In this study, Moroishi et al. show that YAP and TAZ activation induces the negative regulators LATS1 kinase and LATS2 kinase to constitute a negative feedback mechanism to control YAP/TAZ activation duration and cellular response. This study provides new insights into the dynamic regulation and maintenance of Hippo pathway homeostasis.
Keywords: Hippo pathway, YAP, TAZ, negative feedback, phosphorylation, cancer
Abstract
YAP (Yes-associated protein) and TAZ (transcriptional coactivator with PDZ-binding motif) are major downstream effectors of the Hippo pathway that influences tissue homeostasis, organ size, and cancer development. Aberrant hyperactivation of YAP/TAZ causes tissue overgrowth and tumorigenesis, whereas their inactivation impairs tissue development and regeneration. Dynamic and precise control of YAP/TAZ activity is thus important to ensure proper physiological regulation and homeostasis of the cells. Here, we show that YAP/TAZ activation results in activation of their negative regulators, LATS1/2 (large tumor suppressor 1/2) kinases, to constitute a negative feedback loop of the Hippo pathway in both cultured cells and mouse tissues. YAP/TAZ in complex with the transcription factor TEAD (TEA domain family member) directly induce LATS2 expression. Furthermore, YAP/TAZ also stimulate the kinase activity of LATS1/2 through inducing NF2 (neurofibromin 2). This feedback regulation is responsible for the transient activation of YAP upon lysophosphatidic acid (LPA) stimulation and the inhibition of YAP-induced cell migration. Thus, this LATS-mediated feedback loop provides an efficient mechanism to establish the robustness and homeostasis of YAP/TAZ regulation.
Yes-associated protein (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ; also known as WWTR1) play pivotal roles in regulating cell proliferation, survival, and differentiation; organ development; regeneration; and stem cell biology (Barry and Camargo 2013; Johnson and Halder 2014; Mo et al. 2014; Piccolo et al. 2014). YAP and TAZ share ∼50% amino acid sequence identity with a similar domain organization, each containing a TEAD (TEA domain family member)-binding domain, a WW domain, and a C-terminal transactivation domain. YAP and TAZ are transcriptional coactivators that shuttle between the cytoplasm and the nucleus, where they associate with several transcription factors—mainly TEAD transcription factors (Zhao et al. 2008). The YAP/TAZ–TEAD complex activates expression of target genes involved in cell proliferation and survival (Varelas 2014).
The activity of YAP/TAZ is largely regulated through a phosphorylation-dependent inhibition mechanism by the Hippo pathway, which was initially identified through genetic mutant screens for tumor suppressors in Drosophila melanogaster (Pan 2010). In mammals, the core components of the Hippo pathway consist of mammalian STE20-like protein kinase 1 (MST1; also known as STK4) and MST2 (also known as STK3)—the mammalian homologs of Hippo in D. melanogaster—as well as large tumor suppressor 1 (LATS1) and LATS2. When the pathway is activated, MST1/2 phosphorylate and activate the LATS1/2 kinases, which in turn directly phosphorylate YAP/TAZ on multiple serine residues, leading to cytoplasmic retention of YAP/TAZ via a 14-3-3 interaction (Oh and Irvine 2010). Furthermore, phosphorylation of YAP/TAZ by LATS1/2 primes YAP/TAZ for subsequent phosphorylation events by casein kinase 1 (CK1), resulting in recruitment of β-transducin repeat-containing proteins (β-TRCP; the F-box protein subunit of the SCFβ-TRCP ubiquitin–ligase complex) and consequent degradation by the ubiquitin–proteasome pathway (Zhao et al. 2010). Inhibition of YAP/TAZ represents the major functional output of the Hippo pathway.
Although the core components of the Hippo pathway are well characterized, the upstream regulators of this pathway are just beginning to be delineated. Previous studies revealed a general role of G-protein-coupled receptors (GPCRs) as prime regulators of Hippo signaling, where LATS1/2 kinases are acutely inhibited by the extracellular hormones, such as lysophosphatidic acid (LPA) and sphingosine-1-phosphate (S1P) (Park and Guan 2013; Wackerhage et al. 2014). In addition to hormonal regulation, several properties of tissue architecture, such as apical–basal polarity, planar cell polarity, and various types of cell–cell junctions, have been implicated in Hippo pathway regulation (Enderle and McNeill 2013; Thompson et al. 2013). Moreover, studies have revealed that YAP/TAZ are regulated by mechanical cues, such as extracellular matrix (ECM) stiffness and traction forces exerted by neighboring cells (Halder et al. 2012; Low et al. 2014).
Aberrant hyperactivation of YAP/TAZ causes tissue overgrowth and confers principal cancer features, such as epithelial–mesenchymal transition (EMT), increased migration and potential for metastasis, and cancer stem cell properties (Harvey et al. 2013; Moroishi et al. 2015). In contrast, inactivation of YAP/TAZ impairs tissue development, stem cell function, and regeneration (Barry and Camargo 2013; Mo et al. 2014). Therefore, strict and dynamic control of YAP/TAZ activity is essential for proper proliferative cellular response to ensure tissue homeostasis. However, regulatory mechanisms involved in the dynamic control of YAP/TAZ activity following signal perturbation have been poorly understood.
Cell-intrinsic negative feedback loops generally play important roles in establishing robustness to bring systems back to their initial equilibrium states. In D. melanogaster, Yorkie (the fly homolog of YAP/TAZ) was found to induce its negative regulators, including expanded, merlin, four-jointed, and kibra, to provide a possible negative feedback onto itself during developmental growth control (Cho et al. 2006; Hamaratoglu et al. 2006; Genevet et al. 2010). However, the biological significance of this potential negative feedback regulation had not been fully explored. Furthermore, although the evolutionary conservation of the Hippo pathway is extensive, most components of the Hippo pathway have multiple mammalian homologs, conferring additional complexity and divergence in the mammalian Hippo pathway (Bossuyt et al. 2014).
In this study, we reveal a feedback loop between YAP/TAZ and LATS in mammalian cells. YAP/TAZ activation leads to induction of their negative regulators, LATS1/2 kinases, to subsequently restrict YAP/TAZ activity. This feedback regulation plays an important role in terminating LPA-induced YAP activation as well as inhibiting cell migration upon YAP activation. Our results indicate that the LATS-mediated feedback architecture contributes to the robust and dynamic regulations of YAP/TAZ activity.
Results
Mutual inhibition between YAP and TAZ by a mechanism dependent on TEAD-mediated transcription
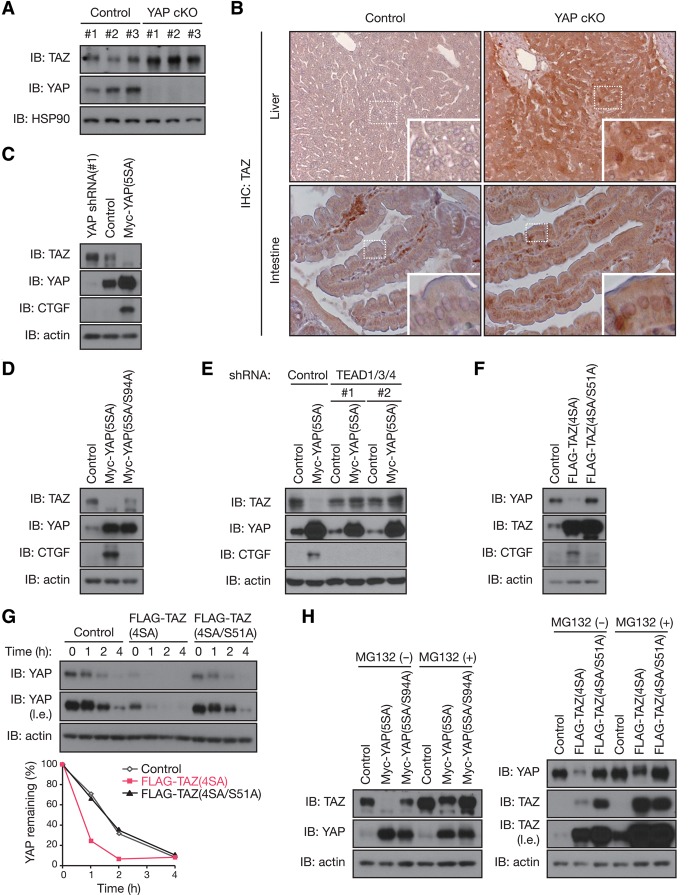
Because the Hippo pathway controls tissue homeostasis and organ size, it must be tightly regulated by extracellular signals as well as internal feedback mechanisms. Convincing studies have revealed that this pathway is regulated by cell density, extracellular signaling, and mechanotransduction. We hypothesized that there may be a mechanism that monitors the total output of YAP/TAZ activity with a feedback control for the pathway. Interestingly, we found that YAP negatively regulates its paralog, TAZ, in vivo. TAZ accumulated in the liver of YAP conditional knockout (cKO) mice (Fig. 1A). We observed significant accumulation as well as nuclear localization of TAZ in the livers and intestines of YAP cKO mice (Fig. 1B). In MCF10A mammary epithelial cells, depletion of endogenous YAP by shRNA increased the amount of endogenous TAZ, whereas overexpression of YAP(5SA) (an active mutant of YAP with all five LATS phosphorylation sites mutated to alanine, thereby unresponsive to inhibition by the LATS kinase) decreased TAZ levels (Fig. 1C). We confirmed the specificity of YAP shRNA, as re-expression of YAP prevented TAZ accumulation in YAP knockdown MCF10A cells (Supplemental Fig. S1A). These observations suggest that cells have an intrinsic mechanism to maintain total output/activity of the Hippo pathway.
