Abstract
Ebola virus (EBOV) belongs to the Filoviridae family and is responsible for a severe disease characterized by the sudden onset of fever and malaise accompanied by other non-specific signs and symptoms; in 30–50% of cases hemorrhagic symptoms are present. Multiorgan dysfunction occurs in severe forms with a mortality up to 90%. The EBOV first attacks macrophages and dendritic immune cells. The innate immune reaction is characterized by a cytokine storm, with secretion of numerous pro-inflammatory cytokines, which induces a huge number of contradictory signals and hurts the immune cells, as well as other tissues. Other highly pathogenic viruses also trigger cytokine storms, but Filoviruses are thought to be particularly lethal because they affect a wide array of tissues. In addition to the immune system, EBOV attacks the spleen and kidneys, where it kills cells that help the body to regulate its fluid and chemical balance and that make proteins that help the blood to clot. In addition, EBOV causes liver, lungs and kidneys to shut down their functions and the blood vessels to leak fluid into surrounding tissues. In this review, we analyze the molecular mechanisms at the basis of Ebola pathogenesis with a particular focus on the cell death pathways induced by the virus. We also discuss how the treatment of the infection can benefit from the recent experience of blocking/modulating cell death in human degenerative diseases.
Facts
The knowledge about Ebola-dependent pathogenesis is limited owing to the need of work into biosafety level 4 (BSL4) laboratories and this represents a significant barrier for experimental study.
Life cycle modeling systems, including minigenome systems and transcription- and replication-competent virus-like particle (VLP) systems, allow modeling of the virus life cycle under BSL2 conditions; however, all current systems model only some aspects of the virus life cycle relying on plasmid-based viral protein expression.
Cytopathic effect have been observed in in vitro filovirus-infected cells, but the mechanisms leading to cell death in EBOV infection are far from being understood.
Electron microscopic analysis of tissues from EBOV-infected animals indicate that infected cells do not undergo apoptosis, but show vacuolization and sign of necrosis.
Open Questions
What are the mechanisms that control cell fate in EBOV-infected cells?
How different steps of EBOV life cycle interct/interfere with cell death machinery (apoptosis and autophagy)?
The modulation of cell death pathways could represent potential therapeutic strategy against EBOV infection?
The virus structure
The Ebola virus (EBOV) belongs to the family Filoviridae. Filoviruses are membrane-enveloped filamentous viruses that contain a negative sense single-stranded RNA. The virus shape is very variable with long tubes and many turns and branches. Morphologically, when studied under the electron microscope, the viral particles look like long stretched filaments with some particles tending to curve into an appearance looking like the number '6'. The long filaments are 80 nm in diameter and either 800–1000 nanometers long. RNA is only 1% of the mass of the virus.1 The large virus structure is composed of three compartments the nucleocapsid, the matrix space and the envelope.
EBOV does an incredible job being composed of only seven genes, coding for eight proteins (Figure 1). The seven genes are for the nucleoprotein (NP), the viral proteins VP24-VP30-VP35-VP40, L (polymerase) and the glycoprotein (GP).2 The surface GP is coded by the GP gene, and is expressed in two molecular forms (GP1 and GP2) that are generated by an RNA editing mechanism; it has important roles in virus infection and pathogenesis, and its expression is tightly regulated during virus replication. It has been recently demonstrated that the level of GP1 and 2 expression regulates the virus production and release.3 The NP embeds the genetic material, forming with proteins VP30 and VP35, a large complex that is involved in synthesizing virus RNAs. Separate genes code for proteins VP40 and VP24 localized in virus matrix space.4, 5
Figure 1.
Overview of EBOV gene products and their interactions with the host cell. There are seven genes in the Ebola virus: the NP, the viral proteins VP24-VP30-VP35-VP40, L (polymerase) and the GP. Figure summarizes the function of genes products within EBOV biology, together with the existing knowledge on host cell factors and functions affected by each EBOV proteins
Five species of EBOV are known all named after the region where has been identified: Bundibugyo, Reston, Sudan, Taï Forest (formerly Côte d'Ivoire ebolavirus) and Zaire.
The two Zaire (EBOV) variants causing human outbreaks in 2014 in West Africa countries (mainly Guinea Konacry, Sierra Leone and Liberia) and in the Democratic Republic of Congo have been demonstrated, using phylogenetic analysis, to be distinct from each other and from variants known from previous EVD outbreaks. The two viruses have been named 'Makona' after the Makona River close to the border between Liberia, Guinea, and Sierra Leone (Ebola virus/H.sapiens/2014/Makona) and 'Lomela' after the Lomela River in Democratic Republic of Congo (Ebola virus/H.sapiens/2014/Lomela).6
Virus entry
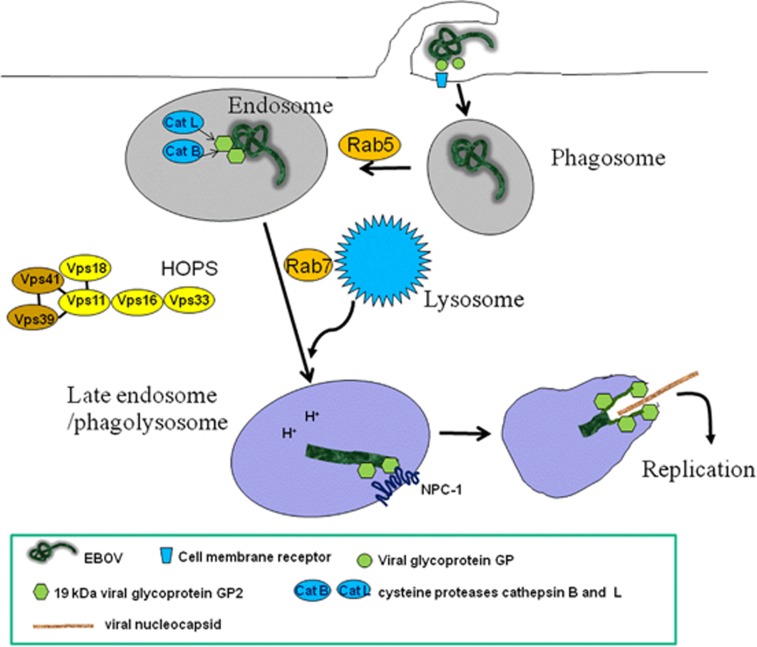
EBOV enter the human body via mucosal surfaces, abrasions and injuries in the skin or by direct parental transmission.7 EBOV then attacks many other organs; in fact, the virus is able to invade almost all human cells using different attachment mechanisms for each cell type, except for lymphocytes. It has been proposed that EBOV can enter the target cells by using different uptake mechanisms including lipid raft, receptor-mediated endocytosis and macropinocytosis (Figure 2).8, 9, 10, 11, 12 Recent reports have shown that cytoskeletal proteins dynamics, and the involvement of the class I phosphatidylinositol-3 kinase–Akt pathway are critical for EBOV uptake.13 However, the size of EBOV particles, which have a uniform diameter of 80 nm, varies dramatically in length ranging from 600–1400 nm, and peak infectivity is associated with 805-nm particles.14 Thus, the size of EBOV particles argues against host cell entry by caveolae (typical for particle sizes ranging from 50–100 nm) or 'canonical' clathrin-coated pits (typical for particles size, 200 nm).15 Another group of proteins involved in Filovirus entry are the β1-integrins,16 which are involved in the uptake of a variety of different viruses.17 Interestingly, detailed study on one of these integrins, the α5β1-integrin, has demonstrated that it is involved not in EBOV internalization, but rather in the regulation of endosomal cathepsin required for EBOV fusion.18
Figure 2.
