Abstract
Purpose
Transfer of genetic material from cancer cells to normal cells occurs via microvesicles. Cell specific phenotypes can be induced in normal cells by the transfer of material in microvesicles, leading to genetic changes. We report the identification and expression of prostate specific genes in normal human marrow cells co-cultured with human prostate cancer cells.
Materials and Methods
We harvested prostate tissue from 11 patients with prostate cancer. In 4 cases prostate tissue was co-cultured across from human marrow for 2 or 7 days but separated from it by a 0.4 μM polystyrene membrane. In 5 cases conditioned medium from patient cancer tissue was collected and ultracentrifuged, and microvesicles were collected for co-culture (3) and vesicle characterization (3). Explanted human marrow was harvested from cultures and RNA extracted. Real-time reverse transcriptase-polymerase chain reaction was done for select prostate specific genes.
Results
Marrow exposed to human prostate tumor or isolated microvesicles in culture in 4 and 3 cases, respectively, showed at least 2-fold or greater prostate gene expression than control marrow. In 1 case in which normal prostate was co-cultured there were no prostate gene increases in normal marrow.
Conclusions
Prostate cancer tumor cells co-cultured with human bone marrow cells induce prostate specific gene expression. The proposed mechanism of transfer of genetic material is via microvesicles. This represents an opportunity for novel therapeutic agents, such as antibodies, to block microvesicle release from cancer cells or for agents that may block cells from accepting microvesicles.
Keywords: prostate, prostatic neoplasms, phenotype, bone marrow, gene expression
Prostate cancer remains a major health problem, affecting more than 186,000 American males in 2008 with 27,000 deaths.1 Therapy has centered on tumor hormone responsiveness and advances have been made but overall results are still discouraging.
The mechanisms of local progression, recurrence and metastasis remain uncertain despite enormous work in this regard. A recent study indicated the secretion of oncosomes or microvesicles from the human prostate cancer cell lines DU145 and LNCap, and showed an association with a region of common chromosomal deletion in metastatic disease.2 This study suggested that microvesicles shed from prostate cancer cells could alter the tumor microenvironment in a manner that may promote disease progression.
Microvesicles released from damaged or stimulated cell populations alter the phenotype of various target cells. They transfer CD41, integrins and CXCR43–5 as well as HIV and prions6,7 between cells. Embryonic stem cell microvesicles reprogram hematopoietic stem/progenitor cells via the horizontal transfer of mRNA and protein.8 Similarly tumor derived microvesicles carry several surface determinants and mRNA, and transfer some of these determinants to monocytes.9 Apoptotic bodies from irradiated EBV carrying cell lines transfer DNA to various co-cultured cells and integrated but not episomal copies of EBV resulted in expression of the EBV encoded genes EBER and EBNAI in recipient cells at high copy number.10 Extracts from T lymphocytes containing transcription factor complexes induced fibroblasts to express lymphoid genes.11 In a recent study Deregibus et al extracted RNA from endothelial progenitor microvesicles and microarray, and performed data analysis.12 They found a total of 298 transcripts, of which 183 were associated with Reference Sequence identifiers and the remaining UniGene, suggesting that the particles did not contain a random sample of cellular mRNA but rather a specific subset.
We previously used a model to investigate the capacity of normal murine lung to alter the genetic phenotype of normal murine marrow cells. In a cross-culture system in which marrow cells are cultured across from normal or irradiated lung we found that marrow cells expressed lung specific mRNA on real-time PCR for surfactants A, B, C and D, aquaporin 5 and Clara cell specific protein.13 CM from lungs mediated similar genetic phenotypes in incubated marrow cells.13 Lung CM, pelleted micro-particles or microvesicles had high levels of lung specific mRNA. Incubation of marrow cells with fluorescence activated cell sorting isolated lung derived microvesicles also induced marked increases in lung specific mRNA and entry of microvesicles into marrow cells. Marrow cells co-cultured across from lung also showed an increased capacity to convert to lung cells after transplantation into irradiated hosts. Recent studies suggest that transcriptional mechanisms were involved in the finally observed genetic phenotype.14 When rat lung was cultured opposite mouse marrow, induced surfactant mRNAs were mouse and rat, indicating that a rat transcription factor was transferred. Further study of murine lung derived microvesicles revealed protein and microRNA. Our working hypothesis is that lung specific mRNA and a protein transcription factor are transferred to cells via microvesicles. Also, microRNA is transferred, which may in turn modulate mRNA levels.