Figure 1.
YAP and TAZ negatively regulate each other via TEAD-mediated transcription. (A) TAZ accumulates in the livers of YAP knockout mice. Immunoblot (IB) analysis of liver extracts from control and liver-specific YAP cKO mice with antibodies to the indicated proteins is shown. n = 3 mice per group. (B) TAZ accumulates in YAP-deficient tissues. Immunohistochemical (IHC) staining of TAZ was performed on paraffin-embedded liver and intestine tissues from control and liver- or intestine-specific YAP cKO mice. Shown at the bottom right are higher magnifications of the indicated boxed regions. (C) The abundance of TAZ is inversely regulated by YAP expression. MCF10A cells stably expressing Myc-YAP(5SA), shRNA specific for YAP, or control vector were subjected to immunoblot analysis. (D) TEAD-binding of YAP is required for YAP-induced TAZ reduction. MCF10A cells with Myc-YAP(5SA), Myc-YAP(5SA/S94A), or control vector overexpression were subjected to immunoblot analysis. (E) TEADs are required for YAP-induced TAZ reduction. MCF10A cells stably expressing Myc-YAP(5SA) (or control vector) were infected with lentiviruses encoding shRNAs specific for TEAD1/3/4. Immunoblot analysis was performed with the indicated antibodies. (F) Active TAZ decreases YAP. MCF10A cells were infected with retroviruses encoding Flag-TAZ(4SA) or Flag-TAZ(4SA/S51A) or with empty retrovirus (control). Cell lysates were immunoblotted with the indicated antibodies. (G) TAZ facilitates YAP degradation. MCF10A cells infected as in F were exposed to 100 μg/mL cycloheximide for the indicated times and then subjected to immunoblot analysis. The percentage of YAP remaining after the various incubation times was quantitated by image analysis. (l.e.) Long exposure. (H) Inhibition of proteasomal degradation blocks mutual regulation between YAP and TAZ. MCF10A cells stably expressing the indicated constructs were exposed to 10 μM proteasome inhibitor MG132 for 10 h and then subjected to immunoblot analysis. See also Supplemental Figure S1.
Next, we investigated whether YAP regulates TAZ in a manner dependent on its transcriptional activity. TEAD is the major YAP-associated transcription factor, and its binding to YAP requires the Ser94 residue in YAP (Zhao et al. 2008). Mutating Ser94 abolished the ability of YAP(5SA) to suppress TAZ (Fig. 1D). Consistently, shRNA-mediated depletion of endogenous TEAD1/3/4 (Supplemental Fig. S1B) attenuated the effect of YAP(5SA) on TAZ (Fig. 1E), suggesting that the protein abundance of TAZ is negatively regulated by YAP-induced TEAD transcriptional activity rather than YAP protein abundance. We confirmed the specificity of TEAD1/3/4 shRNA, as re-expression of TEAD1 restored YAP(5SA)-induced TAZ reduction in TEAD1/3/4 knockdown MCF10A cells (Supplemental Fig. S1C). We examined whether TAZ may reciprocally inhibit YAP. Indeed, overexpression of the active mutant TAZ(4SA), but not the TEAD-binding-defective mutant TAZ(4SA/S51A), in MCF10A cells also repressed endogenous YAP levels (Fig. 1F). Together, these results suggest that YAP and TAZ negatively regulate each other's protein abundance through a mechanism requiring TEAD-mediated transcription.
We explored the mechanism of this mutual inhibition between YAP and TAZ by measuring their mRNA levels. Surprisingly, the mRNA abundance of TAZ showed little change in YAP(5SA)-overexpressing cells (Supplemental Fig. S1D). Similarly, YAP transcription was not altered in TAZ(4SA)-overexpressing cells (Supplemental Fig. S1E). These data suggest that YAP/TAZ negatively regulate each other's protein abundance at a post-transcriptional level. To test this, we measured the stability of endogenous YAP in TAZ(4SA)-overexpressing MCF10A cells cultured in the presence of the protein synthesis inhibitor cycloheximide (Fig. 1G). The half-life of YAP was substantially decreased in cells overexpressing TAZ(4SA) compared with that in control cells. YAP destabilization appears to require TEAD-dependent transcription activity, as YAP was not destabilized in cells expressing TAZ(4SA/S51A). Furthermore, the effect of YAP overexpression on the protein abundance of TAZ and vice versa was attenuated when proteasome function was inhibited by MG132 (Fig. 1H). Thus, these observations indicate that the reciprocal negative regulation between YAP and TAZ requires TEAD-dependent transcription and proteasomal degradation.
YAP and TAZ induce LATS kinase
A prior study in D. melanogaster revealed a codependent regulatory relationship between Yorkie (the fly homolog of YAP/TAZ) and dMyc (encoded by the diminutive gene) in which Yorkie induces dMyc transcription, which in turn represses Yorkie expression (Neto-Silva et al. 2010). However, the mRNA abundance of MYC was not increased, but rather decreased, in YAP(5SA)- or TAZ(4SA)-overexpressing MCF10A cells (Supplemental Fig. S2A), suggesting that the reciprocal inhibitory relationship between YAP and TAZ is unlikely to be mediated by MYC.
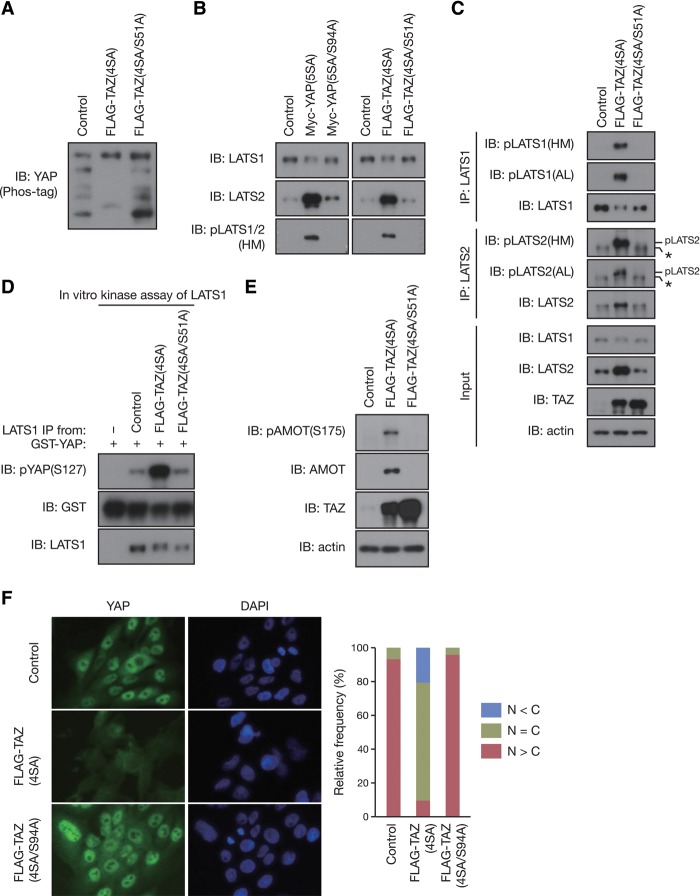
As the protein stability of YAP/TAZ is regulated through phosphorylation by LATS1/2 kinases (Zhao et al. 2010), we examined the phosphorylation status of endogenous YAP in TAZ(4SA)-overexpressing MCF10A cells. YAP phosphorylation was dramatically increased in cells overexpressing TAZ(4SA) compared with those in control or TAZ(4SA/S51A)-overexpressing cells, as indicated by a decreased mobility of YAP on phos-tag SDS-PAGE (Fig. 2A). We further investigated whether YAP/TAZ overexpression affects the protein abundance and/or activity of endogenous LATS1/2. Whereas the amount of LATS1 remained unchanged, the protein abundance of LATS2 as well as phosphorylation levels of LATS1/2 at their hydrophobic motif (HM; an indicator of LATS kinase activity) were dramatically increased by active YAP/TAZ overexpression (Fig. 2B).
Figure 2.