Ebola virus entry. EBOV binds to receptors on the cell surface through the viral spike protein, GP. The virus is then internalized via macropinocytosis and trafficked to endosomal compartments, where the cysteine proteases cathepsin B (CatB) and cathepsin L (CatL) digest GP to a 19 kDa form (GP2). Within the late endosome/lysosome, the viral GP2 interacts with NPC1 allowing fusion between the viral and endosomal membranes. After fusion, the viral nucleocapsid is released into the cytoplasm, where the genome is replicated
The cholesterol-enriched lipid raft microdomains seem to be very important for EBOV entry. In fact, it has been shown that EBOV entry requires functional rafts. In keeping with this assumption, Filoviruses released from infected cells contain raft-associated molecules, suggesting that viral exit occurs at the rafts. There is a consensus to accept that the plasma membrane raft microdomains represent the gateway for the entry and exit of Filoviruses and generation of VLPs.19
The EBOV entry is mediated by the viral spike GP, which docks viral particles to the cell surface. However, additional host factors shuttling from the plasma membrane to the endosomal compartment are required because, subsequent to internalization, the virus utilizes the conventional endolysosomal pathway and is trafficked through early and late endosomes before membrane fusion takes place (Figure 2). Using a genome-wide haploid genetic screen in human cells to identify host factors required for EBOV entry, it has been shown that the membrane fusion mediated by the EBOV GPs, as well as the viral escape from the vesicular compartment require the Niemann-Pick C1 (NPC1) protein, independent of its known function in cholesterol transport. The screen also identified the six members of the homotypic fusion and vacuole protein sorting (HOPS) multisubunit tethering complex, which are involved in the fusion of endosomes to lysosomes, as essential intracellular partners of the virus. In line with these findings, cells defective for the HOPS complex or primary fibroblasts derived from human NPC1 disease patients, are resistant to Ebola infection virus..20, 21, 22
Fusion of the viral and cellular membrane is mediated by GP2,23 which results from proteolytic cleavage of GP1 by the endosomal proteases cathepsin B and cathepsin L.24 Interestingly, the cathepsin dependence of virus entry seems to be cell-type specific. Although virus entry into Vero cells is dependent on the activity of both cathepsin B and cathepsin L, infection of human dendritic cells (DCs) by EBOV does not require active cathepsin L.25 Fusion of the viral and cellular membrane leads to the release of the viral nucleocapsid into the cytoplasm of the infected cell where transcription and replication of the viral genome take place. Viral budding occurs either at intracellular membranes, the multivesicular bodies or at the plasma membrane.
Ebola-induced Major Pathogenetic Events
The complex array of pathogenetic events involved in the severe clinical manifestation of Ebola derives from a number of mechanisms. They include the direct cytopathogenic effects of the virus, that causes the destruction of infected cells, and indirect effects, that represent an amplifying mechanism leading to the destruction/impairment of several crucial body functions, as those played by the innate and adaptive immune system and by the endothelium.
Analyses of human samples obtained from succumbed patients or from experimentally infected animal models indicated that monocytes/macrophages, DCs, fibroblasts, hepatocytes, adrenal cells and epithelial cells can be productively infected by this virus. Furthermore, various studies suggested that monocytes/macrophages and DC are the early replication sites during EBOV infection.26, 27, 28 These cells also have key roles in the dissemination of the virus by migrating out of the spleen and lymph nodes to other tissues.29 Several immunological mechanisms are involved in the pathogenesis of EBOV infection involving both innate and adaptive immune response. In particular, innate immune deregulation (Figure 3) involves inhibition of type-I IFNs response, perturbation of cytokines/chemokines network, functional impairment of DC and natural killer (NK) cells. Adaptive immune deregulation involves both humoral and cell mediated immune arms (Figure 4).
Figure 3.
EBOV infection induces innate immune cell dysfunctions. (a) EBOV infection is able to impair type-I IFNs production by infected cells and to block IFN response in uninfected cells; (b) EBOV infection is able to induce massive cytokines/chemokines production by monocytes/macrophages; (c) EBOV infection is able to impair DC maturation and to deregulate cytokine production. (d) EBOV infection is able to induce massive NK apoptosis, thus avoiding NK function and impairing NK-mediated DC maturation help
Figure 4.
EBOV infection induces adaptive immune cell dysfunctions. (a) Antibodies production represents the best correlate of protection during EBOV infection. Two different forms of EBOV GP, soluble GP (sGP) and glycosylated-GP (GlycGP), are able to drive antibodies shielding and misdirection. (b) EBOV infection of DC results in a deregulated DC/T synapse, characterized by an effective MHC-peptide/TCR interaction (signal 1), in a high inflammatory microenvironment (deregulated signal 3) in the absence of co-stimulatory accessories molecules on DC surface (ineffective signal 2). The inappropriate DC/T-cell interaction induces T-cell apoptosis, avoids CD4 T-cell clonal expansion, thus blocking all CD4 T-cell helper functions such as CD8-mediated cytotoxicity and antibodies-production by B cells
Inhibition of type-I IFNs response
Type-I IFNs response is one of the early and key innate mechanisms involved in the antiviral immune response. A protective role of IFN-α was suggested during EBOV infection, as the early IFN-α production was correlated to survival in a mouse model of EBOV infection30 and in humans.31 Nevertheless, several observations in vitro and in vivo strongly suggest that EBOV is able to evade type-I IFNs response (IFN-α and IFN-β; Figure 3a).32, 33 Inhibition of type-I IFNs was initially described in EBOV-infected endothelial cells,34 and seems to have a key role in filorus pathogenesis. Moreover, EBOV infection of peripheral blood mononuclear cells (PBMC) failed to induce type I IFNs and inhibited IFN-α production induced by double-stranded RNA.35 Several viral proteins are involved in this process. The VP35 has been shown to suppress IFN-β production through multiple inhibitory effects that include the disruption of RIG-1 pathway by preventing IRF-3 phosphorylation,36 the inactivation of IRF-7,37 and the inhibition of activation of IFN-inducible dsRNA and Dicer-dependent protein kinase R.38 In addition, other studies suggest a role of VP24 in disrupting both type-I and type II IFNs signaling, by inhibiting the transcription of antiviral genes. Specifically, VP24 prevents the nuclear accumulation of dimerized phosphorylated STAT-1,39 which participates in both type I (i.e., STAT-1/STAT-2 phosphorylated-dimer) and type II (STAT-1/STAT-1phosphorylated-dimer) signal propagation cascades.40, 41 Finally, recent observations showed that residues within the transmembrane domain of GP contribute to the inhibition of tetherin activity, a type-I IFN-inducible cellular factor able to prevent enveloped virus budding from plasma membranes.42, 43 Several possible mechanisms have been proposed such as interference with tetherin integrity, steric interference between viral and cellular membranes and exclusion of tetherin from the region of plasma membrane from which EBOV bud.44
Cytokines/chemokines deregulation
In vitro studies showed that EBOV infection is able to induce a massive cytokines/chemokines production by PBMC or monocytes/macrophages (Figure 3b).35, 45 Indeed, virion attachment and entry into human macrophages profoundly affects early cellular gene expression. Several inflammatory mediators are induced within the first hour of EBOV exposure, that is, prior to virus gene expression, suggesting a direct role of the GP present on virion surface in inducing an initial inflammatory response.45 Moreover, the ability of shed GP (resulting from the cleavage of surface GP by the cellular metalloprotease TACE) in inducing inflammatory mediators release has been recently shown.46 Shed GP is able to bind and activate non-infected DC and macrophages mainly through TLR4 engagement, inducing the secretion of pro- and anti-inflammatory cytokines. This newly discovered activation mechanism of non-infected immune cells by shed GP could have an important role in the establishment of systemic inflammation during infection, provoking the excessive cytokine storm that appears to be detrimental to survival after infection.