These observations on the transfer of genetic phenotype to marrow cells in mice along with observations of microvesicle derivation from cancer cells formed the basis of our studies of microvesicle evolution in excised human prostate cancer cells.
MATERIALS AND METHODS
Tumor Collection
According to the Rhode Island Hospital committee on protection of human subjects (institutional review board) consent was obtained for each patient. Tumor was surgically removed and processed. The sample was weighed and finely minced into approximately 1 cm2 pieces.
BM Cells
BM was obtained from healthy volunteers with informed consent according to the institutional review board. BM cells (3 × 106 per well) were used 1) as WBM or 2) as Ficoll separated BM cells with mononuclear cells isolated using a Ficoll-Paque™ Premium density gradient according to manufacturer instructions or 3) ACL2 separation. Red blood cells were lysed using BD Pharm Lyse™ lysing buffer according to manufacturer instructions.
Co-Culture
Experiments were done in 3 × 106 BM cells plated per well. Between 50 and 100 mg of minced tumor were placed in a 0.4 μm Millicell® culture plate insert with Dulbecco’s modified Eagle’s medium (Invitrogen™) supplemented with 10% fetal bovine serum (HyClone®), 1% penicillin/streptomycin (Invitrogen) and 20 ng/ml human stem cell factor (R & D Systems®). Control wells were cultured with equal numbers of BM cells plated without tumor. Co-cultures were maintained for 2 or 7 days in 5% CO2 at 37C. For CM the tumor pieces were cultured without BM cells. After 7 days of culture CM was removed and further processed to isolate microvesicles.
Prostate Tumor Microvesicle Isolation
Microvesicles were isolated from CM after 2 or 7 days of culture. CM was centrifuged at 300 × gravity for 10 minutes at 4C. Supernatant was UCF at 28,000 × G for 1 hour at 4C, which was repeated. The resulting pellet was resuspended in PBS and an equal volume of the supravital red fluorescent cell membrane dye PKH26, diluted 1:250 in diluent C (Sigma®) and the cell cytoplasm supravital dye CFSE (Invitrogen) at a final concentration of 0.02 μM. Microvesicles were incubated for 15 minutes at 37C. An equal volume of 10% fetal bovine serum solution in PBS was added and samples were UCF as described. The UCF pellet was resuspended in growth medium and co-cultured with BM cells or further processed for electron microscopy.
Microscopy
Transmission electron
UCF pellets were fixed with 3% glutaraldehyde in 0.15M sodium cacodylate buffer (Electron Microscopy Sciences, Hatfield, Pennsylvania) for several days at 4C. After several rinses the samples were post-fixed in 1% osmium tetroxide (Electron Microscopy Sciences) for 1 hour at 4C. After post-fixation the samples were buffer rinsed, diced into 1.5 mm cubes and covered with 3% agar solution (Electron Microscopy Sciences). After they were hardened excess agar was removed. The sample was dehydrated through a graded series of acetone washes and embedded in Spurr’s epoxy resin (Ladd Research Industries, Williston, Vermont). Semithin sections (1 μm) were prepared using a Reichert Ultracut S microtome (Leica Geosystems, Heerbrugg, Switzerland), stained with methylene blue-azure II and evaluated for areas of interest. Ultrathin sections were prepared, retrieved on 300 mesh thin bar copper grids and contrasted with uranyl acetate and lead citrate (Electron Microscopy Sciences). Sections were examined using a Morgagni™ 268n transmission electron microscope. Images were collected with an AMT Advantage 542 charge coupled device camera system (FEI™).
