Abstract
Cryptococcus gattii is one of the causative agents of human cryptococcosis. Highly virulent strains of serotype B C. gattii have been studied in detail, but little information is available on the pathogenic properties of serotype C isolates. In this study, we analyzed pathogenic determinants in three serotype C C. gattii isolates (106.97, ATCC 24066 and WM 779). Isolate ATCC 24066 (molecular type VGIII) differed from isolates WM 779 and 106.97 (both VGIV) in capsule dimensions, expression of CAP genes, chitooligomer distribution, and induction of host chitinase activity. Isolate WM 779 was more efficient than the others in producing pigments and all three isolates had distinct patterns of reactivity with antibodies to glucuronoxylomannan. This great phenotypic diversity reflected in differential pathogenicity. VGIV isolates WM 779 and 106.97 were similar in their ability to cause lethality and produced higher pulmonary fungal burden in a murine model of cryptococcosis, while isolate ATCC 24066 (VGIII) was unable to reach the brain and caused reduced lethality in intranasally infected mice. These results demonstrate a high diversity in the pathogenic potential of isolates of C. gattii belonging to the molecular types VGIII and VGIV.
Cryptococcosis is a fungal disease that kills approximately 650,000 individuals every year1. Cryptococcus neoformans and C. gattii, the etiological agents of this mycosis2, form the so-called C. neoformans species complex3. C. neoformans is cosmopolitan and commonly causes disease in immunocompromised individuals4, while C. gattii preferentially affects immunocompetent individuals particularly in tropical and sub-tropical regions5. C. gattii outbreaks have also been reported in the Pacific Northwest Region of the United States and in the Vancouver island4,6,7,8,9.
C. gattii is classified into four different molecular types (VGI, VGII, VGIII and VGIV)10,11 and two serotypes (B and C)12. Due to its detection in the US and Canada outbreaks, VGII is the most well characterized molecular type of C. gattii13,14,15. It has been described that virulent C. gattii VGII outbreak lineages derived from mating events in the Brazilian Amazon rainforest and then dispersed to temperate regions16,17,18. VGI isolates can also cause disease in healthy individuals13, while the VGIII molecular type has been associated with clinical syndromes in immunocompromised patients19. Isolation of VGIV isolates is, in general, less frequent, although this molecular type is considered to be endemic in the sub-Saharan Africa11. Importantly, it has been recently demonstrated that virulence is not specifically associated with a particular major molecular type of C. gattii, but rather with still unknown individual strain attributes20.
C. gattii and C. neoformans produce capsules mainly composed of polysaccharides, which are considered to be essential for cryptococcal pathogenesis21. The most abundant component of the capsule is glucuronoxylomannan (GXM), followed by glucuronoxylomannogalactan (GXMGal) and mannoproteins22. Minor, transitory capsular components have also been identified, including heat-shock proteins23, glucans24 and chitooligomers25. Both major and minor capsular components are believed to directly influence the interaction of cryptococci with the host. Recently, chitooligomers have been connected with the ability of C. neoformans to colonize the brain26.
The clinical relevance of VGII isolates of C. gattii has stimulated a number of studies focusing on how the fungus interacts with host cells27,28. On the other hand, the pathogenic properties of VGIII and VGIV isolates are known to a lesser extent. In this study, we have phenotypically characterized three serotype C isolates of C. gattii of the molecular types VGIII and VGIV. Our results suggest an important phenotypic diversity among these three C. gattii isolates, including differences in pigmentation, capsular serology, gene expression in response to capsule-inducing conditions, chitooligomer production in vivo and induction of chitinase activity in mice. These differences were correlated with the ability of C. gattii to kill infected mice.
Results
Phenotypic analysis
Although a revision of the C. neoformans/C. gattii species complex has recently been proposed29, we adopted in this manuscript the standard C. gattii classification into four molecular types, as the acceptance of the new taxonomic proposal in the Cryptococcus community is still uncertain3.
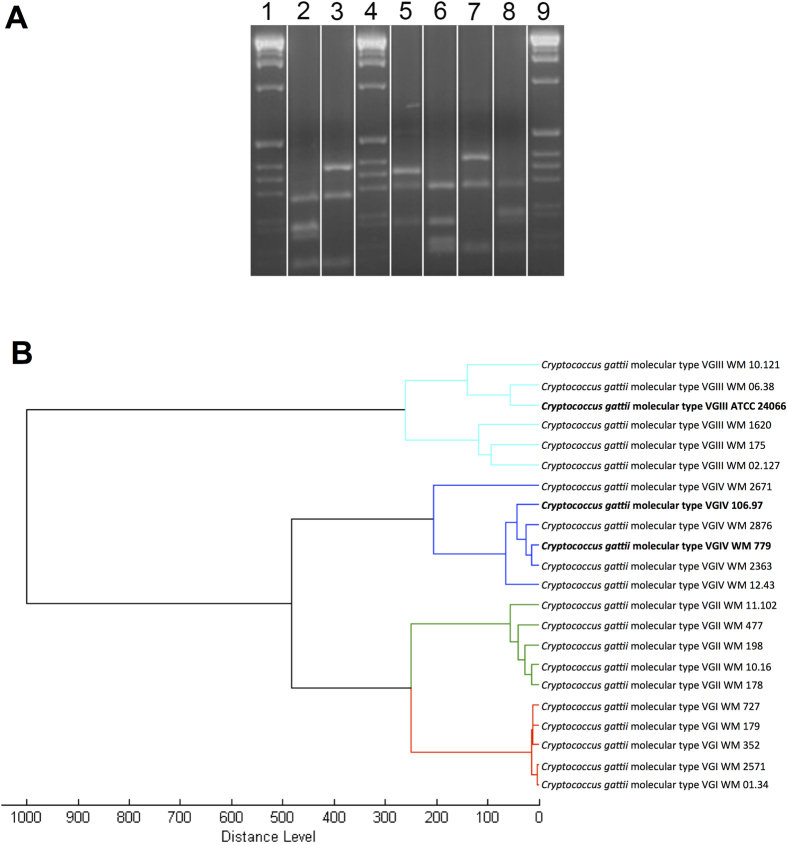
Isolate WM 779 has been characterized before as belonging to the molecular type VGIV30, but the genotypic classification of the isolates 106.97 and ATCC 24066 of C. gattii were not known. That is why, the molecular types of those two isolates were determined via comparative URA5-RFLP analysis with the restriction enzymes HhaI and Sau96I with previously defined molecular types of the reference isolates of the major molecular types of C. gattii (Fig. 1A)31. Isolates ATCC 24066 and 106.97 were classified as molecular types VGIII and VGIV, respectively.
