Abstract
Approximately ten percent of male infertility is caused by non-obstructive azoospermia (NOA), but the etiologies of many NOA remain elusive. Recently, a genome-wide association study (GWAS) of NOA in Han Chinese men was conducted, and only a few genetic variants associated with NOA were found, which might have resulted from genetic heterogeneity. However, those variants that lack genome-wide significance might still be essential for fertility. Functional analysis of genes surrounding these variants in Drosophila identified some spermatogenesis-essential genes. As a complementary method of Drosophila screening, SK1 background Saccharomvces cerevisiae was used as a model to screen meiosis-related genes from the NOA GWAS data in this study. After functional screening, GDA1 (orthologous to humanENTPD6) was found to be a novel meiosis-related gene. The deletion of GDA1 resulted in the failure of yeast sporulation. Further investigations showed that Gda1p was important for pre-meiotic S phase entry. Interestingly, the meiotic role of Gda1p was dependent on its guanosine diphosphatase activity, but not it’s cytoplasmic, transmembrane or stem domains. These yeast data suggest that ENTPD6 may be a novel meiosis-associated NOA-related gene, and the yeast model provides a good approach to analyze GWAS results of NOA.
Infertility, which is a severe threat for the continuation of humans, affects one-sixth of couples worldwide1,2. Approximately half of the infertility cases are attributed to the male2,3. In male infertility, 10–15% of cases are classified as azoospermia, and 60% of these cases are non-obstructive azoospermia (NOA), which generally affects 1% of the male population4,5,6. Recently, some genetic factors, such as single nucleotide polymorphisms (SNPs) and other common structural variants, have been reported to be associated with NOA7,8,9; However, the etiology of most of the NOA remains largely unknown.
The genome-wide association study (GWAS) represents a powerful tool for investigating the genetic architecture of complex human diseases and provides a good approach to study the associations between SNPs and traits such as human diseases10,11. Although GWASs have successfully identified loci that influence a wide variety of human diseases, many of these results are either inconsistent or have failed to be independently validated12,13, possibly due to high genetic heterogeneity. These loci may still indicate genes important for the disease. Systematically functional analyses are expected to help identify genes that are involved in human diseases. Limited by material and financial resources, it is difficult to perform large-scale functional genomic analysis of the pathogenic genes identified by GWASs in mammals. Therefore, an efficient and convenient method is needed to functionally verify the associations of GWAS results with specific diseases in the post-GWAS era.
As a simple single-cell eukaryote, Saccharomvces cerevisiae is widely used as a model organism in biological research and has offered valuable knowledge of the genetics and basic cellular processes that are evolutionarily conserved with higher eukaryotes, such as the cell cycle, DNA replication, recombination, metabolism, aging and meiosis14,15,16,17. Many of these results have been directly extended to mammalian systems, thus providing an important tool in understanding complex human diseases18,19,20. Because of its powerful capacity for genetic manipulation and relative low cost in culturing, yeast has been developed as a very important system for annotating gene function, functional genomics and drug discovery, and it is suitable for uncovering the basic functions of the genes implicated in some human diseases20,21,22,23.
To detect the SNPs associated with NOA, we performed a large-scale genome-wide association study in Han Chinese men, and 103 SNPs were found to be associated with NOA, with p <10−5 24,25,26 in the GWAS scan, but failed in replications. Functional screening of the genes in Drosophila orthologous to those around these SNPs by in vivo RNA interference (RNAi) identified approximately 32% of the analyzed Drosophila genes to be essential for male fertility26. However, because of the lack of chromosome recombination in Drosophila spermatogenesis27, our previous work might have missed some meiosis-related NOA-associated genes. As a classical model for meiotic studies17, functional genomic screening in Saccharomvces cerevisiae provides an efficient and convenient method to identify meiosis-associated genes that might be evolutionarily conserved from yeast to humans. We identified 9 yeast homologs as potential human NOA pathogenic genes by bioinformatics analysis, one of which, MSH5, has been reported to be important for meiosis28,29. After deleting those non-essential genes in SK1 background yeast strains, we found that one gene was required for yeast sporulation. Similar to human NOA, the deletion of GDA1 inhibited gametogenesis. In the GDA1 deletion strain, premeiotic DNA replication was blocked and Sic1p was stabilized, which suggested that Gda1p is primarily required for G1 to pre-meiotic S phase transition. The function of Gda1p in entering the pre-meiotic S phase is dependent on its guanosine diphosphatase activity, but not its glycosylation modification, cytoplasmic, transmembrane or stem domains. Therefore, ENTPD6, the human ortholog of GDA1, may be a NOA pathogenic gene.
Results
Identification of potential non-obstructive azoospermia pathogenic genes by functional screening in yeast
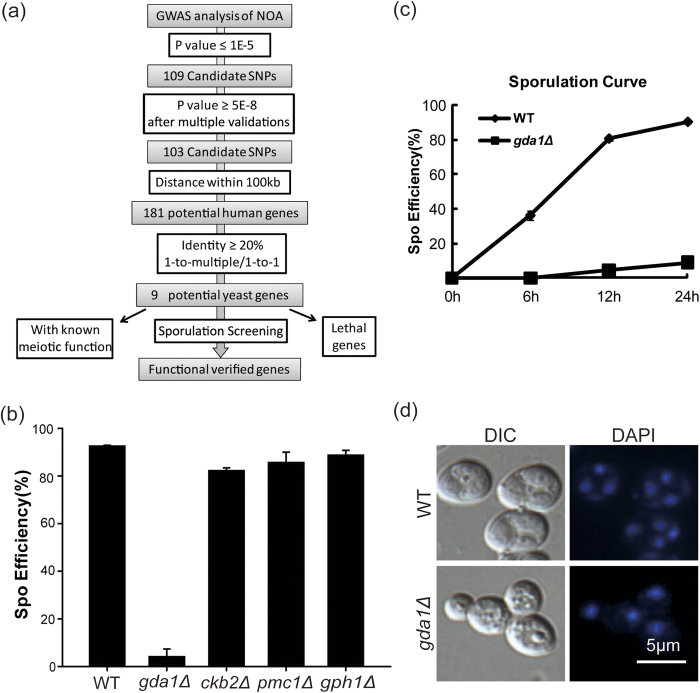
A recent study showed that approximately 32% of GWAS SNPs are located in deoxyribonuclease I hypersensitive sites (DHSs), which are markers of regulatory DNA that can regulate genes within 100 kb30. Thus, we considered genes flanking the tSNPs (Tag Single Nucleotide Polymorphisms) within 100 kb in this study. Our recent work in Drosophila has demonstrated that this approach is effective in identifying genes that are essential for male fertility based on SNPs without genome-wide significant associations with human NOA26. However, Drosophila spermatogenesis does not involve chromosome recombination27, which is an important event for human spermatocyte meiosis. Because yeast is the most powerful model to study meiosis, we used yeast in the present study to screen meiosis-related genes from the NOA GWAS data and used the strategy described in Fig. 1a. In summary, 9 candidate orthologous yeast genes were obtained, corresponding to 11 human genes and 7 susceptible tSNPs (Table S1). Among these, MPP10, RFC5, RPC19 and SLD5 were found to be essential to yeast survival, thus prohibiting our further screening by their deletion. MSH5 was found to be involved in meiosis in yeast28,29. Finally, 4 genes, CKB2, GDA1, GPH1, and PMC1, were selected and underwent functional analysis in the SK1 background yeast strain, which sporulates faster and more synchronously than other strains and is commonly used for the study of sporulation or meiosis31. After deleting these genes by homologous recombination32, wild type (WT) and candidate gene deletion stains were deprived of nitrogen and incubated in sporulation medium for 24 hrs, and the sporulation efficiency was detected by staining with 4’,6-diamidino-2-phenylindole (DAPI). We found that the sporulation efficiency of the gda1Δ strain showed a significant decrease compared with that of the WT strain (Fig. 1b–d), which is similar to some NOAs of humans. ENTPD6 is the orthologous human gene of GDA1, and it is a member of the ENTPD family and localizes in the Golgi apparatus33. Thus, the functional genomic screening of NOA GWAS data in yeast resulted in the identification of ENTPD6 as a potential pathogenic gene of NOA.
