Abstract
This study aimed to determine whether C20orf54 rs13042395 polymorphism modify the risk of esophageal squamous cell carcinoma (ESCC) and gastric cardia adenocarcinomas (GCA) in common population. We conducted a systematic literature review and evaluated the quality of included studies based on Newcastle-Ottawa Scale (NOS). Pooled odds ratios (ORs) and corresponding 95% confidence intervals (95%CIs) were calculated to estimate the strengths of the associations. 9 articles (10 studies) were identified for synthesis analyses. Overall, the results indicated that the C20orf54 rs13042395 genotype was subtly decrease the risk of ESCC (T vs. C: OR = 0.95; 95%CI = 0.90–0.99; P = 0.02) and the rs13042395 polymorphism was associated with a decreased risk of GCA (T vs. C: OR = 0.95; 95%CI = 0.91–0.98; P < 0.01). The subsets were divided by smoking and drinking status, but none of the genetic comparisons reached statistical significance. Subgroup analysis was also stratified by body mass index (BMI), rs13042395 polymorphism was significantly associated with a subtly decreased cancer risk in under-weight group and normal group, but no association was observed in over-weight group. In conclusion, C20orf54 rs13042395 polymorphism was significantly associated with decreased ESCC and GCA risk especially for the subjects with under-weight or normal.
Esophageal cancer and gastric cancer cause more than 400,000 and 700,000 deaths each year, respectively, and represent the sixth and second leading causes of cancer-related death worldwide1. Esophageal squamous cell carcinoma (ESCC) is the most frequent histological subtype of esophageal cancer and accounts for 90% of cases2,3. Synthesis of epidemiological studies indicate that alcohol drinking and tobacco smoking are the major risk factors for esophageal cancer4. Helicobacter pylori (H.pylori) infection is a well-established risk factor for Gastric Cardia Adenocarcinoma (GCA) and has been labeled as a definite human carcinogen by International Agency of Research on Cancer (IARC)5. In western countries, the high body mass index (BMI) has been suggested as a risk factor for GCA and esophageal adenocarcinoma6,7. However, only a subset of individuals exposed to the environmental risk factors would develop ESCC and GCA, it is suggested genetic factors substantially contribute to the ESCC and GCA carcinogenesis8.
In 2010, a large-scale genome-wide association study (GWAS) reported that a new and notable susceptibility locus (rs13042395) located in 5’ flanking region of chromosome 20 open reading frame 54 (C20orf54), it encodes riboflavin transporter 2 protein (RFT2) that was newly identified to play an important role in esophageal and carcinogenesis by modulating riboflavin uptake9. In addition, it has important biological implications for both ESCC and GCA in the Chinese population10,11. C20orf54 is a human riboflavin transporter that has an important role in the intestinal absorption of riboflavin12,13. The deficiency of riboflavin has been documented as a risk factor for ESCC and GCA. Also, riboflavin supplementation has been reported to reduce the risk of ESCC and GCA14.
For C20orf54 rs13042395 genotype and risk of ESCC and GCA, the results were inconsistent. On the basis of the biological and pathologic significance of C20orf54, it is widely shared that functional genetic variations in the C20orf54 may contribute to the development of ESCC and GCA. The objective of the present study was to quantitatively assess the association between C20orf54 rs13042395 polymorphism and risk of ESCC and/or GCA.
Results
Literature search and study characteristics
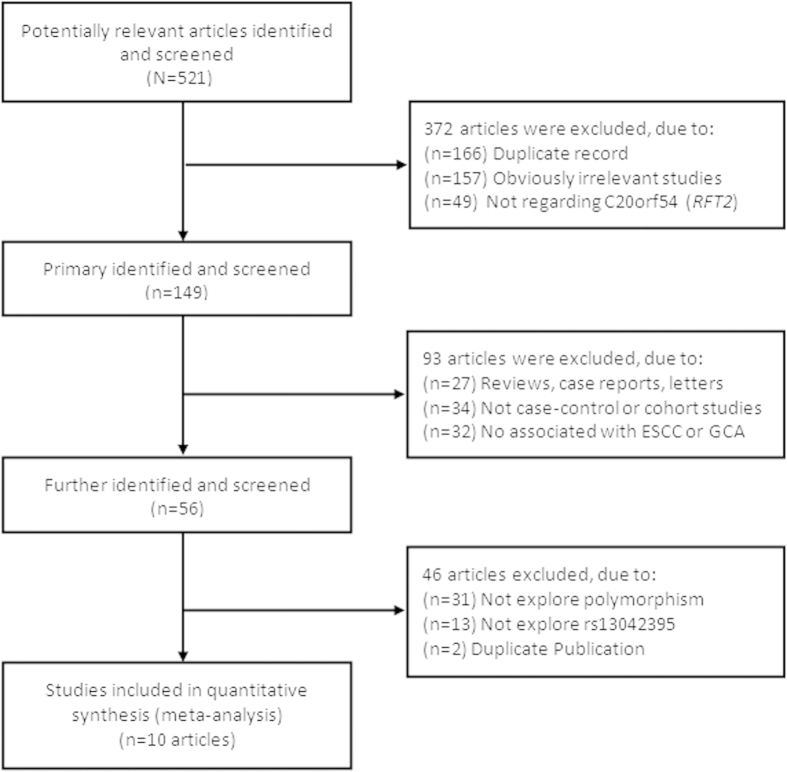
The selection process for relevant studies and a flow diagram are shown in Fig. 1. A computer-assisted search yielded 521 potentially relevant published titles. After primary identified, 149 titles were potentially appropriate, and the corresponding abstracts were reviewed. After further identification and screening individual study, 56 publications underwent full-text review. Finally, producing a total of 10 publications10,15,16,17,18,19,20,21,22,23 (12 studies) for inclusion. Characteristics of included studies are present in Table 1. We identified 12 studies, with a total of 88,547 participants, including 28,765 cases and 59,782 controls. The evidence synthesis included eight studies on ESCC, four GCA. There were 11 studies of Asian and one study of Caucasian. Of the 12 studies, 11 were population-based case-control studies and one was hospital-based case-control study, and eight studies were randomly repeated a portion of samples as quality control while genotyping.