Figure 1. Molecular type and MALDI-TOF analyses of the C. gattii isolates.
A. Molecular type analysis. Determination of the major molecular types via electrophoretic separation of URA5 gene restriction patterns after double digestion with HhaI and Sau96I obtained from isolates 106.97 (VGIV (2)), ATCC 24066 (VGIII (3)), WM179 (VGI reference strain (5)), WM178 (VGII reference strain (6)), WM175 (VGIII reference strain (7)) and WM 779 (VGIV reference strain (8)), 1, 4, 9 = 1 Kb extension ladder (Invitrogen, USA). The gel showed in this panel is representative of different experiments performed under the same experimental conditions. Cropping lines are indicated in white and the unedited gel is available as Supplemental Figure 1. B. MSP dendrograms grouping mass spectra of C. gattii isolates according to their major molecular type. Isolates used in this analysis other than the three currently studied (bold) were obtained from the in-house MALDI_Biotyper BDAL MSP library at Westmead Hospital, Westmead Millennium Culture Collection at Sydney University – Sydney Medical School32.
To verify the molecular type association obtained by URA5-RFLP analysis, Matrix-Assisted Laser Desorption Ionization-Time-of-Flight Mass Spectrometry (MALDI-TOF MS) was used, as it is a simple and reproducible method allowing the separation of the all major molecular types within the C. neoformans/C. gattii species complex29,32,33,34. The MALDI TOF profile of isolate WM 779, the global standard for the VGIV molecular type10, was recently described32,33. The MALDI-TOF MS profiles of isolates ATCC 24066 and 106.97 were generated in the current study. Identification of C. gattii isolates by MALDI-TOF generates consistency score values in the range of 2.3 to 3 for highly probable species identification, 2 to 2.29 for secure genus identification associated with probable species identification, 1.7 to 1.99 for probable genus identification and zero to 1.69 for not reliable identification32. In our study, all isolates were in the range of 2 to 2.29, indicating reliable identification of C. gattii (Table 1). MALDI-TOF spectra confirmed molecular types VGIV and VGIII for isolates 106.97 and ATCC 24066, respectively (Table 1). All studied C. gattii isolates grouped on the basis of MALDI-TOF MS spectra according to molecular types (Fig. 1B).
Table 1. MALDI-TOF MS identification (ID) of the C. gattii isolates analyzed in this study.
| Strain | Organism (best match) | Molecular type | Score valuea |
|---|---|---|---|
| 106.97 | C. gattii | VGIV | 2.276 |
| ATCC 24066 | C. gattii | VGIII | 2.003 |
| WM 779 | C. gattii | VGIV | 2.128 |
aID scores are classified in 2.300 to 3.000 for highly probable species identification; 2.000 to 2.299 for secure genus identification with probable species identification; 1.700 to 1.999 for probable genus identification; and 0.000 to 1.699 for not reliable identification of C. gattii32.
We also analyzed the ability of each isolate to replicate under different conditions and to produce key cryptococcal virulence factors. Growth curves of the three isolates at 30 °C and 37 oC in Sabouraud broth revealed that cells of the isolate 106.97 had slightly faster growth kinetics, although all isolates reached the same cell number after 96 h of cultivation (Fig. 2A). In minimal medium, all isolates had similar growth curves (Fig. 2A). Urease activity was similar in all three isolates (Fig. 2B). Visual analysis of melanin production by the three C. gattii isolates revealed that after 120 h of cultivation in the presence of L-DOPA, the 106.97 and ATCC 24066 isolates had reduced rates of pigmentation at both 30 °C or 37 °C, in comparison with the WM 779 isolate (Fig. 2C). Quantification of pigmentation confirmed the visual analysis of melanin production (Fig. 2D).
Figure 2. General phenotypic analysis of C. gattii.
A. Growth curves of each isolate in Sabouraud broth or in minimal medium at both 30 oC and 37 oC. B. Urease activity from the three serotype C isolates and from S. cerevisiae control strain RSY113. C. Visual analysis of pigmentation after fungal growth in solid media supplemented with L-DOPA in different temperatures. D. Quantitative analysis of pigmentation demonstrated that isolate WM 779 was significantly more efficient in producing pigments than isolates 106.97 and ATCC 24066.
Capsular properties of C. gattii isolates
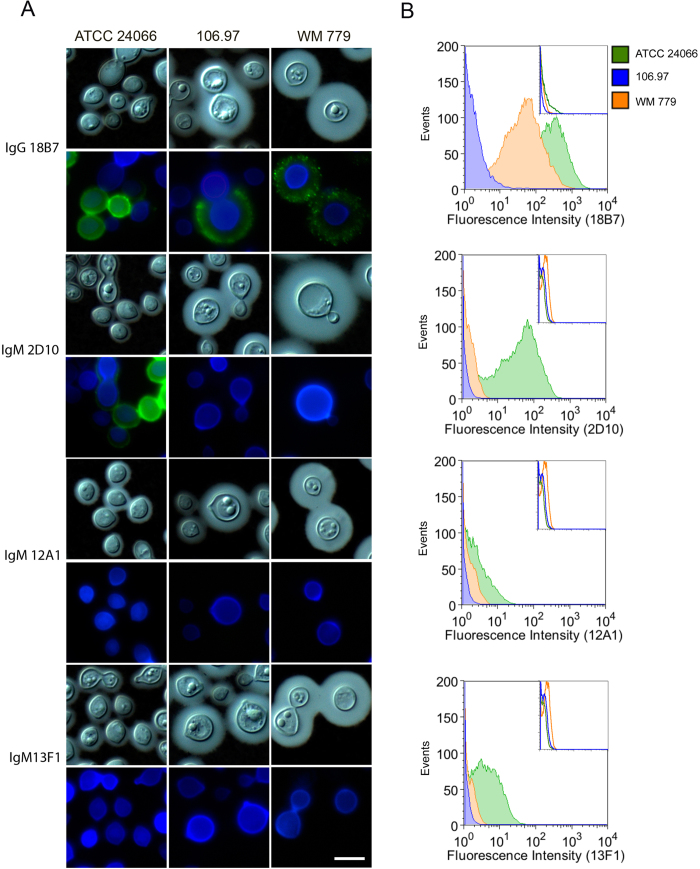
Initially general aspects of morphology and serologic properties of the three C. gattii isolates were characterized (Fig. 3A). All three isolates produced capsule, but ATCC 24066 cells had much smaller capsular dimension. Immunofluorescence tests revealed that isolate 106.97 did not react with any of the antibodies raised against GXM. Assays using mAb 18B7 demonstrated reactivity only with isolates ATCC 24066 and WM 779 resulting annular and punctuate patters, respectively. MAb 2D10 showed annular reactivity with isolate ATCC 24066 and gave negative results for the WM 779 isolate. MAbs 12A1 and 13F1 produced negative serological reactions with all isolates. The results were confirmed in quantitative flow cytometry tests (Fig. 3B), revealing an unexpected diversity in the serological properties of the three C. gattii isolates analyzed here. This led us to investigate the carbohydrate compositions of polysaccharide extracts obtained from each isolate, as well as, to analyze the expression of capsule-related genes.