Massive pro-inflammatory cytokines/chemokines release was confirmed during in vivo EBOV infection both in animal models47 and in humans.31, 48, 49, 50, 51 Different profiles were associated to different clinical outcome, consistent with the idea that systemic inflammation may contribute to a fatal outcome. Survivors of Ebola infection showed an early and short-lived rise in serum cytokines/chemokines, indicative of innate immune response activation, whereas fatal infection is associated to a deregulated inflammatory immune response.48 Delayed elevation in serum viral RNA, concurrent with a delayed inflammatory cytokine and chemokine response seems to be associated with survival In the macaque model.52
In two recent and relatively large studies on human infection, non-survivors develop extremely high levels of pro-inflammatory cytokines (IL-1β, IL-1RA, IL-6, IL-8, IL-15 and IL-16), chemokines (MIP-1α, MIP-1β, MCP-1, MIF, IP-10 GRO-α and eotaxin) that began rising shortly after disease onset and continued to rise until the last sampling within 2–3 days before death.49, 51 In contrast, sCD40L, that may represent ongoing repair of altered endothelium by activated platelets, was detected at high levels in survivors and has been proposed as a novel biomarker of clinical outcome.49 Interestingly, other soluble mediators have been proposed as markers of survival/fatality (IFN-α, IFN-γ, IL-12, IL-17 and TNFα) but different studies showed contrasting results, probably owing to different time of sampling.31, 48, 49, 50, 51 It is interesting to note that IL-10 may have a critical role in modulating the inflammation/regulation profile. Although IL-10 was mildly elevated in survivors, probably as a feedback mechanism to control the inflammatory response, the increase was short lived, as would be expected once cytokine levels returned to normal levels. However, IL-10 was 6- to 10-fold higher in fatal cases and remained elevated until death.31, 48, 49, 50 Thus, suggesting that EBOV-infected macrophages and DC produce inflammatory mediators and chemokines able to recruit additional macrophages and DC to areas of infection, making more target cells available for viral exploitation and further amplifying an already deregulated host response.48
As disease progresses, abnormal production of nitric oxide has been shown,53 inducing several pathological disorders including apoptosis of bystander lymphocytes, tissue damage and loss of vascular integrity, which might contribute to virus-induced shock. Subsequent extensive viral replication leads to increased levels of additional pro-inflammatory cytokines, which then triggers the coagulation cascade. Moreover, death and hemorrhage were associated with elevated thrombomodulin and ferritin levels. An increase of ferritin was also observed in other viral hemorrhagic fever infections such as Dengue54 and Crimean Congo hemorrhagic fevers.55 Other soluble mediators were found associated with hemorrhagic manifestation, such as MCSF, MIP-1α, IP-10 and sICAM. In one possible model, these cytokines could recruit leukocytes to areas of inflammation, and the production of adhesion molecules, such as ICAM, would facilitate leukocyte adhesion, rolling, and diapedesis. This would leave an activated, leukocyte-enriched, procoagulant endothelium, causing deregulated hemostasis, which could manifest clinically as hemorrhage.49 Overall, virus-induced expression of these mediators seems to result in an immunological imbalance, thus contributing to the pathogenesis and disease progression.
DC infection and functional impairment
The ability of EBOV to infect and replicate in DC has been demonstrated in vitro56, 57, 58 and in vivo.29 Interestingly, infected DC exhibited relatively little cell death over 6 days of infection.57 This sustained ability to survive while infected could offer the virus opportunities to disseminate in vivo.
Immature DC function as sentinels of the adaptive immune system. EBOV-infected DC failed to produce cytokines, including type-I IFNs, and were unable to perform a correct maturation process (Figure 1c).30, 56, 57 Specifically, EBOV infection induced 'aberrant' DC maturation, evidenced by upregulation of cell-surface CD40 and CD80, only small increase of CD86 and HLA-DR, absence of CD11c, CD83 upregulation and failure to decrease CCR5, increased expression of cytokine, chemokine, antiviral and anti-apoptotic genes, without significant changes for the expression of lymph node homing receptors or T-cell co-stimulatory molecule genes.56, 59 Aberrant expression of cytokines, chemokines and DC differentiation impairment observed during EBOV infection resulted mainly from the cooperative effect of two different viral proteins VP35 and VP24.60 As expected, the aberrant DC differentiation results in ineffective DC-/T-cell synapses that are unable to induce a correct adaptive immune response (Figure 4). Indeed, EBOV- infected DCs failed to stimulate T-cell proliferation,30, 56, 57 suggesting that EBOV suppression of DC function prevent initiation of adaptive immune responses and facilitate uncontrolled, systemic virus replication. On the other hand, the downstream effects of antigen-presenting cell dysfunction are profound with a marked lack of adaptive immunity noted in fatal cases of filovirus infection.
In the context of innate immune response, a decisive role of NK cells in inducing a protective immunity by EBOV-like particle administration was suggested in a mouse model.61 VLPs directly activated human NK cells in vitro inducing pro-inflammatory cytokine production and CD95L- or perforin-mediated cytolysis of target cells.62 Differently from what happens in wild-type mice, treatment of NK-deficient or -depleted mice with VLPs had no protective effect against EBOV infection and NK cells treated with VLPs protected against EBOV infection when adoptively transferred to naive mice.61 Nevertheless, a massive loss of NK cells was observed in vivo during Ebola infection both in mice63 and in non-human primates29, 64 (Figure 3d). It is well known that NK cells have a crucial role for their ability to mediate direct protective cytotoxicity and to drive adaptive immune response by helping DC maturation.65, 66 Thus, the massive NK cell loss in the peripheral blood may have impact on the failure of infected cells clearance but also be partially responsible for the unbalanced maturation signals for DC (Figure 3d).
Adaptive immune response impairment and lymphocytes loss
An effective immune response needs the coordinate activities of both humoral and cellular arms. In recovered patients, robust immune responses, with early and increasing levels of IgM and IgG, was developed during the acute phase of EBOV infection,67 followed by clearance of circulating viral replication markers, although fatal infections were characterized by impaired humoral responses, with absent virus-specific IgG and barely detectable IgM.48 Interestingly, humoral immune response seems to be long lasting, as survivors of EBOV infection have been recently shown to present serum-neutralizing activity and GP-specific IgG 12 years after infection.68 Several mechanisms have been developed by EBOV to escape humoral immune response (Figure 4). Recently a role of heavy glycosylation of the mucin-like domain of viral GP in shielding the cell-free virus from access to potential virus-neutralizing antibodies was described.69 Moreover, EBOV is able to produce a secreted form of GP that can modulate or misdirect host immune response.70, 71 In particular, soluble GP promote immune evasion by serving as an antibody decoy for GP or by presenting alternative non-neutralizing antibody epitopes.72
During Ebola infection, the antibody titer represents the best correlate of protection,73, 74 however several evidences suggest a key role of T cells in mediating a protective immune response.74, 75, 76 The transfer of both serum and splenocytes from Ebola VLP-vaccinated mice, but not serum or splenocytes alone, conferred protection against lethal-EBOV infection, suggesting that both B and T lymphocytes are absolutely required for VLP-mediated protection against EBOV infection.74 Studies measuring the antigen-specific T-cell response are limited by the difficulty to obtain viable PBMC samples and to perform T-cell functional assay in BSL4 facilities. In recovered patients, early and increasing levels of IgG were followed by a parallel activation of cytotoxic T cells, which was indicated by the upregulation of FasL, perforin, CD28 and IFN-γ mRNA in PBMC.77 Notably, T-cell activation was observed at the time of viral clearance, indicating that cytotoxic responses may also be implicated in the resolution of infection through Fas/FasL and perforin pathways. In contrast, in fatal cases, early expression of IFN-γ was paralleled by a massive increase of the apoptotic marker CD95, and was followed by the disappearance of T-cell related mRNA (including CD3 and CD8) in the days preceding death. Moreover, DNA fragmentation in blood leukocytes was observed, indicating that massive intravascular apoptosis occurred during the days immediately preceding death.77
The ability of EBOV-infected PBMC to induce lymphocyte apoptosis was shown in vitro35 and confirmed in vivo in mice,61 in non-human primate animal models29 and in humans.51 In human fatal cases of EBOV infection, a massive CD4 and CD8 T-lymphocyte loss was observed: CD4 and CD8 lymphocytes represented only 9.2% and 6% of PBMC in fatal cases, compared with >40 and 20% in healthy individuals and survivors. Respectively, 54.1% and 75.8% of these cells expressed CD95, values about 10 times higher than those observed in the healthy individuals, suggesting apoptotic mechanisms largely through Fas/FasL interaction.51 Interestingly, despite the apparent lack of virus-specific IgG in non-human primates and in non-surviving human patients,64, 77 in non-human primates the number of CD20+ B lymphocytes in the blood appears not to be significantly altered after the filovirus infection.
