Abstract
Background:
Minimizing the occurrence of hypoglycemia in patients with type 2 diabetes is a challenging task since these patients typically check only 1 to 2 self-monitored blood glucose (SMBG) readings per day.
Method:
We trained a probabilistic model using machine learning algorithms and SMBG values from real patients. Hypoglycemia was defined as a SMBG value < 70 mg/dL. We validated our model using multiple data sets. In addition, we trained a second model, which used patient SMBG values and information about patient medication administration.
Results:
The optimal number of SMBG values needed by the model was approximately 10 per week. The sensitivity of the model for predicting a hypoglycemia event in the next 24 hours was 92% and the specificity was 70%. In the model that incorporated medication information, the prediction window was for the hour of hypoglycemia, and the specificity improved to 90%.
Conclusions:
Our machine learning models can predict hypoglycemia events with a high degree of sensitivity and specificity. These models—which have been validated retrospectively and if implemented in real time—could be useful tools for reducing hypoglycemia in vulnerable patients.
Keywords: machine learning, hypoglycemia prediction, type 2 diabetes
Hypoglycemia is a significant adverse outcome in patients with type 2 diabetes and has been associated with increased morbidity, mortality, and cost of care.1 In addition, hypoglycemia is a major limiting factor for the optimization of insulin therapy. In patients with frequent self-monitored blood glucose (SMBG) measurements or those who employ continuous glucose monitors, statistical methods may be used to predict hypoglycemia. For example, Rodbard found that hypoglycemia risk can be estimated using mean, standard deviation, coefficient of variation, and the nature of the glucose distribution.2 Kovatchev et al introduced a measure of BG variability called “average daily risk range,” which strongly correlated to hypoglycemia.3 Monnier et al found that the risk of asymptomatic hypoglycemia increases in the presence of increased glucose variability.4 Most patients with type 2 diabetes have only sparse SMBG data, which do not lend themselves to statistical methods. Our goal is to be able to accurately predict an individual’s risk for hypoglycemia using sparse data, and by employing mobile health technology to provide the appropriate preventive actions for patients and caregivers.
For predictions to be clinically useful, the accuracy of the prediction should have significant confidence. Predictions should provide a forecast for a time window that is sufficient to enable meaningful preventive interventions. Predictions should be enabled with BG data alone; other clinical information, when available, can be used if the accuracy of the prediction increases. And finally, the prediction algorithm should require only approximately 1 to 2 SMBG values per day, which is typical for patients with type 2 diabetes
Methods
We employed machine learning methods for our prediction algorithms (see Figure 1). Machine learning is useful when there is a large amount of example data and when the rules for prediction are unclear. In the case of hypoglycemia, we felt that though physicians were able to intuitively estimate the risk of hypoglycemia, they weren’t able to explain specific rules that could be coded in a computer system. In constructing the model, we chose a classification approach rather than a regression approach. If a set of BG values is available for a given week, it can be predicted if the patient will have a hypoglycemic episode in the following week or not. Hence the prediction becomes a binary (yes/no) classification problem. From a computational standpoint, classification problems are easier and more efficient to solve than regression.
Figure 1.
Machine learning methodology.
Data cleansing involves transforming raw data into a form that is easily readable without ambiguity for a machine learning algorithm. This involves identifying any problems such as anomalies, errors, and missing values in a given data set and correcting them. Data cleansing has advantages and disadvantages. Though it may help machine learning algorithms readily understand underlying patterns in the data without ambiguity, it introduces a manual bias of modifying the original pattern in raw data. To account for this in our modeling efforts, we cleansed only the instances of missing value and replaced them with indicators such as “N/A.” Doing so helps the algorithm understand that a specific value may be ignored but that the pattern itself must be considered during learning. Any typographical data entry errors (eg, “3000” instead of “300”) were not corrected. This strategy was chosen to ensure that the model learned to recognize these types of errors and was hence trained to handle such noise in the data. Doing so also makes the model more robust to handle real-world data from virtually any eligible patient, since such errors are common in patient self-reported data.
