells). Zoomed images of two regions are shown below. Conversely, knockdown of N with wor>Ni inhibits abdA expression in rescued NBs in both the central abdA domain and the more terminal AbdB domain of larvae (H abdA red, Dpn green h′ white arrows-no abdA expression in NB, green arrows-abdA expression in NB, yellow arrow- abdA expression in terminal domain). In comparison, most NBs rescued from death in MM3 larvae show abdA expression (g abdA red, dpn green, g′ abdA). (i) Quantification of abdA co-expression in NBs (+ S.D.)
N could regulate abdA expression to activate NB death. We examined this in context of increased and decreased N activity. Expression of Nicd in NBs in the embryo had no effect on early abdA expression in the CNS, but greatly increased the number of abdA expressing NBs at stages 15 and 16 (Figures 5c–f), indicating that N activity is sufficient to induce abdA expression in NBs in late embryogenesis. In the complimentary assay, we knocked down N with wor>Ni, and examined abdA expression in ectopically surviving larval NBs (Figures 5h and h′). We compared the ectopic NBs in N knockdown with those that are rescued by the deletion of the NBRR (MM3, Figure 5i). We found a four-fold decrease in the number of abdA expressing NBs on N knockdown (Figures 5g and i). We found that NBs outside of the normal abdA domain also express abdA before they die (Figure 5g, yellow arrowheads). These NBs also did not express abdA on N knockdown (Figures 5h and h′). Therefore, N is necessary and sufficient for abdA expression in NBs during both the embryonic and larval stages of NB death, placing N upstream of abdA in regulating abdominal NB death.
Delta is required in NB progeny for Enh1 activity and NB death
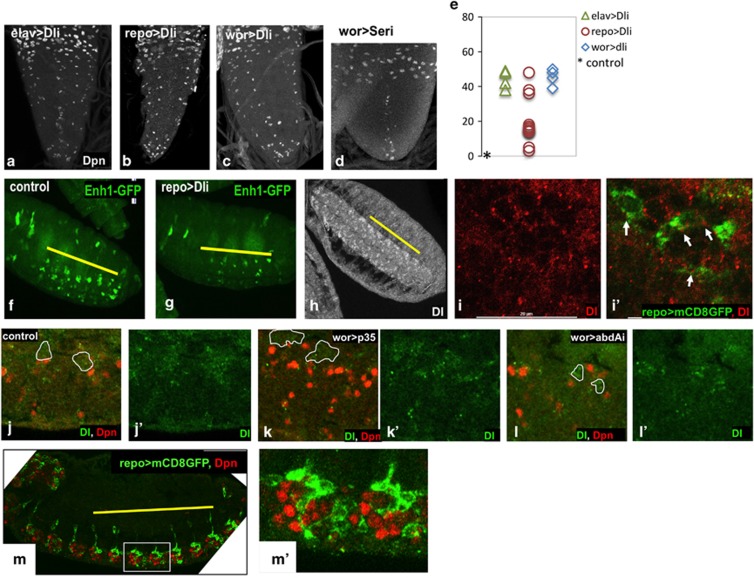
Canonical N signaling is activated by Delta (Dl) or Serrate (Ser) ligands, often presented on neighboring cells.22, 30, 32 The neighbors of the doomed NBs are their own progeny neurons and glia. To test whether N ligands on NB progeny had a role in NB death, we knocked down Dl with repo>Dl RNAi (Dli) (glial) and elav>Dli (predominantly neuronal). Strikingly, knockdown of Dl exclusively in the glial progeny resulted in ectopic NB survival (Figures 6b and e), indicating that Dl expression on glia is required for NB death. Dl knockdown with elav-gal4 also resulted in increased NB survival (Figure 6a), as did knockdown of Dl in NBs and their progeny with wor-gal4 (Figure 6c). In contrast, Ser knockdown did not promote abdominal NB survival (Figure 6d). Taken together, these data indicate that Dl is required on the NB progeny, and possibly also on the stem cell, to activate N-regulated NB death.
Figure 6.