Figure 3. Reactivity of surface GXM with different mAbs.
A. Microscopic analysis of fungal cells after sequential staining with GXM mAbs (green), calcofluor White (blue) and counter-staining with India ink. Each mAb used for GXM detection is indicated on the left. Scale bar: 5 μm. B. Flow cytometry analysis of the reactivity of GXM-binding mAbs with the serotype C C. gattii isolates. Insets denotes the fluorescence of fungal cells in systems where primary antibodies were not used.
Capsular polysaccharides were extracted with DMSO and the resulting fractions had their glycosidic composition determined by GC-MS (Fig. 4). All isolates had similar xylose : glucuronic acid : mannose ratios, which was in accordance with previous studies determining the composition of capsular extracts from isolates belonging to VGIII and VGIV molecular types (serotype C)35. Other sugar units, however, varied in each isolate. Galactose, which was detected in trace amounts in capsular samples from isolate ATCC 24066, was abundant in fractions obtained from isolates 106.97 and WM 779. Glucose was highly concentrated in the samples obtained from isolates ATCC 24066 and 106.97, but not in the glycan fraction extracted from isolate WM 779. Finally, N-acetylglucosamine, which was absent in ATCC 24066 and WM 779 samples, was present at the trace level in fractions extracted from isolate 106.97.
Figure 4. Glycosyl composition of polysaccharide extracts from the C. gattii isolates analyzed in this study.

Monosaccharides were obtained after methanolysis of polysaccharide extracts for further GC-MS analysis, as detailed in the Methods section.
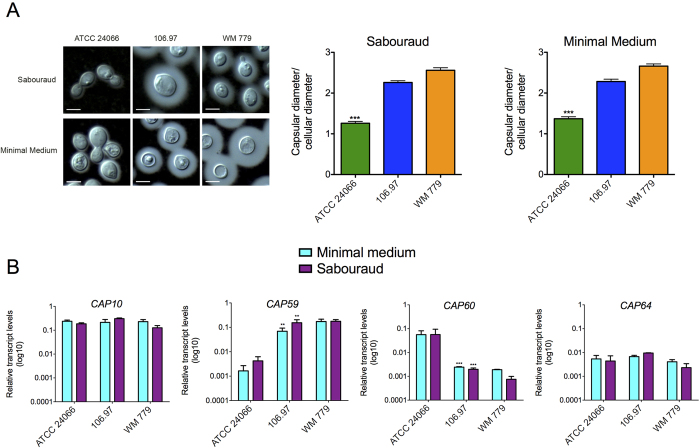
Results obtained from serological, morphological and glycosidic composition analyses clearly indicated a high diversity in capsular structures of the herein analyzed C. gattii isolates. To investigate the possibility of additional diversity at the molecular level, we analyzed the relationship between capsular dimensions and expression of CAP genes in the three isolates after 48 of cultivation in either Sabouraud broth or minimal medium. Cultivation of cryptococci in Sabouraud’s medium is known to repress capsule formation, while large capsules are usually formed by cryptococcal cells in minimal medium36. In comparison with the ATCC 24066 isolate, 106.97 and WM 779 cells showed larger capsular diameters after growth in both Sabouraud and minimal media (Fig. 5A). To further explore these differences, we used q-RT-PCR to determine the relative expression of the CAP family of capsule-related genes (CAP59, CAP10, CAP60 and CAP64; Fig. 5B). CAP64 and CAP10 were similarly expressed in all isolates and conditions. On the other hand, expression of CAP59 was reduced in isolate ATCC 24066, in both Sabouraud and minimal media. The same isolate showed enhanced expression of CAP60 under similar conditions (Fig. 5B).
Figure 5. Capsular dimensions and expression of CAP genes in C. gattii.
A. India ink counter-staining (left panels) and corresponding capsular dimensions (right panels) of C. gattii after growth in Sabouraud or minimal media. Capsular dimensions of the ATCC 24066 isolate were significantly smaller than those observed for 106.97 and WM 779 isolates (***p = <0.0005). Scale bar: 5 μm. B. Expression of CAP genes under the conditions used for determination of capsular dimensions. CAP59 and CAP60 had their expression significantly altered in the ATCC 24066 isolate (***p = <0.0001; **p = <0.005).
