Abstract
Background
Little is known about genes affecting childhood body weight.
Objective
To examine alleles of the mitochondrial uncoupling protein-2 (UCP2) gene for association with obesity, since UCP2 may influence energy expenditure.
Design
We related UCP2 genotype to body composition, and to resting energy expenditure, in 105 children aged 6–10y. Overweight children and non-overweight children of overweight parents were genotyped for a 45 bp deletion/insertion (del/ins) in 3’ UTR of exon 8 and for an exon 4 C to T transition.
Results
89 children were genotyped for the exon 8 allele: 50 children had del/del, 33 del/ins, and 6 ins/ins. Body mass index (BMI) was greater for del/ins (24.1 ± 5.9 kg/m2) than for del/del (20.4 ± 4.8 kg/m2, p<0.001). BMI of ins/ins (23.7 ± 7.8 kg/m2) was not different from del/ins. This effect was independent of race and gender (ANOVAs, p< 0.05). Body composition was also different according to UCP2 genotype. All body circumferences and skin fold thicknesses examined were significantly greater in del/ins than in del/del. DXA body fat mass (p<0.005) was also greater in del/ins than del/del. For 104 children genotyped at exon 4, no significant differences in BMI or body composition were found among the three exon 4 genotypes. Neither resting energy expenditure nor respiratory quotient were different according to UCP2 exon 4 or exon 8 genotype.
Conclusion
The exon 8 ins/del polymorphism of UCP2 appears to be associated with childhood-onset obesity. The UCP2/UCP3 genetic locus may play a role in childhood body weight.
Keywords: Body Mass Index, Weight, Obesity, Polymorphism, Genetics
INTRODUCTION
Body weight in humans appears to be a trait with strong genetic determinants, with heritability estimated to be between 50 and 85 % for adult body weight (1–4). Because obesity is presumed to develop when an imbalance exists between energy intake and expenditure, candidate genes for body weight regulation include those that might be important for the regulation of energy expenditure, such as those that may impact on thermogenesis. The uncoupling proteins (UCPs), which may translocate protons into the mitochondrial matrix, resulting in heat generation without ATP synthesis (5), have therefore been examined for associations with body weight. UCP1, found predominantly in brown adipose tissue and important for shivering thermogenesis in rodents, has been linked in adults to the amount of weight lost during dieting (6), but has not been strongly linked to adult or child body weight (7–11). The human genetic locus containing the recently cloned uncoupling proteins 2 and 3 (11q13 (12–16)), has been linked to a number of factors believed relevant for the regulation of body weight in adults. Adult resting energy expenditure (17–19), weight gain (8); and possibly percentage body fat (18), have all been found to have genetic linkage to the UCP2/UCP3 locus. In one study, a heterozygous polymorphism altering the UCP 3 exon 6-splice donor site that truncates the terminal 37 amino acids of UCP3 was found with higher prevalence in severely obese African American than in lean African American subjects. This same study found that those with this polymorphism had lower basal fat oxidation rates compared with those having the more common allele (20). However, other adult studies have failed to find significant linkage between the UCP2/UCP3 locus and obesity, insulin resistance, or resting energy expenditure (21–28). No previous studies have found linkage of the UCP2/UCP3 locus to body weight in childhood, and little is known about the genes predisposing individuals to become overweight in childhood.
Two independent risk factors for obesity in adulthood are the body mass index (BMI) of school-aged children, and the BMI of their parents (29). Children aged 6–10 y with BMI greater than the 85th centile for age and gender, have a relative risk of adult obesity that is 8.8-fold greater than that of children with a lower BMI. Irrespective of their own BMI, 6–10 year old children with two overweight parents have a relative risk of adult obesity that is 5-fold greater than that of children with two normal weight parents (29). A recent study has suggested that resting energy expenditure may differ in lean and overweight children only when they are grouped according to the degree of their parent’s obesity (30). We hypothesized that children whose BMI was already greater than the 95th centile by age 6–10 y were likely to have genetic factors predisposing them to obesity that were distinct from those that might cause later obesity in the relatively lean children of overweight parents. We further hypothesized that alterations in genes that may regulate energy economy, such as UCP2, would be more likely to contribute to childhood-onset obesity than to obesity that occurs only in adulthood. Therefore, we investigated the association of UCP2 polymorphisms with body weight, body composition, and resting energy expenditure in overweight children and in non-overweight 6–10 year old children with obese parents.