Delta has non-autonomous effects on NB survival. Knockdown of Delta in NB progeny with elav>Dli (a) and repo>Dli (b) results in increased NB survival (Dpn). (Note that panel b is enlarged relative to a, c and d, as the repo>Dli VNCs are very small.) Knockdown of Dl with wor-gal4 (c) also result in ectopic NB survival. Serrate knockdown has no effect (d wor>Seri) e) Dpn positive NBs in the abdA domain were counted in late third instar larvae. No Dpn positive NBs are seen in controls at this stage (*) (f, g) Enh1-GFP expression in the embryo is decreased when Dl is knocked down in glia (f Enh1-GFP/+, g Enh1-GFP/repo-gal4; UAS-Dli). Abdominal neuromeres are marked by a yellow line (h) In stage 16 embryos, Dl expression is seen in the VNC, with higher expression in central abdominal segments (yellow line) (i, i′) Dl puncta (red) can be seen on glial cell membranes, as marked by repo-gal4 UAS-mCD8-GFP (green) j–l) abdA knockdown influences Dl expression in NB progeny in the embryo. In wild-type (j) and wor>p35 embryos (k), Dl expression (green) is seen in many cells neighboring the abdominal NBs (outlines of NB lineages in white, Dpn, red). In contrast, when abdA is knocked down (l, wor>abdAi) Dl expression is largely confined to the NB (j′–l′) Dl expression with brightness adjusted to highlight this difference. (m, m′) The glia cells form a more elaborate network around the NBs in the abdominal neuromeres (green repo>mCD8GFP, red Dpn)
To confirm that glial expression of Dl was required for Enh1 activation, we examined Enh1-GFP expression in repo>Dli embryos (Figures 6f and g). Knockdown of Dl exclusively in glia suppressed Enh1-GFP expression in NBs, as well as NB death. Conversely, Dl overexpression in glia did not increase NB death in thoracic segments (data not shown). Taken together with our data on N, we conclude that Dl/N signaling is permissive for Enh1 expression and death of the NBs, but not sufficient for Enh1 activity outside of the abdA domain.
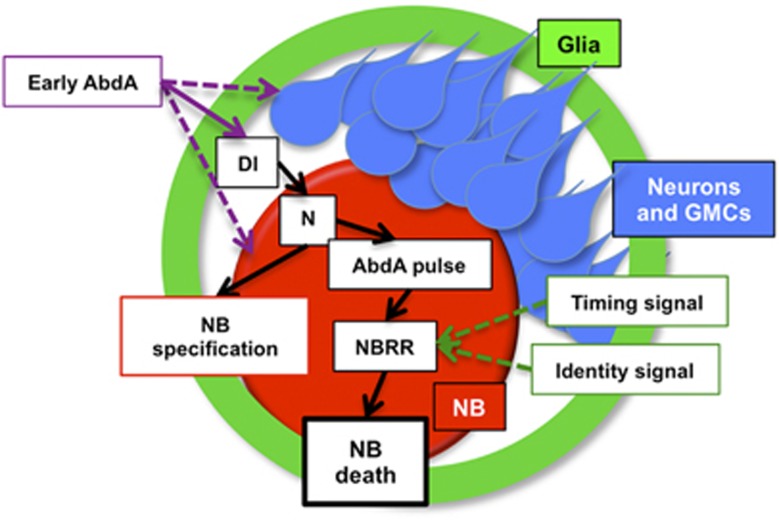
Dl expression can be detected in NBs and their progeny at the time of NB death (Figures 6h and i). The overall expression of Dl is higher in the abdominal segments (Figure 6h), and Dl puncta can clearly be detected on NBs, neurons and glia (Figures 6i and j and data not shown). To test whether abdA might regulate the Dl expression, we knocked down abdA in NBs and their progeny. We found that NB progeny express less Dl on abdA knockdown (Figures 6j and l). In early embryos, abdA specifies the abdominal NB identity, controlling the type of progeny that these NBs produce.36, 37 For example, abdominal NBs produce more glia than their thoracic counterparts, with a more complex morphology (Figure 6m). This could explain the enhanced Dl expression in the abdominal segments. We conclude that abdA acts both upstream and downstream of N signaling to regulate NB death, upstream by regulating Dl expression on NB progeny and downstream to directly or indirectly activate Enh1 (Figure 7).