Fluorescence
BM cells co-cultured for 2 days with isolated microvesicles were harvested by rinsing and collecting cells from a tissue culture flask with 1 × PBS, and centrifugation at 300 × gravity for 10 minutes at 4C. WBM cells containing fluorescent labeled microvesicles were sorted on a BD Influx™ cell sorter equipped with lasers, including a 100 mW 488 nm sapphire, a 100 mW ultraviolet, a 405 nm violet, a 635 nm red and a 561 nm green/yellow laser. PKH26 was excited by the 561 nm laser and emission detected through a 624/40 filter. CFSE was excited by the 488 nm laser and emission was collected through a 528/38 filter. Cytospin slides were made using 1 × 105 cells per slide by spinning cells at 350 rpm for 10 minutes. Slides were cover slipped using Vectashield® with 1.5 μg/ml DAPI. Images were obtained with an Axio Imager Z1 fluorescence microscope and Axiovision 4.6.3 software (Carl Zeiss®). Digital images were acquired with an AxioCam HRm, and 3-dimensional images were obtained using an ApoTome (Carl Zeiss) for structural imaging and a 4-dimensional acquisition module.
RNA Extraction and RT-PCR
RNA from cultured or co-cultured BM cells was isolated using the RNeasy™ Mini Kit or QIAmp® RNA Blood Mini Kit. RNA was measured for quantity and quality (260/280 nm) using a spectrophotometer. Isolated RNA was used to amplify cDNA using a High Capacity cDNA transcription kit (Applied Biosystems®). Equal amounts of RNA were used in all samples in each experiment. Amplification reactions consisted of 1 cycle for 10 minutes at 25C, 2 cycles of 60 minutes each at 37C and 1 cycle of 5 seconds at 85C.
Gene expression was analyzed by RT-PCR in 96-well plates on a 7900 HT-Fast Real Time PCR System (Applied Biosystems). Reactions consisted of 20 × assay mix (Applied Biosystems) for glyceraldehyde-3-dehydrogenase or a target gene, and 2 × TaqMan® PCR Master Mix and predetermined amounts of cDNA. The human prostate assays used were PCA-3, STEAP, PART, acid phosphatase prostate, TMPRSS2, PSA, PSCA, ERG, ETV1 and KLK3. The PCR reaction consisted of an initial enzyme activation step at 95C for 10 minutes, followed by 40 cycles at 95C for 15 seconds and at 60C for 1 minute. We determined a CT for each sample and averaged triplicate sample values. The 2−ΔΔT method was used to calculate the relative expression of each target gene. Briefly, the mean CT of target genes in each sample was normalized to its averaged housekeeping gene CT to determine ΔCT. This value was normalized to control samples (ΔΔCT) and the 2−ΔΔT value was determined. To calculate 2−ΔΔT for target genes with no expression in the control group we assigned a CT of 40 to the control group so that a relative quantity of target gene could be reported. The control group used for all comparisons comprised BM cells co-cultured without prostate tumor, CM or microvesicles for the same duration as experimental groups.
RESULTS
In this study we evaluated 11 patients with prostate cancer and only 1 with normal prostate tissue. Patients had cytological confirmation of prostate cancer. Some patients were studied by co-culturing tumor tissue across from freshly harvested marrow from human volunteers while the tumor tissue of other patients was used to derive microvesicles that were then co-cultured with BM. Prostate cancer tissue from another group of patients was used to isolate microvesicles for electron microscopy or for studies showing microvesicle entry into target marrow cells. The table lists patient demographics. The Appendix shows the 10 prostate specific genes for whose expression target marrow cells were analyzed.
BM co-cultured with prostate cancer tissue was processed in different manners, as described. Patient 1, whose tissue contained no tumor cells, showed no expression of any prostate gene. Control tissue, consisting of normal cultured BM only, expressed low to no prostate gene levels. Thus, we detected quite high, significantly increased gene levels. Data are shown as a structured, color coded, fold difference key (figs. 1 and 2). Values less than a 2-fold increase over the marrow control were considered negative. The different colors represent 2 to 10 to more than 1,000-fold (red areas) increased gene expression detected in marrow. Donor marrow cultured opposite cancer tissue from patient 2 for 7 days showed increases in 8 prostate specific genes, of which 5 showed a 100 to 1,000 to more than 1,000-fold difference (fig. 1). Marrow cultured opposite tumor tissue from patient 3 showed increases in all interrogated prostate specific genes, of which 8 showed greater than 1,000-fold increases. Marrow from patient 4 cultured for only 7 days showed an increase in the 2 prostate specific genes PSA and STEAP at relatively lower levels. Marrow from patient 5 cultured for 7 days showed increases in the 3 prostate specific genes PSA, PSCA and KLK3. Thus, we saw the induction of prostate specific gene expression in normal marrow tissue in each co-culture experiment.