Figure 1. Flow diagram for screening and identification of relevant studies.
Table 1. Characteristics of included studies.
| Author | Year | Study type | Ethnicity (Country) | Cancer type | Source of control | Genotyping | Matching Y/N | Sample size Case/Control | Qualitya control | PHWE |
|---|---|---|---|---|---|---|---|---|---|---|
| Peng[15] | 2014 | Case-control | Asian (China) | ESCC | Population | Sequenom | N | 50/50 | N | NA |
| Peng[15] | 2014 | Case-control | Asian (China) | GCA | Population | Sequenom | N | 50/50 | N | NA |
| Piao [16] | 2014 | Case-control | Asian (Korea) | ESCC | Population | HRM | Y | 321/1700 | Y | 0.724 |
| Gu [17] | 2012 | Case-control | Asian (China) | ESCC | Hospital | MassArray | Y | 379/375 | Y | 0.648 |
| Wei [18] | 2012 | Case-control | Asian (China) | ESCC | Population | PCR-RFLP | Y | 240/198 | Y | 0.354 |
| Dura [19] | 2012 | Case-control | Caucasian (Netherland) | ESCC | Population | Real-Time PCR | Y | 344/580 | N | 0.267 |
| Zhou [20] | 2012 | Case-control | Asian (China) | ESCC | Population | Chips | Y | 4722/4732 | Y | 0.639 |
| Ren [21] | 2011 | Case-control | Asian (China) | GCA | Population | Chips | Y | 2748/10136 | Y | 0.407 |
| Shang [22] | 2011 | Case-control | Asian (China) | ESCC | Population | MALDI-TOF-MS | Y | 190/211 | N | 0.336 |
| Zhang [23] | 2011 | Case-control | Asian (China) | GCA | Population | TaqMan | Y | 1668/1841 | Y | 0.284 |
| Wang [10] | 2010 | GWAS | Asian (China) | ESCC | Population | Chips | Y | 9053/13283 | Y | NA |
| Wang [10] | 2010 | GWAS | Asian (China) | GCA | Population | Chips | Y | 2766/11013 | Y | NA |
ESCC, Esophageal squamous cell carcinoma; GCA, Gastric cardia adenocarcinoma; NA: Not applicable for the lack of C allele;
HRM, High-resolution melting; MALDI-TOF-MS, Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry;
aQuality control was conducted when sample of cases and controls was genotyped.
Assessment of methodological quality
The methodological quality assessment for included studies was summarized in Table 2. According to the NOS, Out of a maximum 9-point score, 4 studies had quality scores of 5–6, 8 studies had high quality scores of 7 or 8. The average scores of case-control studies were 6.67.
Table 2. Methodological quality of studies included in the meta-analysis.
| Selection (score) | Comparability (score) | Exposure (score) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Study | Adequate definition of patient case | Representativeness of patients cases | Selection of controls | Definition of control | Control for important factor or additional factor | Ascertainment of exposure (blinding) | Same method of ascertainment for participants | Non-response Ratea | Total Scoreb |
| Peng[15] | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 5 |
| Peng[15] | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 5 |
| Piao [16] | 1 | 1 | 1 | 1 | 2 | 0 | 1 | 1 | 8 |
| Gu [17] | 1 | 1 | 0 | 1 | 2 | 0 | 1 | 0 | 6 |
| Wei [18] | 1 | 1 | 1 | 1 | 2 | 0 | 1 | 0 | 7 |
| Dura [19] | 1 | 1 | 1 | 1 | 2 | 0 | 1 | 0 | 7 |
| Zhou[20] | 1 | 1 | 1 | 1 | 2 | 0 | 1 | 0 | 7 |
| Ren[21] | 1 | 1 | 1 | 1 | 2 | 0 | 1 | 0 | 7 |
| Shang [22] | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 6 |
| Zhang [23] | 1 | 1 | 1 | 1 | 2 | 0 | 1 | 1 | 8 |
| Wang [10] | 1 | 1 | 1 | 1 | 2 | 0 | 1 | 0 | 7 |
| Wang[10] | 1 | 1 | 1 | 1 | 2 | 0 | 1 | 0 | 7 |
aWhen there was no statistical significance in the response rate between case and control groups by using a chi-squared test (P > 0.05), one point was awarded;
bTotal score was calculated by adding up the points awarded in each item.