Figure 7.
Model for the regulation of abdominal NB death by abdA and N. Early expression of abdA in NBs sets up the abdominal identity of the NBs. This influences the lineages they produce, and possibly their competence to re-express abdA later in embryogenesis in response to N. The abdominal lineages, including neurons and glia produce Dl, which activates N in the NB. N then influences both NB specification and activates a later pulse of abdA. abdA, working together with as yet unidentified timing and cell identity-specific signals, turns on the activity of Enh1, activating rpr, grim and skl to induce cell death
Discussion
Although cell death is an abundant and important aspect of development, particularly in the nervous system, we know relatively little about how cell death is selectively activated in the appropriate cells.3 We have approached this question by examining the developmental pathways that contribute to the activation of a cis-regulatory region required for the death of neural stem cells in Drosophila. We generated a reporter that is specifically expressed in the NBs fated to die, and used this reporter to show that both cell autonomous and non-autonomous regulators activate NB death. We identified a regulatory circuit of N and abdA that regulates NB death, contributing to sculpting the CNS.
Most cells that undergo cell death during Drosophila development turn on the expression of multiple RHG genes, and more than one RHG gene is required for most developmental cell deaths.15 Several non-exclusive models could explain how multiple RHG genes are activated to initiate the timely death of cells. Individual RHG genes could be turned on or off independently, by regulatory sequences specific for each gene.18, 38 The independent activation of multiple RHG genes could kill the cell. Alternatively, temporal and cell identity cues could be integrated at cell-type-specific regulatory regions, to control the expression of multiple RHG genes. Our identification of Enh1 as a regulator of rpr, grim and skl expression in doomed NBs supports this latter model. Similarly, transcription of both hid and rpr are activated by a common regulatory region 5′ of rpr in response to irradiation.20 A combinatorial code of transcriptional regulators could activate cell-type-specific regulatory elements to turn on RHG gene transcription, initiating context-specific cell death in response to a variety of upstream activators.
abdominalA is a critical spatial cue for NB death in the embryo
The homeobox genes of the Antennapedia and bithorax complexes specify the anterior/posterior segmental identity of the Drosophila embryo.11, 34, 37, 39, 40, 41 A role for a pulse of abdA expression to activate the death of post-embryonic NBs has been previously demonstrated,11 as has the involvement of abdA in embryonic NB fate.35 Here, we show that AbdA regulates NB death in the embryo, acting upstream of Enh1 to turn on rpr and grim transcription specifically in abdominal NBs. Based on the data presented here, we conclude that abdA may have two roles in NB fate (Figure 7). Early abdA expression in the CNS regulates NB identity, and the type of progeny produced by the NBs.35 The broad early expression of abdA does not activate Enh1, suggesting that there are temporal constraints on the ability of abdA to activate apoptosis. However, abdominal identity regulates the number of glia produced in NB lineages.36 We find that levels of Dl on the NB progeny are influenced by abdA, possibly due to the increase in number and complexity of the glia produced in this region.
Later expression of abdA in response to N activity is necessary and sufficient for Enh1 activation and NB death. However, not all lineages were competent to express Enh1-GFP on abdA overexpression. This suggests that Enh1 is a cell death regulatory region for a specific subset of NBs, and expression is constrained both temporally and by other cell identity factors. The spatial regulation of cell death by proteins involved in developmental patterning is likely to be important in other developmental contexts.
Enh1 contains several potential hox binding sites, suggesting that abdA might directly regulate Enh1 activity. However, the low level of abdA expression at the time of NB death precluded the detection of abdA binding to the Enh1 region by ChIP (L Zhou, personal communication).