Figure 1.
Normal human BM cells co-cultured with prostate tumor samples. BM cells used were Ficoll-Paque Premium (Ficol) separated BM cells. Red blood cells were lysed using BD Pharm Lyse. Colors indicate prostate specific RNA fold increases in BM cells on RT-PCR vs BM cells cultured without prostate tumor. A, 2-day culture. B, 7-day culture. ACL2, ACL2 separation.
Figure 2.
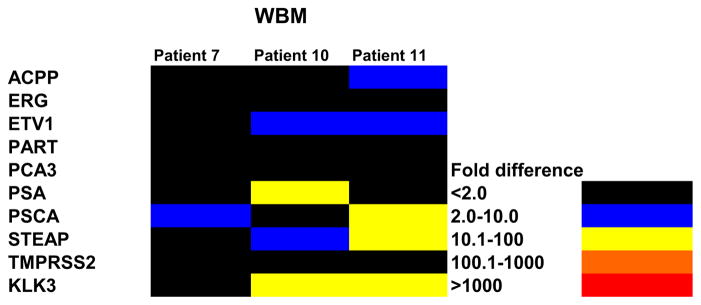
Isolated prostate tumor microvesicles from prostate tumor sample cultures were co-cultured with human BM cells for 7 days. Colors indicate fold increases in prostate specific RNA on RT-PCR vs cells cultured without CM.
Patient demographics
| Pt No.—Age | Race | PSA (ng/ml) | Gleason Grade | Medications |
|---|---|---|---|---|
| 1—65* | White | 3.7 | 3 + 3 | Flecainide, pravastatin |
| 2—57 | White | 4.7 | 4 + 5 | Atenolol, simvastatin |
| 3—58 | Black | 27 | 4 + 5 | Lisinopril, glucophage, Glucotrol®, Lantus® |
| 4—56 | White | 10 | 3 + 3 | None |
| 5—64 | White | 6.7 | 3 + 4 | None |
| 6—55 | White | 9.0 | 3 + 4 | None |
| 7—65 | White | 9.3 | 4 + 4 | None |
| 8—61 | White | 3.3 | 4 + 3 | Balsalozide, Diovan®, omeprazole, sertraline, zolipidezem |
| 9—75 | White | 7.5 | 4 + 3 | None |
| 10—61 | White | 4.2 | 3 + 4 | Atacand®, aspirin |
| 11—71 | White | 8.2 | 3 + 4 | Advil®, fluticasone |
Normal control prostate tissue.
In another experiment prostate cancer tissue from patient 7 was established in liquid culture for 7 days without BM and CM was collected. Pelleted CM contained microvesicles, which were evaluated by electron microscopy (fig. 3). Residual material was incubated with normal human BM for 7 days. Microvesicles from patients 10 and 11 were also incubated with marrow for 7 days. Increased prostate specific genes were seen in each case (fig. 2).
Figure 3.