The C. gattii isolates differ in chitooligomer detection and induction of host chitinase
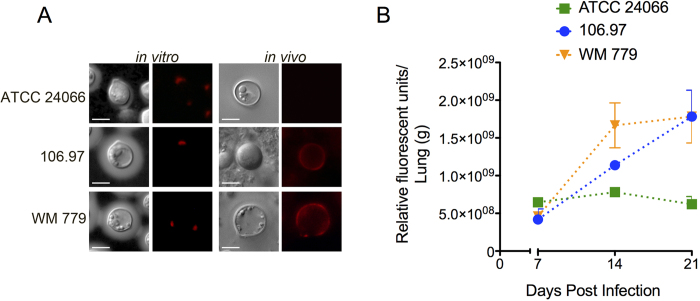
Chitooligomers are readily detectable components of the cell wall-capsule interface in both C. neoformans and C. gattii (molecular type VGII)25. These structures participate in the architecture37 and biological functions38 of capsular components. We therefore evaluated whether chitooligomer detection varied among the three isolates studied here. Microscopic analysis of C. gattii cells grown in vitro revealed that all three isolates had the staining profile that was previously observed in C. gattii serotype B and C. neoformans serotype A isolates25, consisting of polarized regions of the cell wall recognized by WGA (Fig. 6A). Considering that the profile of chitooligomer detection in vivo varies depending on the induction of pulmonary chitinases37, we also tested the distribution of GlcNAc oligomers in these three isolates after infection of mice. In this analysis, 106.97 and WM 779 cells obtained from the lungs of infected animals had an annular pattern of chitooligomer staining. However, similar experiments with the ATCC 24066 isolate gave negative results for chitooligomer staining. We also tested the levels of chitinase activity in the lungs of infected mice (Fig. 6B). C. gattii 106.97 and WM 779 (annular detection of chitooligomers) induced chitinase activity in a time-dependent fashion. In contrast, C. gattii ATCC 24066 isolate (no chitooligomer detection) induced chitinase activity at the lowest levels.
Figure 6. Analysis of C. gattii chitooligomers and host-derived chitinase activity.
A. In vitro and in vivo detection of chitooligomers using TRITC-WGA. The typical polarized pattern of chitooligomer detection was observed in vitro. In vivo analysis revealed annular patterns of oligomer detection in isolates 106.97 and WM 779 and negative staining for the ATCC 24066 isolate. Scale bar, 5 μm. B. Pulmonary chitinase activity after infection with C. gattii. From day 14 to 21 post infection, chitinase activity was significantly higher when mice was infected with isolates 106.97 or WM 779, in comparison to the ATCC 24066 isolate (P < 0.05).
C. gattii isolates of the VGIII and VGIV molecular types have different pathogenic potential
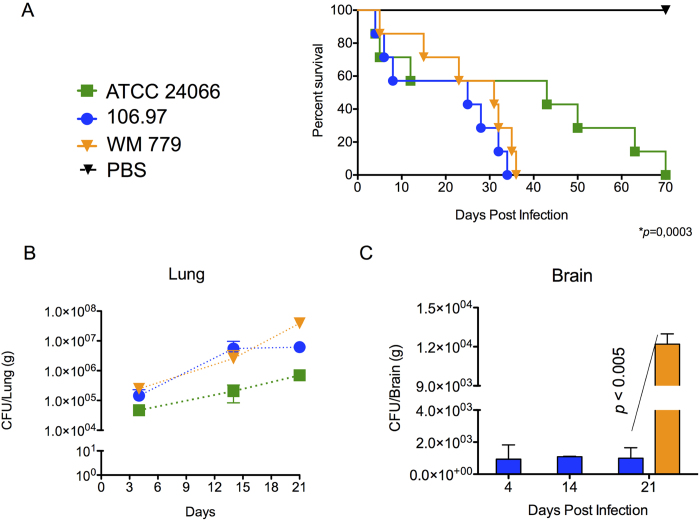
At this point, our results indicated that the C. gattii isolates studied herein differed in key determinants of the pathogenic potential observed within C. gattii. In summary, isolate ATCC 24066 differed from WM 779 and 106.97 cells in molecular type, capsule dimensions, expression of CAP genes, chitooligomer distribution, and induction of host chitinase, but additional diversity among the three isolates was also detected. For instance, isolate WM 779 was more efficient than the others in producing pigments, as already reported20, and all three isolates had distinct patterns of reactivity with antibodies to GXM. To evaluate whether this great diversity was translated into differential pathogenicity, we infected mice with the three isolates separately and compared host survival and fungal burden. Mortality curves (Fig. 7A) revealed that isolate ATCC 24066 was less efficient than the others in killing mice (P = 0.0003). Isolates WM 779 and 106.97 were similar in their ability to cause lethality and, in fact, they produced higher pulmonary fungal burden (Fig. 7B). Isolate ATCC 24066 was unable to reach the brain (data not shown), in contrast with 106.97 and WM 779 isolates (Fig. 7C). The later was only detected in the brain 21 days after infection, which contrasted with the cerebral detection of isolate 106.97 in days 4, 14 and 21 post-infection. However, after three weeks of infection, isolate WM 779 was approximately 100-fold more abundant in the brains of infected mice than 106.97.
Figure 7. Mortality curves.
(A) and fungal burden (B,C) after infection of mice with serotype C isolates of C. gattii. A. Mice infected with the ATCC 24066 isolate lived longer than those infected with isolates 106.97 and WM 779 (P = 0.0003). B. Pulmonary burden after infection with isolate ATCC 24066 was always smaller (P<0.05) in comparison with systems where animals were inoculated with isolates 106.97 or WM 779. C. Brain colonization after infection of mice with isolates 106.96 and WM 779. Isolate WM 779 took longer to infect the brain of mice, but fungal burden 21 days post-infection was significantly higher for this isolate. Negative results were obtained when fungal colonization was analyzed after infection with isolate ATCC 24066. Isolate 106.97, blue bars; isolate WM 779, orange bars.
Discussion
Pathogenic diversity within microbial species has been known for decades39, but the molecular determinants regulating this phenomenon are multiple and, in most species, poorly known. In the C. neoformans/C. gattii complex, pathogenic diversity is important for the outcome of human disease, which has been correlated with cryptococcal-phagocyte interactions and laccase-dependent melanin pathways in vitro40. A recent study using an invertebrate model of infection revealed a formidable pathogenic diversity in 40 isolates of C. gattii, although the molecular determinants associated with this high variability remained undetermined20. The results described in the present study indicated remarkable differences in the pathogenicity of three isolates belonging to the same serotype (C), but to different molecular types (VGIII and VGIV) of C. gattii. The pathogenic determinants linked to these differences were mainly related to the surface architecture of C. gattii and included melanin formation, capsular structure and chitooligomer distribution.