Notch signaling links progeny to the elimination of the parental stem cell
The N signaling pathway is involved in many binary cell fate decisions in development, and can behave as both an oncogene and a tumor suppressor.23 N signaling can be both pro- or anti-apoptotic in neurons of the developing CNS in flies and mammals,27, 28, 42, 43, 44 and is involved in the non-autonomous regulation of cell survival by hedgehog signaling in the developing eye.29
We find that the N pathway is required for NB death in the embryo, in contrast to previous data showing that N is not involved in pNB death later in development.45 Importantly, we find that N is necessary and sufficient for Enh1 expression in abdominal NBs. Epistasis and expression data indicate that N regulates embryonic NB death by influencing a pulse of abdA expression in the abdominal NBs. Interestingly, N has been reported to be important for transient Hox gene expression during mouse somitogenesis.46, 47 The ability of N to activate the late pulse of abdA in NBs is constrained to the segments that normally express abdA earlier in development. This suggests that early abdA expression, or chromatin conformation around the abdA gene,48 may be permissive for later abdA activation by N.
Activation of canonical N signaling requires the binding of N to the Dl or Ser ligands, displayed in trans on neighboring cells or in cis on the signal receiving cell.30 We find that Dl knockdown in glia and neurons inhibits Enh1 activity and NB death. As all neurons and glia in the embryonic CNS are the progeny of the NBs, this finding implicates the NB progeny in signaling the death of their parental NBs. This could provide a timing mechanism for NBs to be eliminated only after they produced the proper array of progeny. Interestingly, there is evidence that glia are important in limiting the neural precursor pool in mammalian and zebrafish neurogenesis.49, 50
During development, neural stem cells live in intimate contact with their progeny neurons and glia. Thus, feedback from the progeny might be part of the combinatorial code that regulates NB death. A role for neurons and glia in regulating the death of their parental stem cells has important implications for how we think about the composition and role of the stem cell niche, and may influence the design and implementation of stem cell-based therapies for disease.51
Materials and methods
Fly stocks and genotypes
Flies were raised at 25 °C. The fly lines used in this study are as follows: wild-type flies were yw67c23, Df(3L)MM3,13 Df (3L)H999 and wor-Gal4,25 and UAS-dcr2 was obtained from TRiP. The following lines were obtained from the Bloomington Stock Center (Bloomington, IN, USA): repo-Gal4 (BL7415), Notch-RNAi (BL33616, BL 33611, BL31383, BL27988, BL35640, BL35213, BL31180, BL33621), Delta-RNAi (BL34322), mam-RNAi (BL28046), Ser-RNAi (BL34700), UAS-abdA (BL912), UAS-AbdB (BL913), UAS-Ubx (BL911) UAS-mcd8-GFP (BL5137), elav-Gal4 and abdA-RNAi (BL35644). abdA-RNAi was also obtained from VDRC (106155). UAS-abdAHA, UAS-UbxHA, UAS-HoxA9 and UAS-HoxB852 were provided by Y Graba. UAS-NICD 33 was provided by S Artavanis-Tsakonas and Kazuya Hori. PCNA-GFP53 was provided by IK Hariharan. Enh1- GFP construction is described below.
Fluorescent in situ hybridization (FISH)
Fluorescent in situ hybridization (FISH) or RNA-protein double labeling was carried out as previously described.13 Digoxigenin (DIG)-labeled probes for grim, rpr, and GFP were used. Biotin-labeled-GFP was used for double FISH. When expression levels are compared, in situs were processed in parallel, and imaged with matched confocal settings and image processing.
For RNA-TUNEL double labeling, the RNA in situ was carried out as described above. After probe detection, TUNEL labeling (TMR red, Roche, Indianapolis, IN, USA) was performed as follows: label solution and enzyme (9 : 1) were added to each sample and incubated overnight at 4 °C.54 After washes in PBS with 0.1% Triton X-100 the samples were mounted either in Vectorshield (Vector Labs, Burlingame, CA, USA) or Fluoromount-G (SouthernBiotech, Birmingham, AL, USA). Images were obtained using a Nikon A1SiR confocal (Melville, NY, USA).