YAP and TAZ activate LATS1/2 kinases. (A) TAZ increases YAP phosphorylation. Cell lysates from the same experiment shown in Figure 1F were analyzed with the phos-tag electrophoresis for assessment of YAP phosphorylation status. (B) YAP/TAZ increase the protein abundance of LATS2 and phosphorylation levels of LATS1/2 at their HM. Cell lysates from the same experiment shown in Figure 1D (left) or 1F (right) were subjected to immunoblot (IB) analysis. (C) YAP/TAZ increase the activating phosphorylation of LATS1/2 at their HM and activation loop (AL). MCF10A cells were infected with retroviruses encoding Flag-TAZ(4SA) or Flag-TAZ(4SA/S51A) or with empty retrovirus (control). Cell lysates were then subjected to immunoprecipitation (IP) and immunoblot analysis. The asterisk indicates a nonspecific band. (D) TAZ activates LATS1 kinase activity. Endogenous LATS1 was immunoprecipitated from MCF10A cells infected as in C. The resulting immunoprecipitates were subjected to in vitro kinase assay. (E) Angiomotin (AMOT) is phosphorylated and accumulated by TAZ overexpression. MCF10A cells infected as in C were subjected to immunoblot analysis. (F) Active TAZ promotes YAP cytoplasmic localization. MCF10A cells infected as in C were subjected to immunostaining analysis. YAP subcellular localization was determined by immunofluorescence staining for endogenous YAP along with DAPI for DNA. Representative images are presented in the left panel. (Right panel) Cells in five random views (∼100 cells) were selected for the quantification of YAP localization. See also Supplemental Figure S2.
Because human LATS1 and LATS2 have similar apparent molecular weights, they were not resolved by the SDS-PAGE gel. Therefore the phospho-LATS antibody detected the combined phosphorylation signals of both LATS1 and LATS2. To further evaluate activating phosphorylation of LATS1/2 individually, we immunoprecipitated endogenous LATS1 or LATS2 from MCF10A cells and examined the phosphorylation levels of their activation loop (AL) and HM, both of which correlate with LATS kinase activity (Hergovich 2013). Immunoblot analysis revealed that activating phosphorylation of both LATS1 and LATS2 were markedly increased in cells overexpressing TAZ(4SA) (Fig. 2C), suggestive of increased LATS1/2 kinase activity in these cells. Indeed, in vitro kinase assay revealed that the kinase activity of endogenous LATS1 was substantially increased in TAZ(4SA)-overexpressing MCF10A cells (Fig. 2D). These results indicate that YAP and TAZ not only increase the amount of LATS2 but also stimulate the intrinsic kinase activity of both LATS1 and LATS2.
Consistent with the observed LATS1 and LATS2 activation, phosphorylation of angiomotin (AMOT) at Ser175, a known direct substrate site for LATS1/2 kinases (Adler et al. 2013; Chan et al. 2013; Dai et al. 2013; Hirate et al. 2013), was increased in TAZ(4SA)-overexpressing MCF10A cells (Fig. 2E). Moreover, the protein abundance of AMOT was also increased without a corresponding effect on the amount of AMOT mRNA (Supplemental Fig. S2B). These results are consistent with the previous observation that LATS-dependent phosphorylation stabilizes AMOT (Adler et al. 2013). We also determined endogenous YAP localization in TAZ(4SA)-expressing cells. As expected, TAZ(4SA) overexpression caused a dramatic alteration of YAP subcellular distribution in MCF10A cells (Fig. 2F). There was much more cytoplasmic YAP and less nuclear YAP in the TAZ(4SA)-expressing cells than the control or TAZ(4SA/S51A)-expressing cells. Together, our data show that TAZ(4SA) overexpression not only increases endogenous LATS1/2 kinase activity but also elevates phosphorylation of the physiological LATS1/2 substrates YAP and AMOT. Based on the above data, we conclude that YAP/TAZ–TEAD-mediated transcription increases LATS2 protein abundance as well as the kinase activity of both LATS1 and LATS2, therefore constituting a potential negative feedback loop.
YAP/TAZ and TEAD directly regulate LATS2 transcription
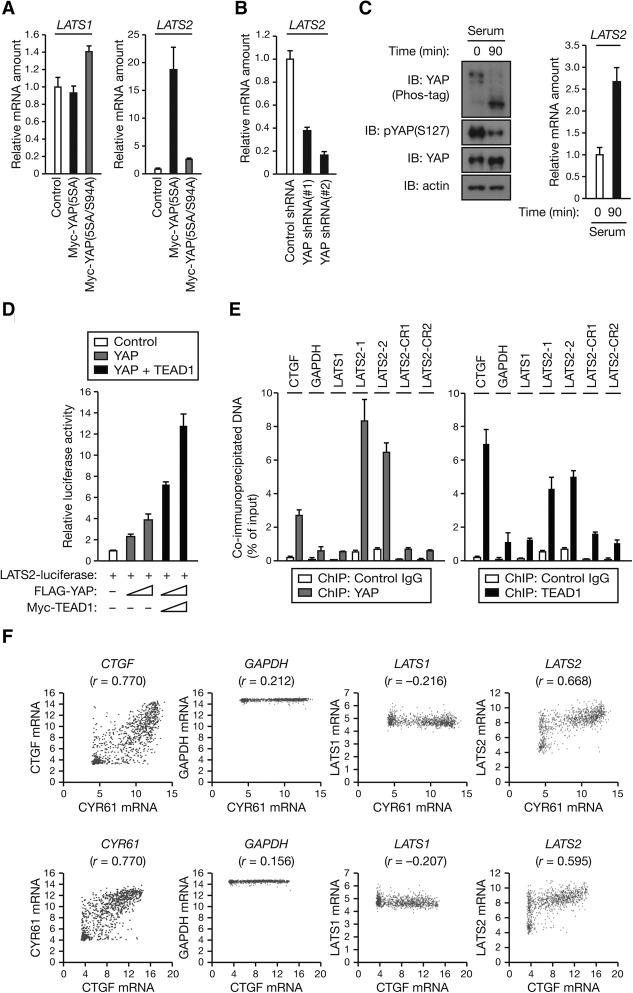
As YAP/TAZ–TEAD-mediated transcription is found to increase the amount of LATS2, we hypothesized that YAP/TAZ and TEAD might induce LATS2 transcription. Indeed, the amount of LATS2 mRNA, but not that of LATS1 mRNA, was substantially increased in MCF10A cells overexpressing active YAP or TAZ (Fig. 3A; Supplemental Fig. S3A). Conversely, shRNA-mediated depletion of endogenous YAP resulted in down-regulation of LATS2 transcript (Fig. 3B; Supplemental Fig. S3B).
Figure 3.
YAP and TAZ directly induce LATS2 expression. (A) YAP induces the transcription of LATS2. Total RNA extracted from MCF10A cells stably expressing the indicated constructs were subjected to RT and real-time PCR analysis. Data are means ± SD of triplicates from a representative experiment. (B) Loss of YAP decreases the mRNA abundance of LATS2. Total RNA extracted from MCF10A cells stably expressing shRNA specific for YAP (or control) were subjected to RT and real-time PCR analysis of LATS2 mRNA. Data are means ± SD of triplicates from a representative experiment. (C) Serum stimulation activates YAP and increases LATS2 mRNA levels. MCF10A cells were starved in serum-free medium for 8 h and then stimulated with 5% horse serum for 90 min. (Left) Cell lysates were subjected to immunoblotting (IB) with the indicated antibodies. (Top left) Where indicated, gels containing phos-tag were employed for assessment of YAP phosphorylation status. (Right) Total RNA were subjected to RT and real-time PCR analysis of LATS2 mRNA. Data are means ± SD of triplicates from a representative experiment. (D) Activation of LATS2 reporter by YAP and TEAD1. HEK293A cells were transfected with LATS2 luciferase construct and with (or without) increasing amounts (1× and 2×) of an expression vector for Flag-YAP or Myc-TEAD1. Luciferase activities were assayed 30 h after transfection. Data are means ± SD from three independent experiments. (E) Endogenous YAP and TEAD1 both bind to the LATS2 promoter. MCF10A cells starved in serum-free medium for 18 h were stimulated with 5% horse serum for 2 h. The cells were then subjected to chromatin immunoprecipitation (ChIP) with antibodies to endogenous YAP or TEAD1 (or control IgG), and the precipitated DNA was quantitated by real-time PCR analysis with primers specific for a promoter region or a control region (CR) of the indicated genes. Data are means ± SD of triplicates from a representative experiment. (F) Dot plots showing the positive correlation between LATS2 and CYR61 or CTGF mRNA expression in cell lines from 967 subjects. R-values were calculated for each correlation; P < 0.0001; Pearson's correlation coefficient. See also Supplemental Figure S3.
Both YAP and TAZ can be potently dephosphorylated and activated by serum (Yu et al. 2012). As expected, serum stimulation induced YAP dephosphorylation as determined by immunoblotting using an antibody specific to YAP phosphorylated at Ser127 (a direct LATS1/2 phosphorylation site responsible for cytoplasmic localization of YAP) and differential migration on phos-tag-containing gels (Fig. 3C, left). Consistent with a positive role of YAP in LATS2 transcription, we found that the amount of LATS2 mRNA was increased by serum (Fig. 3C, right), supporting a role for endogenous YAP in LATS2 transcription regulation. Thus, these data suggest a physiological role of YAP in promoting LATS2 transcription.