Figure 1. Identification of potential non-obstructive azoospermia pathogenic genes by functional genomic screening in yeast.
(a) Flow chart of the screening strategy. The selection criteria for candidate genes in Saccharomvces cerevisiae included a tSNP with an association P-value < 10−5 and a P-value ≥ 5*10−8 after multiple validations, and human genes flanking the tSNPs within 100 kb, homology type (one to many or one to one) and orthology identity >20% were considered. After eliminating the well-studied genes in meiosis and lethal genes, the candidate genes were screened for their sporulation efficiency after deletion. (b) The sporulation efficiency of the yeast in which candidate genes were deleted. Wild type and candidate gene deletion stains were incubated in sporulation medium for 24 hrs. Sporulation efficiency was the percentage of cells induced to sporulate that became dyads and tetrads by staining with DAPI. (c) The gda1Δ strain showed a decrease in sporulation efficiency compared with the WT strain. A sporulation time course indicated the percentage of cells/asci with dyads and tetrads in the gda1Δ and WT strains. Diploid yeast cells were deprived of nutrients, induced to enter sporulation synchronously, and stained with DAPI at different times post-induction. (d) WT and gda1Δ spores were stained with DAPI to show the decrease of sporulation efficiency.
GDA1 is required for pre-meiotic S phase entry
Because the deletion of GDA1 inhibited sporulation, we next determined which phases of sporulation were affected in the GDA1 deleted strain. Meiosis in yeast is initiated by the expression of Ime1p, which serves as the master regulatory switch for meiosis34,35. First, we detected the expression of Ime1p in the GDA1 deletion strain by generating a 3×Myc tag on the C-terminus of IME1. During the sporulation processes, we found that the expression of Ime1p in the GDA1 deletion strain was only delayed by approximately 1 hr compared with the WT strain (Fig. 2a). Real-time PCR analysis of IME1 mRNA in WT and the GDA1 deletion strain showed similar results to the protein level (Fig. 2b). These results suggest that GDA1 is not the major regulator of meiosis initiation, even though it is involved in this process to some extent. We next detected the pre-meiotic DNA synthesis by flow cytometry analysis to test whether the GDA1-null mutant influenced pre-meiotic DNA replication. We found that the pre-meiotic DNA replication was repressed in the GDA1 deletion strain (Fig. 2c).
Figure 2. GDA1 is required for pre-meiotic S phase entry.
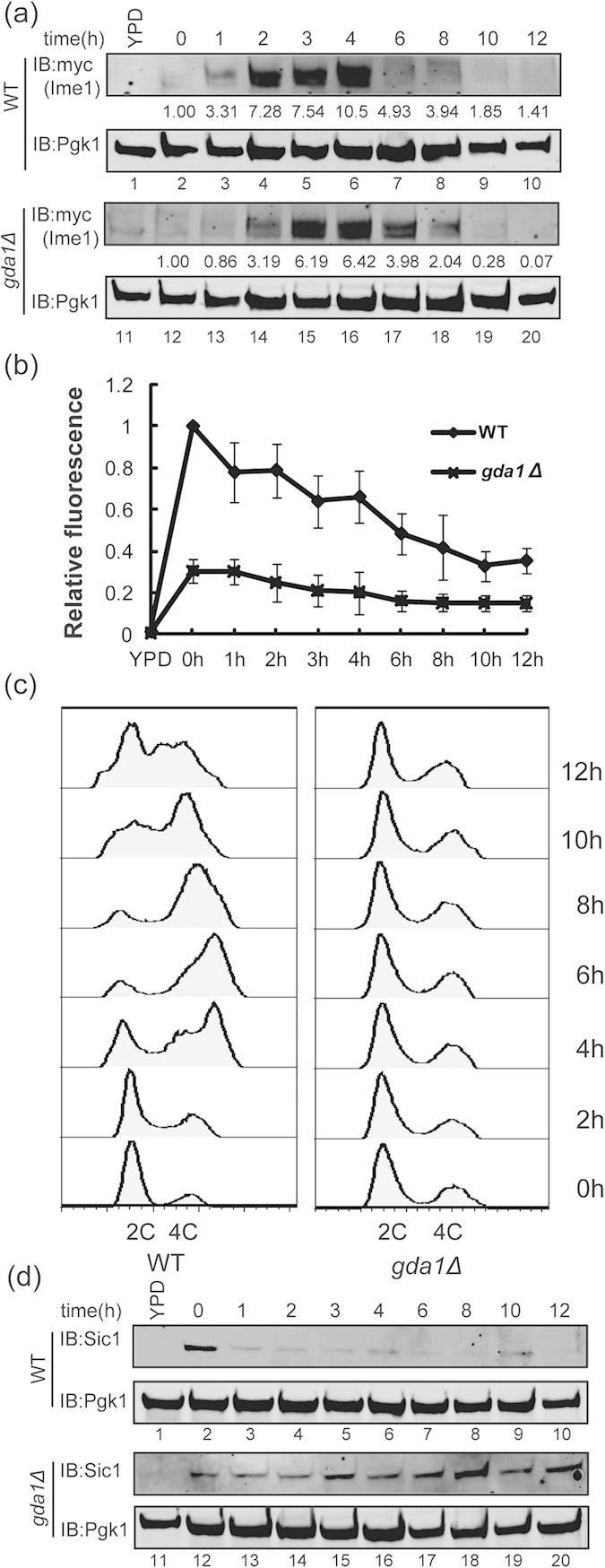
The expression of Ime1p was only slightly delayed in the gda1Δ strain during sporulation. WT or gda1Δ strains expressing the IME1-3×myc allele were incubated in sporulation medium and samples were collected at different times. The expression of Ime1p-3×myc over time was analyzed by immunoblotting with anti-Myc antibody. Pgk1p served as a loading control. Full-length blots/gels are presented in Supplementary Figure 5. (b) Real-time PCR analysis of the IME1 expression level in WT and GDA1 deletion strains. (c) The pre-meiotic DNA replication was inhibited in the gda1Δ strain during sporulation. WT or gda1Δ strains were incubated in sporulation medium and samples were collected at different times after induction. DNA content was analyzed by flow cytometry to detect pre-meiotic DNA replication (2C to 4C transition). (d) The stabilization of Sic1p in the gda1Δ strain during sporulation. WT or gda1Δ strains were incubated in sporulation medium and samples were collected at different times after induction. The expression of Sic1p over time was analyzed by immunoblotting with specific anti-Sic1 antibody. Pgk1p served as a loading control. Full-length blots/gels are presented in Supplementary Figure 5.