One key virulence factor in C. neoformans and C. gattii is the ability to produce a polysaccharide capsule41. Mutants with defective capsule formation, in general, have avirulent phenotypes21,42,43. In our study, the less virulent isolate (ATCC 24066) manifested the lowest capsular dimensions, independently on the medium used for fungal growth. Accordingly, this phenotype was accompanied by decreased expression of CAP59, a gene that has been predicted to encode a protein required for GXM export44. Unexpectedly, CAP60, which has been described in seminal studies as one of the genes required for capsule formation45, was overexpressed in these cells. These results suggest that, during synthesis of capsular components and assembly of the capsule, the product of CAP59 is required for the functions of CAP60. This hypothesis is further supported by the observation that CAP60 and CAP59, despite their similarity in sequence and chromosome localization, cannot be functionally substituted by each other45.
Antibody reactivity with capsular components is essential for the classification of Cryptococcus species in different serotypes46. Our study demonstrated that serotype C isolates of C. gattii manifest an unexpected variability in their ability to react with GXM antibodies. These results support the notion that serotype classification of cryptococci does not reflect the phenotypic properties of different isolates. The less pathogenic isolate, ATCC 24066, was effectively recognized by two protective monoclonal antibodies, namely 18B7 and 2D1047,48. Isolate WM 779 was only recognized by mAb 18B7 and the 106.97 isolate had no reactivity with any of the antibodies tested here. This serologic variability imply that capsular structures and / or epitope distribution are diverse in at least two molecular types of C. gattii belonging to the same serotype (C) suggesting that the traditional classification of the C. neoformans species complex into four different serotypes may include still unknown capsular, serological determinants. In our study, carbohydrate analysis of capsular extracts revealed similar GXM composition, but the presence of secondary monosaccharides was different in each isolate. Therefore, the possibility that capsular components associate with other surface components resulting in modified serological reactivity cannot be ruled out. This hypothesis is supported by the recent demonstration that glycan association in the surface of C. neoformans resulted in hybrid polysaccharides with diverse immunological functions38.
The presence of fungal cells in brain tissue also varied in a murine model of cryptococcosis after infection with each isolate studied here. Only isolates 106.97 and WM 779, both VGIV isolates, disseminated to the brain, an observation that was linked to their ability to kill mice. A recent study by our group suggested that recognition of chitooligomers by macrophages is involved in dissemination of C. neoformans to the central nervous system26. In the present study, brain infection was in fact associated with increased chitooligomer distribution at the surface of fungal cells in vivo and elevated chitinase activity in the lung of infected mice. Pulmonary chitinase activity was linked with cryptococcal infection49 and increased exposure of surface fungal chitooligomers37. In addition, it has been recently demonstrated that chitinase-mediated chitin recognition induced pathologic responses to cryptococcal infection50. Therefore, we hypothesize that the increased chitooligomer exposure in isolates WM 779 and 106.97 results from their ability to induce pulmonary chitinase, in contrast to isolate ATCC 24066 (VGIII). This enhanced chitooligomer exposure could result in a more effective recognition of the fungus by macrophages, facilitating dissemination26. Finally, in our model, the highest brain fungal burden was observed 21 days after infection of mice with isolate WM 779, the most efficient in the production of melanin, as similarly reported among VGIV isolates20. Pigment synthesis has been linked with neurotropism, which is likely related to the high availability of diphenolic substrates used for pigment synthesis in the brain51. Melanin and fungal virulence are linked in many ways, since this pigment is required for protection of fungal cells against the antimicrobial arsenal produced by the host52,53. Importantly, the hypothetic importance of melanin formation for the virulence of cryptococci during human infection has been recently supported by the observation that laccase-dependent melanin pathways in different isolates are related to human clinical presentation and outcome40.
Variable virulence in C. gattii isolates has been suggested in a number of studies3,16,20,54. In a recent report with 40 globally selected strains including all molecular types, it has been demonstrated that virulence was not specifically associated with a particular molecular type of C. gattii, but rather with still undetermined attributes of each isolate analyzed20. Independently on the niche of isolation, some strains of the molecular types VGI, VGIII and VGIV were shown to be as virulent or even more virulent than the strains of the highly virulent subtype VGIIa, responsible for several fatal cases in the ongoing Vancouver Island outbreak9,55. The VGIV isolate WM 779, the one producing the highest fungal burden in the present study, was recently classified as a highly virulent strain20. In fact, we observed in our study that the two VGIV strains were more virulent than a VGIII isolate. The clinical relevance of this observation is still unknown, but human cases of meningitis caused by serotype C, VGIV, C. gattii have been reported in Mexico56 and India57. The molecular type VGIV also predominates in AIDS patients in Sub-Saharan Africa58.
Our work supports the notion that C. gattii isolates belonging to molecular types VGIII and VGIV are highly diverse in pathogenicity and phenotype. Although the molecular regulators of this potential diversity are still obscure, we propose that differences in melanin production, expression of CAP genes, capsular structure and surface architecture are involved in pathogenic diversity. These results reinforce the notion that even the serotype C VGIII and VGIV C. gattii isolates manifest hypervirulent phenotypes and open new venues for the investigation on how C. gattii cells cause damage to the host.
Methods
Methods described in this section were carried out in accordance with the approved guidelines available in http://www.nature.com/srep/policies/index.html.