We then investigated whether LATS2 is a direct transcriptional target of the YAP–TEAD complex. We identified three putative TEAD-binding sequences in the human LATS2 promoter region and cloned the LATS2 promoter into a luciferase reporter plasmid to evaluate its responsiveness to YAP and TEAD. Expression of YAP in HEK293A cells markedly increased LATS2 luciferase reporter activity in a concentration-dependent manner, and this activation was further enhanced by coexpression of TEAD1 (Fig. 3D). Chromatin immunoprecipitation (ChIP) using anti-YAP or anti-TEAD1 antibodies revealed that endogenous YAP and TEAD1 both specifically associated with the LATS2 promoter in MCF10A cells (Fig. 3E). No significant association of YAP or TEAD1 with a nonpromoter control region (CR) of the LATS2 gene or the LATS1 promoter was detected. CTGF (connective tissue growth factor), a known direct target gene of the YAP–TEAD complex (Zhao et al. 2008), and GAPDH (glyceraldehyde 3-phosphate dehydrogenase) were used as a positive and negative control, respectively. Together, our data suggest that YAP–TEAD directly binds to the promoter of LATS2 to induce its expression.
We analyzed the gene expression data from the Cancer Cell Line Encyclopedia (CCLE) (Fig. 3F; Barretina et al. 2012). As expected, there is a strong positive correlation between the two YAP/TAZ target genes CYR61 (cysteine-rich angiogenic inducer 61) and CTGF but no correlation between GAPDH (a negative control) and either CYR61 or CTGF. Interestingly, we also found a strong positive correlation between LATS2 mRNA and either CYR61 or CTGF expression. In contrast, no significant correlation was observed between LATS1 and either CYR61 or CTGF. These data provide further in vivo evidence supporting that YAP/TAZ promote LATS2 transcription.
YAP/TAZ stimulate the kinase activity of LATS1/2 through NF2 (neurofibromin 2)
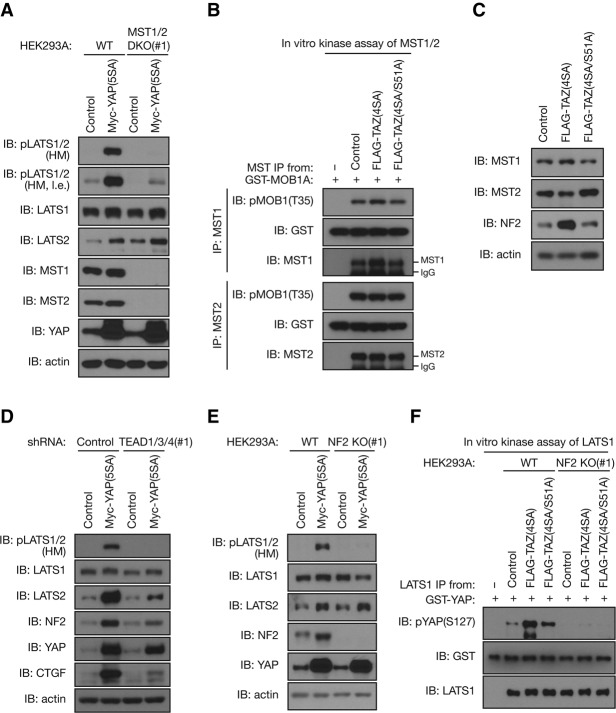
The YAP/TAZ-induced feedback mechanism includes induction of LATS2 protein expression as well as activation of both LATS1 and LATS2 kinase activity. We next explored the mechanism of LATS1/2 kinase activation by YAP/TAZ. As we observed an increased phosphorylation of LATS1/2 at the HM, a site known to be phosphorylated by MST1/2 (Chan et al. 2005), we investigated whether MST1/2 are responsible for this activation. To test this possibility, we deleted MST1/2 in HEK293A cells using CRISPR/Cas9 genome-editing technology (Ran et al. 2013). Consistent with a positive role for MST1/2 in YAP phosphorylation, MST1/2 deletion reduced the basal phosphorylation of YAP (Supplemental Fig. S4A). To avoid the influences from off-target effects, we obtained multiple clones and confirmed that re-expression of MST1/2 prevented YAP dephosphorylation in MST1/2-deficient HEK293A cells (Supplemental Fig. S4A). Similar to the observation in MCF10A cells (Fig. 2B,C), YAP(5SA) or TAZ(4SA) overexpression increased the amount of LATS2 as well as LATS1/2 HM phosphorylation in HEK293A cells (Fig. 4A; Supplemental Fig. S4B). Importantly, loss of endogenous MST1/2 attenuated the effect of YAP(5SA) on LATS1/2 phosphorylation (Fig. 4A; Supplemental Fig. S4C). However, MST1/2 depletion failed to completely block YAP-induced LATS1/2 HM phosphorylation, possibly due to the presence of other LATS-activating kinases. We then measured the kinase activity of endogenous MST1 and MST2. However, TAZ(4SA) overexpression had no effect on MST1/2 kinase activity, as indicated by their substrate (MOB1) phosphorylation (Fig. 4B). Immunoblot analysis revealed that the amount of MST1 and MST2 remained unchanged in TAZ(4SA)-overexpressing MCF10A cells (Fig. 4C). Although MST1/2 appeared to be largely required for YAP/TAZ-induced LATS1/2 activation, the above results suggest that the protein abundance and intrinsic kinase activity of MST1/2 are not the key steps stimulated by YAP/TAZ overexpression.
Figure 4.
YAP and TAZ stimulate the intrinsic kinase activity of LATS1/2 through NF2. (A) MST1/2 are largely required for YAP-induced LATS1/2 activation. Wild-type (WT) and MST1/2 double-knockout (DKO) HEK293A cells infected with retroviruses encoding Myc-YAP(5SA) or with control empty retrovirus were subjected to immunoblot (IB) analysis. (l.e.) Long exposure. (B) TAZ does not stimulate MST1/2 kinase activity. Endogenous MST1 or MST2 was immunoprecipitated (IP) from MCF10A cells infected with retroviruses encoding Flag-TAZ(4SA) or Flag-TAZ(4SA/S51A) or with the empty retrovirus (control). The resulting immunoprecipitates were subjected to in vitro kinase assay. (C) TAZ increases NF2 abundance. MCF10A cells infected as in B were subjected to immunoblot analysis. (D) TEADs are required for NF2 induction and LATS1/2 activation by YAP. MCF10A cells stably expressing Myc-YAP(5SA) or control vector were infected with lentiviruses encoding shRNAs specific for TEAD1/3/4. Immunoblot analysis was performed with the indicated antibodies. (E) NF2 is required for YAP-induced activating phosphorylation of LATS1/2. Wild-type and NF2 knockout (KO) HEK293A cells infected with retroviruses encoding Myc-YAP(5SA) or with control empty retrovirus were subjected to immunoblot analysis. (F) NF2 is required for TAZ-induced LATS1 activation. Endogenous LATS1 was immunoprecipitated from wild-type or NF2 knockout HEK293A cells stably expressing Flag-TAZ(4SA), Flag-TAZ(4SA/S51A), or control vector. The resulting immunoprecipitates were subjected to in vitro kinase assay. See also Supplemental Figure S4.
Recently, the tumor suppressor NF2 (Merlin in D. melanogaster) was revealed to promote LATS1/2 kinase activation without activating the intrinsic kinase activity of MST1/2 (Yin et al. 2013). Furthermore, Merlin is already an established transcriptional target of Yorkie, the fly homolog of YAP/TAZ (Hamaratoglu et al. 2006). Indeed, we found that mRNA and protein abundance of NF2 both were markedly increased by YAP(5SA) or TAZ(4SA) overexpression in MCF10A cells (Fig. 4C,D; Supplemental Fig. S4D). ChIP assay revealed that both endogenous YAP and TEAD1 specifically associated with the NF2 promoter (Supplemental Fig. S4E). Moreover, shRNA-mediated depletion of endogenous TEAD1/3/4 attenuated the effect of YAP(5SA) on the induction of NF2 and LATS2 as well as the activating phosphorylation of the LATS1/2 HM (Fig. 4D). We thus hypothesized that the increased NF2 protein abundance may be responsible for the YAP/TAZ-mediated LATS activation. To test this possibility, we deleted NF2 in HEK293A cells using CRISPR/Cas9 genome-editing technology. Consistent with a positive role of NF2 in YAP phosphorylation, NF2 deletion reduced the basal phosphorylation of YAP (Supplemental Fig. S4F). We obtained multiple clones and confirmed that re-expression of NF2 prevented YAP dephosphorylation in NF2-deficient HEK293A cells (Supplemental Fig. S4F). Strikingly, loss of endogenous NF2 diminished the effect of YAP(5SA) on LATS1/2 phosphorylation (Fig. 4E; Supplemental Fig. S4G). A similar function of NF2 in TAZ-induced LATS activation was observed (Supplemental Fig. S4H). Moreover, in vitro kinase assay revealed that TAZ(4SA) overexpression increased the kinase activity of endogenous LATS1 in NF2 wild-type cells, yet LATS1 activation was largely abolished in NF2 knockout cells (Fig. 4F). Consistent with a positive role of NF2 in LATS1/2 activation, NF2 deletion also reduced basal LATS1 kinase activity. We confirmed that overexpression of NF2 and MST1/2 increased LATS1/2 phosphorylation but did not affect LATS2 induction by YAP(5SA), which is a mutant unresponsive to inhibition by the Hippo pathway, suggesting that the negative feedback loop could still be operational via LATS2 induction even under condition of high Hippo pathway activity (Supplemental Fig. S4I). Together, our observations indicate that YAP/TAZ directly induce NF2 and LATS2 expression to increase the kinase activity of LATS1/2.