The early phase of sporulation begins when cells make the decision to differentiate into spores, they then exit the mitotic cycle in G1 and enter the pre-meiotic S phase17. To determine whether the GDA1-null mutant influenced the transition from G1 to the pre-meiotic S phase, we detected the expression of Sic1p, which acts as a central G1 to pre-meiotic S phase transition regulator by inhibiting the Clb5,-6/Cdk1 activity36,37, presents early in meiosis and subsequently disappears when cells enter the pre-meiotic S phase38,39. Consistent with other reports38,39, Sic1p was detected at 1–2 hrs after the induction of sporulation and then disappeared in the following stages in the WT yeast strain (Fig. 2d, lanes 2–10). However, in the GDA1 deletion strain, Sic1p was detected throughout the process of the induction of sporulation (Fig. 2d, lanes 12–20), which indicated that the GDA1-null mutant influenced the G1 to pre-meiotic S phase transition. Therefore, we concluded that the GDA1-null mutant mainly arrests before the pre-meiotic S phase and GDA1 is required for pre-meiotic S phase entry.
The pre-meiotic S phase entry defect could be rescued by the expression of GDA1 in the GDA1 deletion strain
To further confirm the effect of GDA1 in entering the pre-meiotic S phase, a GDA1 expression vector under the control of its own promoter was generated and transformed into a GDA1 deletion strain. After being incubated in sporulation medium for 24 hrs, we detected the sporulation efficiency of WT vector (containing the empty vector), gda1Δ vector (containing the empty vector) and gda1Δ GDA1 (containing the GDA1 expression vector under the control of its own promoter) strains, and we found that GDA1 expression in the gda1Δ strain could partially rescue its sporulation defect up to more than 60% compared with the gda1Δ vector strain, whose sporulation efficiency remained less than 10% (Fig. 3a). By contrast, the gda1Δ GDA1 strain could produce spores (Fig. 3b) and complete pre-meiotic DNA replication (Fig. 3c); however, the gda1Δ vector strain failed to do so. We then detected the Sic1p expression pattern during sporulation processes in these three strains and found that the Sic1p expression in the gda1Δ GDA1 strain was similar to that of the WT vector strain (Fig. 3d). These results suggested that the defect of sporulation could be rescued by the expression of GDA1 in its deletion strain, and GDA1 indeed played very important roles in entering the pre-meiotic S phase.
Figure 3. The pre-meiotic S phase entry defect could be rescued by GDA1 in the gda1Δ strain.
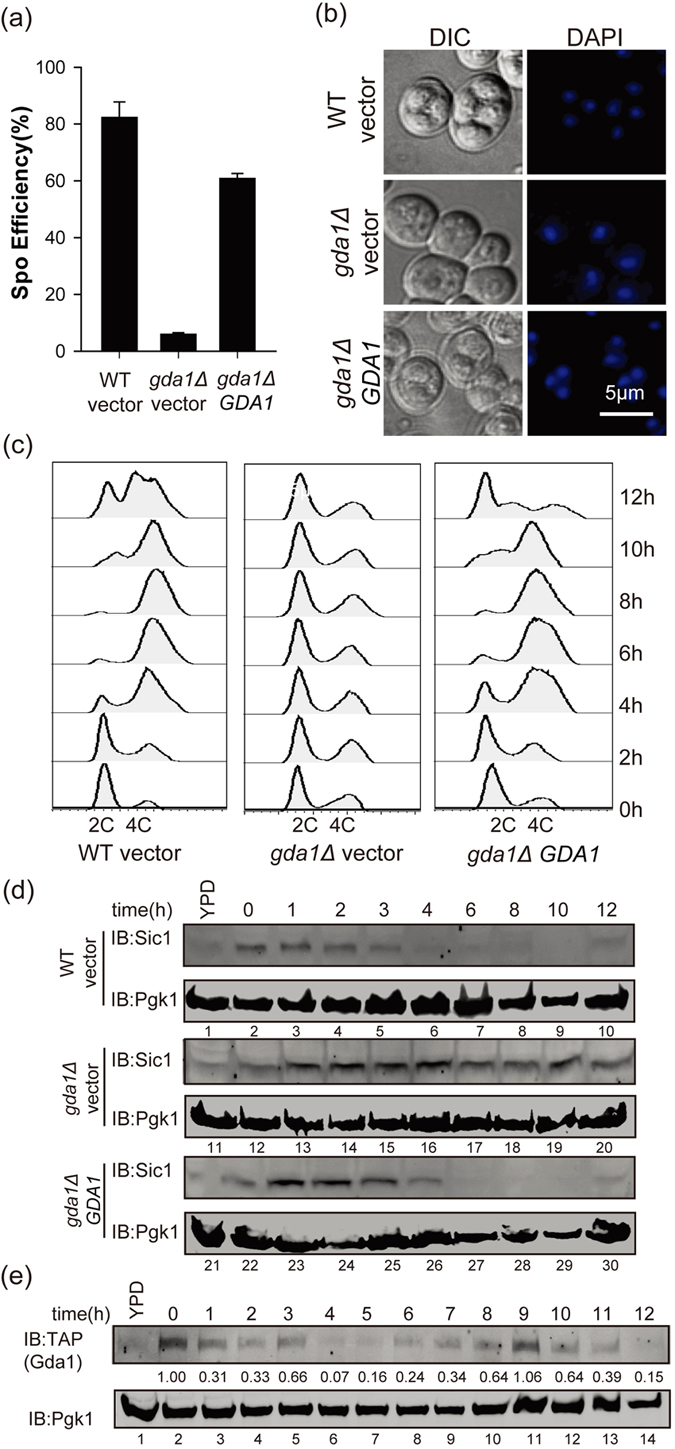
(a) The expression of GDA1 could rescue the sporulation process in the gda1Δ strain. The WT strain harbored the empty vector, and the gda1Δ strains harbored either the empty vector or GDA1 under the control of its own promoter. The strains were incubated in sporulation medium for 24 hrs. Sporulation efficiency was assessed by staining with DAPI. (b) Microscopic observation of the WT strain harboring the empty vector and the gda1Δ strains harboring either the empty vector or GDA1 under the control of its own promoter after sporulation induction for 24 hrs. (c) The expression of GDA1 could rescue the pre-meiotic DNA replication defect of the gda1Δ strain. The WT strain harbored the empty vector, and the gda1Δ strains harbored either the empty vector or GDA1 under the control of its own promoter. The strains were incubated in sporulation medium, and samples were collected at different times after induction. DNA content was analyzed by flow cytometry to detect pre-meiotic DNA replication (2C to 4C transition). (d) The expression of of GDA1 could rescue the stabilization of Sic1p in the gda1Δ strain during sporulation. The WT strain harbored empty vector, and the gda1Δ strains harbored either empty vector or GDA1 under the control of its own promoter. The strains were incubated in sporulation medium, and samples were collected at different times after induction. The expression of Sic1p over time was analyzed by immunoblotting with specific anti-Sic1 antibody. Pgk1p served as a loading control. Full-length blots/gels are presented in Supplementary Figure 5. (e) The expression of Gda1p during sporulation. The WT strain expressing the GDA1-TAP allele was incubated in sporulation medium, and samples were collected at different times after sporulation induction. The expression of Gda1-TAP over time was analyzed by immunoblotting with anti-TAP antibody. Pgk1p served as a loading control. Full-length blots/gels are presented in Supplementary Figure 5.