METHODS
Subjects
Subjects were recruited through notices mailed to 6–10 year-old children in the Montgomery County, MD school district, and in the case of overweight children, by referral from local physicians. We studied a total of 105 African American, Asian, and Caucasian children (ages 6–10 y, 63 girls and 42 boys) for whom we obtained samples for genomic DNA analysis (Tables 1 and 2). Children were recruited for studies of physiology and metabolism in three groups: A) 58 overweight children (children with a body mass index > 95th percentile for age, gender, and race (31) who did not have a discernable medical cause of their obesity); B) 32 children with a body mass index between the 15th and 95th percentiles, both of whose parents had a history of overweight or obesity, defined as a body mass index > 25 kg/m2 (32); and C) 15 children with a body mass index between the 15th and 85th percentiles with one parent having a history of overweight or obesity. Of the 58 overweight children, 38 had two parents, and 18 had one parent, with a body mass index > 25 kg/m2.
TABLE 1.
Study subjects grouped by exon 8 del/ins genotype
| del/del (n = 50) | del/ins (n = 33) | ins/ins (n = 6) | |
|---|---|---|---|
| Age (y)Δ | 8.3 ± 1.3 | 8.6 ± 1.2 | 8.3 ± 1.7 |
| Weight (kg) | 36.5 ± 11.4 | 46.6 ± 16.7*** | 47.5 ± 23.4* |
| Height (cm) | 134.2 ± 9.6 | 136.6 ± 9.7 | 139.4 ± 14.3 |
| Body Mass Index (kg/m2) | 20.4 ± 4.8 | 24.1 ± 5.9*** | 23.7 ± 7.8 |
| Body Mass Index SDS | 2.0 ± 2.5 | 3.3 ± 2.8** | 3.1 ± 3.2 |
| Percent Overweight (%)¶ | 33.7 ± 33.0 | 56.3 ± 38.9*** | 49.7 ± 44.4 |
| Gender (%) | |||
| Girls | 57% | 65% | 60% |
| Boys | 43% | 35% | 40% |
| Race (%) | |||
| Asian | 10% | 7% | 0% |
| African American | 30% | 41% | 20% |
| Caucasian | 60% | 51% | 80% |
| Bone Age (y) | 9.0 ± 1.7 | 9.4 ± 2.0 | 9.8 ± 2.7 |
| Boy’s Testis Volume (cc) | 2.4 ± 1.1 | 2.7 ± 1.6 | 2.7 ± 1.2 |
| Girls’ Breast Stage (%) | |||
| I | 75% | 59% | 60% |
| II | 25% | 41% | 40% |
| Pubic Hair Stage (%) | |||
| I | 81% | 73% | 60% |
| II | 13% | 10% | 40% |
| III | 6% | 17% | 0% |
| Plasma Estradiol, pg/mL (girls) | 10.9 ± 8.9 | 9.4 ± 5.1 | 10.0 ± 3.6 |
| Plasma Testosterone, ng/dL (boys) | 12.4 ± 5.7 | 14.1 ± 6.7 | 11.0 ± 1.9 |
| Plasma DHEA-S (ug/dL) | 47.2 ± 36.2 | 53.1 ± 43.1 | 64.5 ± 36.8 |
Mean ± SD.
p<0.05;
p<0.005;
p<0.001 versus del/del group.
Percent overweight is defined as percentage by which body mass index exceeds the 50th centile for age, gender, and race (31).
TABLE 2.