YAP induces negative feedback in mouse livers
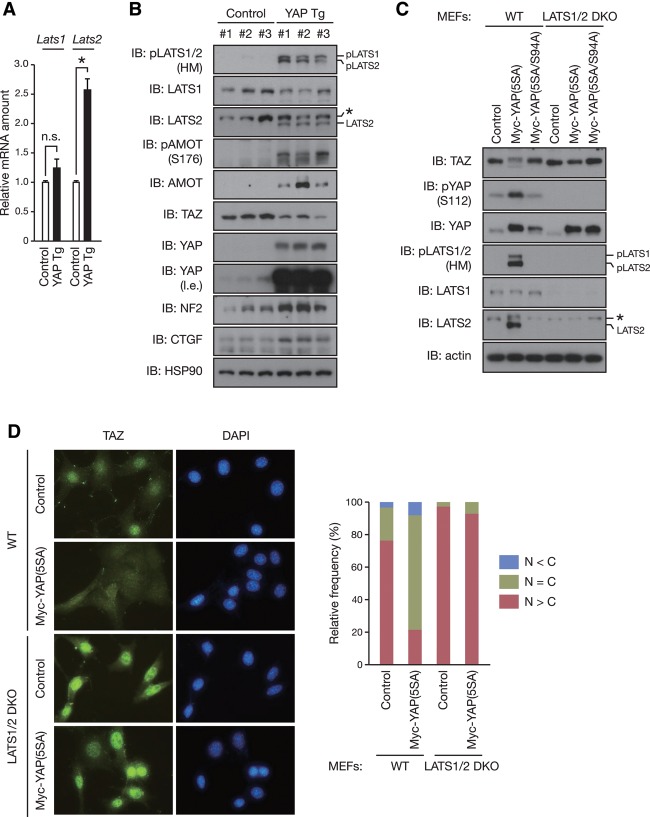
We then investigated whether this feedback mechanism operates in vivo using YAP transgenic (Tg) mice (Dong et al. 2007). Consistent with what we observed with the cell lines in vitro, a significant increase of Lats2 mRNA, but not Lats1 mRNA, was detected in the livers of YAP Tg mice (Fig. 5A). Furthermore, we observed a marked increase in the activating phosphorylation of the LATS1/2 HM as well as the protein abundance of LATS2 (Fig. 5B). As observed with the cultured cells, NF2 accumulated in the livers of YAP Tg mice. Activation of LATS1/2 kinases was associated with the decreased protein abundance of TAZ, which has two phosphodegrons and is strongly destabilized by the Hippo pathway. To further confirm LATS activation, we examined the phosphorylation of AMOT at Ser176 (corresponding to Ser175 in humans). Again, the AMOT phosphorylation was increased, indicating increased LATS activity. Notably, the protein abundance of AMOT was also elevated in the YAP Tg liver, consistent with the previous observation that the phosphorylation stabilizes AMOT (Adler et al. 2013). Collectively, these data support a model in which YAP/TAZ induce LATS activation to constitute a feedback mechanism in the Hippo pathway.
Figure 5.
LATS1/2 mediate negative feedback regulation of YAP/TAZ activity. (A) YAP induces the transcription of Lats2 in vivo. RT and real-time PCR analysis of the indicated mRNA in the livers of 2-mo-old YAP Tg and non-Tg (control) mice fed 0.2 mg/mL doxycycline in drinking water for 2 wk. Normalized data are expressed relative to the corresponding value for non-Tg littermates and are mean ± SD. n = 3 mice per group. (n.s.) Not significant. (*) P < 0.05 (Student's t test). (B) YAP stimulates negative feedback in vivo. Immunoblot (IB) analysis of liver extracts from mice treated as in A with antibodies to the indicated proteins. n = 3 mice per group. The asterisk indicates a nonspecific band. (l.e.) Long exposure. (C) LATS1/2 are required for YAP-induced negative feedback regulation of YAP and TAZ. Wild-type (WT) or Lats1–/–Lats2Δ/Δ (LATS1/2 double-knockout [DKO]) mouse embryonic fibroblasts (MEFs) were infected with retroviruses encoding Myc-YAP(5SA) or Myc-YAP(5SA/S94A) or with empty retrovirus. Cell lysates were immunoblotted with the indicated antibodies. The asterisk indicates a nonspecific band. (D) LATS1/2 are required for YAP-induced cytoplasmic localization of TAZ. MEFs infected as in C were subjected to immunostaining analysis. TAZ subcellular localization was determined by immunofluorescence staining for endogenous TAZ along with DAPI for DNA. Representative images are presented in the left panel. (Right panel) Cells in five random views (∼100 cells) were quantified for TAZ localization. See also Supplemental Figure S5.
LATS1/2 are required for feedback regulation of YAP/TAZ activity
To test our hypothesis that LATS1/2 mediate this feedback regulation of YAP/TAZ activity, we examined the effect of LATS1/2 depletion on YAP-mediated TAZ inhibition. Depletion of endogenous LATS1/2 by siRNA attenuated the effect of YAP(5SA) on endogenous TAZ reduction in MCF10A cells (Supplemental Fig. S5A), suggesting that LATS1/2 are required for this regulation. To further confirm the role of LATS1/2 in feedback regulation, we generated LATS1/2-deficient mouse embryonic fibroblasts (MEFs) by infecting Lats1–/–Lats2F/F MEFs (Kim et al. 2013) with adenoviruses encoding Cre recombinase. We confirmed that almost all floxed alleles were deleted by Cre recombinase in Lats1–/–Lats2F/F MEFs, giving rise to LATS1/2 double-knockout MEFs (Supplemental Fig. S5B). In wild-type MEFs, overexpression of YAP(5SA) increased the activating phosphorylation of LATS1/2 at the HM as well as the mRNA and protein abundance of LATS2 (Fig. 5C; Supplemental Fig. S5B). LATS activation was associated with the decreased protein abundance of TAZ as well as the increased phosphorylation of endogenous YAP at Ser112 [corresponding to Ser127 in humans; the phospho-YAP antibody does not recognize the exogenous YAP(5SA)]. However, the effects of YAP(5SA) on endogenous YAP/TAZ were almost completely abolished in LATS1/2 double-knockout MEFs (Fig. 5C). We next determined TAZ localization, which is tightly regulated by LATS-dependent phosphorylation. As predicted, YAP(5SA) expression increased the cytoplasmic and decreased the nuclear localization of endogenous TAZ in wild-type MEFs, but this change of TAZ subcellular localization was not observed in LATS1/2 double-knockout MEFs (Fig. 5D). We also found that loss of LATS1/2 blocked the effect of YAP(5SA) on the phosphorylation and protein levels of AMOT (Supplemental Fig. S5C). Thus, collectively, our observations demonstrate that LATS1/2 are involved in cross-regulation among YAP, TAZ, and AMOT.
Feedback regulation maintains the transient nature of LPA-induced YAP activation
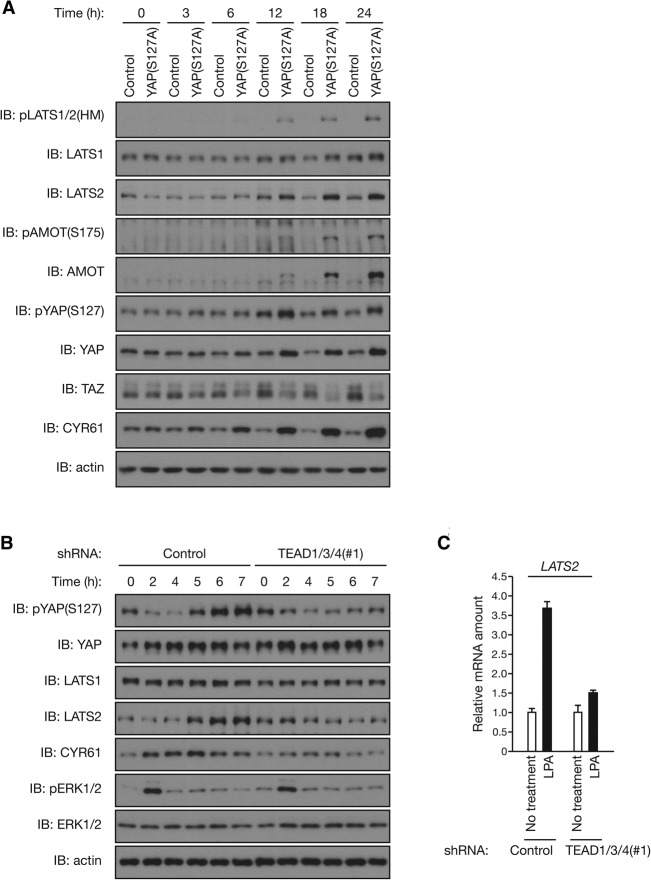
As LATS induction is found to provide a feedback mechanism for YAP/TAZ regulation, we investigated the dynamics of this regulation. To this end, we generated MCF10A cells in which a tetracycline-inducible construct encoding YAP(S127A) was stably integrated. We also introduced the S127A mutation into the YAP construct to distinguish between endogenous and exogenous YAP; therefore, the phospho-YAP antibody detects only endogenous YAP phosphorylated at Ser127. Immunoblot analysis revealed a significant YAP induction in cells inducibly expressing YAP(S127A) at 12 h of doxycycline treatment (Fig. 6A). Remarkably, a substantial increase in the activating phosphorylation of LATS1/2 and the protein abundance of LATS2 was also observed as early as 12 h after doxycycline addition. LATS activation was associated with the increased phosphorylation of endogenous YAP and AMOT as well as decreased TAZ protein abundance, indicative of activation of negative feedback. Given that protein expression of YAP(S127A) and its effects on downstream targets, as indicated by YAP target gene CYR61 expression, were apparent 6–12 h after the onset of doxycycline treatment, these results indicate a prompt induction of LATS-mediated negative feedback following YAP activation.