We then detected the expression of Gda1p during the process of sporulation by generating a TAP tag on its C-terminus, and we found that Gda1p accumulated in the early phase of sporulation and subsequently decreased in the middle phases at approximately 4–6 hrs (Fig. 3d), which is consistent with its function in the meiotic early phase. Gda1p could also be detected in the late phase of sporulation (Fig. 3e), which may be related to its role in spore wall biogenesis40.
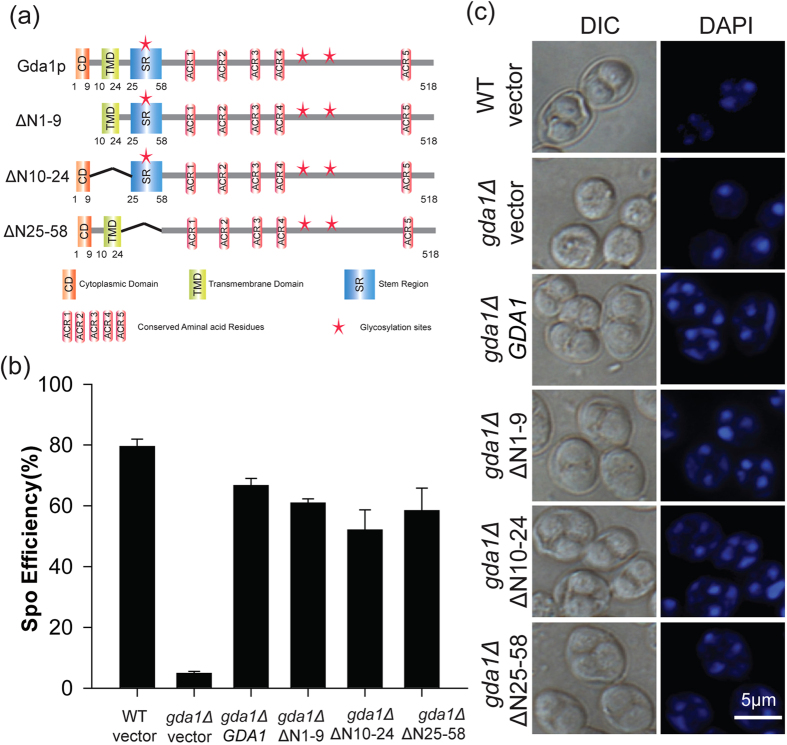
The cytoplasmic, transmembrane and stem domains of Gda1p are not necessary for its role in meiosis
Gda1p is a guanosine diphosphatase that is located in the Golgi, and it is involved in the transport of GDP-mannose into the Golgi lumen by converting guanosine diphosphate (GDP) to guanosine monophosphate (GMP) after mannose is transferred to its substrate. The GDA1 deletion strain has severe defects in the N- and O-mannosylation of proteins and glycosphingolipids41,42. Gda1p has a short cytoplasmic domain in its N-terminus. The next domain is a single transmembrane domain, followed by a stem region and a large catalytic luminal domain (Fig. 4a). To further study the functional role of Gda1p in sporulation, we generated three truncations: ΔN1-9 (Gda1 Δ1–9aa, in which the cytoplasmic domain is deleted), ΔN10–24 (Gda1 Δ10–24aa, in which the transmembrane domain is deleted) and ΔN25–58 (Gda1 Δ25–58aa, in which the stem region is deleted) (Fig. 4a). These were transformed into the gda1Δ strain. We found that none of the mutants affected the sporulation efficiency compared with the WT GDA1 (Fig. 4b,c). These results suggested that the function of Gda1p in meiosis is not dependent on these three domains. Additionally, in agreement with our results, it has been reported that the cytoplasmic and transmembrane domains are not necessary for the Golgi localization of Gda1p43.
Figure 4. N-terminal domains of Gda1p are not necessary for its role in meiosis.
(a) Schematic representation of the domains of Gda1p and some mutants, including ΔN1–9 (Δ1–9aa), ΔN10–24 (Δ10–24aa), and ΔN25–58 (Δ25–58aa). CD indicates the cytoplasmic domain; TMD indicates the transmembrane domain; SR indicates the stem region; ACR1–5 indicates the conserved amino acids related to guanosine diphosphatase activity; stars indicate the glycosylation sites. (b) The function of GDA1 in meiosis was independent of its localization-terminal domains. The WT strain harboring the empty vector and the gda1Δ strains harboring either the empty vector or GDA1, ΔN1–9, ΔN10–24, ΔN25–58 under the control of its own promoter were incubated in sporulation medium for 24 hrs. Sporulation efficiency was determined by staining with DAPI. (c) Microscopic observation of the WT strain harboring the empty vector and the gda1Δ strains harboring either the empty vector or GDA1 and its variants under the control of its own promoter after sporulation induction for 24 hrs.
Glycosylation of Gda1p is not necessary for sporulation
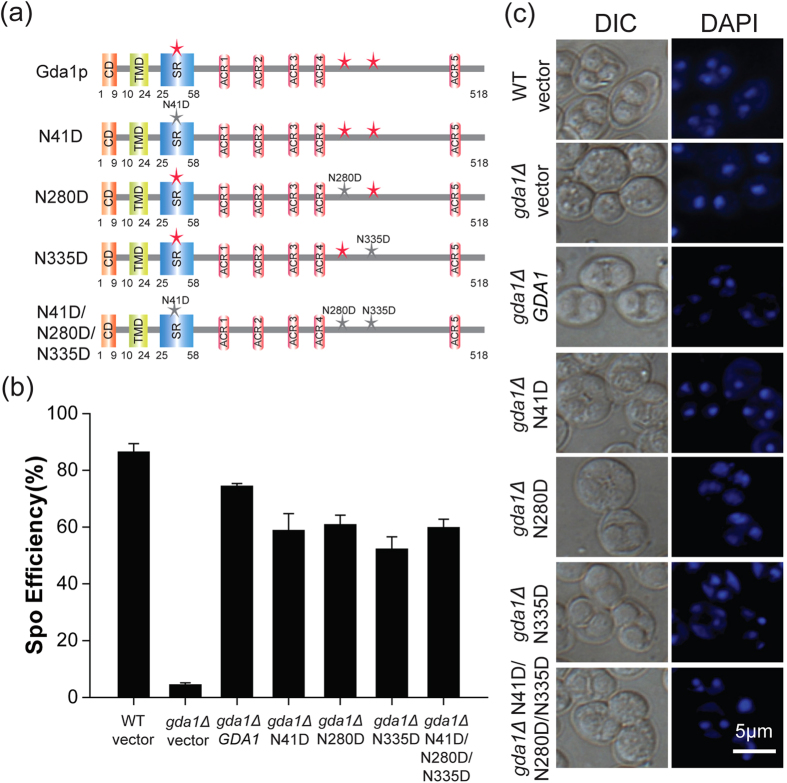
The nucleoside triphosphate diphopshohydrolases (NTPDases) often contain glycosylation sites, and the glycosylation of NTPDase is important for correct protein folding, membrane targeting and activity44. As an NTPDase, Gda1p contains three glycosylation sites, which are N41, N280 and N33545. To detect whether the glycosylation of Gda1p is involved in its function in sporulation, we abolished its glycosylation by mutating these glycosylation sites to aspartic acids (N41D, N280D and N335D), either separately or together (N41D/N280D/N335D) (Fig. 5a), and transformed these four mutants into the gda1Δ strain. By detecting the sporulation efficiency, we found that these mutants did not affect yeast sporulation compared with the control groups (Fig. 5b,c), which indicated that the glycosylation modification of Gda1p is not necessary for its role in meiosis.