Evidence synthesis
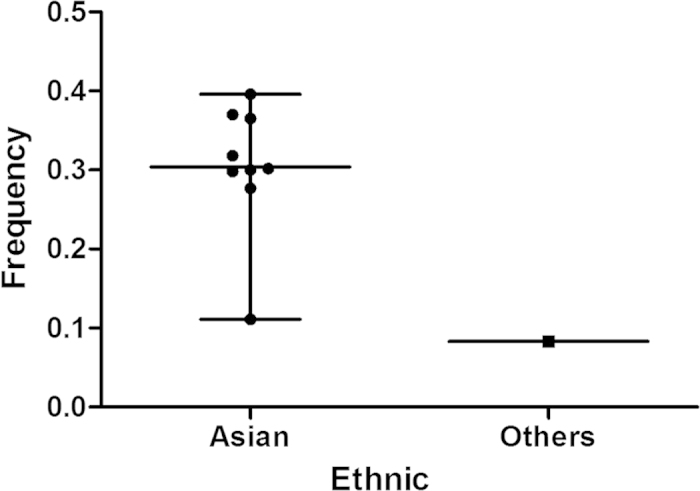
For all of 12 data sets, the frequencies of risk T allele in rs13042395 are presented in Fig. 2. The T allele frequencies for Asians and other populations were 30.41% and 8.30%, respectively.
Figure 2. Frequencies of T allele in rs13042395 among controls stratified by ethnicity.
The evaluation of the association between the C20orf54 rs13042395 polymorphism and the susceptibility to ESCC and GCA is presented in Table 3. Overall analysis indicated that the variant T allele of rs13042395 could significantly decrease the risk of ESCC and/or GCA in all genetic models (T vs. C: OR = 0.95, 95% CI = 0.92–0.97, P < 0.01; CT vs. CC: OR = 0.94, 95% CI = 0.88–0.99, P = 0.04; TT vs. CC: OR = 0.91, 95% CI = 0.83–0.99, P = 0.04; CT + TT vs. CC: OR = 0.94, 95% CI = 0.89–0.99, P = 0.01) except recessive model (TT vs. CT + CC: OR = 0.93, 95% CI = 0.86–1.02, P = 0.12) (Fig. 3).
Table 3. Main results of pooled ORs in the meta-analysis.
| Cases | Controls | Heterogeneity test | Hypothesis test | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Comparisons | n/N | n/N | Q | P | I2(%) | Summary OR (95% CI) | Z | P | Studies |
| Total | |||||||||
| T vs C | 10039/36106 | 23195/79834 | 10.32 | 0.33 | 13 | 0.95(0.92,0.97) | 3.74 | <0.01 | 10 |
| CT vs CC | 4304/9515 | 8381/17945 | 9.68 | 0.21 | 28 | 0.94(0.88,0.99) | 2.04 | 0.04 | 8 |
| TT vs CC | 953/6308 | 1820/11536 | 8.57 | 0.29 | 18 | 0.91(0.83,0.99) | 2.08 | 0.04 | 8 |
| CT + TT vs CC | 5306/10712 | 10115/19865 | 9.25 | 0.41 | 3 | 0.94(0.89,0.99) | 2.50 | 0.01 | 10 |
| TT vs CT + CC | 953/10612 | 1820/19765 | 9.92 | 0.19 | 29 | 0.93(0.86,1.02) | 1.55 | 0.12 | 8 |
| ESCC | |||||||||
| T vs C | 8444/29922 | 16973/58402 | 6.90 | 0.33 | 13 | 0.95(0.90,0.99) | 2.24 | 0.02 | 7 |
| CT vs CC | 2416/6140 | 2657/7147 | 7.21 | 0.21 | 31 | 0.95(0.89,1.03) | 1.24 | 0.22 | 6 |
| TT vs CC | 489/3780 | 649/4706 | 3.21 | 0.67 | 0 | 0.91(0.80,1.04) | 1.38 | 0.17 | 6 |
| CT + TT vs CC | 2931/6246 | 3768/7846 | 4.38 | 0.63 | 0 | 0.95(0.88,1.02) | 0.49 | 0.14 | 7 |
| TT vs CT + CC | 489/6196 | 649/7796 | 5.87 | 0.32 | 15 | 0.94(0.83,1.07) | 0.96 | 0.34 | 6 |
| GCA | |||||||||
| T vs C | 1595/6584 | 6222/21432 | 3.42 | 0.18 | 42 | 0.95(0.91,0.98) | 3.00 | <0.01 | 3 |
| CT vs CC | 1888/3375 | 5724/10798 | 2.25 | 0.13 | 55 | 0.93(0.85,1.01) | 1.69 | 0.09 | 2 |
| TT vs CC | 464/2582 | 1171/6830 | 5.35 | 0.02 | 81 | 0.93(0.69,1.26) | 0.45 | 0.65 | 2 |
| CT + TT vs CC | 2375/4466 | 6347/12019 | 4.75 | 0.09 | 58 | 0.93(0.87,1.00) | 2.00 | 0.05 | 3 |
| TT vs CT + CC | 464/4416 | 1171/11969 | 4.02 | 0.05 | 75 | 0.93(0.82,1.05) | 1.22 | 0.22 | 2 |
Figure 3. Forest plot of cancer risk associated with C20orf54 rs13042395 for the allele comparison (T vs. C).