Microorganisms and phenotypic characterization
The three C. gattii serotype C isolates used in this study included 106.97 (clinical isolate35), ATCC 24066 (clinical, reference isolate; American Type Culture Collection) and WM 779 (animal isolate, Westmead Millennium Culture Collection, The University of Sydney at Westmead Millennium Institute, Australia). Yeast cells were cultivated for 48 h at 30 °C with shaking in Sabouraud dextrose liquid medium (2% glucose, 1% peptone) and in a chemically defined minimal medium (pH 5.5) containing glucose (15 mM), MgSO4 (10 mM), KH2PO4 (29.4 mM), glycine (13 mM), and thiamine-HCl (3 μM). Growth rates were determined by counting the cell number in a Neubauer chamber in 24 h intervals. Yeast cells were harvested by centrifugation and washed in phosphate-buffered saline (PBS). Molecular typing was performed as described previously31. Analysis of the three isolates by MALDI-TOF MS was performed as recently described32,33. Three combined mass spectra (MSP) per isolate were generated on the MALDI-TOF Biotyper (BRUKER, Germany) using the MALDI Biotyper Automation Control software version 2.0.43.8 (BRUKER, Germany). For all morphological analyses, the cells were fixed with paraformaldehyde 4% for 30 min at room temperature and washed again in PBS, for further counter staining with India ink. Capsule to cell body diameter ratios (n = 100 cells) were determined using the Image J software (http://imagej.nih.gov). For determination of urease activity, the cells were grown for 24 h in liquid Sabouraud medium, washed three times with PBS and tested as described before59. Saccharomyces cerevisae (strain RSY113) was used as a negative control in these assays. Melanin production was determined visually after the cells were spotted on solid minimal medium supplemented with 1 mM l-dopa and cultivated for 120 h either at 30 oC or 37 oC60,61. Quantification of pigment formation was determined densitometrically after digitalization of the images generated for visual analysis.
Analysis of expression of CAP genes
Each isolate was cultivated in liquid Sabouraud and in minimal media for 48 h as described above. Cells were collected by centrifugation for RNA extraction using the Trizol reagent (Invitrogen) according to the manufacturer’s protocol. Briefly, after DNAse treatment, RNA preparations were submitted to cDNA synthesis using M-MLV reverse transcriptase enzyme (Promega). Real-time PCR reactions were performed in an Applied Biosystems StepOnePlus Real-Time PCR System. Thermal cycling conditions consisted of an annealing step at 95 °C for 15 s followed by an elongation step at 60 °C for 1 min along for 40 cycles. Platinum SYBR Green qPCR SuperMix-UDG with ROX (Invitrogen) was used with 4 pmol of each primer and 2 μl of the cDNA template in a final reaction volume of 20 μl. Each sample was analyzed in triplicate. Melting curve analyses were performed at the end of each reaction. Results were normalized by using yeast ß-actin gene amplification. Relative expression was determined by the 2−ΔCT method described by Livak & Schmittgen, 200162.
Determination of glycosidic composition in cellular glycans
Cellular polysaccharides were extracted with dimethyl sulfoxide (DMSO), using protocols previously established for efficient removal of GXM from C. neoformans cells63. Glycosidic composition was performed by combined gas chromatography/mass spectrometry (GC/MS) of the per-O-trimethylsilyl (TMS) derivatives of the monosaccharide methyl glycosides produced from the sample by acidic methanolysis according to previously described methods64.
Fluorescence-based analysis of the cell surface
General properties of the cell surface of C. gattii were analyzed by fluorescence microscopy. Yeast cells (107) were fixed in 4% paraformaldehyde (1 h, room temperature), blocked with 1% (w/v) BSA in PBS (1 h, 37 °C) and subsequently incubated with four different monoclonal antibodies (mAbs) raised to GXM, which were kindly donated by Dr. Arturo Casadevall (Baltimore, USA). The mAbs used in this study were: IgG1 18B747 and IgMs 2D10, 13F1 and 12A144,47. All mAbs were incubated with C. gattii at a final concentration of 10 μg/ml (1 h, 37 °C). The cells were subsequently incubated with Alexa Fluor™ 488-conjugated secondary antibodies (Invitrogen; 10 μg/ml, 1 h, 37 °C) and washed with PBS. Stained cells were analyzed by flow cytometry using a FACScan apparatus. Data was analyzed using the FlowJo7 software (FlowJo© Tree Star, Inc.). Alternatively, stained cells were probed with Calcofluor White at 5 μg/ml for 30 min (37 °C). The cells were finally counter-stained with India ink for analysis of capsular dimensions. After further washing with PBS, the cells were observed under a fluorescence optical microscope Axioplan 2 (Zeiss, Germany). Images were processed using the analysis software (Zen 2.0) and Adobe Photoshop CS5 was used for preparing figure panels.
In vivo assays
BALB/c (female) mice (n = 7) approximately 4–6 weeks old were used for determination of mortality curves, fungal burden, pulmonary microscopic examination and chitinase activity after infection with C. gattii. The animals were anesthetized with ketamine (10 mg/kg) and xylazine (4 mg/kg) and inoculated intranasally with a fungal suspension of 105 yeast cells in a final volume of 50 μl PBS. Groups were observed twice a day for 70 days for determination of mortality rates. Alternatively, groups of 12 mice were infected as described above with each of the isolates studied here. After 4, 14 and 21 days post infection, animals (n = 4) infected with each isolate were sacrificed and the lungs and the brains were excised and homogenized. These macerates were plated on Sabouraud medium for colony forming units (CFU) determination as previously described65. Lung macerates were also used for determination of chitinase activity. The homogenized pulmonary tissues were clarified by centrifugation for removal of cells and debris. Pelleted cells were stained with the wheat germ lectin (WGA) conjugated to tetramethylrhodamine (TRITC) at 5μg/ml for 30min (37°C). Supernatants were assessed for chitinase activity using the Chitinase Assay Kit Fluorimetric (CS1030 - Sigma Aldrich) according to the manufacturer instructions. Briefly, 5 μl of each sample were diluted to 100 μl with the commercial working substrate, for further incubation for 1 h at 37 °C. Reactions were read fluorimetrically as indicated by the manufacturer. The use of animals in this study was carried out with the approval of the Ethics Committees for animal use at the Federal Universities of Rio Grande do Sul and Rio de Janeiro (project number 093/14). The animals were kept in groups of up to 4 individuals, in cages with food and water ad libitum. Animals were maintained following the rules of the National Council of Animal Experimentation Control (CONCEA) and the Brazilian College of Animal Experimentation (COBEA).
Statistical Tests
Student’s t test for comparison of sample data and the Kaplan-Meier survival statistics were carried out using the GraphPad Prism 6.0 software (GraphPad Software).
Additional Information
How to cite this article: Rodrigues, J. et al. Pathogenic diversity amongst serotype C VGIII and VGIV Cryptococcus gattii isolates. Sci. Rep. 5, 11717; doi: 10.1038/srep11717 (2015).