Figure 5. Glycosylation modification of Gda1p is not necessary for yeast sporulation.
(a) Schematic representation of the mutant glycosylation sites, which included GDA1 (Gda1p 1–518aa), N41D (Gda1p N41D), N280D (Gda1p N280D), N335D (Gda1p N335D), and N41D/N280D/N335D (Gda1p. N41D/N280D/N335D). (b) Glycosylation site mutations of GDA1 were not necessary for yeast sporulation. The WT strain harboring empty vector and the gda1Δ strains harboring the empty vector or GDA1, N41D, N280D, N335D, N41D/N280D/N335D under the control of its own promoter were incubated in sporulation medium for 24 hrs. Sporulation efficiency was determined by staining with DAPI. (c) Microscopic observation of the WT strain harboring empty vector and the gda1Δ strains harboring either the empty vector, WT GDA1 or glycosylation site mutants under the control of their own promoter after sporulation induction for 24 hrs.
The function of Gda1p in entering the pre-meiotic S phase is dependent on its guanosine diphosphatase activity
The NTPDase enzyme activity depends on several strictly conserved motifs, called apyrase conserved regions (ACRs), and some key residues were found to be essential to the activity of these NTPDases44,46. There are five ACRs in Gda1p, called ACR1–5, and these ACRs are distributed in the lumenal domain of Gda1p44. To identify whether the guanosine diphosphatase activity of Gda1p is necessary for its function in sporulation, we selected and mutated R176, E216 and D245/G247 to abolish its enzyme activity, which were named as R176A, E216D and D245A/G247A (Fig. 6a). The expression of these mutants was superior to that of the WT proteins (Fig. S1a), which might have been due to the complementary demand of the activity of this enzyme. After transforming these mutants into the gda1Δ strain, we detected the sporulation efficiency and pre-meiotic DNA synthesis. We found that these three mutant strains failed to exhibit pre-meiotic DNA replication and sporulation, similar to the gda1Δ strain (Fig. 6b,c, Fig. S1b). We then detected Sic1p expression in these strains, and similar to the gda1Δ strain, we found that the Sic1p was stabilized throughout the sporulation process in the enzyme activity-abolished strains (gda1ΔR176A, gda1ΔE216D and gda1ΔD245A/G247A) (Fig. 6d). Therefore, we concluded that the function of Gda1p in entering the pre-meiotic S phase is dependent on its guanosine diphosphatase activity.
Figure 6. The function of Gda1p in entering the pre-meiotic S phase is dependent on its guanosine diphosphatase activity.
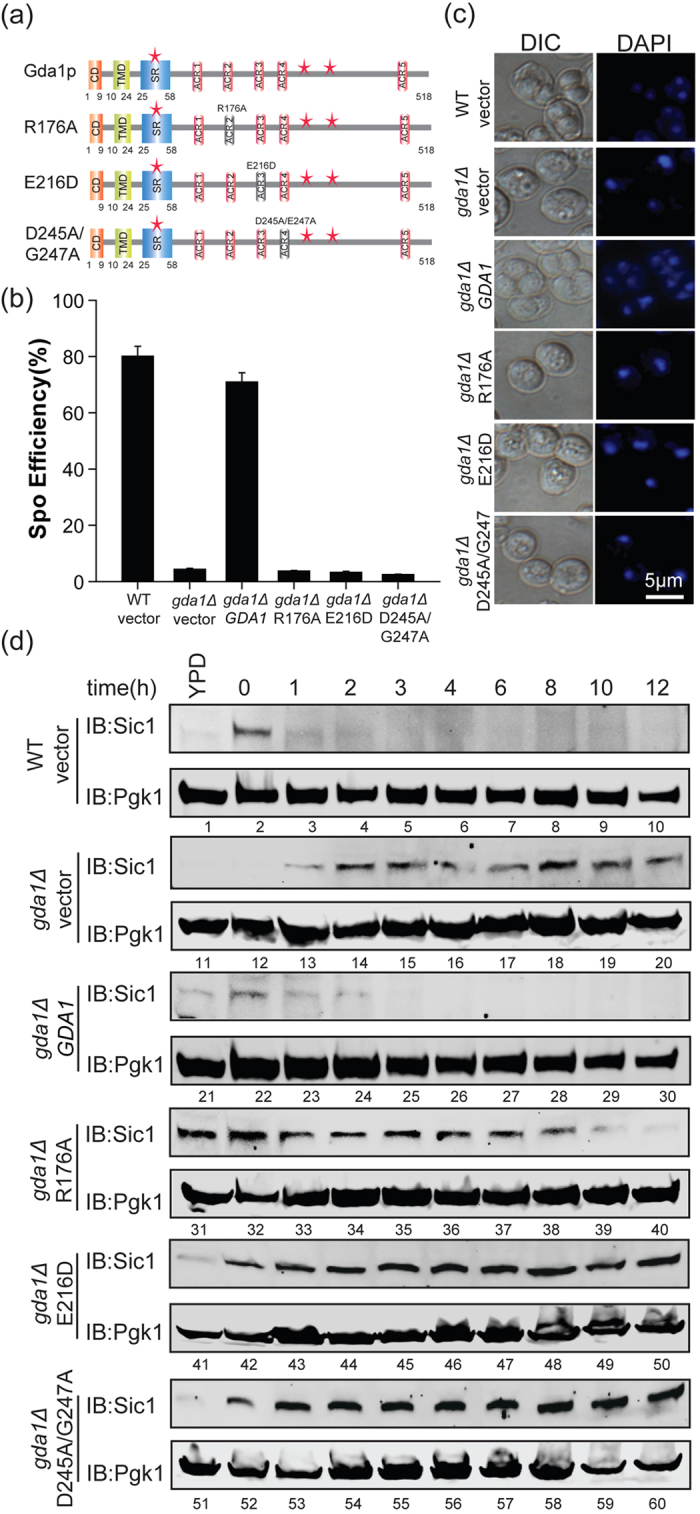
(a) Schematic representation of the key guanosine diphosphatase activity mutants, which include GDA1 (Gda1p 1–518aa), R176A (Gda1p R176A), E216D (Gda1p E216D), and D245A/G247A (Gda1p D245A/G247A). (b) The disruption of Gda1p guanosine diphosphatase activity induced a decrease in sporulation efficiency compared with the WT strain. The WT strain harboring the empty vector and the gda1Δ strains harboring either the empty vector or GDA1, R176A, E216D, D245A/G247A under the control of its own promoter were incubated in sporulation medium for 24 hrs. Sporulation efficiency was determined by staining with DAPI. (c) Microscopic observation of the WT strain harboring empty vector and the gda1Δ strains harboring either the empty vector, WT GDA1 or key guanosine diphosphatase activity mutants under the control of their own promoter after sporulation induction for 24 hrs. (d) The disruption of GDA1 guanosine diphosphatase activity stabilized Sic1p during sporulation. The WT strain harboring empty vector and the gda1Δ strains harboring either empty vector or GDA1, R176A, E216D, D245A/G247A mutants under the control of their own promoter were incubated in sporulation medium and samples were collected at different times after induction. The expression of Sic1p over time was analyzed by immunoblotting with specific anti-Sic1 antibodies. Pgk1p served as a loading control. Full-length blots/gels are presented in Supplementary Figure 5.