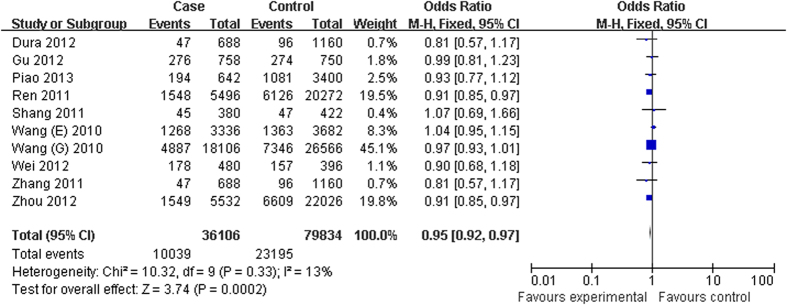
The association between C20orf54 rs13042395 polymorphism and ESCC risk was explored in eight studies. The results indicated that the C20orf54 rs13042395 genotype subtly decreased the risk of ESCC, as revealed by the allele genetic model (T vs. C: OR = 0.95, 95% CI = 0.90–0.99, P = 0.02) (Table 3). GCA was defined by tumor site in four studies. The C20orf54 rs13042395 polymorphism was associated with a decreased risk of GCA (T vs. C: OR = 0.95, 95% CI = 0.91–0.98, P < 0.01) (Table 3).
We performed subgroup analyses stratified by smoking status, all the genetic comparisons did not reach statistical significance in smokers (T vs. C: OR = 0.98, 95% CI = 0.89–1.08, P = 0.73; CT vs. CC: OR = 0.90, 95% CI = 0.66–1.22, P = 0.49; TT vs. CC: OR = 1.00, 95% CI = 0.79–1.27, P = 0.99; CT + TT vs. CC: OR = 0.96, 95% CI = 0.85–1.08, P = 0.48; TT vs. CT + CC: OR = 1.06, 95% CI = 0.84–1.33, P = 0.62) and never smokers (T vs. C: OR = 1.00, 95% CI = 0.94–1.06, P = 0.91; CT vs. CC: OR = 0.95, 95% CI = 0.87–1.02, P = 0.18; TT vs. CC: OR = 0.97, 95% CI = 0.84–1.11, P = 0.66; CT + TT vs. CC: OR = 0.94, 95% CI = 0.87–1.02, P = 0.12; TT vs. CT + CC: OR = 1.13, 95% CI = 0.76–1.67, P = 0.54) (Table 4).
Table 4. Stratified analyses of the C20orf54 rs13042395 polymorphism on cancer risk.
| Heterogeneity test | Hypothesis test | ||||||
|---|---|---|---|---|---|---|---|
| Comparisons | Q | P | I2(%) | Summary OR (95%CI) | Z | P | Studies |
| Smoking status | |||||||
| Smokers | |||||||
| T vs C | 0.95 | 0.81 | 0 | 0.98(0.89,1.08) | 0.34 | 0.73 | 4 |
| CT vs CC | 11.38 | 0.01 | 74 | 0.90(0.66,1.22) | 0.68 | 0.49 | 4 |
| TT vs CC | 1.92 | 0.59 | 0 | 1.00(0.79,1.27) | 0.02 | 0.99 | 4 |
| CT + TT vs CC | 4.79 | 0.19 | 37 | 0.96(0.85,1.08) | 0.70 | 0.48 | 4 |
| TT vs CT + CC | 5.62 | 0.13 | 47 | 1.06(0.84,1.33) | 0.50 | 0.62 | 4 |
| Never smokers | |||||||
| T vs C | 3.47 | 0.33 | 13 | 1.00(0.94,1.06) | 0.11 | 0.91 | 4 |
| CT vs CC | 0.30 | 0.96 | 0 | 0.95(0.87,1.02) | 0.35 | 0.18 | 4 |
| TT vs CC | 1.13 | 0.77 | 0 | 0.97(0.84,1.11) | 0.44 | 0.66 | 4 |
| CT + TT vs CC | 0.52 | 0.91 | 0 | 0.94(0.87,1.02) | 1.56 | 0.12 | 4 |
| TT vs CT + CC | 14.63 | <0.01 | 79 | 1.13(0.76,1.67) | 0.61 | 0.54 | 4 |
| Drinking status | |||||||
| Drinkers | |||||||
| T vs C | 0.36 | 0.95 | 0 | 0.98(0.88,1.10) | 0.27 | 0.19 | 4 |
| CT vs CC | 12.86 | 0.01 | 77 | 0.83(0.56,1.22) | 0.96 | 0.34 | 4 |
| TT vs CC | 0.59 | 0.90 | 0 | 0.99(0.75,1.31) | 0.06 | 0.96 | 4 |
| CT + TT vs CC | 4.62 | 0.20 | 35 | 0.95(0.82,1.10) | 0.65 | 0.51 | 4 |
| TT vs CT + CC | 4.75 | 0.19 | 37 | 1.07(0.82,1.40) | 0.53 | 0.60 | 4 |
| Never drinkers | |||||||
| T vs C | 1.02 | 0.80 | 0 | 0.96(0.90,1.01) | 1.61 | 0.11 | 4 |
| CT vs CC | 0.88 | 0.83 | 0 | 0.95(0.88,1.02) | 1.38 | 0.17 | 4 |
| TT vs CC | 0.67 | 0.88 | 0 | 0.92(0.81,1.05) | 1.22 | 0.22 | 4 |
| CT + TT vs CC | 1.05 | 0.79 | 0 | 0.94(0.88,1.01) | 1.59 | 0.11 | 4 |
| TT vs CT + CC | 0.43 | 0.93 | 0 | 0.94(0.83,1.07) | 0.87 | 0.39 | 4 |
| BMI | |||||||
| Under weight | |||||||
| T vs C | 0.58 | 0.75 | 0 | 0.87(0.77,0.98) | 2.26 | 0.02 | 3 |
| CT vs CC | 0.38 | 0.83 | 0 | 0.91(0.77,1.08) | 1.06 | 0.29 | 3 |
| TT vs CC | 0.66 | 0.72 | 0 | 0.70(0.52,0.93) | 2.42 | 0.02 | 3 |