Supplementary Material
Acknowledgments
We thank Carolina Pereira Silveira for help with in vivo experimentation. We are also grateful to Leonardo Nimrichter and Jorge José-Jó Bastos Ferreira for helpful suggestions. Carbohydrate analyses were performed at the Complex Carbohydrate Research Center, University of Georgia (Atlanta), which is supported in part by the Department of Energy-funded Center for Plant and Microbial Complex Carbohydrates (DE-FG-9-93ER-20097). This work was supported by grants from the Brazilian agencies CNPq, CAPES, and FAPERJ. The authors also acknowledge support from the Instituto Nacional de Ciência e Tecnologia de Inovação em Doenças Negligenciadas (INCT-IDN). MLR is the recipient of a Pathfinder Award from the Wellcome Trust (UK). WM is supported by an NHMRC (Australia) grant #APP1031943.
Footnotes
Author Contributions J.R., R.M.C.G. and F.L.F. Phenotypic, capsule, chitinase and chitooligomer analyses. C.F. MALDI-TOF analysis. R.M.C.G., K.M. and W.M. molecular typing. J.R., R.O.S. and C.C.S. Gene expression. J.R. and L.K. Animal experimentation. J.R., R.M.C.G., W.M., M.H.V., A.S. and M.L.R. preparation of the manuscript. All authors discussed the results, wrote and approved the final manuscript.
References
- Park B. J. et al. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23, 525–30 (2009). [DOI] [PubMed] [Google Scholar]
- Chayakulkeeree M. & Perfect J. R. Cryptococcosis. Infect Dis Clin North Am 20, 507–44, v-vi (2006). [DOI] [PubMed] [Google Scholar]
- Ngamskulrungroj P. et al. Genetic diversity of the Cryptococcus species complex suggests that Cryptococcus gattii deserves to have varieties. PLoS One 4, e5862 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huston S. M. & Mody C. H. Cryptococcosis: an emerging respiratory mycosis. Clin Chest Med 30, 253–64, vi (2009). [DOI] [PubMed] [Google Scholar]
- Lin X. Cryptococcus neoformans: morphogenesis, infection, and evolution. Infect Genet Evol 9, 401–16 (2009). [DOI] [PubMed] [Google Scholar]
- Byrnes E. J. 3rd et al. First reported case of Cryptococcus gattii in the Southeastern USA: implications for travel-associated acquisition of an emerging pathogen. PLoS One 4, e5851 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J. R. et al. Cryptococcus gattii in the United States: clinical aspects of infection with an emerging pathogen. Clin Infect Dis 53, 1188–95 (2011). [DOI] [PubMed] [Google Scholar]
- Kidd S. E. et al. Characterization of environmental sources of the human and animal pathogen Cryptococcus gattii in British Columbia, Canada, and the Pacific Northwest of the United States. Appl Environ Microbiol 73, 1433–43 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd S. E. et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci U S A 101, 17258–63 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer W. et al. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med Mycol 47, 561–70 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. C., Meyer W. & Sorrell T. C. Cryptococcus gattii infections. Clin Microbiol Rev 27, 980–1024 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E. E. An immunologic comparison of 12 strains of Cryptococcus neoformans (Torula histolytica). Proc Soc Exp Biol Med 71, 644–6 (1949). [DOI] [PubMed] [Google Scholar]
- Byrnes E. J. 3rd, Bartlett K. H., Perfect J. R. & Heitman J. Cryptococcus gattii: an emerging fungal pathogen infecting humans and animals. Microbes Infect 13, 895–907 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carriconde F. et al. Clonality and alpha-a recombination in the Australian Cryptococcus gattii VGII population–an emerging outbreak in Australia. PLoS One 6, e16936 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen F., Chowdhary A., Prakash A., Yntema J. B. & Meis J. F. Molecular characterization of Cryptococcus gattii genotype AFLP6/VGII isolated from woody debris of divi-divi (Caesalpinia coriaria), Bonaire, Dutch Caribbean. Rev Iberoam Micol 31, 193–6 (2014). [DOI] [PubMed] [Google Scholar]
- Hagen F. et al. Ancient dispersal of the human fungal pathogen Cryptococcus gattii from the Amazon rainforest. PLoS One 8, e71148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelthaler D. M. et al. Cryptococcus gattii in North American Pacific Northwest: whole-population genome analysis provides insights into species evolution and dispersal. MBio 5, e01464–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billmyre R. B. et al. Highly recombinant VGII Cryptococcus gattii population develops clonal outbreak clusters through both sexual macroevolution and asexual microevolution. MBio 5, e01494–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi S., Dyavaiah M., Larsen R. A. & Chaturvedi V. Cryptococcus gattii in AIDS patients, southern California. Emerg Infect Dis 11, 1686–92 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firacative C., Duan S. & Meyer W. Galleria mellonella model identifies highly virulent strains among all major molecular types of Cryptococcus gattii. PLoS One 9, e105076 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. C. & Kwon-Chung K. J. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol Cell Biol 14, 4912–9 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartivarian S. E. et al. Localization of mannoprotein in Cryptococcus neoformans. J Bacteriol 171, 6850–2 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira C. P. et al. The heat shock protein (Hsp) 70 of Cryptococcus neoformans is associated with the fungal cell surface and influences the interaction between yeast and host cells. Fungal Genet Biol 60, 53–63 (2013). [DOI] [PubMed] [Google Scholar]
- Cordero R. J. et al. Chronological aging is associated with biophysical and chemical changes in the capsule of Cryptococcus neoformans. Infect Immun 79, 4990–5000 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues M. L., Alvarez M., Fonseca F. L. & Casadevall A. Binding of the wheat germ lectin to Cryptococcus neoformans suggests an association of chitinlike structures with yeast budding and capsular glucuronoxylomannan. Eukaryot Cell 7, 602–9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca F. L. et al. Binding of the wheat germ lectin to Cryptococcus neoformans chitooligomers affects multiple mechanisms required for fungal pathogenesis. Fungal Genet Biol 60, 64–73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P. Y., Sham A. & Kronstad J. W. Cryptococcus gattii isolates from the British Columbia cryptococcosis outbreak induce less protective inflammation in a murine model of infection than Cryptococcus neoformans. Infect Immun 77, 4284–94 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngamskulrungroj P., Chang Y., Sionov E. & Kwon-Chung K. J. The primary target organ of Cryptococcus gattii is different from that of Cryptococcus neoformans in a murine model. MBio 3, e00103–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen F. et al. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol, 10.1016/j.fgb.2015.02.009 (2015). [DOI] [PubMed] [Google Scholar]