Discussion
To date, hundreds of tag single nucleotide polymorphisms (tSNPs) have been found to be associated with human diseases by genome-wide association studies10, and GWAS provides a good approach to study the associations between tSNPs and traits such as major diseases10,11. However, the results of many GWASs are either inconsistent or have failed to be independently replicated due to the high genetic heterogeneity of the population. Nevertheless, the identified tSNPs may still indicate important genes for the diseases, but most of them still lack systematically functional studies12,13. The yeast model has been widely used in large-scale functional genomic screenings to reveal the functions of some genes implicated in many human diseases15,18,20,21,22. Therefore, for NOA, functional genomic screening in yeast may be an efficient and convenient approach to identify genes that are essential for gametogenesis using NOA GWAS results, especially for meiosis, defects in which are important causes of NOA.
Using a strategy to identify meiosis-essential genes in yeast based on SNPs associated with human NOA without genome-wide significance (Fig. 1a), we identified 9 homologs of human genes that were indicated by NOA SNPs in yeast. After eliminating the lethal genes, we found that 1 (25%) of the 4 were required for sporulation, which indicated the usefulness of NOA GWAS data to find meiosis-essential genes. It has been found that ~340 genes (~6% of the yeast genome) are required for sporulation47, so our hit rate was higher than that obtained in simple large-scale functional genomic analysis.
In this study, we found that the deletion of GDA1 resulted in the abolishment of the sporulation process in the SK1 background yeast strain. The GDA1-null mutant failed to produce gametes, which is similar to the phenotype of human NOA. Gda1p is a guanosine diphosphatase that is involved in the transport of GDP-mannose into the Golgi lumen by converting GDP to GMP after mannose is transferred to its substrate41,42. To test whether the products of Gda1p were essential for sporulation, we directly added the product analog of Gda1p, Guanosine 5’-monophosphate disodium salt hydrate (GMP-Na2) or Uridine 5’-monophosphosphate disodium salt (UMP-Na2), into the YPA medium and sporulation medium. After sporulation, we found that the addition of GMP-Na2 and UMP-Na2 could not rescue the sporulation defect of the GDA1 deletion strain (Fig. S2). KRE2 and its paralog KTR6 have been reported to be Golgi α1, 2- mannosyltransferases, and they can add the second and third mannose on O-linked glycans and release Gda1p’s substrate GDP from GDP- mannose in the Golgi48,49,50. Therefore, considering the concentration of GDP in the Golgi, KRE2 and its paralog KTR6 should be upstream of GDA1. Unexpectedly, both the KRE2-null mutant and the KRE2/KTR6 double deletion mutant sporulated as efficiently as the control strain (Fig. S3). All of these results suggested that the nucleoside metabolism-related function of Gda1p may not be essential for sporulation.
It was reported that Gda1p also participates in the N- and O-mannosylation of proteins and glycosphingolipids41,42. We found that the guanosine diphosphatase activity of Gda1p was essential to its role in the entry into the pre-meiotic S phase by affecting the stability of Sic1p (Fig. 6). Gda1p may affect Sic1p stability in the following ways: 1) Gda1p may directly regulate the glycosylation of Sic1p to promote its degradation, or 2) Gda1p may influence the glycosylation of some other proteins, which are essential for pre-meiotic S phase entry. As far as we know, there is no report on the glycosylation of Sci1p. Thus, we further considered the second indirect possibility; it was reported that the G1-cyclin/Cdk1 complexes catalyzed the phosphorylation of Sic1p during the mitotic G1/S transition, therefore the phosphorylated Sic1p could be recognized and further ubiquitinated by the ubiquitin ligase SCFCdc4 to promote its degradation51,52. Any molecule related to the above-mentioned process might be targeted by Gda1p to influence the stability of Sicp1. Consistent with this hypothesis, it was reported that Skp1p could be glycosylated to modulate the E3 activity of SCFCdc4 in Dictyostelium53. Therefore, Gda1p may influence the glycosylation of either Skp1p or any other unknown proteins to promote Sic1p destruction during pre-meiotic S phase entry.
ENTPD6 is the human ortholog of GDA1; ENTPD6 is a member of the ENTPD family, which shows UDPase activity and localizes in the Golgi apparatus33, and is highly expressed in the testis54. It has been reported that protein glycosylation plays a very important role during mammalian reproduction55. Therefore, ENTPD6 may be a NOA pathogenic gene and might be involved in meiosis. The mechanistic study and functional analysis of GDA1 in yeast have provided important clues for further exploring the role of ENTPD6 in human spermatogenesis.
In contrast to our results, it was reported that GDA1 deletion results in the increase of the sporulation efficiency in the S288C background yeast strain56. To test their results, we created a GDA1 deletion strain in the BY4743 background and tested its sporulation efficiency (Fig. S4). We found that the GDA1-null mutant showed an increase in sporulation efficiency compared with the WT strain, which was entirely different from the SK1 background strain used in this study. This result suggests that there might have been a redundancy of the function of Gda1p in the S288C background. This result is very similar to the pathology of some human diseases and hints that ENTPD6 mutations may result in NOA in some people but not in other populations. The detailed mechanism underlying this phenomenon needs to be further investigated in future work.
Materials and Methods
Screening of candidate functional yeast gene orthologs based on SNPs associated with human NOA
The tSNPs associated with human NOA (with P values less than 10−5) were extracted from previous GWAS of NOA performed in Han Chinese men24. To obtain candidate fertility-related genes, we considered human genes located within 100 kb upstream or downstream of the SNPs. We then selected the corresponding unique homologous yeast genes (orthologous type: one-to-many or one-to-one) of the candidate human genes for potential targets to be verified. We only considered yeast genes with at least 20% sequence identity with their human orthologs. The genome backgrounds for humans and yeast were GRCh37 and R64-1-1, respectively. SNPs localizations were batch-obtained from the UCSC genome browser (http://genome.ucsc.edu/). Gene descriptions, genome localizations, and orthologous relationships of the human and yeast genes were annotated via BioMart (http://www.biomart.org/).
Antibodies
The Myc and FLAG antibodies were purchased from Abmart (Shanghai, China), the Sic1 antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, United States), and the TAP antibody was purchased from Thermo Scientific (Waltham, MA USA). The Pgk1 polyclonal antibody was generated in rabbits using the corresponding recombinant proteins as antigens.
Strains and Plasmids
All plasmids and yeast strains used in this study are described in Tables S2 and S3.
Growth and sporulation
Cells were grown in YPD medium (1% yeast extract, 2% peptone, 2% glucose), or YPA medium (1% yeast extract, 2% peptone, 2% potassium acetate). The sporulation was performed as previously described57. Cells were grown overnight in liquid YPD medium and diluted in liquid YPA medium to an OD600 of 0.3 and cultured for 10 hr at 30 °C. Cells were harvested and resuspended in sporulation medium (2% potassium acetate) to OD600 of 1.9 and sporulated at 30 °C for different lengths of time. In the Gda1p product addition experiments, various concentrations of guanosine 5’-monophosphate disodium salt hydrate (CAS:5550-12-9) or Uridine 5’-monophosphosphate disodium salt (CAS:3387-36-8) were added into the YPA medium and sporulation medium to test their effects on sporulation.