Figure 6.
YAP/TEAD-induced feedback mechanism maintains the transient YAP activation on stimulation. (A) YAP induction promptly stimulates the LATS-mediated feedback loop. Tetracycline repressor-expressing MCF10A cells were infected with retroviruses encoding YAP(S127A) or with empty retrovirus, incubated with 1 μg/mL doxycycline for the indicated times, and then subjected to immunoblot (IB) analysis. (B) YAP/TEAD-induced negative feedback is required for the transient activation of YAP upon LPA stimulation. MCF10A cells infected with lentiviruses encoding shRNA specific for TEAD1/3/4 (or control) were starved in serum-free medium for 15 h and then stimulated with 5 μM LPA for the indicated times. Cell lysates were subjected to immunoblotting with the indicated antibodies. (C) LPA induces LATS2 transcription in a TEAD-dependent manner. MCF10A cells infected and starved as in B were stimulated with 5 μM LPA for 2 h. Total RNA were subjected to RT and real-time PCR analysis of LATS2 mRNA. Data are means ± SD of triplicates from a representative experiment. See also Supplemental Figure S6.
We propose that the LATS-mediated feedback mechanism is involved in the dynamic regulation of the Hippo pathway, such as the duration of YAP activation upon upstream signal stimulation. LPA acts through GPCR to induce YAP activation (Yu et al. 2012). In control MCF10A cells, LPA treatment induced an acute YAP dephosphorylation, as determined by immunoblotting using the phospho-YAP antibody (Fig. 6B). The LPA-induced YAP activation is supported by the increased CTGF and CYR61 transcription (Supplemental Fig. S6A). We also found that LPA increased LATS2 expression, which temporally followed YAP dephosphorylation (Fig. 6B,C), suggesting that YAP activation induces LATS2 expression upon LPA stimulation. Notably, LATS2 protein accumulation was slower than CYR61 (Fig. 6B), although mRNAs of both genes were induced at the early time point (2 h) (Fig. 6C; Supplemental Fig. S6A), indicating a possible additional level of regulation. Importantly, the effect of LPA on YAP dephosphorylation was transient, as YAP phosphorylation was recovered ∼5 h after LPA stimulation and even increased above the prestimulated level at later time points (Fig. 6B). Because LATS2 accumulation was correlated with this “rephosphorylation” of YAP, we propose that the YAP-induced and transcription-dependent feedback machinery is responsible for terminating YAP activity. Therefore, YAP activation by a given signal will not be prolonged forever but rather will be temporally controlled.
To test our model, we suppressed the induction of negative feedback by knocking down TEAD, thus blocking YAP-induced transcription. As expected, depletion of endogenous TEAD1/3/4 by shRNA in MCF10A cells (Supplemental Fig. S1B) attenuated LPA-induced YAP target gene expression, including CTGF, CYR61, and LATS2 (Fig. 6B, C; Supplemental Fig. S6A). TEAD depletion had no effect on the initial YAP dephosphorylation by LPA. However, strikingly, TEAD knockdown abolished YAP “rephosphorylation” following LPA-induced YAP dephosphorylation (Fig. 6B). In contrast, no substantial difference was observed in an ERK (extracellular signal-regulated kinase) phosphorylation–dephosphorylation pattern following LPA stimulation between control and TEAD1/3/4 knockdown MCF10A cells (Fig. 6B), suggesting that TEAD-mediated negative feedback specifically regulates YAP inactivation after LPA stimulation. Therefore, blockade of this negative feedback resulted in a sustained YAP activation/dephosphorylation by LPA (Fig. 6B). Together, our observations indicate that the YAP–LATS negative feedback mechanism plays a critical role in modulating signal duration.
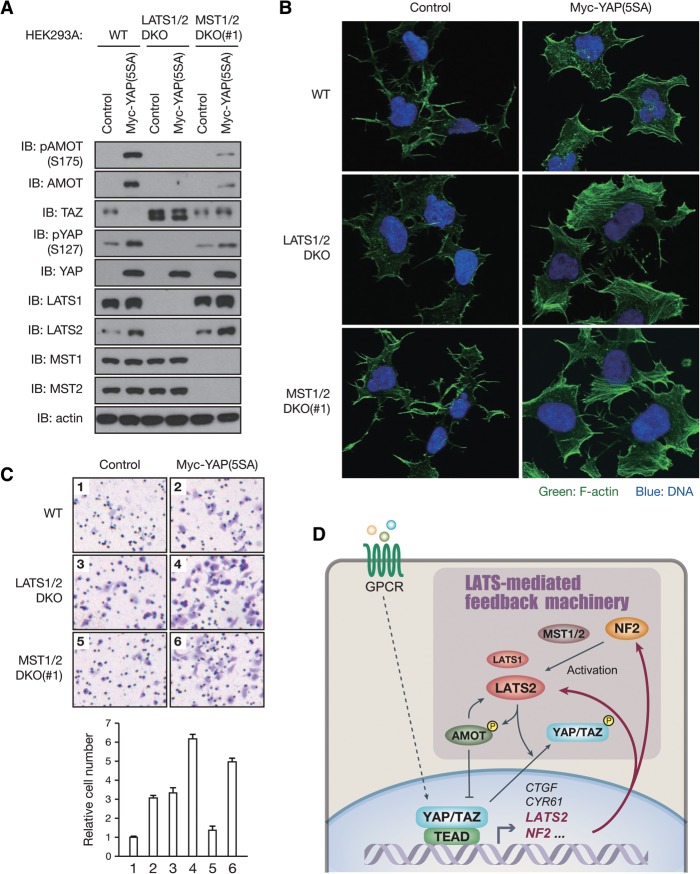
LATS-mediated negative feedback inhibits cell migration
As the LATS-mediated feedback is found to influence the dynamic regulation of YAP activity following Hippo pathway perturbation by LPA–GPCR signaling, we investigated whether this feedback mechanism regulates YAP/TAZ-mediated biology, such as migration. To confer transient YAP activation, we generated LATS1/2- or MST1/2-deficient HEK293A cells in which a tetracycline-inducible construct encoding YAP(5SA) was stably integrated. We confirmed the specificity of CRISPR/Cas9-mediated LATS1/2 depletion as re-expression of LATS1/2 prevented YAP dephosphorylation in LATS1/2 double-knockout HEK293A cells (Supplemental Fig. S7A). In wild-type HEK293A cells, YAP induction initiates the feedback machinery to activate LATS1/2, resulting in phosphorylation and degradation of endogenous TAZ as well as phosphorylation and accumulation of AMOT (Fig. 7A). These feedback regulations on TAZ and AMOT were significantly attenuated in MST1/2 double-knockout cells and almost completely abolished in LATS1/2 double-knockout cells (Fig. 7A). AMOT is a filamentous actin (F-actin)-binding protein. While YAP and TAZ are known to promote cell migration (Chan et al. 2008; Haskins et al. 2014), recent studies revealed that the phosphorylation of AMOT by LATS1/2 inhibits AMOT binding to F-actin (Chan et al. 2013; Dai et al. 2013; Hirate et al. 2013), reduces actin stress fiber formation, and thereby inhibits migration (Dai et al. 2013). We thus hypothesized that the LATS-mediated feedback mechanism limits cell migration induced by YAP activation. To test this possibility, we investigated the effect of LATS1/2 or MST1/2 depletion on YAP(5SA) overexpression. Immunofluorescence analysis revealed that YAP(5SA) overexpression promoted F-actin formation in wild-type cells (Fig. 7B), yet additional ablation of LATS1/2 or MST1/2 dramatically enhanced actin stress fiber formation (Fig. 7B). Consistent with this, deletion of LATS1/2 or MST1/2 on top of YAP(5SA) overexpression further promoted HEK293A cell migration (Fig. 7C). Together, these observations indicate that LATS-mediated feedback mechanism limits cell migration through cross-regulation among YAP, TAZ, and AMOT.
Figure 7.