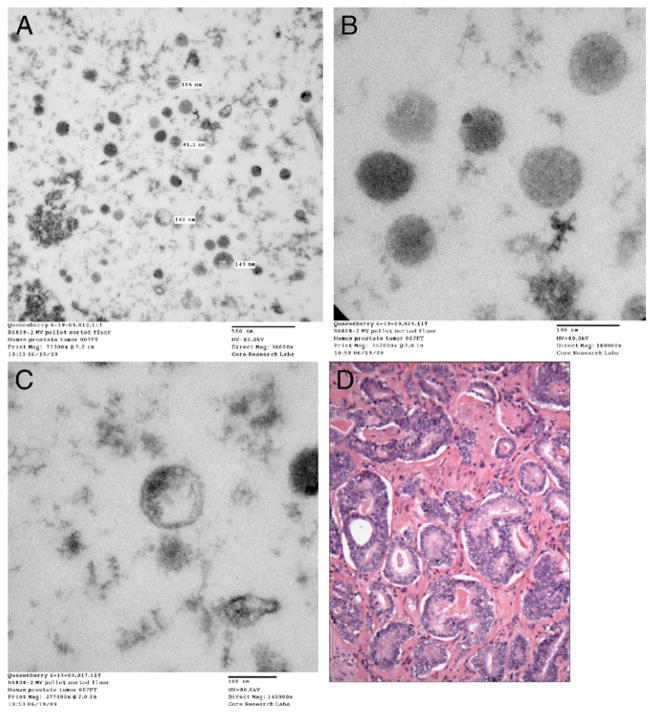
Transmission electron microscopy reveals microvesicles from prostate tumor samples cultured without BM cells for 7 days. CM was removed and microvesicles were isolated by high speed centrifugation. Sections were examined by transmission electron microscope and images were collected with charge coupled device camera. A, microvesicles. Reduced from ×36,000. B, microvesicles. Reduced from ×180,000. C, microvesicles. Reduced from ×140,000. D, pathology slide shows prostate tumor cells from same patient. H & E, reduced from ×600.
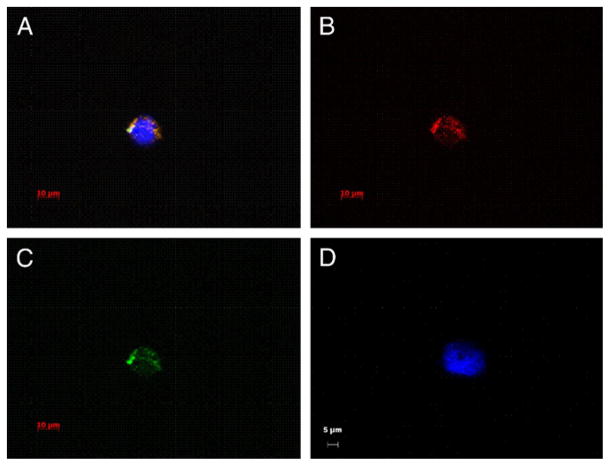
Figure 4 shows fluorescence microscopy images of BM cells containing fluorescent labeled prostate tumor microvesicles. Tumor tissue from patient 9 was isolated and stained. The resulting microvesicles were co-cultured with normal human marrow cells for 2 days. Approximately 6% of marrow cells absorbed fluorescent microvesicles (fig. 4). Figure 4, A shows a cell with 3 fluorescent colors. We used a Texas Red filter to reveal fluorescence from PKH26 and a FITC filter for green CFSE labeled microvesicles (fig. 4, B and C). Figure 4, D shows a cell counterstained with DAPI that did not take up microvesicles. These data clearly indicate uptake of prostate cancer derived microvesicles by marrow cells.
Figure 4.
Fluorescence microscopy shows digital images of BM cells containing fluorescent labeled prostate tumor microvesicles. A to C, representative PKH26 and CFSE labeled prostate tumor derived microvesicles in BM cell. Scale bar indicates 10 μm. A, composite of images using DAPI, FITC and Texas Red filters. Yellow-orange areas indicate double positive PKH26 and CFSE label in microvesicles. Blue area indicates nuclear stain DAPI. B, image using Texas Red filter. C, image using FITC filter. D, representative DAPI stained BM cell negative for fluorescently labeled microvesicles. Reduced from 63×, scale bar indicates 5 μm.
The existence of microvesicles was established using electron microscopy. A patient sample was processed. Figure 3, D shows a photomicrograph of prostate cancer morphology in this patient.