| CT + TT vs CC | 1.95 | 0.38 | 0 | 0.89(0.76,1.05) | 1.41 | 0.16 | 3 |
| CC vs CT + TT | 1.48 | 0.48 | 0 | 0.74(0.56,0.98) | 2.07 | 0.04 | 3 |
| Normal weight | |||||||
| T vs C | 0.69 | 0.71 | 0 | 0.85(0.80,0.91) | 4.76 | <0.01 | 3 |
| CT vs CC | 3.95 | 0.14 | 0 | 0.82(0.75,0.89) | 4.49 | <0.01 | 3 |
| TT vs CC | 0.74 | 0.69 | 0 | 0.76(0.69,0.85) | 3.35 | <0.01 | 3 |
| CT + TT vs CC | 1.49 | 0.47 | 0 | 0.81(0.75,0.88) | 4.90 | <0.01 | 3 |
| CC vs CT + TT | 0.55 | 0.76 | 0 | 0.93(0.74,1.16) | 0.67 | 0.50 | 3 |
| Brought forward | |||||||
| Over weight | |||||||
| T vs C | 2.62 | 0.27 | 24 | 1.03(0.88,1.21) | 0.38 | 0.70 | 3 |
| CT vs CC | 0.70 | 0.71 | 0 | 1.02(0.82,1.26) | 0.19 | 0.85 | 3 |
| TT vs CC | 3.46 | 0.18 | 42 | 1.06(0.73,1.54) | 0.31 | 0.76 | 3 |
| CT + TT vs CC | 1.17 | 0.56 | 0 | 1.04(0.85,127) | 0.37 | 0.71 | 3 |
| CC vs CT + TT | 5.54 | 0.06 | 64 | 1.08(0.76,1.54) | 0.45 | 0.66 | 3 |
In the subsets divided by drinking status, whereas no significant associations were detected among the drinkers (T vs. C: OR = 0.98, 95% CI = 0.88–1.10, P = 0.19; CT vs. CC: OR = 0.83, 95% CI = 0.56–1.22, P = 0.34; TT vs. CC: OR = 0.99, 95% CI = 0.75–1.31, P = 0.96; CT + TT vs. CC: OR = 0.95, 95% CI = 0.82–1.10, P = 0.51; TT vs. CT + CC: OR = 1.13, 95% CI = 0.76–1.67, P = 0.54) and never drinkers (T vs. C: OR = 0.96, 95% CI = 0.90–1.01, P = 0.11; CT vs. CC: OR = 0.95, 95% CI = 0.88–1.02, P = 0.17; TT vs. CC: OR = 0.92, 95% CI = 0.81–1.05, P = 0.22; CT + TT vs. CC: OR = 0.94, 95% CI = 0.88–1.01, P = 0.11; TT vs. CT + CC: OR = 0.94, 95% CI = 0.83–1.07, P = 0.39) (Table 4).
Furthermore, the subgroup analysis was stratified by BMI, C20orf54 rs13042395 polymorphism was significantly associated with a subtly decreased cancer risk in under-weight group (T vs. C: OR = 0.87, 95% CI = 0.77–0.98, P = 0.02; TT vs. CC: OR = 0.70, 95% CI = 0.52–0.93, P = 0.02; TT vs. CT + CC: OR = 0.74, 95% CI = 0.56–0.98, P = 0.04) and normal weight group (T vs. C: OR = 0.85, 95% CI = 0.80–0.91, P < 0.01; TC vs. CC: OR = 0.82, 95% CI = 0.75–0.89, P < 0.01; CT + TT vs. CC: OR = 0.81, 95% CI = 0.75–0.88, P < 0.01), but no association was observed in over weight group (Table 4).
Test of heterogeneity and sensitivity analysis
Our data sets indicated that there was no significant heterogeneity between studies among all comparisons in the overall analysis (Pheterogeneity > 0.05, I2 ≦ 50%). One-way sensitivity analyses were performed to assess the influence of the results by the systematic omission of the individual studies from the analyses. The dataset showed that the corresponding pooled ORs were not materially altered, indicating that our results were statistically robust (data not shown).
Publication bias
There was no evidence for publication bias using either Begg’s rank correction. Begg’s funnel plot and Egger’s linear regression test were performed to assess the publication bias of the quantitative synthesis literature. The shape of the funnel plot (Begg’s rank correction) did not reveal any evidence of obvious asymmetry (Fig. 4), and no evidence for publication bias using Egger’s linear regression test (Table 5).
Figure 4. Funnel plot of C20orf54 rs13042395 polymorphism and susceptibility to ESCC and GCA for dominant model (CT + TT vs. CC).