Immunostaining
Staining of whole embryos and larval CNS was carried out as previously described.13, 55 The following primary antibodies in various combinations were used in the study: rat anti-Dpn (1 : 1.5, a gift from C. Doe, University of Oregon), goat anti-abdA (1 : 500, dH17, Santa Cruz, Santa Cruz, CA, USA), rabbit or mouse anti-GFP antibody (1 : 1000, rabbit, Invitrogen, Grand Island, NY, USA or mouse jl8, Clontech, Mountain View, CA, USA), mouse-anti-Delta (1 : 30, DSHB, Iowa City, IA, USA), rabbit anti-cleaved Dcp1 (1 : 100, Cell Signaling, Danvers, MA, USA). Secondary antibodies (Molecular Probes, Eugene, OR, USA) were used at 1 : 200 dilutions. Antibody/TUNEL co-labeling was done as previously described.54 Images were obtained on a Nikon A1 confocal (Melville, NY, USA), and were processed in Nikon Elements or Photoshop (CS5 v 12.1, Adobe Systems, San Jose, CA, USA) for brightness and contrast.
DNA constructs and transgenic flies
The Enh1-GFP reporter was generated by Gateway cloning (Invitrogen). Genomic sequences 3L:18,351,841to18,356,841 (PCR'd from CH322-130A2056) were inserted into a GFP-destination vector57 (primers: Enh1-attb1-F, GGGGACAAGTTTGTACAAAAAAGCAGGCTGTAGGCAAATAGTAATGAAAG, Enh1-attb2-R,GGGGACCACTTTGTACAAGAAAGCTGGGTGTCTGCTTTCCAAATTCC). Transgenic flies were generated by Genetic Services (Cambridge, MA, USA). The attP40 Site on the second chromosome was selected for insertion. Two independent insertions were analyzed in this study. Of note, one of the insertions was at the targeted attP40 site while another is a random insertion on the third chromosome. Expression of GFP in the two insertions is very similar.
Acknowledgments
This work was funded in part by R01GM55568 (KW) and an MGH Fund for Medical Discovery fellowship (RA). RA and KW designed the experiments, RA produced all of the data, with assistance from TS. YT conducted some of the original screen. KW directed the research and prepared the manuscript. We thank Ritu Tomar, Francisca Rodriguez and Hsiao-Yu Huang for technical assistance, Chris Doe and the Developmental Studies Hybridoma Bank for antibodies, Iswar Hariharan, Yacine Graba, Spyros Artavanis-Tsakonas and Kazuya Hori and the Bloomington Stock Center for fly strains, Liz Perkins and the TRiP (Boston, MA, USA) for RNAi lines, Ila Joshi and Katia Georgopoulos for comments on the manuscript and Lei Zhou for information about unpublished ChIP analyses.
Glossary
- NBs
neuroblasts
- pNBs
post-embryonic neuroblasts
- CNS
central nervous system
- VNC
ventral nerve cord
- rpr
reaper
- hid
head involution defective
- skl
sickle
- RHG
reaper, hid and grim
- NBRR
neuroblast regulatory region
- N
Notch
- abdA
abdominalA
- Dl
Delta
- HOT
high occupancy target
- Enh1
enhancer1
- Dpn
Deadpan
- PCNA
proliferating cell nuclear antigen
- wor
worniu
- RNAi
RNA interference
- mam
mastermind
- Nicd
Notch intracellular domain
- elav
embryonic lethal abnormal vision
- repo
reversed polarity
- Ser
Serrate
- TUNEL
terminal deoxynucleotidyl transferase nick end labeling
- Ubx
Ultrabithorax
- GMC's
ganglion mother cells
- L3
third larval instar
- LL3
late third larval instar
- cDcp1
cleaved Dcp1 caspase
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by H Steller
Supplementary Material
References
- Baehrecke EH. How death shapes life during development. Nat Rev Mol Cell Biol. 2002;3:779–787. doi: 10.1038/nrm931. [DOI] [PubMed] [Google Scholar]
- Nonomura K, Yamaguchi Y, Hamachi M, Koike M, Uchiyama Y, Nakazato K, et al. Local apoptosis modulates early mammalian brain development through the elimination of morphogen-producing cells. Dev Cell. 2013;27:621–634. doi: 10.1016/j.devcel.2013.11.015. [DOI] [PubMed] [Google Scholar]