DISCUSSION
Cancer derived microvesicles were first noted in the 1970s. Subsequently the effects of normal or cancer cell derived microvesicles on cancer cells or their environment were noted to promote survival, immune surveillance escape,15–18 extracellular matrix degradation,19 –21 angiogenesis22,23 and metastasis.24 Tumor derived microvesicles can enhance the metastatic potential of melanoma cells in vivo.25 Studies in human cancer cell lines showed the delivery of oncogenic epidermal growth factor receptor via microvesicles to cultured endothelial cells,24 and derivation of microvesicles from human prostate2 and colorectal cell lines.26 Skog et al reported that human glioblastoma tissues obtained from surgical resection showed release of microvesicles containing mRNA, miRNA and angiogenic proteins.27 These microvesicles were imbibed by normal host cells, such as brain microvascular endothelial cells. They detected mRNA mutant/variants and miRNA characteristic of glioma in serum microvesicles in patients with glioblastoma. Wysoczynski and Ratajczak noted that human and murine lung cancer cell lines secrete microvesicles, and tumor microvesicles enhanced the metastatic potential of murine and human lung cancer cells in vivo.28
Our series indicates that prostate cancer cells in close proximity to human marrow cells induce the expression of prostate specific mRNA in the marrow cells and shows the exchange of the prostate specific phenotype from tumor tissue to marrow cells. Also, we noted that isolated microvesicles entered marrow cells and induced prostate specific mRNA in BM cells. Some study limitations are its small sample size and possible origin of microvesicles from normal prostate tissue. However, our data indicate induced genetic changes in marrow cells toward a prostate specific phenotype.
CONCLUSIONS
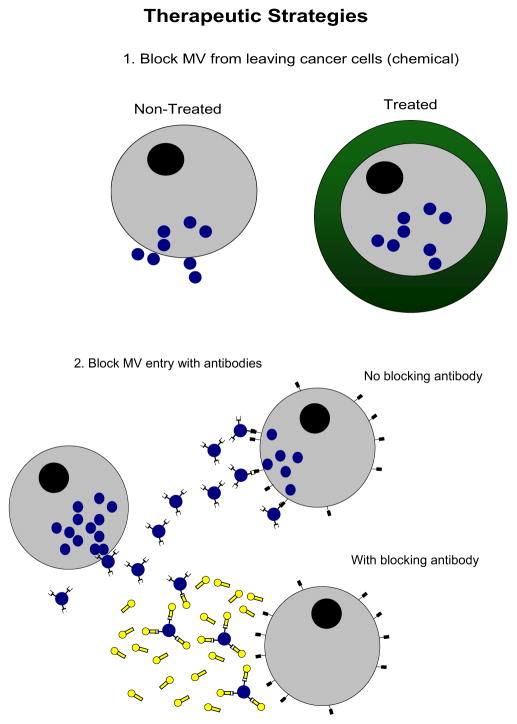
These observations suggest that microvesicles derived from prostate cancer cells could enter circulating monocytes, stem cells or other cells, altering their phenotype toward that of a prostate cancer cell. Results indicate significantly increased gene expression in BM cells co-cultured with prostate tumor cells (Gleason grades 6–9). Our study establishes a base on which to begin evaluating the significance of microvesicle mediated genetic transfer, mechanisms of transfer (ie surface epitope profiles) and therapeutic options for blocking (ie antibodies to microvesicle surface epitopes) or manipulating such transfer to influence the disease process. Future studies will determine whether there is transfer of genetic or transcriptional factors via microvesicles from human prostate cancer cells to fresh human BM cells, whether transfer of genetic material impacts tumorigenicity and metastasis, and whether this process can be inhibited to prevent disease progression (fig. 5). Understanding the role of endogenous intracellular factors involved in the regulation of prostate cancer progression has the potential to lead to the development and selective application of novel, mechanism directed chemotherapeutic agents.
Figure 5.
Therapeutic strategies to prevent microvesicle (MV) transfer, including chemical treatment to block microvesicles from leaving cells (1) and antibody treatment of cells, which result in blocking sites where microvesicles bind to cells (2).
Acknowledgments
Supported by NCRR 1P20RR025179-01.