DAPI staining
Sporulation was assayed by the microscopic examination of cultures that had been incubated in SPM (sporulation medium) for 24 hrs. Approximately two hundred cells per culture were counted and the percentage of cells that had formed Asci was scored. The nuclear DNA was stained by 4’,6-diamidino-2-phenylindole (DAPI) as previously described58. The nuclei were visualized and counted using a Nikon Eclipse Ti microscope.
Yeast whole-cell extract preparation and immunoblotting analysis
The yeast whole-cell extract preparation was performed as previously described59. The samples were collected and resuspended in 30 μl distilled water, and an equal volume of 0.2 M NaOH was added. Next, cells were collected after 10 min of incubation at room temperature, and the supernatant was carefully removed. Approximately 30 μl of SDS-sample buffer (100 mM Tris-HCL, pH6.8, 200 mM DTT, 4% SDS, 0.2% BPB, and 20% glycerol) was added to the pellet, and the cells were resuspended and boiled for 10 min and centrifuged briefly. The extract was loaded onto an SDS-PAGE gel and detected by immunoblotting with primary antibodies. The immunoblotting was performed using a fluorescent dye-labeled secondary antibody (Invitrogen), and the blots were scanned using an Odyssey infrared imager.
Isolation of RNA from yeast
The RNA isolation was performed as previously described60. The samples were collected and resuspended in 400 μl AE buffer (50 mM Na acetate pH 5.3, 10 mM EDTA), and 40 μl 10% SDS was added. The suspension was vortexed for 5 min, and 400 μl fresh phenol was added. The mixture was again vortexed for 5 min and incubated at 65 °C for 4 min; the mixture was rapidly chilled on ice for 5 min and then centrifuged for 2 min at 12000 rpm. The upper aqueous phase was transferred to a fresh tube. Then, phenol and chloroform (1:1) was added and extracted for 5 min at room temperature. After being centrifuged for 5 min at 12000 rpm, the upper aqueous phase was again transferred to a fresh tube, and 40 μl 3 M Na acetate and 2.5 volumes ethanol were added to precipitate RNA. After washing with 80% ethanol, the pellet was dried for 5 min, resuspended in 20 μl DEPC-treated water and stored at −80 °C.
Real-time PCR for IME1 mRNA
Real-time PCR was carried out with the Roche Light Cycler® 480II System. cDNA was synthesized by the PrimeScriptTM RT Reagent Kit (TaKaRa, RR037A), A 10 μL volume of the system with 5 μL of 2×EvaGreen mix (Applied Biological Materials Inc., MasterMix-S), 0.8 μL of each primer (10 nmol/L), 2 μL of sample cDNA, and 2.2 μL of ddH2O was prepared for amplification. Primer sets for IME1 (5’-GACACAACCACCGATCAAGAAG-3’ and 5’-GATGAGTGGAACGTAGATGCG-3’) and ACT1 (5’-CCTACGTTGGTGATGAAGCT-3’ and 5’-GTCAGTCAAATCTCTACCGG-3’) were used. The real-time PCR was initiated at 95 °C for 10 min, followed by 40 cycles of denaturation for 5 sec at 95 °C, annealing for 30 sec at 60 °C, and elongation for 60 sec at 72 °C. Fluorescence signals were collected at 72 °C during the elongation step. Each DNA template was performed in triplicate. The results were analyzed using the LightCycle480 SW 1.5.1.
FACS analysis of DNA replication
One-milliliter samples from meiotic cultures were pelleted and resuspended in 70% ethanol. Samples were stored at −20 °C until FACS analysis. Before FACS analysis, cells were washed with 50 mM sodium citrate and resuspended in 0.5 ml of 50 mM sodium citrate containing 0.1 mg/ml RNase A at 30 °C for 2 hrs. Alternatively, 0.5 ml of 50 mM sodium citrate containing 2 μM Sytox Green (for final concentration 1 μM) was added. Samples were briefly sonicated and analyzed on a Becton-Dickinson FACScan analyzer.
Additional Information
How to cite this article: Wang, Q. et al. Yeast model identifies ENTPD6 as a potential non-obstructive azoospermia pathogenic gene. Sci. Rep. 5, 11762; doi: 10.1038/srep11762 (2015).
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 31401159, 31171374, 81222006), the National Basic Research Program of China (Grant No. 2011CB944303, 2012CB944404, 2013CB911400), the One Hundred Talents Program of the Chinese Academy of Sciences.
Footnotes
Author Contributions Q.W. and C.L. performed most of the experiments and helped to write the manuscript. C.T., H.G., Y.J. and L.W. performed the yeast knocked out experiments. H.Z. and Y.S. performed the flow cytometry analysis. Y.W., Y.L., T.Z., W.D. and Z.Z. designed and performed some experiments. W.L., J.S., X.G. and Z.H. designed the experiments and wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
References
- Maduro M. R. & Lamb D. J. Understanding new genetics of male infertility. J Urol 168, 2197–205 (2002). [DOI] [PubMed] [Google Scholar]
- Hirsh A. Male subfertility. BMJ 327, 669–72 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarow J. P. et al. Best practice policies for male infertility. J Urol 167, 2138–44 (2002). [PubMed] [Google Scholar]
- Jarow J. P., Espeland M. A. & Lipshultz L. I. Evaluation of the azoospermic patient. J Urol 142, 62–5 (1989). [DOI] [PubMed] [Google Scholar]
- Jarvi K. et al. CUA Guideline: The workup of azoospermic males. Can Urol Assoc J 4, 163–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosnitzer M., Goldstein M. & Hardy M. P. Review of Azoospermia. Spermatogenesis 4, e28218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H. et al. Genome-wide expression of azoospermia testes demonstrates a specific profile and implicates ART3 in genetic susceptibility. PLoS Genet 4, e26 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston K. I. & Carrell D. T. Genome-wide study of single-nucleotide polymorphisms associated with azoospermia and severe oligozoospermia. J Androl 30, 711–25 (2009). [DOI] [PubMed] [Google Scholar]
- Navarro-Costa P., Goncalves J. & Plancha C. E. The AZFc region of the Y chromosome: at the crossroads between genetic diversity and male infertility. Hum Reprod Update 16, 525–42 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D. & O’Donnell C. J. An open access database of genome-wide association results. BMC Med Genet 10, 6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J. & Singleton A. Genomewide association studies and human disease. N Engl J Med 360, 1759–68 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sijde M. R., Ng A. & Fu J. Systems genetics: From GWAS to disease pathways. Biochim Biophys Acta 1842, 1903–1909 (2014). [DOI] [PubMed] [Google Scholar]
- Chen D. & Gyllensten U. Lessons and implications from association studies and post-GWAS analyses of cervical cancer. Trends Genet 31, 41–54. (2014). [DOI] [PubMed] [Google Scholar]
- Kaeberlein M. Lessons on longevity from budding yeast. Nature 464, 513–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., Chervitz S. A. & Cherry J. M. Yeast as a model organism. Science 277, 1259–60 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guide to yeast genetics and molecular biology. Methods Enzymol 194, 1–863 (1991). [PubMed] [Google Scholar]
- Neiman A. M. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 189, 737–65 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Nobel Lecture. Yeast and cancer. Biosci Rep 22, 373–94 (2002). [DOI] [PubMed] [Google Scholar]
- Mirzaei H., Suarez J. A. & Longo V. D. Protein and amino acid restriction, aging and disease: from yeast to humans. Trends Endocrinol Metab 25, 558–566 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. G. & Snyder M. Yeast as a model for human disease. Curr Protoc Hum Genet Chapter 15, Unit 15 6 (2006). [DOI] [PubMed] [Google Scholar]