Study subjects grouped by exon 4 T/C Genotype
| C/C (n = 42) | C/T (n = 47) | T/T (n = 14) | |
|---|---|---|---|
| Age (y) Δ | 8.3 ± 1.5 | 8.4 ± 1.2 | 8.8 ± 1.4 |
| Weight (kg) | 38.8 ± 13.8+ | 40.4 ± 13.5 | 50.8 ± 19.5 |
| Height (cm) | 133.1 ± 9.2+ | 133.6 ± 9.5 | 139.8 ± 12.3 |
| Body Mass Index (kg/m2) | 21.9 ± 5.3 | 22.1 ± 5.7 | 24.2 ± 7.0 |
| Body Mass Index SDS | 2.3 ± 2.5 | 2.5 ± 2.8 | 3.6 ± 3.3 |
| Percent Overweight (%)¶ | 38.4 ± 35.7 | 42.4 ± 37.4 | 60.7 ± 41.8 |
| Gender (%) | |||
| Girls | 62% | 57% | 60% |
| Boys | 38% | 43% | 40% |
| Race (%) | |||
| Asian | 14% | 19% | 0% |
| African American | 29% | 35% | 54% |
| Caucasian | 57% | 53% | 46% |
| Bone Age (y) | 9.2 ± 1.7 | 9.2 ± 1.8 | 9.9 ± 2.3 |
| Boy’s Testis Volume (cc) | 2.1 ± 0.8 | 2.5 ± 0.9 | 2.9 ± 1.2 |
| Girls’ Breast Stage (%) | |||
| I | 79% | 53% | 75% |
| II | 13% | 33% | 25% |
| III | 8% | 4% | 0% |
| Pubic Hair Stage (%) | |||
| I | 81% | 83% | 41% |
| II | 13% | 7% | 44% |
| III | 6% | 12% | 15% |
| Plasma Estradiol, pg/mL (girls) | 12.4 ± 10.4 | 9.9 ± 5.7 | 10.7 ± 4.8 |
| Plasma Testosterone, ng/dL (boys) | 13.8 ± 6.8 | 12.1 ± 3.6 | 14.2 ± 2.8 |
| Plasma DHEA-S (ug/dL) | 50.9 ± 42.0 | 47.0 ± 36.1 | 68.1 ± 37.8 |
Mean ± SD.
p<0.05 versus T/T group.
Percent overweight is defined as percentage by which body mass index exceeds the 50th centile for age, gender, and race (31).
All African Americans reported having black parents and grandparents, all Asians had Asian parents and grandparents and all Caucasians had white parents and grandparents. For analysis, only one child from each family was studied. All subjects were free of significant medical disease. All had normal physical examinations and normal hepatic, renal, and thyroid function. Each subject underwent a detailed medical history, and was examined for clinical signs of adrenarche or gonadarche. The study was approved by the National Institutes of Health Intramural Clinical Research Subpanel. Each child gave written assent, and a parent gave written consent, for protocol participation.
Protocol
Participants were studied in the morning, more than 12 hours after their last meal. Anthropometric measurements were obtained as recommended (33). These measurements included weight, stature, skin fold thicknesses and body circumferences obtained with a flexible, non-stretching, tape measure. Waist circumference was measured at the smallest horizontal circumference between the 12th rib and the iliac crest. Hip circumference was measured around the buttocks at the maximum circumference. The waist circumference divided by the hip circumference was calculated to determine the waist-to-hip ratio. Measurement of height (measured three times) was performed using a stadiometer (Holtain Ltd., Crymych, Wales) calibrated before each height to the nearest 1 mm. Weight was obtained to the nearest 0.1 kg using a calibrated digital scale (Scale-Tronix, Wheaton, IL). A roentgenogram of the left hand and wrist for bone maturation was also obtained (34).
Resting energy expenditure (REE) and body composition using dual energy x-ray absorptiometry (DXA) were measured at the NIH Warren Grant Magnuson Clinical Center as previously described (35). Subjects were at bed-rest for at least 2 hours before measurement of REE, and had nothing to eat or drink except water after midnight on the day of the study. Subjects were awake , lay supine in bed, and watched non-commercial children’s television shows during measurement. Oxygen consumption and carbon dioxide production were measured at one-minute intervals by respiratory exchange using a ventilated hood system (DeltaTrac, SensorMedics, Yorba Linda, CA). REE was calculated from the rates of oxygen consumption and carbon dioxide production (36). Breath-by-breath measurements of VO2 and VCO2 were automatically averaged and recorded at 1-minute intervals for at least 15 minutes, until more than 10 minutes with coefficient of variation in energy expenditure <5% was present. The mean of the energy expenditure and respiratory quotient (RQ) measurements was determined for each subject and used for further analysis.
All subjects had blood samples drawn after an overnight fast. Total testosterone, estradiol, and DHEA-S were measured at Covance Laboratories (Vienna, VA) as previously described (37, 38).