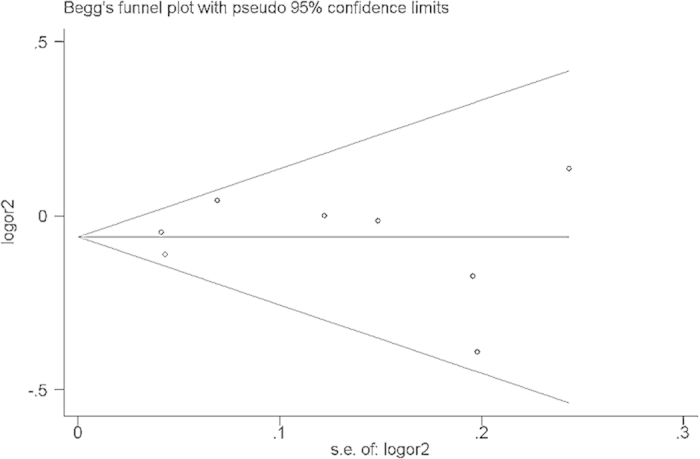
Table 5. Publication bias of C20orf54 rs13042395 for Egger’s test.
| Comparisons | t | p | 95% CI |
|---|---|---|---|
| T vs C | −0.54 | 0.604 | −2.066 ~ 1.283 |
| CT vs CC | −0.09 | 0.933 | −1.995 ~ 1.857 |
| TT vs CC | −0.71 | 0.505 | −2.163 ~ 1.191 |
| CT + TT vs CC | −0.01 | 0.993 | −1.768 ~ 1.756 |
| CC vs CT + TT | −0.52 | 0.619 | −2.239 ~ 1.450 |
Discussion
Results from previous individual published studies investigating the associations between C20orf54 rs13042395 polymorphism and cancer risk (ESCC and/or GCA) were inconclusive. The present study is considered to be the first quantitative meta-analysis concerning the effect of C20orf54 rs13042395 polymorphism on risks of ESCC and GCA and specific stratified analysis (smoking status, drinking status and BMI). By analyzing the data that extracted from 10 published studies, we revealed that C20orf54 rs13042395 polymorphism might be associated with decreased ESCC and GCA risk especially for under-weight and normal weight groups.
The genetic basis of ESCC and GCA between a large number of SNPs and disease predisposition has been explored, and the rs13042395 in C20orf54 was significantly associated with ESCC and GCA risk in the GWAS among Chinese population10. However, other two Chinese population-based GWASs both failed to expore a significant association of rs13042395 with the risk of ESCC and GCA2,11. In the present study, we identified a significant association of rs13042395 with the risk of ESCC and GCA. This indicated that the finding of GWAS need independent replication studies to verify.
ESCC and GCA are complex diseases likely resulting from multiple interacting genetic polymorphisms and gene-environment interactions. Both in the western countries and Asian especially in China, heavy smoking and alcohol consumption were identified as the main environmental risk factors for ESCC and GCA24,25. C20orf54 has a high homology with rat C20orf54, a transmembrane protein involved in the uptake of riboflavin in the small intestine10. The C20orf54 genotypes modulated the risk of ESCC in smokers, drinkers, or in individuals with a negative family history18. These findings suggest that C20orf54 may alter environmental risk factors. Interestingly, our results indicated that smoking and drinking did not significantly alter the effects of C20orf54 rs13042395 polymorphism on the risk of ESCC and/or GCA. However, on this point, our meta-analysis obtained the consistent conclusions came up with Wang et al.10.
In the present study, the C20orf54 rs13042395 T allele significantly decreased the risk of ESCC and/or GCA in the subjects with BMI less than 24 especially between 18.5 to 24. Overweight and obesity have been consistently related to gastric and esophageal adenocarcinoma, but not to squamous cell carcinoma26,27,28. The influence of obesity on gastric and esophageal adenocarcinoma may be related to higher incidence of gastroesophageal reflux in obese individuals29, and the risk of gastroesophageal reflux is strongly associated with the risk for Barrett’s esophagus30,31.
The following limitations should be acknowledged in our studies. First, the present meta-analysis only included design of case-control studies, some of which were hospital based studies. Thus, the controls may not reflect the representative element of the source population. Second, although all eligible studies were summarized, the relatively small study number may lead to reduced statistical power when stratified according to the cancer type, ethnicity, smoking status, drinking status and BMI. Third, the pooled datasets without excluding the studies with inefficient points based on NOS. In addition, Large-scale studies will be needed for high-risk population screening, individualized prevention, treatment and exposure rating in the future.
In summary, current data suggest that C20orf54 rs13042395may be associated with a significantly decreased risk of ESCC and GCA, especially for the subjects with BMI less than 24 particularly between 18.5 to 24. Notably, based on the well-designed studies at multicenters with large sample size will be needed for further validate our results.
Materials and Methods
Data source and search strategy
We comprehensively identified studies through searching PubMed, Embase, Web of Science, Chinese National Knowledge Infrastructure (CNKI) and Wanfang database using terms “C20orf54”, “RFT2” and “rs13042395” for both case–control and cohort studies, which evaluated the association between C20orf54 rs13042395 polymorphism and the risk of ESCC and/or GCA (last search update: March 24, 2015). The search was limited to papers published in English or Chinese language. In addition, Reference lists of retrieved articles were examined manually to further identify potentially relevant studies.