T-cell apoptosis can be the result of a deregulated DC/T synapse during EBOV infection. It is well known that T-cell activation needs a coordination of three different signals (Figure 4): (i) TCR recognition of MHC-peptide; (ii) binding of several co-stimulatory molecules between DC and T cells; and (iii) balanced ensemble of soluble factors in the microenvironment. A well-orchestrated DC/T-cell interaction of all three signals is necessary to effectively activate CD4 T cells that, in turn, exploit all their help activities, such as clonal expansion of specific T-cell clones, driving CD8 T-cell cytotoxicity, and sustaining antibody-producing B cells. During EBOV infection, we can speculate that T/DC synapsis is ineffective as it is characterized by TCR/MHC-peptide recognition (signal 1) in a high inflammatory microenvironment (deregulated signal 3) but in absence of co-stimulatory accessory molecules on DC surface (ineffective signal 2). As well known, this inappropriate interaction induces T-cell apoptosis, thus blocking all T-cell helper functions on CD8-mediated cytotoxicity and the production of antibodies by B cells (Figure 4). The final result is a marked collapse of adaptive immune response.
Notably, in a mice model, a residual T-cell function is observed in the remaining cells despite their massive loss in numbers.63 The number of functional T cells that are generated during the late phase of infection is likely too low to control high viral titers although they are sufficient upon transfer to newly infected animals to induce disease control.78 Studies in mouse model indicate that although immediate control of EBOV infection may be achieved by CD8+ T cells, B and CD4+ T cells are important for long-term control and clearance of virus replication.75 During Ebola infection, lymphoid depletion and necrosis have also been reported in spleen, thymus and lymph nodes of dying patients, as well as and in experimentally infected non-human primates. Studies carried out, after the 2000 Ebola outbreak in Uganda, showed a drastic decrease in the number of circulating T lymphocytes in succumbing people but not in survivors. Interestingly, despite the large loss of lymphocytes occurring during the infection, no signs of virus infection in lymphocytes could be detected, suggesting a bystander mechanism of apoptosis. The molecular mechanism leading to the apoptosis induction in bystander lymphocytes during the Ebola infection is not defined, however it has been proposed that it results from the activation of several different cell death modalities. These might include the death receptors pathways mediated by both TNF-related apoptosis-inducing ligand and Fas, stimulated by soluble mediators or possibly by direct interactions between lymphocytes and EBOV proteins. In addition, recent studies have shown that EBOV does not induce apoptosis in infected cells but rather leads to a non-apoptotic form of cell death, that ultrastructural analysis indicates to represent necrotic cell death.79
These findings highlight a key pathogenetic role for the TNF-α in this process, shedding some light on possible molecular mechanisms involved and indicating some potential therapeutic targets.
Cytopathogenesis of Non-immune Cells
Although the classic severe EBOV disease presentation is characterized by hemorrhagic events (petechiae, ecchymosis, mucosal hemorrhages and visceral hemorrhagic effusions), studies defining the molecular mechanisms of endothelial impairment are elusive. The major pathogenetic events for the endothelial cells seem to be determined by the EBOV GP. The GP has been suggested to have a key role in the induction of cytotoxicity and injury in endothelial cells, which is characterized by cell rounding and detachment associated by downregulating cell-adhesion molecules typical of anoikis.80
It has been shown that VLPs consisting of the EBOV matrix protein VP40 and GP (1,2) can activate endothelial cells and induce a decrease of their barrier function. In contrast, the soluble GP does not activate endothelial cells or change the endothelial barrier function. Interestingly, the VLP-induced decrease in barrier function is further enhanced by TNF-α, which is known to induce a long-lasting decrease in endothelial-cell barrier function and is hypothesized to have multiple roles in EBOV pathogenesis.81
Over the last decade, the knowledge of cell death signals involved in disease pathogenesis totally changed. Indeed, in addition to apoptosis, multiple forms of regulated necrosis have been shown to have a key role in pathologies such as sepsis, inflammatory diseases and infectious disorders.82, 83 In particular, the regulation of necroptosis in vivo is currently under the focus of many laboratories.84, 85 Considering the essential role of TNFα in modulating necroptosis/apoptosis or cell survival, it would be very interesting to determine whether during the EBOV infection the reported necrosis is because of the activation of this cell death modality.
The liver is another important target for EBOV,86, 87, 88, 89 probably having an important role in the disease pathogenesis and hepatocellular necrosis have been reported both in patients and in experimental animal models.90 Indeed, the hemorrhagic events typical of the classic Ebola infection could be related to impaired synthesis of blood coagulation protein/enzymes owing to the severe hepatocellular necrosis.91 In a recent study, 399 microRNAs were identified by deep sequencing of tissues of the Black flying fox (a confirmed natural reservoir of the human hemorrhagic fever-inducing pathogens), which has a key role in protecting these animals from developing the disease. Of the microRNAs identified, several were predicted to target genes involved in the DNA damage response, apoptosis and autophagy.92 These findings underlie the important link between the hemorrhagic fever pathogenesis and the role played by autophagy in the organism homeostasis. Autophagy aids in the removal of pathogens (a process called xenophagy) by working in conjunction with the innate immune system.93, 94, 95, 96 However, several microorganisms have evolved unique mechanisms to circumvent, suppress or exploit autophagic machinery to ensure their own survival and replication.97, 98 For instance, HSV-1 and HIV-1 block autophagy to abrogate their degradation through this pathway by blunting autophagosome formation or interrupting autophagosome–lysosome fusion, respectively.99, 100, 101, 102 By contrast, pathogens such as polio and dengue activate autophagy to enhance their own replication.103 Ebola VLP containing VP40, GP and NP protect rodents and non-human primates from lethal-EBOV infection, thus representing as good candidates for vaccine. Various findings indicate that eVLP stimulate early innate immune responses through TLRs and type-I IFNs signaling pathways to protect the host from EBOV infection. Interestingly, both TLRs and type-I IFNs mediate induction of autophagy promoting the autophagosome fusion with pathogen-containing phagosomes leading to the elimination of intracellular pathogens.104 In addition, proteins that regulate transport and fusion events between vesicles are important in autophagosome formation and maturation. Rab 7, a protein involved in transport to late endosomes and in the biogenesis of the perinuclear lysosome compartment, is required for the normal progression of autophagosomes to autophagolysosomes.105 EBOV VP24 has been shown by a mass spectrometry approach to interact with many proteins which modulate endosomal trafficking such as Rab-11B and Rab-7a.106
A very recent study identified a role for two-pore channels (TPCs) in EBOV infection.104 The blockade of the TPC function by the selective inhibitor bis-benzylisoquinoline alkaloid, tetrandrine, prevents EBOV from escaping the endosomal network into the cell cytoplasm, and consequentely the virus spreading. TPCs are localized in endosomes and lysosomes and their stimulation by nicotinic acid adenine dinucleotide phosphate (NAADP) mediates the release of cytosolic Ca++.105 Interestingly we reported that the activation of TPCs by NAADP leads to an induction of autophagy.105 In keeping with these findings, it has been shown that tetrandrine is a potent autophagy agonist. In fact, low dose of this compound induce the formation of autophagolysosomes and the accumulation of GFP-LC3 puncta.107 These findings suggest an important role of autophagy in the host response to EBOV infection. Future studies should address this hypothesis.