Genomic DNA was isolated from peripheral blood leukocytes and amplified by the polymerase chain reaction (PCR). For exon 8, a sense primer (hUCP2e8f, 5’ CAG TGA GGG AAG TGG GAG G 3’) and an antisense primer (hUCP2e8r, 5’ GGG GCA GGA CGA AGA TTC 3’) that flank the region containing a 45 bp insertion/deletion in the 3'-untranslated region of exon 8 of UCP2 were used as previously described (17). These PCR primers produce either 457 or 412 bp products, depending upon whether the insert is present. Subjects were identified on the basis of PCR products as being either homozygous for the 3’ UTR exon 8 deletion (del/del), heterozygous (ins/del), or homozygous for the 3’ UTR exon 8 insertion (ins/ins). For exon 4, the sense primerEx4F1 (5’ TGT CTA CTC TGT TCC CTC CCC 3’) and antisense primer Ex4R1 (5’ GGC CTA CAC CCT TGC TCC 3’) were used to amplify genomic DNA. A second set of PCR primers (Ex4A/VF: 5’ GGG CCA GTG CGA CCT ACA G 3’ and Ex4A/VR: 5’ ATG CGG ACA GAG GCA AAG C 3’) were then used to introduce an EclHk1 restriction site that produces 167 bp (for T) or 151 bp products (for C), allowing subjects to be identified as being either homozygous C/C, heterozygous C/T, or homozygous T/T (39).
Data Analysis
Parametric data were analyzed on a Macintosh PowerPC using SuperAnova 1.11 and StatView 4.5 software (Abacus Concepts, Inc., Berkeley, CA). Results of anthropometry and DXA body composition from all three genotypes were compared with analysis of variance. Logarithmic transformation was performed before analysis for all anthropometric circumference measurements, for plasma DHEA-S, and for DXA lean body mass and body fat mass measurements. Significant ANOVAs were followed by post-hoc Fisher protected least significant difference tests. All tests were two-tailed. Gender, race, and UCP2 polymorphism status were factors for analyses of body mass index and DXA percentage body fat. Trunk lean body mass, arm %fat mass, and trunk %fat mass could not be successfully normalized by transformation, and were compared with the nonparametric Mann Whitney test. Statistical results were interpreted with Bonferoni adjustment for multiple comparisons. Nominal p-values are reported. Categorical data were examined with contingency table analysis. The presence of linkage disequilibrium between exon 4 and exon 8 alleles was tested using the chi-square statistic. There were 88 children with results available from both exon 4 and 8 polymorphisms for this analysis. Analysis of covariance, with the log of DXA lean body mass, the log of DXA body fat mass, and gender as the covariates, were performed to evaluate the effect of UCP2 genotype on the log of resting energy expenditure. Because some study participants had evidence of early puberty or adrenarche, analyses were also performed using evidence of pubertal or adrenarchal sex hormone production as a factor. The presence of puberty or adrenarche, either on physical examination (presence of axillary or pubic hair, or Tanner breast stage > I) or laboratory testing (estradiol > 36.7 pmol/L [15 pg/mL]; testosterone > 0.7 nmol/L; dehydroepiandrosterone sulfate > 0.95 μmol/L [35 μg/dL]). These hormone cut-points represent the mean plus two standard deviations for each hormone in the assays employed for children less than age 6 years (40–42). There were no significant differences between results from those with and without evidence of early puberty or adrenarche; therefore, results for the entire group of 105 children are presented.
RESULTS
Genotypes were obtained in 89 children for the exon 8 insertion/deletion (ins/del) allele (Table 1), including 50 children with homozygous 45 bp deletion alleles in the 3'-untranslated region of exon 8 of UCP2 (del/del), 33 children heterozygous for the insertion allele (del/ins), and 6 children with homozygous insertions (ins/ins). Because so few ins/ins subjects were identified, there was insufficient power to determine significance of differences found in ins/ins subjects. For the 104 children genotyped at exon 4 (Table 2), there were 42 homozygous C/C, 47 heterozygous T/C, and 15 children with homozygous T/T. In the 88 children for whom genotypes at both exons were available, polymorphisms in exons 4 and 8 were in linkage disequilibrium (Table 3, χ2 = 50.2, p< 0.001).
TABLE 3.
Linkage of exon 8 and exon 4 polymorphisms
| C/C | C/T | T/T | Totals | |
|---|---|---|---|---|
| Del/Del | 30 (34%) | 17 (19%) | 2 (2%) | 49 (56%) |
| Del/Ins | 2 (2%) | 25 (29%) | 6 (7%) | 33 (37%) |
| Ins/Ins | 0 (0%) | 1 (1%) | 5 (6%) | 6 (7%) |
| Totals | 32 (36%) | 43 (49%) | 13 (15%) | 88 (100%)* |
Polymorphisms in exons 4 and 8 were in linkage disequilibrium, χ2 = 50.2, p< 0.001. Percentage of all subjects is given in parentheses.