Inclusion and exclusion criteria
Studies were included in the analysis if following criteria were met: (i) based on case-control studies (including cohort studies and GWASs) examined the associations between the C20orf54 rs13042395 and ESCC or GCA; (ii) sufficient allele or genotype data for estimating an odds ratio (OR) with corresponding 95% confidence intervals (95%CIs); (iii) genotype distribution of control groups must be in accordance with the assumptions of Hardy-Weinberg equilibrium (HWE). Case-control studies based on the esophageal adenocarcinomas and/or gastric non-cardia adenocarcinoma were excluded. In case of redundant publications, only the studies with the largest sample size and/or latest published date were included.
Data extraction and quality assessment
Two independent authors (Fujiao Duan and Shuli Cui) extracted the data from the eligible publications. Data for analyses, including first author, publication year, study design, ethnicity, cancer type, source of control, detection methods of C20orf54 rs13042395 polymorphism and quality control or not, characteristics of cases and controls. If discrepancies existed, consensus would be finally reached on discussion.
We assessed quality of included studies by a modified checklist based on the Newcastle–Ottawa Scale (NOS)32, with discrepancies resolved by consensus. A nine-point scale of the NOS (range, 0–9 points) has been developed for the evaluation. A high-quality study was defined as one with great than or equal to 7 points.
Quantitative data synthesis and analyses
We utilized RevMan 5.0 (Cochrane Collaboration, Oxford, UK) and STATA 12.0 (StataCorp, College Station, TX, USA) to perform all the statistical analysis.
RevMan 5.0 was used to estimate the association between C20orf54 rs13042395 polymorphism and cancer risk by the pooled ORs with corresponding to 95%CIs. The stratified analysis was conducted by ethnicity (Asian, Caucasian), smoking status (smokers, never smokers), drinking status (drinkers, never drinkers) and BMI (under weight <18.5, normal weight: 18.5–24, over weight >24).
Heterogeneity was explored by the chi-squared test (χ2) of heterogeneity and the inconsistency index (I2) between each individual study. By heterogeneity test, if P-value for heterogeneity test (Pheterogeneity) < 0.05 or I2 > 50%, the sources of heterogeneity would be used for meta regression in STATA 12.033. Random- or fixed-effects models were used depending on Pheterogeneity. If Pheterogeneity ≥ 0.05, we used the fixed effect model (the Mantel-Haenszel method)34. Otherwise, random effects model (DerSimonian and Laird method) was selected35. The significance of merged OR was dependent on the Z-test, P < 0.05 was considered significant.
Sensitivity analysis, in which one study is omitted at a time, was performed to assess the quality and consistency of the results.
Publication bias was evaluated by Begg’s test (rank correlation test)36 and then statistically using Egger’s test (weighted linear regression test)37. This analysis was performed using the STATA 12.0 procedure of ‘Metabias’.
Additional Information
How to cite this article: Duan, F. et al. Esophageal Squamous Cell Carcinoma and Gastric Cardia Adenocarcinoma Shared Susceptibility Locus in C20orf54: Evidence from Published Studies. Sci. Rep. 5, 11961; doi: 10.1038/srep11961 (2015).
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China (No.81202278). The authors would like to thank the people who participated in this study.
Footnotes
Author Contributions All authors contributed significantly to this work. F.J.D. designed and drafted the manuscript. S.L.C. and L.P.D. collected studies, summarized data and copyedited this article. X.Z., C.H.S. and Y.S. collected articles, summarized data, did statistical work. All authors reviewed this manuscript and approved the final draft.
References
- Parkin D. M., Bray F., Ferlay J. & Pisani P. Global cancer statistics, 2002. CA Cancer J Clin 55, 74–108 (2005). [DOI] [PubMed] [Google Scholar]
- Wu C. et al. Genome-wide association study identifies three new susceptibility loci for esophageal squamous-cell carcinoma in Chinese populations. Nat Genet 43, 679–684 (2011). [DOI] [PubMed] [Google Scholar]
- Metzger R., Schneider P. M., Warnecke-Eberz U., Brabender J. & Holscher A. H. Molecular biology of esophageal cancer. Onkologie 27, 200–206 (2004). [DOI] [PubMed] [Google Scholar]
- Hashibe M. et al. Esophageal cancer in Central and Eastern Europe: tobacco and alcohol. Int J Cancer 120, 1518–1522 (2007). [DOI] [PubMed] [Google Scholar]
- Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7–14 June 1994. IARC Monogr Eval Carcinog Risks Hum 61, 1–241 (1994). [PMC free article] [PubMed] [Google Scholar]
- Chow W. H. et al. Body mass index and risk of adenocarcinomas of the esophagus and gastric cardia. J Natl Cancer Inst 90, 150–155 (1998). [DOI] [PubMed] [Google Scholar]
- Hongo M., Nagasaki Y. & Shoji T. Epidemiology of esophageal cancer: Orient to Occident. Effects of chronology, geography and ethnicity. J Gastroenterol Hepatol 24, 729–735 (2009). [DOI] [PubMed] [Google Scholar]
- Lichtenstein P. et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 343, 78–85 (2000). [DOI] [PubMed] [Google Scholar]
- He Y. et al. Effect of riboflavin-fortified salt nutrition intervention on esophageal squamous cell carcinoma in a high incidence area, China. Asian Pac J Cancer Prev 10, 619–622 (2009). [PubMed] [Google Scholar]
- Wang L. D. et al. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet 42, 759–763 (2010). [DOI] [PubMed] [Google Scholar]
- Shi Y. et al. A genome-wide association study identifies new susceptibility loci for non-cardia gastric cancer at 3q13.31 and 5p13.1. Nat Genet 43, 1215–1218 (2011). [DOI] [PubMed] [Google Scholar]
- Yamamoto S. et al. Identification and functional characterization of rat riboflavin transporter 2. J Biochem 145, 437–443 (2009). [DOI] [PubMed] [Google Scholar]
- Fujimura M. et al. Functional characteristics of the human ortholog of riboflavin transporter 2 and riboflavin-responsive expression of its rat ortholog in the small intestine indicate its involvement in riboflavin absorption. J Nutr 140, 1722–1727 (2010). [DOI] [PubMed] [Google Scholar]
- Siassi F. & Ghadirian P. Riboflavin deficiency and esophageal cancer: a case control-household study in the Caspian Littoral of Iran. Cancer Detect Prev 29, 464–469 (2005). [DOI] [PubMed] [Google Scholar]
- Peng S. Q. Analysis on clinicopathologic features influencing factors of survival and the variation of rs13042395 loci for patients with concurrent esophageal and gastric cardia cancers, Master thesis, Henan University of Science and Technology (2014).