Conclusions and Future Perspectives
The ongoing Ebola epidemics, determined by a separate clade of EBOV, from the previously identified strains,108 has led to >10 000 deaths so far, significantly highliting the need for specific therapies. At the moment, no approved vaccine or drug is available for Ebola. Experimental vaccines and treatments for Ebola are under development, but they have not yet been fully tested for safety or effectiveness. Current experimental approaches for treatment, or post-exposure prophylaxis of EBOV diseases, are based on: antivirals directly targeting the virus (i.e., small molecules inhibiting viral polymerase, phosphor-oligonucleotides to block viral protein production, single or multiple small interfering RNAs to silence viral genes); compounds targeting host functions required for viral replication and spread (i.e., multi-ion channel inhibitor and adrenoceptor antagonist as inhibitor of filovirus cell entry; selective estrogen receptor modulators to control late viral entry); immune-modulating drugs, aiming at promoting of host defense and modulate the harmful host immune responses (i.e., compounds addressing coagulation factors and/or cytokines activity, multiple interferons and anti-opioid peptides); single or multiple mono or polyclonal antibodies for viral neutralization and killing of infected cells; passive transfers of immunity using convalescent plasma; vaccines for post-exposure treatment; finally, advanced life support is recognized as a key intervention to sustain and restore perturbed vital functions in infected patients.109, 110, 111
EBOV is able to evade innate and adaptive (both humoral and cellular) responses by encoding for multiple viral proteins that inhibit both type-I IFNs synthesis and response, by masking viral epitopes by glycosylation processes, by deregulating inflammatory response, by preventing DC maturation, thus resulting in a catastrophic failure of innate and adaptive immunity. Thus host factors have a key role for viral replication and release, and may represent good targets for therapeutic strategies. Among host factors representing potentially promising targets for anti-Ebola strategies, newly discovered mechanisms may provide a new perspective for elaborating innovative strategies. In particular, we can take some advantage from the knowledge of cell death in the Ebola pathogenesis to open up the way to new strategies toward the development of antiviral therapeutic approaches. Considering the similarities between the role played by TNF in the pathogenesis of bacterial sepsis and the EBOV infection, it might be possible to envisage the treatment of hemorrhagic fevers with anti-TNF antibodies which are known to protect from sepsis.108 TNF inhibition can be achieved with several commercially available monoclonal antibodies or with a circulating receptor fusion protein.108 The anti-TNF monoclonal antibody biologics are all currently approved by the US Food and Drug Administration for human use and there are no major side effects for short-term treatments as in the case of the EBOV infections. Studies in patients with sepsis have shown that acute statin treatment reduces the risk of developing severe sepsis (multi-organ failure) by 83%, and multi-organ failure is what kills people with EBOV infection. Moreover, acute treatment with statins and other immunomodulatory agents (e.g., ACE inhibitors, ARBs etc.) significantly improves the 30-day survival in patients hospitalized with pneumonia and sepsis.112
Another possible attempt would be to prevent cell death and in particular necroptosis. For this it would be very important to test in animal models the effects of necrostatin1 which has been shown to be a potent inhibitor of this form of programmed necrosis. In connection with this, when the EBOV-infected cells were treated with dsRNA-dependent caspase recruiter (dsCARE) virus titers were strongly reduced.79
Finally, on the basis of our hypothesis that autophagy can help the host's innate immune response to fight the EBOV infection, the treatment with autophagy inducers such as rapamycin, resveratrol and other compounds should be tested in animal models.113 Thus from these considerations it is possible to conclude that alternative strategies to combat the hemorrhagic viral infections exist, although the development of a specific vaccine is for sure the best approach to prevent these pandemic infections.114, 115
Acknowledgments
This work was supported by grants from AIRC, the Italian Ministry of University and Research (FIRB Accordi di Programma 2011) and from the Italian Ministry of Health (Ricerca Finalizzata and Ricerca Corrente).
Glossary
- EBOV
Ebola virus
- BSL
biosafety level
- NP
nucleoprotein
- GP
glycoprotein
- PI3K
phosphatidylinositol-3 kinase
- NPC1
Niemann-Pick C1 protein
- HOPS
homotypic fusion and vacuole protein sorting
- DC
dendritic cell
- IFN
interferon
- NK
natural killer
- RIG-1
retinoic acid-inducible gene 1
- IRF
interferon regulatory factor
- STAT-1
signal transducer and activator of transcription protein 1
- PBMC
peripheral blood mononuclear cells
- TLR4
toll-like receptor 4
- IL
interleukin
- MIP
macrophage inflammatory protein
- MCP-1
monocyte chemotactic protein 1
- MIF
macrophage migration inhibitory factor
- IP-10
interferon-inducible protein 10
- GRO-α
growth-related oncogene alpha
- MCSF
macrophage colony-stimulating factor
- ICAM
intercellular adhesion molecule 1
- VLP
virus-like particle
- FasL
Fas ligand
- TCR
T-cell receptor
- MHC
major histocompatibility complex
- TRAIL
TNF-related apoptosis-inducing ligand
- TNF
tumor necrosis factor
- HSV-1
Herpes simplex virus 1
- TPCs
two-pore channels
- NAADP
nicotinic acid adenine dinucleotide phosphate
- ACE
angiotensin-converting enzyme
- ARBs
angiotensin receptor blockers
- dsCARE
dsRNA-dependent caspase recruiter
The authors declare no conflict of interest.
Footnotes
Edited by RA Knight
References
- Goeijenbier M, van Kampen JJ, Reusken CB, Koopmans MP, van Gorp EC. Ebola virus disease: a review on epidemiology, symptoms, treatment and pathogenesis. Neth J Med. 2014;72:442–448. [PubMed] [Google Scholar]
- Hoenen T, Groseth A, Falzarano D, Feldmann H. Ebola virus: unravelling pathogenesis to combat a deadly disease. Trends Mol Med. 2006;12:206–215. doi: 10.1016/j.molmed.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Mohan GS, Ye L, Li W, Monteiro A, Lin X, Sapkota B, et al. Less is more: Ebola surface glycoprotein expression levels regulate virus production and infectivity. J Virol. 2014;89:1205–1217. doi: 10.1128/JVI.01810-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Noda T, Halfmann P, Jasenosky L, Kawaoka Y. Ebola virus (EBOV) VP24 inhibits transcription and replication of the EBOV genome. J Infect Dis. 2007;196:S284–S290. doi: 10.1086/520582. [DOI] [PubMed] [Google Scholar]
- Hoenen T, Biedenkopf N, Zielecki F, Jung S, Groseth A, Feldmann H, et al. Oligomerization of Ebola virus VP40 is essential for particle morphogenesis and regulation of viral transcription. J Virol. 2010;84:7053–7063. doi: 10.1128/JVI.00737-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JH, Andersen KG, Baize S, Bào Y, Bavari S, Berthet N, et al. Nomenclature- and database-compatible names for the two Ebola virus variants that emerged in Guinea and the Democratic Republic of the Congo in 2014. Viruses. 2014;6:4760–4799. doi: 10.3390/v6114760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann H, Geisbert TW. Ebola haemorrhagic fever. Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Empig CJ, Goldsmith MA. Association of the caveola vesicular system with cellular entry by filoviruses. J Virol. 2002;76:5266–5270. doi: 10.1128/JVI.76.10.5266-5270.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A. Analysis of filovirus entry into vero e6 cells, using inhibitors of endocytosis, endosomal acidification, structural integrity, and cathepsin (B and L) activity. J Infect Dis. 2007;196:S251–S258. doi: 10.1086/520597. [DOI] [PubMed] [Google Scholar]