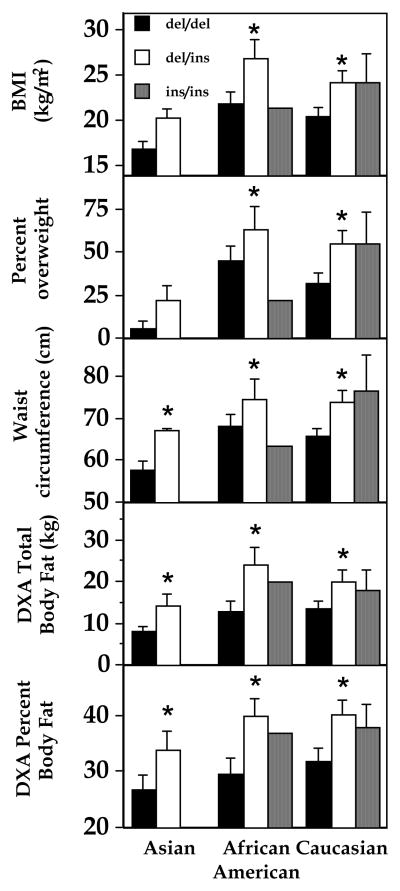
For both exon 4 and exon 8 polymorphisms, gender and racial distributions were not different in each group, and the groups were also not different in age, bone age, pubertal maturation, and plasma adrenal and gonadal hormones (Tables 1 and 2). However, both weight (p < 0.001) and body mass index (p < 0.001) were significantly greater in del/ins than in del/del (Table 1). These effects were confirmed when BMI was expressed either as standard deviation score or as percentage overweight (Table 1) and the effects on BMI and DXA body fat mass were independent of race (Fig. 1) and gender (ANOVA, p< 0.01). BMI was not significantly different for children with different exon 4 genotypes (T/T 24.2 ± 7.0, T/C 22.1 ± 5.7, and C/C 21.9 ± 5.3 kg/m2, p = 0.12, ANOVA, Table 2).
Figure 1.
BMI, percent overweight, waist circumference, DXA total body fat mass, and DXA total percent boy fat in African American, Caucasian, and Asian subjects. *p<0.05, del/del vs. del/ins. For Asians’ BMI, del/del vs. del/ins p = 0.07. There were no Asians and only one African American with the ins/ins genotype. Percent overweight is defined as percentage by which body mass index exceeds the 50th centile for age, gender, and race (31).
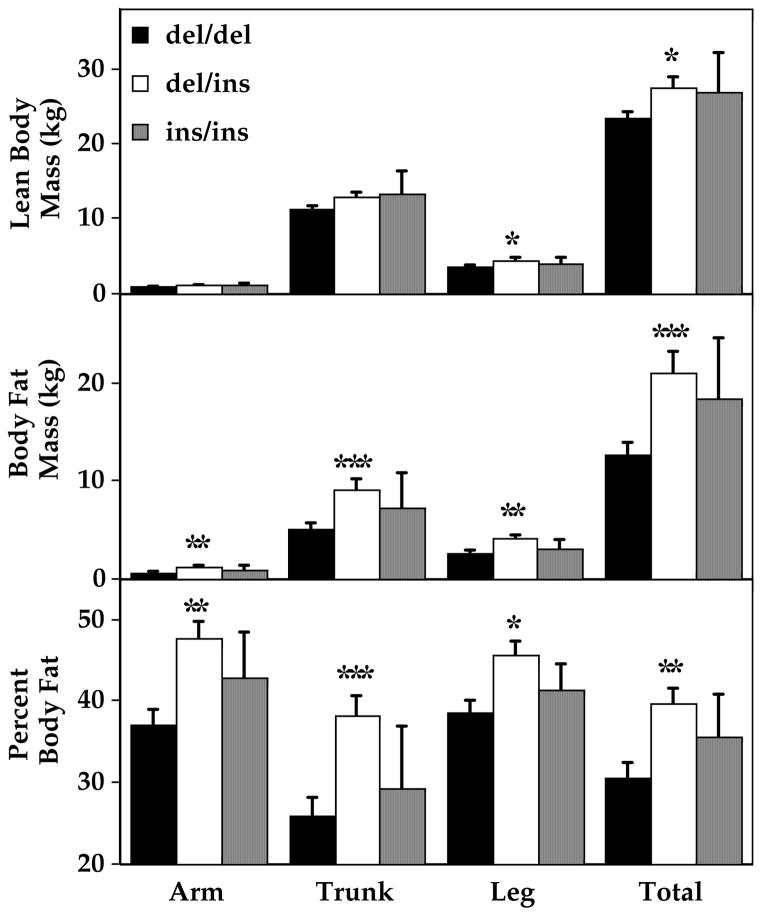
Body composition was also different according to UCP2 exon 8 genotype. Anthropometric measurements were significantly greater in del/ins than in del/del for all circumferences and skin fold thicknesses examined (Table 4). Waist:hip ratio was the only anthropometric variable not different among the groups, and similar results were found for all three racial groups studied (Fig. 1). These results were confirmed by DXA (Figure 2): Body fat mass and percentage of total body mass that was fat were significantly greater in del/ins than in del/del, and these effects were independent of race and gender (ANOVAs, p<0.01). As would be expected given their greater body fat mass, total lean body mass was also greater in del/ins than in del/del subjects (p<0.05). There were no significant differences in body composition, assessed either by DXA or by anthropometrics, for children with the three exon 4 genotypes (Table 5).