Based on the clinical objective, it is clear that a BG value and its respective timestamp are the only 2 data variables that are consistently available. By using the time stamp, other variables can be derived such as time of the day, day of the week, month, and so on. Also, it is known that differences between successive BG values (ie, variability) may have a strong relationship with the occurrence of hypoglycemia events.3,4 Hence, these relationships were taken into consideration. Once the relationships were identified and data were preprocessed, we introduced the data to the algorithms, which learn patterns within and between factors as well as the relationship of those patterns with the prediction variable. Every algorithm has parametric values which define a configuration of the algorithm which best predicts the prediction variable.
We used 3 criteria for selecting among the numerous machine learning algorithms available. First, the algorithm must be widely accepted in the data science community for better support during modeling. Second, the algorithm should have a proven track record for accuracy and efficiency based on the type of data (mixed measure—numerical predictors and categorical response). Third, the algorithm should have a history of successful commercial usage in scoring data in a production level system for a large user base.5,6 We selected 4 algorithms to test: random forest, support vector machine (SVM), k-nearest neighbor, and naïve Bayes.
To test the effectiveness of the model, different data sets were used. The training data set of SMBG values came from deidentified patient data from a clinical trial of patients with type 2 diabetes.7 The data had been deidentified following procedures approved by the University of Maryland Baltimore’s Institutional Review Board. The original data set contained 56 000 SMBG data points collected in a 1-year prospective study. In this study, patients were treated with a variety of medications, including oral antihyperglycemic agents and insulin.
Validation testing of the model was carried out using a segment of the SMBG data not used in the model training. Further cross-validation testing utilized 3 other large data sets, from which we were able to generate 10 814 test samples. The testing procedure involves removing the prediction variable (SMBG value from day 8) from the test set. Hence, the model will have data only on the identified factors. The values predicted by the model during testing were then matched with the reference values to identify cases of data when the model did and did not predict correctly. The various cases of the model’s success and failure in prediction are expressed in the form of a confusion matrix.
We used 2 sets of input variables. First, for the 1-day time window prediction model, we used the 11 most recent SMBG values and respective timestamp (hour of the day) in the previous 7 days from a given day. We used the BG value and timestamp for a given day as well as the difference between each chosen SMBG value and the immediately previous SMBG value. Second, for the hour of the day prediction model that used medication information, in addition to the those variables, we used the 11 most recent medication administration data points, and the respective timestamp (hour of the day) in the previous 7 days from a given day. We used the medication class (ie, oral agent, long-acting insulin, etc) and respective dosage as well.
Results
We concluded that a 1-day (24-hour) prediction window would be the most useful time frame for delivering an intervention. We fixed this time window and studied the effect of sample size on the accuracy of our models. Accuracy peaked with a sample size of 10 to 11 SMBG values per week (see Figure 2). Subsequent models were then designed to have a 1-day prediction window and a sample size requirement of 11 SMBG values per week.
Figure 2.
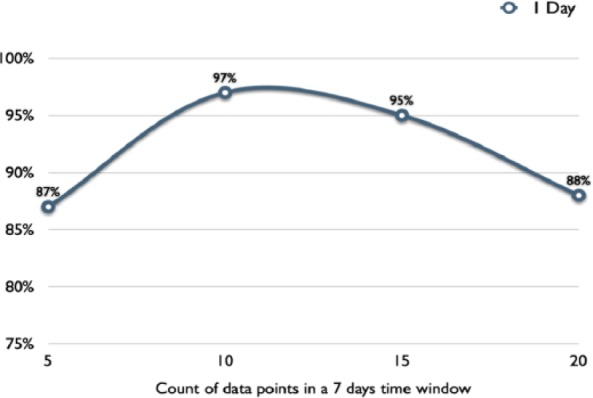
Performance (accuracy) of model across various sample sizes for 1-day prediction of hypoglycemia event.
Next, we trained different models using the algorithms described in the methods section; random forest, SVM, k-nearest neighbor, and naïve Bayes. The accuracy of predicting hypoglycemia using the various models is shown in Table 1. We further cross-validated the model with distinct data sets (also shown in Table 1). Note that models 1 and 3 performed best across all data sets with a prediction accuracy of over 90%.
Table 1.