- Dekkers MP, Nikoletopoulou V, Barde YA. Cell biology in neuroscience: death of developing neurons: new insights and implications for connectivity. J Cell Biol. 2013;203:385–393. doi: 10.1083/jcb.201306136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell DG, Paredes MF, Galvao RP, Jones DL, Froemke RC, Sebe JY, et al. Intrinsically determined cell death of developing cortical interneurons. Nature. 2012;491:109–113. doi: 10.1038/nature11523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum B, Benvenisty N. The tumorigenicity of human embryonic stem cells. Adv Cancer Res. 2008;100:133–158. doi: 10.1016/S0065-230X(08)00005-5. [DOI] [PubMed] [Google Scholar]
- Peterson C, Carney GE, Taylor BJ, White K. reaper is required for neuroblast apoptosis during Drosophila development. Development. 2002;129:1467–1476. doi: 10.1242/dev.129.6.1467. [DOI] [PubMed] [Google Scholar]
- Sousa-Nunes R, Cheng LY, Gould AP. Regulating neural proliferation in the Drosophila CNS. Curr Opin Neurobiol. 2010;20:50–57. doi: 10.1016/j.conb.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–863. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- Truman JW, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev Biol. 1988;125:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- Bello BC, Hirth F, Gould AP. A pulse of the Drosophila Hox protein abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron. 2003;37:209–219. doi: 10.1016/s0896-6273(02)01181-9. [DOI] [PubMed] [Google Scholar]
- Maurange C, Gould AP. Brainy but not too brainy: starting and stopping neuroblast divisions in Drosophila. Trends Neurosci. 2005;28:30–36. doi: 10.1016/j.tins.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Tan Y, Yamada-Mabuchi M, Arya R, St, Pierre S, Tang W, Tosa M, et al. Coordinated expression of cell death genes regulates neuroblast apoptosis. Development. 2011;138:2197–2206. doi: 10.1242/dev.058826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornbluth S, White K. Apoptosis in Drosophila: neither fish nor fowl (nor man, nor worm) J Cell Sci. 2005;118:1779–1787. doi: 10.1242/jcs.02377. [DOI] [PubMed] [Google Scholar]
- Bangs P, White K. Regulation and execution of apoptosis during Drosophila development. Dev Dyn. 2000;218:68–79. doi: 10.1002/(SICI)1097-0177(200005)218:1<68::AID-DVDY6>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- White K, Tahaoglu E, Steller H. Cell killing by the Drosophila gene reaper. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- Chen P, Nordstrom W, Gish B, Abrams JM. grim, A novel cell death gene in Drosophila. Genes Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- Kurada P, White K. Ras promotes cell survival in Drosophila by downregulating hid expression. Cell. 1998;95:319–329. doi: 10.1016/s0092-8674(00)81764-x. [DOI] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lin N, Carroll PM, Chan G, Guan B, Xiao H, et al. Epigenetic blocking of an enhancer region controls irradiation-induced proapoptotic gene expression in Drosophila embryos. Dev Cell. 2008;14:481–493. doi: 10.1016/j.devcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link N, Kurtz P, O'Neal M, Garcia-Hughes G, Abrams JM. A p53 enhancer region regulates target genes through chromatin conformations in cis and in trans. Genes Dev. 2013;27:2433–2438. doi: 10.1101/gad.225565.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierfelice T, Alberi L, Gaiano N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron. 2011;69:840–855. doi: 10.1016/j.neuron.2011.02.031. [DOI] [PubMed] [Google Scholar]
- Dotto GP. Notch tumor suppressor function. Oncogene. 2008;27:5115–5123. doi: 10.1038/onc.2008.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson R, Chabu C, Sheehan A, Doe CQ. Scribble protein domain mapping reveals a multistep localization mechanism and domains necessary for establishing cortical polarity. J Cell Sci. 2004;117:6061–6070. doi: 10.1242/jcs.01525. [DOI] [PubMed] [Google Scholar]
- Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman JW, Moats W, Altman J, Marin EC, Williams DW. Role of Notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster. Development. 2010;137:53–61. doi: 10.1242/dev.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertet C, Li X, Erclik T, Cavey M, Wells B, Desplan C. Temporal patterning of neuroblasts controls Notch-mediated cell survival through regulation of Hid or reaper. Cell. 2014;158:1173–1186. doi: 10.1016/j.cell.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen AE, Ding T, Fan Y, Graves HK, Herz HM, Lindblad JL, et al. Non-cell autonomous control of apoptosis by ligand-independent Hedgehog signaling in Drosophila. Cell Death Differ. 2013;20:302–311. doi: 10.1038/cdd.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13:654–666. doi: 10.1038/nrg3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C, Renner S, Luer K, Technau GM. The commonly used marker ELAV is transiently expressed in neuroblasts and glial cells in the Drosophila embryonic CNS. Dev Dyn. 2007;236:3562–3568. doi: 10.1002/dvdy.21372. [DOI] [PubMed] [Google Scholar]
- Zacharioudaki E, Magadi SS, Delidakis C. bHLH-O proteins are crucial for Drosophila neuroblast self-renewal and mediate Notch-induced overproliferation. Development. 2012;139:1258–1269. doi: 10.1242/dev.071779. [DOI] [PubMed] [Google Scholar]
- Go MJ, Eastman DS, Artavanis-Tsakonas S. Cell proliferation control by Notch signaling in Drosophila development. Development. 1998;125:2031–2040. doi: 10.1242/dev.125.11.2031. [DOI] [PubMed] [Google Scholar]
- Miller DF, Rogers BT, Kalkbrenner A, Hamilton B, Holtzman SL, Kaufman T. Cross-regulation of Hox genes in the Drosophila melanogaster embryo. Mech Dev. 2001;102:3–16. doi: 10.1016/s0925-4773(01)00301-x. [DOI] [PubMed] [Google Scholar]
- Prokop A, Bray S, Harrison E, Technau GM. Homeotic regulation of segment-specific differences in neuroblast numbers and proliferation in the Drosophila central nervous system. Mech Dev. 1998;74:99–110. doi: 10.1016/s0925-4773(98)00068-9. [DOI] [PubMed] [Google Scholar]
- Beckervordersandforth RM, Rickert C, Altenhein B, Technau GM. Subtypes of glial cells in the Drosophila embryonic ventral nerve cord as related to lineage and gene expression. Mech Dev. 2008;125:542–557. doi: 10.1016/j.mod.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Rogulja-Ortmann A, Technau GM. Multiple roles for Hox genes in segment-specific shaping of CNS lineages. Fly (Austin) 2008;2:316–319. doi: 10.4161/fly.7464. [DOI] [PubMed] [Google Scholar]
- Bergmann A, Agapite J, McCall K, Steller H. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell. 1998;95:331–341. doi: 10.1016/s0092-8674(00)81765-1. [DOI] [PubMed] [Google Scholar]
- Marin EC, Dry KE, Alaimo DR, Rudd KT, Cillo AR, Clenshaw ME, et al. Ultrabithorax confers spatial identity in a context-specific manner in the Drosophila postembryonic ventral nervous system. Neural Dev. 2012;7:31. doi: 10.1186/1749-8104-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suska A, Miguel-Aliaga I, Thor S. Segment-specific generation of Drosophila capability neuropeptide neurons by multi-faceted Hox cues. Dev Biol. 2011;353:72–80. doi: 10.1016/j.ydbio.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkholz O, Vef O, Rogulja-Ortmann A, Berger C, Technau GM. Abdominal-B and caudal inhibit the formation of specific neuroblasts in the Drosophila tail region. Development. 2013;140:3552–3564. doi: 10.1242/dev.096099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundell MJ, Lee HK, Perez E, Chadwell L. The regulation of apoptosis by Numb/Notch signaling in the serotonin lineage of Drosophila. Development. 2003;130:4109–4121. doi: 10.1242/dev.00593. [DOI] [PubMed] [Google Scholar]