Abbreviations and Acronyms
- BM
bone marrow
- CFSE
carboxyfluorescein diacetate succinimidyl ester
- CM
conditioned medium
- CT
cycle threshold
- DAPI
4,6-diamidino-2-phenylindole
- EBV
Epstein Barr virus
- FITC
fluorescein isothiocyanate
- KLK3
kallikrein 3
- PART
prostate androgen regulated transcript 1
- PBS
phosphate buffered saline
- PCA-3
prostate cancer antigen 3
- PCR
polymerase chain reaction
- PSA
prostate specific antigen
- PSCA
prostate stem cell antigen-A
- RT
reverse transcriptase
- STEAP
6-transmembrane epithelial antigen of prostate
- TMPRSS2
transmembrane protease, serine 2–
- UCF
ultracentrifuged
- WBM
whole BM
APPENDIX
Genes Investigated
| Gene | Ca Implications |
|---|---|
| Acid phosphatase prostate | Up-regulated in pts with prostate Ca |
| ERG (transcription regulator) | Abnormal gene infusion associated with prostate Ca occurs when ERG or ETV fuses with TMPRSS2 (involved with Ewing’s sarcoma) |
| ETV1 (transcription activator) | See ERG |
| PART | Androgen dependent prostate Ca marker |
| PCA-3 | Prostate Ca specificity |
| PSCA | Up-regulated in pts with prostate Ca |
| PSA | Up-regulated in pts with prostate Ca |
| STEAP | Up-regulated in pts with prostate Ca |
| TMPRSS2 | See ERG |
| KLK3 | Product of KLK3 gene is PSA |
Footnotes
Study received approval from Rhode Island Hospital committee on protection of human subjects.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Di Vizio D, Kim J, Hager MH, et al. Oncosome formation in prostate cancer: association with a region of frequent chromosomal deletion in metastatic disease. Cancer Res. 2009;69:5601. doi: 10.1158/0008-5472.CAN-08-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janowska-Wieczorek A, Majka M, Kijowski J, et al. Platelet-derived microparticles bind to hematopoietic stem/progenitor cells and enhance their engraftment. Blood. 2001;98:3143. doi: 10.1182/blood.v98.10.3143. [DOI] [PubMed] [Google Scholar]
- 4.Baj-Krzyworzeka M, Majka M, Pratico D, et al. Platelet-derived microparticles stimulate proliferation, survival, adhesion, and chemotaxis of hematopoietic cells. Exp Hematol. 2002;30:450. doi: 10.1016/s0301-472x(02)00791-9. [DOI] [PubMed] [Google Scholar]
- 5.Rozmyslowicz T, Majka M, Kijowski J, et al. Platelet- and megakaryocyte-derived microparticles transfer CXCR4 receptor to CXCR4-null cells and make them susceptible to infection by X4-HIV. AIDS. 2003;17:33. doi: 10.1097/00002030-200301030-00006. [DOI] [PubMed] [Google Scholar]
- 6.Fackler OT, Peterlin BM. Endocytic entry of HIV-1. Curr Biol. 2000;10:1005. doi: 10.1016/s0960-9822(00)00654-0. [DOI] [PubMed] [Google Scholar]
- 7.Fevrier B, Vilette D, Archer F, et al. Cells release prions in association with exosomes. Proc Natl Acad Sci U S A. 2004;101:9683. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ratajczak J, Miekus K, Kucia M, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 9.Baj-Krzyworzeka M, Szatanek R, Weglarczyk K, et al. Tumor-derived microvesicles carry several surface determinants and mRNA of tumor cells and transfer some of these determinants to monocytes. Cancer Immunol Immunother. 2006;55:808. doi: 10.1007/s00262-005-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmgren L, Szeles A, Rajnavolgyi E, et al. Horizontal transfer of DNA by the uptake of apoptotic bodies. Blood. 1999;93:3956. [PubMed] [Google Scholar]
- 11.Hakelien AM, Landsverk HB, Robl JM, et al. Reprogramming fibroblasts to express T-cell functions using cell extracts. Nature Biotechnol. 2002;20:460. doi: 10.1038/nbt0502-460. [DOI] [PubMed] [Google Scholar]
- 12.Deregibus MC, Cantaluppi V, Calogero R, et al. Endothelial progenitor cell-derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood. 2007;110:2440. doi: 10.1182/blood-2007-03-078709. [DOI] [PubMed] [Google Scholar]