- Nanbo A, Imai M, Watanabe S, Noda T, Takahashi K, Neumann G, et al. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010;6:e1001121. doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 2010;6:e1001110. doi: 10.1371/journal.ppat.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt CL, Kolokoltsov AA, Davey RA, Maury W. The Tyro3 receptor kinase Axl enhances macropinocytosis of Zaire ebolavirus. J Virol. 2011;85:334–347. doi: 10.1128/JVI.01278-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed MF, Kolokoltsov AA, Freiberg AN, Holbrook MR, Davey RA. Phosphoinositide-3 kinase-Akt pathway controls cellular entry of Ebola virus. PLoS Pathog. 2008;4:e1000141. doi: 10.1371/journal.ppat.1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beniac DR, Melito PL, Devarennes SL, Hiebert SL, Rabb MJ, Lamboo LL, et al. The organisation of Ebola virus reveals a capacity for extensive, modular polyploidy. PLoS One. 2012;7:e29608. doi: 10.1371/journal.pone.0029608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleksandrowicz P, Marzi A, Biedenkopf N, Beimforde N, Becker S, Hoenen T, et al. Ebola virus enters host cells by macropinocytosis and clathrin-mediated endocytosis. JID. 2011;204:S957–S967. doi: 10.1093/infdis/jir326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada A, Watanabe S, Ito H, Okazaki K, Kida H, Kawaoka Y. Downregulation of beta1 integrins by Ebola virus glycoprotein: implication for virus entry. Virology. 2000;278:20–26. doi: 10.1006/viro.2000.0601. [DOI] [PubMed] [Google Scholar]
- Siljamäki E, Rintanen N, Kirsi M, Upla P, Wang W, Karjalainen M, et al. Cholesterol dependence of collagen and echovirus 1 trafficking along the novel α2β1 integrin internalization pathway. PLoS One. 2013;8:e55465. doi: 10.1371/journal.pone.0055465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt CL, Lennemann NJ, Maury W. Filovirus entry: a novelty in the viral fusion world. Viruses. 2012;4:258–275. doi: 10.3390/v4020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavari S, Bosio CM, Wiegand E, Ruthel G, Will AB, Geisbert TW, et al. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J Exp Med. 2002;195:593–602. doi: 10.1084/jem.20011500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EH, Obernosterer G, Raaben M, Herbert AS, Deffieu MS, Krishnan A, et al. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J. 2012;31:1947–1960. doi: 10.1038/emboj.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam B, Lins L, Stroobant V, Thomas A. Brasseur R Distribution of hydrophobic, residues is crucial for the fusogenic properties of the Ebola virus GP2 fusion peptide. J Virol. 2004;78:2131–2136. doi: 10.1128/JVI.78.4.2131-2136.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecher M, Schornberg KL, Delos SE, Fusco ML, Saphire EO, White JM. Cathepsin cleavage potentiates the Ebola virus glycoprotein to undergo a subsequent fusion-relevant conformational change. J Virol. 2012;86:364–372. doi: 10.1128/JVI.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez O, Ndungo E, Tantral L, Miller EH, Leung LW, Chandran K, et al. A mutation in the Ebola virus envelope glycoprotein restricts viral entry in a host species- and cell-type-specific manner. J Virol. 2013;87:3324–3334. doi: 10.1128/JVI.01598-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M, Hatfill S, Hensley L, Huggins JW. Haematological, biochemical and coagulation changes in mice, guinea-pigs and monkeys infected with a mouse-adapted variant of Ebola Zaire virus. J Comp Pathol. 2001;125:243–253. doi: 10.1053/jcpa.2001.0503. [DOI] [PubMed] [Google Scholar]
- Bray M, Mahanty S. Ebola hemorrhagic fever and septic shock. J Infect Dis. 2003;188:1613–1617. doi: 10.1086/379727. [DOI] [PubMed] [Google Scholar]
- Geisbert TW, Young HA, Jahrling PB, Davis KJ, Kagan E, Hensley LE. Mechanisms underlying coagulation abnormalities in ebola hemorrhagic fever: overexpression of tissue factor in primate monocytes/macrophages is a key event. J Infect Dis. 2003;188:1618–1629. doi: 10.1086/379724. [DOI] [PubMed] [Google Scholar]
- Geisbert TW, Hensley LE, Larsen T, Young HA, Reed DS, Geisbert JB, et al. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am J Pathol. 2003;163:2347–2370. doi: 10.1016/S0002-9440(10)63591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanty S, Gupta M, Paragas J, Bray M, Ahmed R, Rollin PE. Protection from lethal infection is determined by innate immune responses in a mouse model of Ebola virus infection. Virology. 2003;312:415–424. doi: 10.1016/s0042-6822(03)00233-2. [DOI] [PubMed] [Google Scholar]
- Hutchinson KL, Rollin PE. Cytokine and chemokine expression in humans infected with Sudan Ebola virus. J Infect Dis. 2007;196:S357–S363. doi: 10.1086/520611. [DOI] [PubMed] [Google Scholar]
- Ansari AA. Clinical features and pathobiology of Ebolavirus infection. J Autoimmun. 2014;55C:1–9. doi: 10.1016/j.jaut.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Misasi J, Sullivan NJ. Camouflage and misdirection: the full-on assault of Ebola virus disease. Cell. 2014;159:477–486. doi: 10.1016/j.cell.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt BH, Sanchez A, Offermann MK. Ebola virus selectively inhibits responses to interferons, but not to interleukin-1beta, in endothelial cells. J Virol. 1999;73:3491–3496. doi: 10.1128/jvi.73.4.3491-3496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M, Mahanty S, Ahmed R, Rollin PE. Monocyte-derived human macrophages and peripheral blood mononuclear cells infected with ebola virus secrete MIP-1alpha and TNF-alpha and inhibit poly-IC-induced IFN-alpha in vitro. Virology. 2001;284:20–25. doi: 10.1006/viro.2001.0836. [DOI] [PubMed] [Google Scholar]
- Basler CF, Wang X, Muhlberger E, Volchkov V, Paragas J, Klenk HD, et al. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc Natl Acad Sci USA. 2000;97:12289–12294. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins KC, Cardenas WB, Basler CF. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKKepsilon and TBK-1. J Virol. 2009;83:3069–3077. doi: 10.1128/JVI.01875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann M, Gantke T, Muhlberger E. Ebola virus VP35 antagonizes PKR activity through its C-terminal interferon inhibitory domain. J Virol. 2009;83:8993–8997. doi: 10.1128/JVI.00523-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SP, Valmas C, Martinez O, Sanchez FM, Basler CF. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J Virol. 2007;81:13469–13477. doi: 10.1128/JVI.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC. Introduction: interferon signals: what is classical and what is nonclassical. J Interferon Cytokine Res. 2005;25:732. doi: 10.1089/jir.2005.25.732. [DOI] [PubMed] [Google Scholar]
- Jouvenet N, Neil SJ, Zhadina M, Zang T, Kratovac Z, Lee Y, et al. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J Virol. 2009;83:1837–1844. doi: 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ, Sandrin V, Sundquist WI, Bieniasz PD. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe. 2007;2:193–203. doi: 10.1016/j.chom.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletsky RL, Francica JR, Agrawal-Gamse C, Bates P. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc Natl Acad Sci USA. 2009;106:2886–2891. doi: 10.1073/pnas.0811014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl-Jensen V, Kurz S, Feldmann F, Buehler LK, Kindrachuk J, DeFilippis V, et al. Ebola virion attachment and entry into human macrophages profoundly effects early cellular gene expression. PLoS Negl Trop Dis. 2011;5:e1359. doi: 10.1371/journal.pntd.0001359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero-Perez B, Volchkova VA, Dolnik O, Lawrence P, Volchkov VE. Shed GP of Ebola virus triggers immune activation and increased vascular permeability. PLoS Pathog. 2014;10:e1004509. doi: 10.1371/journal.ppat.1004509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley LE, Young HA, Jahrling PB, Geisbert TW. Proinflammatory response during Ebola virus infection of primate models: possible involvement of the tumor necrosis factor receptor superfamily. Immunol Lett. 2002;80:169–179. doi: 10.1016/s0165-2478(01)00327-3. [DOI] [PubMed] [Google Scholar]