The LATS-mediated negative feedback limits actin stress fiber formation and cell migration upon YAP activation. (A) Inducible expression of YAP triggers the negative feedback machinery via LATS1/2 and MST1/2. Tetracycline repressor-expressing HEK293A cells were infected with retroviruses encoding Myc-YAP(5SA) or with control empty retrovirus, incubated with 200 ng/mL doxycycline for 24 h, and then subjected to immunoblot (IB) analysis. (B) Loss of LATS1/2 or MST1/2 enhances actin stress fiber formation by YAP(5SA) overexpression. HEK293A cells infected and induced as in A were seeded on fibronectin-coated coverslips for 1 h in serum-free medium. F-actin was stained with Alexa fluor 488 phalloidin (green) along with DAPI for DNA (blue). (C) LATS1/2 or MST1/2 depletion enhances cell migration by YAP(5SA) overexpression. HEK293A cells infected and induced as in A were subjected to transwell migration assay. Cells on the bottom sides of the transwells were stained with crystal violet (top) and quantified (bottom). (D) A proposed model of negative feedback regulation in the mammalian Hippo pathway. In response to YAP/TAZ-activating stimulations, YAP/TAZ in complex with TEAD directly induces LATS2 expression. YAP/TAZ also stimulate the kinase activity of LATS1/2 through NF2 induction. YAP/TAZ activation therefore results in activation of their negative regulators, LATS1/2, to constitute a negative feedback loop of the Hippo pathway. See also Supplemental Figure S7.
Discussion
Despite the critical role of YAP/TAZ in regulating cell fate, tissue homeostasis, and tumorigenesis, mechanisms involved in the dynamic control of YAP/TAZ activity following perturbation, particularly the feedback regulation conferring transient activation/deactivation in response to stimuli and preventing overactivation, have not been elucidated. In this study, we show that YAP/TAZ activation stimulates their negative regulators, LATS1/2 kinases, to restrict YAP/TAZ activity. YAP/TAZ in complex with TEAD directly stimulate LATS2 transcription and indirectly activate LATS1/2 kinase activity through NF2 induction and AMOT accumulation, therefore constituting a YAP/TAZ–LATS negative feedback loop (Fig. 7D). This negative feedback regulation is also observed in vivo by the Tg overexpression of YAP or deletion of the Yap gene in mouse tissues. Consistent with our findings, Chen et al. (2015) report a robust homeostatic mechanism maintaining YAP activity in vivo; mice harboring a YAP(S112A) knock-in mutation in the endogenous Yap locus were phenotypically normal due to a compensatory decrease in YAP protein levels. This negative feedback regulation is mediated by increased LATS activity and YAP phosphorylation in the phosphodegron (Chen et al. 2015). Together, our findings show that the LATS-mediated feedback mechanism enables the robustness, dynamics, and homeostasis of YAP/TAZ regulation.
The negative feedback mechanism employed by the mammalian Hippo pathway appears to differ significantly from that in D. melanogaster, where Yorkie induces expression of expanded, merlin, four-jointed, or kibra (Cho et al. 2006; Hamaratoglu et al. 2006; Genevet et al. 2010), but not warts (the fly homolog of LATS1/2), in dividing epithelial cells of the wing or eye disc (Jukam et al. 2013). On the contrary, Yorkie indirectly suppresses Warts transcription to establish positive feedback regulation in combination with tissue-specific transcription factors in the context of Drosophila eye development (Jukam et al. 2013). This is in a dramatic contrast to mammalian cells in which YAP/TAZ activate LATS. It is possible that the mechanism observed in D. melanogaster might be employed in some mammalian cell types. Although the evolutionary conservation of the Hippo pathway is extensive, most of the mammalian homologs of the pathway components are encoded by more than one gene. As mammals acquire two orthologs of Drosophila Warts, it is interesting that one (LATS1) is not induced by YAP/TAZ, similar to the Drosophila Warts, while the other (LATS2) is induced by YAP/TAZ. This evolutionary divergence in pathway components adds an extra layer of complexity and contributes to the dynamic regulation of the mammalian Hippo pathway (Bossuyt et al. 2014).
We also showed that LATS-mediated feedback regulation plays an important role in terminating LPA-induced YAP activation signal. As LATS-mediated negative feedback requires transcription and translation, it takes more time to fully induce the negative regulatory effects than other well-characterized negative feedback loops mediated by protein modifications, such as receptor phosphorylation by GPCR kinases (GRKs) followed by β-arrestin-mediated uncoupling of the GPCR from the G proteins (Pierce et al. 2002) or feedback phosphorylation of upstream components of a signaling cascade (e.g., inhibitory phosphorylation of RAF by ERK in the MAPK signaling) (Dougherty et al. 2005). Thus, the LATS-mediated feedback architecture provides enough time for YAP/TAZ to mediate their biological functions as transcriptional coactivators by taking advantage of the time delay of transcription and translation. When the feedback loop is disrupted by TEAD knockdown, YAP dephosphorylation/activation by LPA is sustained. Therefore, this feedback mechanism is essential for proper duration of YAP activation in response to stimuli.
Besides YAP/TAZ, AMOT is another physiological substrate of LATS. We demonstrate that YAP/TAZ-induced LATS activation can indeed lead to increased AMOT phosphorylation and protein accumulation. Notably, the increase of AMOT may also contribute to the negative feedback because AMOT can directly bind to and inhibit YAP/TAZ as well as indirectly activate LATS (Zhao et al. 2011; Hirate et al. 2013). One can predict with certainty that LATS must have many additional physiological substrates. Sequence analyses of the human proteome show that there are >1000 peptides with the LATS phosphorylation consensus motif (HXRXXS). Therefore, the negative YAP/TAZ–LATS feedback loop is likely to provide a mechanism by which YAP/TAZ may modulate phosphorylation of other LATS substrates. One may further speculate that YAP/TAZ could exert some of their biological functions through LATS-mediated phosphorylation of other LATS substrates.
Given the observed frequent YAP/TAZ activation in human cancers, the general lack of cancer-relevant somatic or germline mutations in core members of the Hippo pathway is a major conundrum to the field (Harvey et al. 2013; Moroishi et al. 2015; Plouffe et al. 2015). One possibility is that negative feedback control of YAP/TAZ activity suppresses tumorigenesis following small perturbation of the pathway. Inactivation of LATS, though, can increase YAP/TAZ activity; however, the YAP/TAZ activation would be limited by the YAP/TAZ-mediated LATS activation, which could also phosphorylate other substrates to limit cell growth. Single mutation in the Hippo pathway components might not be enough—rather, it might require network-level strong perturbation—to drive tumorigenesis. In support of this idea, genetic mutations inhibiting the functions of key negative feedback regulators are found in human cancer: NF2 is mutated with high frequency in neurofibromatosis (Asthagiri et al. 2009), and loss of NF2 as well as LATS2 is also frequently observed in malignant mesothelioma (Murakami et al. 2011), indicative of their functions as bona fide tumor suppressor genes. Further investigations delineating the feedback regulation will have important implications in understanding both the basic biology of the Hippo pathway and tumorigenesis in cancer with alteration of this pathway.
Materials and methods
Full experimental procedures are shown in the Supplemental Material.
Gene deletion of HEK293A cells by the CRISPR/Cas9 system
LATS1/2-, MST1/2-, or NF2-deficient HEK293A cells were created through the CRISPR/Cas9 system (Ran et al. 2013). HEK293A cells were transfected with a Cas9 expression plasmid together with single-guide RNA (sgRNA) expression plasmids (PX459; Addgene plasmid no. 48139). The guide sequences were designed using the CRISPR design tool at http://www.genome-engineering.org/crispr or were taken from archived guide sequences from the genome-scale CRISPR knockout (GeCKO2) library (Sanjana et al. 2014). The guide sequences used were 5′-CGTGCAGCTCTCCGCTCTAA-3′ for human LATS1, 5′-TACGCTGGCACCGTAGCCCT-3′ for human LATS2, 5′-ATACACCGAGATATCAAGGC-3′ for human MST1, 5′-AGTACTCCATAACAATCCAG-3′ for human MST2, and 5′-GTCCATGGTGACGATCCTCA-3′ for human NF2. Following transfection and selection with puromycin, cells were single-cell-sorted by fluorescence-activated cell sorting (FACS) into a 96-well plate format. Knockout clones were selected by immunoblot analysis, and genomic mutations were confirmed by genome sequencing. Two independent clones were analyzed as indicated.
Gene deletion of MEFs by adenoviral infection
SV40 large T-immortalized Lats1–/–Lats2F/F MEFs were described previously (Murakami et al. 2011). For Lats2 deletion, MEFs were infected with an adenovirus encoding Cre recombinase (no. 1700, Vector Biolabs).
Kinase assay
The immunoprecipitated LATS1 was subjected to a kinase assay in 30 μL of kinase assay buffer supplemented with 500 μM ATP and 1 μg of GST-YAP as substrates. The reaction mixtures were incubated for 30 min at 30°C and subjected to immunoblot analysis. The same procedure was used for the MST1 and MST2 kinase assay, except that GST-MOB1A was used as the substrate.
Luciferase assay
A 1000-base-pair (bp) fragment of human genomic DNA containing the promoter lesion of LATS2 was cloned into the pGL3-Basic vector. HEK293A cells were transfected with pGL3 LATS2 luciferase reporter, pCMV Flag-YAP, pRK7 Myc-TEAD1, and Renilla plasmids. Luciferase assay was performed 30 h after transfection using the Dual Glo luciferase system (Promega). All luciferase activities were normalized to Renilla.