- 13.Aliotta JM, Sanchez-Guijo FM, Dooner GJ, et al. Alteration of marrow cell gene expression, protein production, and engraftment into lung by lung-derived microvesicles: a novel mechanism for phenotype modulation. Stem Cells. 2007;9:2245. doi: 10.1634/stemcells.2007-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aliotta JM, Pereira M, Johnson KW, et al. Microvesicle entry into marrow cells mediates tissue-specific changes in mRNA by direct delivery of mRNA and induction of transcription. Exp Hematol. 2010;38:233. doi: 10.1016/j.exphem.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitlow MB, Klein LM. Response of SCC-12F, a human squamous cell carcinoma cell line, to complement attack. J Invest Dermatol. 1997;109:39. doi: 10.1111/1523-1747.ep12276459. [DOI] [PubMed] [Google Scholar]
- 16.Carney DF, Hammer CH, Shin ML. Elimination of terminal complement complexes in the plasma membrane of nucleated cells: influence of extra-cellular CA2+ and association with ca2+ J Immunol. 1986;137:263. [PubMed] [Google Scholar]
- 17.Hakulinen J, Junnikkala S, Sorsa T, et al. Complement inhibitor membrane cofactor protein (MCP; CD46) is constitutively shed from cancer cell membranes and converted by a maetalloproteinase to a functionally active soluble form. Eur J Immunol. 2004;34:2620. doi: 10.1002/eji.200424969. [DOI] [PubMed] [Google Scholar]
- 18.Andreola G, Rivoltini L, Castelli C, et al. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. J Exp Med. 2002;195:1303. doi: 10.1084/jem.20011624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hotary KB, Allen ED, Brooks PC, et al. Membrane type1 matrix metalloproteinase usurps tumor growth control imposed by the three dimensional extracellular matrix. Cell. 2003;114:33. doi: 10.1016/s0092-8674(03)00513-0. [DOI] [PubMed] [Google Scholar]
- 20.Ginestra A, Miceli D, Dolo V, et al. Membrane vesicles in ovarian cancer fluids: a new potential marker. Anticancer Res. 1998;19:3439. [PubMed] [Google Scholar]
- 21.Ginestra A, La P, Saladino F, et al. The amount and proteolytic content of vesicles shed by human cancer cell lines correlates with their in vivo invasiveness. Anticancer Res. 1998;18:3433. [PubMed] [Google Scholar]
- 22.Rauch U, Antoniak S. Tissue factor-positive microparticles in blood associated with coagulopathy in cancer. Thromb Haemost. 2007;97:9. [PubMed] [Google Scholar]
- 23.Polgar J, Matuskova J, Wagner DD. The P-selectin, tissue factor, coagulation triad. J Tromb Haemost. 2005;3:1590. doi: 10.1111/j.1538-7836.2005.01373.x. [DOI] [PubMed] [Google Scholar]
- 24.Al-Nedawi K, Meehan B, Micallef J, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumor cells. Nat Cell Biol. 2008;10:619. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 25.Lima LG, Chammas R, Monteiro RQ, et al. Tumor-derived microvesicles modulate the establishment of metastatic melanoma in a phosphatidyserine-dependent manner. Cancer Lett. 2009;283:168. doi: 10.1016/j.canlet.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 26.Choi DS, Lee JM, Park GW, et al. Proteomic analysis of microvesicles derived from human colorectal cancer cells. J Proteome Res. 2007;6:4646. doi: 10.1021/pr070192y. [DOI] [PubMed] [Google Scholar]
- 27.Skog J, Würdinger T, van Rijn S. Glioblastoma microvesicles transport RNA and proteins that promote tumor growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wysoczynski M, Ratajczak MZ. Lung cancer secreted microvesicles: Under appreciated modulators of microenvironment in expanding tumors. Int J Cancer. 2009;125:1595. doi: 10.1002/ijc.24479. [DOI] [PMC free article] [PubMed] [Google Scholar]