- Meyer W. et al. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA-a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis 20, 1790–9 (1999). [DOI] [PubMed] [Google Scholar]
- Meyer W. et al. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg Infect Dis 9, 189–95 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firacative C., Trilles L. & Meyer W. MALDI-TOF MS enables the rapid identification of the major molecular types within the Cryptococcus neoformans/C. gattii species complex. PLoS One 7, e37566 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posteraro B. et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based method for discrimination between molecular types of Cryptococcus neoformans and Cryptococcus gattii. J Clin Microbiol 50, 2472–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTaggart L. R. et al. Rapid identification of Cryptococcus neoformans and Cryptococcus gattii by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 49, 3050–3 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden D. C., Fries B. C., Wang F. & Casadevall A. Capsule structural heterogeneity and antigenic variation in Cryptococcus neoformans. Eukaryot Cell 6, 1464–73 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O. & Casadevall A. Experimental modulation of capsule size in Cryptococcus neoformans. Biol Proced Online 6, 10–15 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca F. L. et al. Role for chitin and chitooligomers in the capsular architecture of Cryptococcus neoformans. Eukaryot Cell 8, 1543–53 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos C. L. et al. Chitin-like molecules associate with Cryptococcus neoformans glucuronoxylomannan to form a glycan complex with previously unknown properties. Eukaryot Cell 11, 1086–94 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald J. R. & Musser J. M. Evolutionary genomics of pathogenic bacteria. Trends Microbiol 9, 547–53 (2001). [DOI] [PubMed] [Google Scholar]
- Sabiiti W. et al. Efficient phagocytosis and laccase activity affect the outcome of HIV-associated cryptococcosis. J Clin Invest 124, 2000–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O. et al. The capsule of the fungal pathogen Cryptococcus neoformans. Adv Appl Microbiol 68, 133–216 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho R. M. et al. The vacuolar-sorting protein Snf7 is required for export of virulence determinants in members of the Cryptococcus neoformans complex. Sci Rep 4, 6198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza C. A. et al. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol Cell Biol 21, 3179–91 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rivera J., Chang Y. C., Kwon-Chung K. J. & Casadevall A. Cryptococcus neoformans CAP59 (or Cap59p) is involved in the extracellular trafficking of capsular glucuronoxylomannan. Eukaryot Cell 3, 385–92 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. C. & Kwon-Chung K. J. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans. Infect Immun 66, 2230–6 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dromer F., Gueho E., Ronin O. & Dupont B. Serotyping of Cryptococcus neoformans by using a monoclonal antibody specific for capsular polysaccharide. J Clin Microbiol 31, 359–63 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A. et al. Characterization of a murine monoclonal antibody to Cryptococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob Agents Chemother 42, 1437–46 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee J., Scharff M. D. & Casadevall A. Protective murine monoclonal antibodies to Cryptococcus neoformans. Infect Immun 60, 4534–41 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicencio A. G. et al. Pulmonary cryptococcosis induces chitinase in the rat. Respir Res 9, 40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner D. L. et al. Chitin Recognition via Chitotriosidase Promotes Pathologic Type-2 Helper T Cell Responses to Cryptococcal Infection. PLoS Pathog 11, e1004701 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosanchuk J. D., Rosas A. L., Lee S. C. & Casadevall A. Melanisation of Cryptococcus neoformans in human brain tissue. Lancet 355, 2049–50 (2000). [DOI] [PubMed] [Google Scholar]
- Doering T. L., Nosanchuk J. D., Roberts W. K. & Casadevall A. Melanin as a potential cryptococcal defence against microbicidal proteins. Med Mycol 37, 175–81 (1999). [PubMed] [Google Scholar]
- Nosanchuk J. D. & Casadevall A. Impact of melanin on microbial virulence and clinical resistance to antimicrobial compounds. Antimicrob Agents Chemother 50, 3519–28 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. & May R. C. Virulence in Cryptococcus species. Adv Appl Microbiol 67, 131–90 (2009). [DOI] [PubMed] [Google Scholar]
- Fraser J. A. et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437, 1360–4 (2005). [DOI] [PubMed] [Google Scholar]
- Olivares L. R. et al. Genotyping of Mexican Cryptococcus neoformans and C. gattii isolates by PCR-fingerprinting. Med Mycol 47, 713–21 (2009). [DOI] [PubMed] [Google Scholar]
- Cogliati M. et al. Cryptococcus gattii serotype-C strains isolated in Bangalore, Karnataka, India. Mycoses 55, 262–8 (2012). [DOI] [PubMed] [Google Scholar]
- Litvintseva A. P., Thakur R., Reller L. B. & Mitchell T. G. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in Sub-Saharan Africa. J Infect Dis 192, 888–92 (2005). [DOI] [PubMed] [Google Scholar]
- Kwon-Chung K. J., Wickes B. L., Booth J. L., Vishniac H. S. & Bennett J. E. Urease inhibition by EDTA in the two varieties of Cryptococcus neoformans. Infect Immun 55, 1751–4 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmetzsch L. et al. Role for Golgi reassembly and stacking protein (GRASP) in polysaccharide secretion and fungal virulence. Mol Microbiol 81, 206–18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker L. G., Specht C. A., Donlin M. J. & Lodge J. K. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot Cell 6, 855–67 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–8 (2001). [DOI] [PubMed] [Google Scholar]
- Bryan R. A. et al. Radiological studies reveal radial differences in the architecture of the polysaccharide capsule of Cryptococcus neoformans. Eukaryot Cell 4, 465–75 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle R. K. & Poppe I. Carbohydrate composition analysis of glycoconjugates by gas-liquid chromatography/mass spectrometry. Methods Enzymol 230, 1–15 (1994). [DOI] [PubMed] [Google Scholar]
- Rodrigues M. L. et al. Monoclonal antibody to fungal glucosylceramide protects mice against lethal Cryptococcus neoformans infection. Clin Vaccine Immunol 14, 1372–6 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.