ChIP assay
MCF10A cells were dual-cross-linked consecutively with 2 mM disuccinimidyl glutarate (DSG) for 45 min and 1% formaldehyde for 10 min, and chromatin DNA was digested with micrococcal nuclease (MNase). Immunoprecipitation reactions were carried out with chromatin extracts overnight at 4°C. Precipitated DNA was quantitated by real-time PCR analysis. All ChIP signals were normalized to the input (labeled as percentage of input on the vertical axis).
YAP mutant mice
To achieve liver-specific or intestine-specific gene deletion, YapF/F mice were bred to Albumin-Cre (Zhang et al. 2010) or Villin-Cre (Taniguchi et al. 2015) Tg mice, respectively. For liver-specific YAP Tg mice, 2-mo-old YAP Tg and non-Tg littermates were fed 0.2 mg/mL doxycycline (Sigma) in drinking water supplemented with 2.5% sucrose, as described previously (Dong et al. 2007). Two-month-old to 3-mo-old sex-matched mice were used for all experimental procedures.
More detailed experimental procedures and other methods are described in the Supplemental Material.
Supplementary Material
Acknowledgments
We thank Dr. Dae-Sik Lim for Lats1–/–Lats2F/F MEFs; Dr. Feng Zhang for PX459 plasmid; Dr. Bin Zhao for pAMOT antibody; and Dr. Jung-Soon Mo, Dr. Carsten Gram Hansen, Dr. Audrey W. Hong, and Dr. Guangbo Liu for insightful discussions. This work was supported by grants from the National Institutes of Health (EY022611 and CA132809) to K.-L.G. T.M. is supported by the Japan Society for the Promotion of Science (JSPS) Post-doctoral Fellowships for Research Abroad and a grant from the Yasuda Medical Foundation. S.W.P is supported by a University of California at San Diego Cellular and Molecular Pharmacology training grant (T32 GM007752).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.262816.115.
References
- Adler JJ, Johnson DE, Heller BL, Bringman LR, Ranahan WP, Conwell MD, Sun Y, Hudmon A, Wells CD. 2013. Serum deprivation inhibits the transcriptional co-activator YAP and cell growth via phosphorylation of the 130-kDa isoform of angiomotin by the LATS1/2 protein kinases. Proc Natl Acad Sci 110: 17368–17373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asthagiri AR, Parry DM, Butman JA, Kim HJ, Tsilou ET, Zhuang Z, Lonser RR. 2009. Neurofibromatosis type 2. Lancet 373: 1974–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. 2012. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483: 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry ER, Camargo FD. 2013. The Hippo superhighway: signaling crossroads converging on the Hippo/Yap pathway in stem cells and development. Curr Opin Cell Biol 25: 247–253. [DOI] [PubMed] [Google Scholar]
- Bossuyt W, Chen CL, Chen Q, Sudol M, McNeill H, Pan D, Kopp A, Halder G. 2014. An evolutionary shift in the regulation of the Hippo pathway between mice and flies. Oncogene 33: 1218–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EH, Nousiainen M, Chalamalasetty RB, Schafer A, Nigg EA, Sillje HH. 2005. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24: 2076–2086. [DOI] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Guo K, Ng CP, Lee I, Hunziker W, Zeng Q, Hong W. 2008. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res 68: 2592–2598. [DOI] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Guo F, Tan I, Leung T, Hong W. 2013. Actin-binding and cell proliferation activities of angiomotin family members are regulated by Hippo pathway-mediated phosphorylation. J Biol Chem 288: 37296–37307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Zhang N, Xie R, Wang W, Cai K-S, David KK, Huang B, Yabuta N, Nojima H, et al. 2015. Homeostatic control of Hippo signaling activity revealed by an endogenous activating mutation in YAP. Genes Dev (this issue) 10.1101/gad.264234.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. 2006. Delineation of a Fat tumor suppressor pathway. Nat Genet 38: 1142–1150. [DOI] [PubMed] [Google Scholar]
- Dai X, She P, Chi F, Feng Y, Liu H, Jin D, Zhao Y, Guo X, Jiang D, Guan KL, et al. 2013. Phosphorylation of angiomotin by Lats1/2 kinases inhibits F-actin binding, cell migration, and angiogenesis. J Biol Chem 288: 34041–34051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. 2007. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130: 1120–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty MK, Muller J, Ritt DA, Zhou M, Zhou XZ, Copeland TD, Conrads TP, Veenstra TD, Lu KP, Morrison DK. 2005. Regulation of Raf-1 by direct feedback phosphorylation. Mol Cell 17: 215–224. [DOI] [PubMed] [Google Scholar]
- Enderle L, McNeill H. 2013. Hippo gains weight: added insights and complexity to pathway control. Sci Signal 6: re7. [DOI] [PubMed] [Google Scholar]
- Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. 2010. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell 18: 300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Dupont S, Piccolo S. 2012. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat Rev Mol Cell Biol 13: 591–600. [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. 2006. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol 8: 27–36. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Zhang X, Thomas DM. 2013. The Hippo pathway and human cancer. Nat Rev Cancer 13: 246–257. [DOI] [PubMed] [Google Scholar]
- Haskins JW, Nguyen DX, Stern DF. 2014. Neuregulin 1-activated ERBB4 interacts with YAP to induce Hippo pathway target genes and promote cell migration. Sci Signal 7: ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A. 2013. Regulation and functions of mammalian LATS/NDR kinases: looking beyond canonical Hippo signalling. Cell Biosci 3: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirate Y, Hirahara S, Inoue K, Suzuki A, Alarcon VB, Akimoto K, Hirai T, Hara T, Adachi M, Chida K, et al. 2013. Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr Biol 23: 1181–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Halder G. 2014. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov 13: 63–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukam D, Xie B, Rister J, Terrell D, Charlton-Perkins M, Pistillo D, Gebelein B, Desplan C, Cook T. 2013. Opposite feedbacks in the Hippo pathway for growth control and neural fate. Science 342: 1238016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Kim M, Lee S, Kuninaka S, Saya H, Lee H, Lee S, Lim DS. 2013. cAMP/PKA signalling reinforces the LATS–YAP pathway to fully suppress YAP in response to actin cytoskeletal changes. EMBO J 32: 1543–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low BC, Pan CQ, Shivashankar GV, Bershadsky A, Sudol M, Sheetz M. 2014. YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Lett 588: 2663–2670. [DOI] [PubMed] [Google Scholar]
- Mo JS, Park HW, Guan KL. 2014. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep 15: 642–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroishi T, Hansen CG, Guan KL. 2015. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer 15: 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Mizuno T, Taniguchi T, Fujii M, Ishiguro F, Fukui T, Akatsuka S, Horio Y, Hida T, Kondo Y, et al. 2011. LATS2 is a tumor suppressor gene of malignant mesothelioma. Cancer Res 71: 873–883. [DOI] [PubMed] [Google Scholar]
- Neto-Silva RM, de Beco S, Johnston LA. 2010. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev Cell 19: 507–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. 2010. Yorkie: the final destination of Hippo signaling. Trends Cell Biol 20: 410–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. 2010. The hippo signaling pathway in development and cancer. Dev Cell 19: 491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HW, Guan KL. 2013. Regulation of the Hippo pathway and implications for anticancer drug development. Trends Pharmacol Sci 34: 581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo S, Dupont S, Cordenonsi M. 2014. The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev 94: 1287–1312. [DOI] [PubMed] [Google Scholar]
- Pierce KL, Premont RT, Lefkowitz RJ. 2002. Seven-transmembrane receptors. Nat Rev Mol Cell Biol 3: 639–650. [DOI] [PubMed] [Google Scholar]
- Plouffe SW, Hong AW, Guan KL. 2015. Disease implications of the Hippo/YAP pathway. Trends Mol Med 21: 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11: 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Wu LW, Grivennikov SI, de Jong PR, Lian I, Yu FX, Wang K, Ho SB, Boland BS, Chang JT, et al. 2015. A gp130–Src–YAP module links inflammation to epithelial regeneration. Nature 519: 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BJ, Pichaud F, Roper K. 2013. Sticking together the Crumbs—an unexpected function for an old friend. Nat Rev Mol Cell Biol 14: 307–314. [DOI] [PubMed] [Google Scholar]
- Varelas X. 2014. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development 141: 1614–1626. [DOI] [PubMed] [Google Scholar]
- Wackerhage H, Del Re DP, Judson RN, Sudol M, Sadoshima J. 2014. The Hippo signal transduction network in skeletal and cardiac muscle. Sci Signal 7: re4. [DOI] [PubMed] [Google Scholar]
- Yin F, Yu J, Zheng Y, Chen Q, Zhang N, Pan D. 2013. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell 154: 1342–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al. 2012. Regulation of the Hippo–YAP pathway by G-protein-coupled receptor signaling. Cell 150: 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, Giovannini M, Liu P, Anders RA, Pan D. 2010. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell 19: 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. 2008. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22: 1962–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. 2010. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(β-TRCP). Genes Dev 24: 72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL. 2011. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev 25: 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.