Comparison of Performance (Accuracy) of 4 First-Generation Models Across 4 Distinct Data Sets.
| Data set (sample size) | Model 1 (RF) (%) | Model 2 (KNN) (%) | Model 3 (SVM) (%) | Model 4 (naïve Bayes) (%) |
|---|---|---|---|---|
| Set 1 (1037) | 91.0 | 24.7 | 93.3 | 2.3 |
| Set 2 (6686) | 95.2 | 53.3 | 96.0 | 0.6 |
| Set 3 (1091) | 94.0 | 41.2 | 97.0 | 0.5 |
| Set 4 (2000) | 97.0 | 63.0 | 97.5 | 48.5 |
The sample size used for each data set is shown. Note that the sample size indicates the number of 7-day samples, not the total number of SMBG values in the data set. All data are on file at WellDoc, Inc. Note that the methodology for collection of data set 4 is in Quinn et al.7 KNN, k nearest neighbor; RF, random forest; SVM, support vector machine.
We subsequently used model 1 for further optimization because we felt that a random forest model may be easier to study and analyze as compared to the “black box” SVM model. As we attempted to optimize the models based on these 4 algorithms, we found that when the model was optimized for high sensitivity, specificity was reduced, and vice versa (see Table 2). A model which would predict most hypoglycemia but that would produce many false positives, would not be able to deliver meaningful interventions to patients. We also examined how the distribution of SMBGs affects the predictions. In our data sets, it was clear that patients do not always collect SMBG values at a regular frequency. In a given week, some patients may check more frequently earlier in the week; others may check more frequently later in the week. Table 2 shows how the distribution of SMBG values affects the predictions. Our third-generation model, which we call model 1.3, was optimized for a good balance between sensitivity (approximately 90%) and sensitivity (approximately 70%).
Table 2.
Comparison of Performance (Specificity and Sensitivity) of Models for Different Distribution of BG Data Across a Week.
| Model number | Week segment | Specificity (%) | Sensitivity (%) |
|---|---|---|---|
| Model 1.1 | Most BGs toward the beginning of week | 12 | 86 |
| Most BGs toward end of week | 5 | 92 | |
| Model 1.2 | Most BGs toward the beginning of week | 99 | 3 |
| Most BGs toward end of week | 100 | 6 |
These models were optimized from the first-generation model 1. Note that optimizing the model for high sensitivity resulted in low specificity and vice versa. BG, blood glucose.
In our testing scenarios, we present the model with 11 SMBG values over 7 days and ask for a prediction of hypoglycemia (Y/N) on the eighth day. We wanted to know how the model would perform in comparison to human experts. We presented 200 samples (11 SMBG values per sample) to model 1.3 and to 3 endocrinologists. Model 1.3 had a higher degree of sensitivity than the humans (91.7% vs 52.7 ± 16%) but a lower specificity than the human experts (69.5% vs 79.8 ± 5%).
Because of the concern for low specificity and the possibility of using false positive predictions to drive interventions, we wanted to examine other variables which could improve the performance of our models. Potential variables might include physical activity (eg, exercise), medication use (eg, insulin dose), and nutritional data (eg, carbohydrate intake). Unfortunately, our data sets did not include sufficient values from these other variables except for medication use. We then used data samples with 11 SMBG values during the week and at least 11 instances of medication use data during the same week. To be included in the analysis, we required that the medication data point be paired with an SMBG value (within 1 hour). We then trained a model using these inputs and tested it and cross-validated it using distinct data sets as we did before with our SMBG values-only models. The model was designed to make a prediction for the hour of hypoglycemia on the eighth day. The results are shown summarized in Table 3. Note that with medication information, the specificity of the predictions increased by 32%. Both sensitivity and specificity were about 90% as calculated using 2 distinct data sets.
Table 3.