- Yang X, Klein R, Tian X, Cheng HT, Kopan R, Shen J. Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev Biol. 2004;269:81–94. doi: 10.1016/j.ydbio.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Udolph G, Rath P, Tio M, Toh J, Fang W, Pandey R, et al. On the roles of Notch, Delta, Kuzbanian, and inscuteable during the development of Drosophila embryonic neuroblast lineages. Dev Biol. 2009;336:156–168. doi: 10.1016/j.ydbio.2009.09.030. [DOI] [PubMed] [Google Scholar]
- Almeida MS, Bray SJ. Regulation of post-embryonic neuroblasts by Drosophila grainyhead. Mech Dev. 2005;122:1282–1293. doi: 10.1016/j.mod.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Cordes R, Schuster-Gossler K, Serth K, Gossler A. Specification of vertebral identity is coupled to Notch signalling and the segmentation clock. Development. 2004;131:1221–1233. doi: 10.1242/dev.01030. [DOI] [PubMed] [Google Scholar]
- Zakany J, Kmita M, Alarcon P, de la Pompa JL, Duboule D. Localized and transient transcription of Hox genes suggests a link between patterning and the segmentation clock. Cell. 2001;106:207–217. doi: 10.1016/s0092-8674(01)00436-6. [DOI] [PubMed] [Google Scholar]
- Bowman SK, Deaton AM, Domingues H, Wang PI, Sadreyev RI, Kingston RE, et al. H3K27 modifications define segmental regulatory domains in the Drosophila bithorax complex. eLife. 2014;3:e02833. doi: 10.7554/eLife.02833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CL, Martinez-Cerdeno V, Noctor SC. Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J Neurosci. 2013;33:4216–4233. doi: 10.1523/JNEUROSCI.3441-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma EY, Rubel EW, Raible DW. Notch signaling regulates the extent of hair cell regeneration in the zebrafish lateral line. J Neurosci. 2008;28:2261–2273. doi: 10.1523/JNEUROSCI.4372-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corty MM, Freeman MR. Cell biology in neuroscience: Architects in neural circuit design: glia control neuron numbers and connectivity. J Cell Biol. 2013;203:395–405. doi: 10.1083/jcb.201306099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banreti A, Hudry B, Sass M, Saurin AJ, Graba Y. Hox proteins mediate developmental and environmental control of autophagy. Dev Cell. 2014;28:56–69. doi: 10.1016/j.devcel.2013.11.024. [DOI] [PubMed] [Google Scholar]
- Thacker SA, Bonnette PC, Duronio RJ. The contribution of E2F-regulated transcription to Drosophila PCNA gene function. Curr Biol. 2003;13:53–58. doi: 10.1016/s0960-9822(02)01400-8. [DOI] [PubMed] [Google Scholar]
- Sarkissian T, Timmons A, Arya R, Abdelwahid E, White K. Detecting apoptosis in Drosophila tissues and cells. Methods. 2014;68:89–96. doi: 10.1016/j.ymeth.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Parthasarathy N, Krause HM. Fluorescent in situ hybridization protocols in Drosophila embryos and tissues. Methods Mol Biol. 2008;420:289–302. doi: 10.1007/978-1-59745-583-1_18. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boy AL, Zhai Z, Habring-Muller A, Kussler-Schneider Y, Kaspar P, Lohmann I. Vectors for efficient and high-throughput construction of fluorescent Drosophila reporters using the PhiC31 site-specific integration system. Genesis. 2010;48:452–456. doi: 10.1002/dvg.20637. [DOI] [PubMed] [Google Scholar]
- Rogulja-Ortmann A, Luer K, Seibert J, Rickert C, Technau GM. Programmed cell death in the embryonic central nervous system of Drosophila melanogaster. Development. 2007;134:105–116. doi: 10.1242/dev.02707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.