- Baize S, Leroy EM, Georges AJ, Georges-Courbot MC, Capron M, Bedjabaga I, et al. Inflammatory responses in Ebola virus-infected patients. Clin Exp Immunol. 2002;128:163–168. doi: 10.1046/j.1365-2249.2002.01800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy AK, Erickson BR, Flietstra TD, Rollin PE, Nichol ST, Towner JS, et al. Ebola hemorrhagic Fever: novel biomarker correlates of clinical outcome. J Infect Dis. 2014;210:558–566. doi: 10.1093/infdis/jiu088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villinger F, Rollin PE, Brar SS, Chikkala NF, Winter J, Sundstrom JB, et al. Markedly elevated levels of interferon (IFN)-gamma, IFN-alpha, interleukin (IL)-2, IL-10, and tumor necrosis factor-alpha associated with fatal Ebola virus infection. J Infect Dis. 1999;179:S188–S191. doi: 10.1086/514283. [DOI] [PubMed] [Google Scholar]
- Wauquier N, Becquart P, Padilla C, Baize S, Leroy EM. Human fatal zaire ebola virus infection is associated with an aberrant innate immunity and with massive lymphocyte apoptosis. PLoS Negl Trop Dis. 2010;4:e837. doi: 10.1371/journal.pntd.0000837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins K, Cooper C, Warren T, Wells J, Bell T, Raymond J, et al. Characterization of clinical and immunological parameters during Ebola virus infection of rhesus macaques. Viral Immunol. 2015;28:32–41. doi: 10.1089/vim.2014.0085. [DOI] [PubMed] [Google Scholar]
- Sanchez A, Lukwiya M, Bausch D, Mahanty S, Sanchez AJ, Wagoner KD, et al. Analysis of human peripheral blood samples from fatal and nonfatal cases of Ebola (Sudan) hemorrhagic fever: cellular responses, virus load, and nitric oxide levels. J Virol. 2004;78:10370–10377. doi: 10.1128/JVI.78.19.10370-10377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiyaratana W, Chuansumrit A, Atamasirikul K, Tangnararatchakit K. Serum ferritin levels in children with dengue infection. Southeast Asian J Trop Med Public Health. 2008;39:832–836. [PubMed] [Google Scholar]
- Barut S, Dincer F, Sahin I, Ozyurt H, Akkus M, Erkorkmaz U. Increased serum ferritin levels in patients with Crimean-Congo hemorrhagic fever: can it be a new severity criterion. Int J Infect Dis. 2010;14:e50–e54. doi: 10.1016/j.ijid.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Bosio CM, Aman MJ, Grogan C, Hogan R, Ruthel G, Negley D, et al. Ebola and Marburg viruses replicate in monocyte-derived dendritic cells without inducing the production of cytokines and full maturation. J Infect Dis. 2003;188:1630–1638. doi: 10.1086/379199. [DOI] [PubMed] [Google Scholar]
- Mahanty S, Hutchinson K, Agarwal S, McRae M, Rollin PE, Pulendran B. Cutting edge: impairment of dendritic cells and adaptive immunity by Ebola and Lassa viruses. J Immunol. 2003;170:2797–2801. doi: 10.4049/jimmunol.170.6.2797. [DOI] [PubMed] [Google Scholar]
- Martinez O, Johnson J, Manicassamy B, Rong L, Olinger GG, Hensley LE, et al. Zaire Ebola virus entry into human dendritic cells is insensitive to cathepsin L inhibition. Cell Microbiol. 2010;12:148–157. doi: 10.1111/j.1462-5822.2009.01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melanson VR, Kalina WV, Williams P. Ebola virus infection induces irregular dendritic cell gene expression. Viral Immunol. 2015;28:42–50. doi: 10.1089/vim.2014.0091. [DOI] [PubMed] [Google Scholar]
- Lubaki NM, Ilinykh P, Pietzsch C, Tigabu B, Freiberg AN, Koup RA, et al. The lack of maturation of Ebola virus-infected dendritic cells results from the cooperative effect of at least two viral domains. J Virol. 2013;87:7471–7485. doi: 10.1128/JVI.03316-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield KL, Perkins JG, Swenson DL, Deal EM, Bosio CM, Aman MJ, et al. Role of natural killer cells in innate protection against lethal Ebola virus infection. J Exp Med. 2004;200:169–179. doi: 10.1084/jem.20032141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller CL, Ruthel G, Warfield KL, Swenson DL, Bosio CM, Aman MJ, et al. NKp30-dependent cytolysis of filovirus-infected human dendritic cells. Cell Microbiol. 2007;9:962–976. doi: 10.1111/j.1462-5822.2006.00844.x. [DOI] [PubMed] [Google Scholar]
- Bradfute SB, Braun DR, Shamblin JD, Geisbert JB, Paragas J, Garrison A, et al. Lymphocyte death in a mouse model of Ebola virus infection. J InfectDis. 2007;196:S296–S304. doi: 10.1086/520602. [DOI] [PubMed] [Google Scholar]
- Reed DS, Hensley LE, Geisbert JB, Jahrling PB, Geisbert TW. Depletion of peripheral blood T lymphocytes and NK cells during the course of ebola hemorrhagic fever in cynomolgus macaques. Viral Immunol. 2004;17:390–400. doi: 10.1089/vim.2004.17.390. [DOI] [PubMed] [Google Scholar]
- Marcenaro E, Carlomagno S, Pesce S, Moretta A, Sivori S. NK/DC crosstalk in anti-viral response. Adv ExpMed Biol. 2012;946:95–308. doi: 10.1007/978-1-4614-0106-3_17. [DOI] [PubMed] [Google Scholar]
- Wehner R, Dietze K, Bachmann M, Schmitz M. The bidirectional crosstalk between human dendritic cells and natural killer cells. J Innate Immun. 2011;3:258–263. doi: 10.1159/000323923. [DOI] [PubMed] [Google Scholar]
- McElroy AK, Akondy RS, Davis CW, Ellebedy AH, Mehta AK, Kraft CS, et al. Human Ebola virus infection results in substantial immune activation. Proc Natl Acad Sci USA. 112:4719–4724. doi: 10.1073/pnas.1502619112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobarzo A, Ochayon DE, Lutwama JJ, Balinandi S, Guttman O, Marks RS, et al. Persistent immune responses after Ebola virus infection N Engl J Med 2013. 369492–493. [DOI] [PubMed] [Google Scholar]
- Cook JD, Lee JE. The secret life of viral entry glycoproteins: moonlighting in immune evasion. PLoS Pathog. 2013;9:e1003258. doi: 10.1371/journal.ppat.1003258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindzelskii AL, Yang Z, Nabel GJ, Todd RF, Petty HR. Ebola virus secretory glycoprotein (sGP) diminishes Fc gamma RIIIB-to-CR3 proximity on neutrophils. J Immunol. 2000;164:953–958. doi: 10.4049/jimmunol.164.2.953. [DOI] [PubMed] [Google Scholar]
- Yang Z, Delgado R, Xu L, Todd RF, Nabel EG, Sanchez A, et al. Distinct cellular interactions of secreted and transmembrane Ebola virus glycoproteins. Science. 1998;279:1034–1037. doi: 10.1126/science.279.5353.1034. [DOI] [PubMed] [Google Scholar]
- Mohan GS, Li W, Ye L, Compans RW, Yang C. Antigenic subversion: a novel mechanism of host immune evasion by Ebola virus. PLoS Pathog. 2012;8:e1003065. doi: 10.1371/journal.ppat.1003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan NJ, Martin JE, Graham BS, Nabel GJ. Correlates of protective immunity for Ebola vaccines: implications for regulatory approval by the animal rule. Nat Rev Microbiol. 2009;7:393–400. doi: 10.1038/nrmicro2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baize S, Leroy EM, Georges-Courbot MC, Capron M, Lansoud-Soukate J, Debre P, et al. Defective humoral responses and extensive intravascular apoptosis are associated with fatal outcome in Ebola virus-infected patients. Nat Med. 1999;5:423–426. doi: 10.1038/7422. [DOI] [PubMed] [Google Scholar]
- Warfield KL, Olinger G, Deal EM, Swenson DL, Bailey M, Negley DL, et al. Induction of humoral and CD8+ T cell responses are required for protection against lethal Ebola virus infection. J Immunol. 2005;175:1184–1191. doi: 10.4049/jimmunol.175.2.1184. [DOI] [PubMed] [Google Scholar]