Cross-Validation Results for BG and Medication Model Across 2 Distinct Data Sets.
| Data set A (accuracy 90.6%) |
Data set B (accuracy 91.1%) |
|||||
|---|---|---|---|---|---|---|
| Hypo event | No hypo event | Specificity (%) | Hypo event | No hypo event | Specificity (%) | |
| Predicted hypo | 227 | 20 | 92.5 | 387 | 34 | 91.7 |
| Predicted no hypo | 29 | 248 | 41 | 377 | ||
| Sensitivity (%) | 88.7 | 90.4 | ||||
Discussion
The challenge of hypoglycemia prediction in type 2 diabetes is that these patients typically collect fewer SMBG values than patients with type 1 diabetes. In patients with type 1 diabetes who use continuous glucose monitoring, a regression approach to predicting future blood glucose values seems reasonable.8 The sparse data in the type 2 diabetes situation make a mathematical approach unfeasible. We decided to make the problem a classification one (ie, hypoglycemia y/n?) rather than a regression one (ie, what is the future value?). We also wanted to mimic the approach used by human experts, who do not clearly define the rules they use when making a prediction. Machine learning algorithms are quite useful in situations such as these. After testing multiple models and cross-validating them with multiple data sets, we have demonstrated the usefulness of this approach. As expected, the addition of more variables (in our case, medication information) improved the performance of the models.
Since these models have a high degree of sensitivity and specificity and compare favorably to human experts, employing them in an automated manner to drive interventions in real time may be useful in preventing hypoglycemia. One design, which could utilize this approach, is a mobile health platform. Patients enter their SMBG values in a cellular-enabled device. The SMBG values are transmitted to a server, where the model is running in real time. When a hypoglycemia event is predicted with a high degree of certainty, a self-management message can be generated to the user. Furthermore, clinical decision support regarding the risk of hypoglycemia could be sent to health care providers.
Conclusion
We developed hypoglycemia prediction models using machine learning methods. These models performed with a high degree of sensitivity and specificity using sparse SMBG data from distinct data sets. Models which used medication information had improved specificity, which would reduce false positive predictions. Future studies comparing the accuracy of machine learning models and traditional statistical methods for predicting hypoglycemia may be useful. Further testing should be conducted to confirm that the models can be deployed in real time and could extend beyond predicting hypoglycemia to also deliver an intervention message, which could further reduce the occurrence of this serious diabetes complication.
Footnotes
Abbreviations: SMBG, self-monitored blood glucose; SVM, support vector machine.
Declaration of Conflicting Interests: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: All authors were employees of WellDoc, Inc at the time this work was conducted.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Zhang Y, Wieffer H, Modha R, Balar B, Pollack M, Krish-narajah G. The burden of hypoglycemia in type 2 diabetes: a systematic review of patient and economic perspectives. Clin Outcomes Manag. 2010;17(12):547-557. [Google Scholar]
- 2. Rodbard D. Hypo- and hyperglycemia in relation to the mean, standard deviation, coefficient of variation, and nature of the glucose distribution. Diabetes Technol Ther. 2012;14(10):868-876. [DOI] [PubMed] [Google Scholar]
- 3. Kovatchev BP, Otto E, Cox D, Gonder-Frederick L, Clarke W. Evaluation of a new measure of blood glucose variability in diabetes. Diabetes Care. 2006;29(11):2433-2438. [DOI] [PubMed] [Google Scholar]
- 4. Monnier L, Wojtusciszyn A, Colette C, Owens D. The contribution of glucose variability to asymptomatic hypoglycemia in persons with type 2 diabetes. Diabetes Technol Ther. 2011;13(8):813-818. [DOI] [PubMed] [Google Scholar]
- 5. Kotsiantis SB. Supervised machine learning: a review of classification techniques. Informatica. 2007;31:249-268. [Google Scholar]
- 6. Bishop CM, Nasrabadi NM. Pattern Recognition and Machine Learning. Vol. 1 New York, NY: Springer; 2006. [Google Scholar]
- 7. Quinn CC, Shardell MD, Terrin ML, Barr EA, Ballew SH, Gruber-Baldini AL. Cluster-randomized trial of a mobile phone personalized behavioral intervention for blood glucose control. Diabetes Care. 2011;34(9):1934-1942. Erratum in: Diabetes Care. 2013;36(11):3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cameron F, Niemeyer G, Gundy-Burlet K, Buckingham B. Statistical hypoglycemia prediction. J Diabetes Sci Technol. 2008;2(4):612-621. [DOI] [PMC free article] [PubMed] [Google Scholar]