- Styles E., Youn J. Y., Mattiazzi Usaj M. & Andrews B. Functional genomics in the study of yeast cell polarity: moving in the right direction. Philos Trans R Soc Lond B Biol Sci 368, 20130118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff D. F. et al. Yeast reveal a “druggable” Rsp5/Nedd4 network that ameliorates alpha-synuclein toxicity in neurons. Science 342, 979–83 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiazzi M., Petrovic U. & Krizaj I. Yeast as a model eukaryote in toxinology: a functional genomics approach to studying the molecular basis of action of pharmacologically active molecules. Toxicon 60, 558–71 (2012). [DOI] [PubMed] [Google Scholar]
- Hu Z. et al. A genome-wide association study in Chinese men identifies three risk loci for non-obstructive azoospermia. Nat Genet 44, 183–6 (2012). [DOI] [PubMed] [Google Scholar]
- Hu Z. et al. Association analysis identifies new risk loci for non-obstructive azoospermia in Chinese men. Nat Commun 5, 3857 (2014). [DOI] [PubMed] [Google Scholar]
- Yu J. et al. Identification of seven genes essential for male fertility through a genome-wide association study of non-obstructive azoospermia and RNA interference-mediated large-scale functional screening in Drosophila. Hum Mol Genet 24, 1493–503 (2014). [DOI] [PubMed] [Google Scholar]
- McKee B. D., Yan R. & Tsai J. H. Meiosis in male Drosophila. Spermatogenesis 2, 167–184 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N. M., Ponte L. & Halsey C. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev 9, 1728–39 (1995). [DOI] [PubMed] [Google Scholar]
- Pochart P., Woltering D. & Hollingsworth N. M. Conserved properties between functionally distinct MutS homologs in yeast. J Biol Chem 272, 30345–9 (1997). [DOI] [PubMed] [Google Scholar]
- Maurano M. T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane S. M. & Roth R. Carbohydrate metabolism during ascospore development in yeast. J Bacteriol 118, 8–14 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol 194, 3–21 (1991). [DOI] [PubMed] [Google Scholar]
- Wang T. F. & Guidotti G. Golgi localization and functional expression of human uridine diphosphatase. J Biol Chem 273, 11392–9 (1998). [DOI] [PubMed] [Google Scholar]
- Kassir Y., Granot D. & Simchen G. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell 52, 853–62 (1988). [DOI] [PubMed] [Google Scholar]
- Vershon A. K. & Pierce M. Transcriptional regulation of meiosis in yeast. Curr Opin Cell Biol 12, 334–9 (2000). [DOI] [PubMed] [Google Scholar]
- Barberis M. Sic1 as a timer of Clb cyclin waves in the yeast cell cycle—design principle of not just an inhibitor. FEBS J 279, 3386–410 (2012). [DOI] [PubMed] [Google Scholar]
- Schwob E., Bohm T., Mendenhall M. D. & Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79, 233–44 (1994). [DOI] [PubMed] [Google Scholar]
- Benjamin K. R., Zhang C., Shokat K. M. & Herskowitz I. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev 17, 1524–39 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirick L., Goetsch L., Ammerer G. & Byers B. Regulation of meiotic S phase by Ime2 and a Clb5,6-associated kinase in Saccharomyces cerevisiae. Science 281, 1854–7 (1998). [DOI] [PubMed] [Google Scholar]
- Gonzalez M. et al. A screen for deficiencies in GPI-anchorage of wall glycoproteins in yeast. Yeast 27, 583–96 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninsone P., Miret J. J. & Hirschberg C. B. The Golgi guanosine diphosphatase is required for transport of GDP-mannose into the lumen of Saccharomyces cerevisiae Golgi vesicles. J Biol Chem 269, 207–11 (1994). [PubMed] [Google Scholar]
- Lopez-Avalos M. D., Uccelletti D., Abeijon C. & Hirschberg C. B. The UDPase activity of the Kluyveromyces lactis Golgi GDPase has a role in uridine nucleotide sugar transport into Golgi vesicles. Glycobiology 11, 413–22 (2001). [DOI] [PubMed] [Google Scholar]
- Vowels J. J. & Payne G. S. A role for the lumenal domain in Golgi localization of the Saccharomyces cerevisiae guanosine diphosphatase. Mol Biol Cell 9, 1351–65 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles A. F. The GDA1_CD39 superfamily: NTPDases with diverse functions. Purinergic Signal 7, 21–45 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeijon C. et al. Guanosine diphosphatase is required for protein and sphingolipid glycosylation in the Golgi lumen of Saccharomyces cerevisiae. J Cell Biol 122, 307–23 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirley T. L., Crawford P. A. & Smith T. M. The structure of the nucleoside triphosphate diphosphohydrolases (NTPDases) as revealed by mutagenic and computational modeling analyses. Purinergic Signal 2, 379–89 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyenihi A. H. & Saunders W. S. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163, 47–54 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausler A. & Robbins P. W. Glycosylation in Saccharomyces cerevisiae: cloning and characterization of an alpha-1,2-mannosyltransferase structural gene. Glycobiology 2, 77–84 (1992). [DOI] [PubMed] [Google Scholar]
- Abeijon C., Orlean P., Robbins P. W. & Hirschberg C. B. Topography of glycosylation in yeast: characterization of GDPmannose transport and lumenal guanosine diphosphatase activities in Golgi-like vesicles. Proc Natl Acad Sci U S A 86, 6935–9 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier M., Sdicu A. M. & Bussey H. The KTR and MNN1 mannosyltransferase families of Saccharomyces cerevisiae. Biochim Biophys Acta 1426, 323–34 (1999). [DOI] [PubMed] [Google Scholar]
- Verma R. et al. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278, 455–60 (1997). [DOI] [PubMed] [Google Scholar]
- Feldman R. M., Correll C. C., Kaplan K. B. & Deshaies R. J. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91, 221–30 (1997). [DOI] [PubMed] [Google Scholar]
- West C. M. et al. Purification and characterization of an alpha1,2,-L-fucosyltransferase, which modifies the cytosolic protein FP21,from the cytosol of Dictyostelium. J Biol Chem 271, 12024–35 (1996). [DOI] [PubMed] [Google Scholar]
- Tada Y. et al. Ectonucleoside triphosphate diphosphohydrolase 6 expression in testis and testicular cancer and its implication in cisplatin resistance. Oncol Rep 26, 161–7 (2011). [DOI] [PubMed] [Google Scholar]
- Yonezawa N. Posttranslational modifications of zona pellucida proteins. Adv Exp Med Biol 759, 111–40 (2014). [DOI] [PubMed] [Google Scholar]
- Deutschbauer A. M., Williams R. M., Chu A. M. & Davis R. W. Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 99, 15530–5 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile T. M. & Amon A. Meiosis I is established through division-specific translational control of a cyclin. Cell 133, 280–91 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D. & Wittenberg C. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev 12, 2698–710 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y., Asakawa K., Toda T. & Katayama S. A rapid method for protein extraction from fission yeast. Biosci Biotechnol Biochem 70, 1992–4 (2006). [DOI] [PubMed] [Google Scholar]
- Schmitt M. E., Brown T. A. & Trumpower B. L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18, 3091–2 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.