- Piao J. M. et al. Replication of results of genome-wide association studies on esophageal squamous cell carcinoma susceptibility loci in a Korean population. Dis Esophagus 27, 798–801 (2014). [DOI] [PubMed] [Google Scholar]
- Gu H. et al. Replication study of PLCE1 and C20orf54 polymorphism and risk of esophageal cancer in a Chinese population. Mol Biol Rep 39, 9105–9111 (2012). [DOI] [PubMed] [Google Scholar]
- Wei W. et al. Functional single nucleotide polymorphism in C20orf54 modifies susceptibility to esophageal squamous cell carcinoma. Dis Esophagus 26, 97–103 (2013). [DOI] [PubMed] [Google Scholar]
- Dura P. et al. GWAS-uncovered SNPs in PLCE1 and RFT2 genes are not implicated in Dutch esophageal adenocarcinoma and squamous cell carcinoma etiology. Eur J Cancer Prev 22, 417–419 (2013). [DOI] [PubMed] [Google Scholar]
- Fuyou Z. et al. Correlation of C20orf54 gene polymorphism with the smoking, drinking and BMI in esophageal squamous cell carcinoma. Journal of Henan University (Medical science) 31, 6 (2012). [Google Scholar]
- Ren J. Family history and genetic susceptibility to gastric cardia adenocarcinoma of Chinese han population, Doctor thesis, Zhengzhou University, (2011).
- Shang X. Association of the single nucleotide polymorphisms of hRFT genes with susceptibility to Hazakh’s esophageal cancer in XinJiang, Master thesis, Shihezi University, (2011).
- Zhang H. et al. Genetic variants at 1q22 and 10q23 reproducibly associated with gastric cancer susceptibility in a Chinese population. Carcinogenesis 32, 848–852 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan T. L., Davis S., Kristal A. & Thomas D. B. Obesity, alcohol, and tobacco as risk factors for cancers of the esophagus and gastric cardia: adenocarcinoma versus squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 4, 85–92 (1995). [PubMed] [Google Scholar]
- Fan Y., Yuan J. M., Wang R., Gao Y. T. & Yu M. C. Alcohol, tobacco, and diet in relation to esophageal cancer: the Shanghai Cohort Study. Nutr Cancer 60, 354–363 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacInnis R. J., English D. R., Hopper J. L. & Giles G. G. Body size and composition and the risk of gastric and oesophageal adenocarcinoma. Int J Cancer 118, 2628–2631 (2006). [DOI] [PubMed] [Google Scholar]
- Merry A. H., Schouten L. J., Goldbohm R. A. & van den Brandt P. A. Body mass index, height and risk of adenocarcinoma of the oesophagus and gastric cardia: a prospective cohort study. Gut 56, 1503–1511 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagergren J., Bergstrom R., Adami H. O. & Nyren O. Association between medications that relax the lower esophageal sphincter and risk for esophageal adenocarcinoma. Ann Intern Med 133, 165–175 (2000). [DOI] [PubMed] [Google Scholar]
- Edelstein Z. R., Bronner M. P., Rosen S. N. & Vaughan T. L. Risk factors for Barrett’s esophagus among patients with gastroesophageal reflux disease: a community clinic-based case-control study. Am J Gastroenterol 104, 834–842 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson C., Johansson A. L., Nyren O. & Lagergren J. Socioeconomic factors and risk of esophageal adenocarcinoma: a nationwide Swedish case-control study. Cancer Epidemiol Biomarkers Prev 14, 1754–1761 (2005). [DOI] [PubMed] [Google Scholar]
- Liu X. et al. Gastro-esophageal reflux disease symptoms and demographic factors as a pre-screening tool for Barrett’s esophagus. PLoS One 9, e94163 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells G. et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. (2011). Date of access: 25/11/2012. http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp
- Thompson S. G. & Higgins J. P. How should meta-regression analyses be undertaken and interpreted? Stat Med 21, 1559–1573 (2002). [DOI] [PubMed] [Google Scholar]
- Mantel N. & Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22, 719–748 (1959). [PubMed] [Google Scholar]
- DerSimonian R. & Laird N. Meta-analysis in clinical trials. Control Clin Trials 7, 177–188 (1986). [DOI] [PubMed] [Google Scholar]
- Begg C. B. & Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 50, 1088–1101 (1994). [PubMed] [Google Scholar]
- Egger M., Davey Smith G., Schneider M. & Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]