- Warfield KL, Olinger GG. Protective role of cytotoxic T lymphocytes in filovirus hemorrhagic fever. J Biomed Biotechnol. 2011;2011:984241. doi: 10.1155/2011/984241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong G, Richardson JS, Pillet S, Patel A, Qiu X, Alimonti J, et al. Immune parameters correlate with protection against Ebola virus infection in rodents and nonhuman primates. Sci Transl Med. 2012;4:158ra146. doi: 10.1126/scitranslmed.3004582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradfute SB, Warfield KL, Bavari S. Functional CD8+ T cell responses in lethal Ebola virus infection. J Immunol. 2008;180:4058–4066. doi: 10.4049/jimmunol.180.6.4058. [DOI] [PubMed] [Google Scholar]
- Olejnik J, Alonso J, Schmidt KM, Yan Z, Wang W, Marzi A, et al. Ebola virus does not block apoptotic signaling pathways. J Virol. 2013;87:5384–5396. doi: 10.1128/JVI.01461-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray RB, Basu A, Steele R, Beyene A, McHowat J, Meyer K, et al. Ebola virus glycoprotein-mediated anoikis of primary human cardiac microvascular endothelial cells. Virology. 2004;321:181–188. doi: 10.1016/j.virol.2003.12.014. [DOI] [PubMed] [Google Scholar]
- Wahl-Jensen VM, Afanasieva TA, Seebach J, Ströher U, Feldmann H, Schnittler HJ. Effects of Ebola virus glycoproteins on endothelial cell activation and barrier function. J Virol. 2005;79:10442–10450. doi: 10.1128/JVI.79.16.10442-10450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect Immun. 2005;73:1907–1916. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H, Mimuro H, Ogawa M, Kobayashi T, Sanada T, Kim M, et al. Cell death and infection: a double-edged sword for host and pathogen survival. J Cell Biol. 2011;195:931–942. doi: 10.1083/jcb.201108081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Kroemer G. Necroptosis: a specialized pathway of programmed necrosis. Cell. 2008;135:1161–1163. doi: 10.1016/j.cell.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol. 2010;11:700–714. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- Fisher-Hoch SP, Brammer TL, Trappier SG, Hutwagner LC, Farrar BB, Ruo SL, et al. Pathogenic potential of filoviruses: role of geographic origin of primate host and virus strain. J Infec Dis. 1992;166:753–763. doi: 10.1093/infdis/166.4.753. [DOI] [PubMed] [Google Scholar]
- Johnson E, Jaax N, White J, Jahrling P. Lethal experimental infections of rhesus monkeys by aerosolized Ebola virus. Int J Exp Pathol. 1995;76:227–236. [PMC free article] [PubMed] [Google Scholar]
- Ryabchikova EI, Kolesnikova LV, Luchko SV. An analysis of features of pathogenesis in two animal models of Ebola virus infection. J Infect Dis. 1999;179:S199–S202. doi: 10.1086/514293. [DOI] [PubMed] [Google Scholar]
- Martines RB, Ng DL, Greer PW, Rollin PE, Zaki SR. Tissue and cellular tropism, pathology and pathogenesis of Ebola and Marburg viruses. J Pathol. 2015;235:153–174. doi: 10.1002/path.4456. [DOI] [PubMed] [Google Scholar]
- Kindrachuk J, Wahl-Jensen V, Safronetz D, Trost B, Hoenen T, Arsenault R, et al. Ebola virus modulates transforming growth factor β signaling and cellular markers of mesenchyme-like transition in hepatocytes. J Virol. 2014;88:9877–9892. doi: 10.1128/JVI.01410-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paessler S, Walker DH. Pathogenesis of the viral hemorrhagic fevers. Annu Rev Pathol. 2013;8:411–440. doi: 10.1146/annurev-pathol-020712-164041. [DOI] [PubMed] [Google Scholar]
- Cowled C, Stewart CR, Likic VA, Friedländer MR, Tachedjian M, Jenkins KA, et al. Characterisation of novel microRNAs in the Black flying fox (Pteropus alecto) by deep sequencing. BMC Genomics. 2014;15:682. doi: 10.1186/1471-2164-15-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauckman KA, Owusu-Boaitey N, Mysorekar IU. Selective autophagy: xenophagy. Methods. 2014;75:120–127. doi: 10.1016/j.ymeth.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumura K, Choi AM, Ryter SW. Emerging role of selective autophagy in human diseases. Front Pharmacol. 2014;5:244. doi: 10.3389/fphar.2014.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulus GL, Xavier RJ. Autophagy and checkpoints for intracellular pathogen defense. Curr Opin Gastroenterol. 2015;31:14–23. doi: 10.1097/MOG.0000000000000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joven J, Guirro M, Mariné-Casadó R, Rodríguez-Gallego E, Menéndez JA. Autophagy is an inflammation-related defensive mechanism against disease. Adv Exp Med Biol. 2014;824:43–59. doi: 10.1007/978-3-319-07320-0_6. [DOI] [PubMed] [Google Scholar]
- Sir D, Ou JH. Autophagy in viral replication and pathogenesis. Mol Cells. 2010;29:1–7. doi: 10.1007/s10059-010-0014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev. 2011;240:92–104. doi: 10.1111/j.1600-065X.2010.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, Alexander D, Tallóczy Z, Sun Q, Wei Y, Zhang W, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol. 2009;186:255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiramel AI, Brady NR, Bartenschlager R. Divergent roles of autophagy in virus infection. Cells. 2013;2:83–104. doi: 10.3390/cells2010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YR, Wang PS, Wang JR, Liu HS. Enterovirus 71-induced autophagy increases viral replication and pathogenesis in a suckling mouse model. J Biomed Sci. 2014;21:80. doi: 10.1186/s12929-014-0080-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudchodkar SB, Levine B. Viruses and autophagy. Rev Med Virol. 2009;19:359–378. doi: 10.1002/rmv.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- García-Dorival I, Wu W, Dowall S, Armstrong S, Touzelet O, Wastling J, et al. Elucidation of the Ebola virus VP24 cellular interactome and disruption of virus biology through targeted inhibition of host-cell protein function. J Proteome Res. 2014;13:5120–5135. doi: 10.1021/pr500556d. [DOI] [PubMed] [Google Scholar]
- et al. Ebola virus. Two-pore channels control Ebola virus host cell entry and are drug targets for disease treatment. Science. 2015;347:995–998. doi: 10.1126/science.1258758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira GJ, Hirata H, Fimia GM, do Carmo LG, Bincoletto C, Han SW, et al. Nicotinic acid adenine dinucleotide phosphate (NAADP) regulates autophagy in cultured astrocytes. J Biol Chem. 2011;286:27875–27881. doi: 10.1074/jbc.C110.216580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong K, Chen C, Zhan Y, Chen Y, Huang Z, Li W. Autophagy-related gene 7 (ATG7) and reactive oxygen species/extracellular signal-regulated kinase regulate tetrandrine-induced autophagy in human hepatocellular carcinoma. J Biol Chem. 2012;287:35576–35588. doi: 10.1074/jbc.M112.370585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baize S, Pannetier D, Oestereich L, Rieger T, Koivogui L, Magassouba N, et al. Emergence of Zaire Ebola virus disease in Guinea. N Engl J Med. 2014;371:1418–1425. doi: 10.1056/NEJMoa1404505. [DOI] [PubMed] [Google Scholar]
- Yazdanpanah Y, Arribas JR, Malvy D. Treatment of Ebola virus disease.Intensive. Care Med. 2015;41:115–117. doi: 10.1007/s00134-014-3529-8. [DOI] [PubMed] [Google Scholar]
- Ippolito G, Feldmann H, Lanini S, Vairo F, Di Caro A, Capobianchi MR, et al. Viral hemorrhagic fevers: advancing the level of treatment. BMC Med. 2012;10:31. doi: 10.1186/1741-7015-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring G, Rohrmann K, Atenchong N, Mittler E, Becker S, Dahlmann F, et al. The clinically approved drugs amiodarone, dronedarone and verapamil inhibit filovirus cell entry. J Antimicrob Chemother. 2014;69:2123–2131. doi: 10.1093/jac/dku091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P, Cui X, Barochia A, Li Y, Natanson C, Eichacker PQ. The evolving experience with therapeutic TNF inhibition in sepsis: considering the potential influence of risk of death. Expert Opin Investig Drugs. 2011;20:1555–1564. doi: 10.1517/13543784.2011.623125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Wen X, Peng J, Lu Y, Guo Z, Lu J. Systematic review and meta-analysis on the association between outpatient statins use and infectious disease-related mortality. PLoS One. 2012;7:e51548. doi: 10.1371/journal.pone.0051548. [DOI] [PMC free article] [PubMed] [Google Scholar]