TABLE 4.
Body composition according to exon 8 genotype
| del/del (n = 50) | del/ins (n = 33) | ins/ins (n = 6) | |
|---|---|---|---|
| Circumference (cm)Δ | |||
| Neck | 29.1 ± 2.6 | 30.9 ± 3.0** | 32.3 ± 5.9 |
| Upper Arm | 23.7 ± 4.6 | 27.0 ± 5.4** | 28.0 ± 6.7 |
| Forearm | 20.4 ± 2.7 | 22.4 ± 3** | 22.5 ± 3.7 |
| Axilla | 27.2 ± 4.5 | 30.4 ± 5.1** | 31.5 ± 7.2 |
| Chest | 73.3 ± 10.3 | 80.0 ± 10.7** | 82.9 ± 17.6 |
| Waist | 65.1 ± 10.7 | 72.2 ± 12.0** | 77.7 ± 21.2 |
| Hip | 75.9 ± 10.6 | 83.9 ± 11.7** | 86.5 ± 16.2 |
| Thigh | 41.0 ± 6.5 | 46.8 ± 8.1** | 49.2 ± 10.8 |
| Calf | 29.7 ± 4.3 | 32.5 ± 5.1* | 33.0 ± 6.1 |
| Waist:hip ratio | 0.85 ± 0.04 | 0.86 ± 0.04 | 0.87 ± 0.08 |
| Skin fold thickness (mm) | |||
| Triceps | 19.1 ± 8.8 | 24.1 ± 11.0* | 26.7 ± 13.0 |
| Biceps | 9.5 ± 5.9 | 12.1 ± 5.5* | 14.9 ± 9.1 |
| Subscapular | 14.5 ± 9.3 | 19.5 ± 10.1* | 21.4 ± 11.7 |
| Suprailiac | 17.1 ± 10.6 | 22.9 ± 10.5* | 22.9 ± 11.7 |
| Sum of 4 skin folds | 60.2 ± 32.7 | 78.6 ± 35.6* | 85.9 ± 45.0 |
| DXA Lean Mass (kg) | 23.4 ± 5.1 | 27.3 ± 7.8* | 26.8 ± 10.6 |
| DXA Fat Mass (kg) | 12.6 ± 8.8 | 21.1 ± 11.5** | 18.4 ± 12.2 |
| DXA % Fat Mass | 30.6 ± 11.5 | 39.7 ± 10.2** | 35.6 ± 10.5 |
Mean ± SD.
p<0.05;
p<0.005 versus del/del group.
Figure 2.
Regional body composition by DXA. *p<0.05, **p<0.005, ***p<0.001, del/del vs. del/ins.
TABLE 5.
Body composition according to exon 4 genotype
| C/C (n = 42) | C/T (n = 47) | T/T (n = 14) | |
|---|---|---|---|
| Circumference (cm)Δ | |||
| Neck | 29.9 ± 3.3 | 29.9 ± 3.0 | 31.2 ± 4.5 |
| Upper Arm | 24.8 ± 5.1 | 25.1.0 ± 5.5 | 27.3 ± 6.2 |
| Forearm | 21.1 ± 2.7 | 21.3 ± 3.1 | 22.1 ± 3.5 |
| Axilla | 28.3 ± 4.8 | 28.6 ± 5.2 | 30.8 ± 6.1 |
| Chest | 75.5 ± 11.5.0 | 76.4 ± 11.3 | 80.9 ± 14.3 |
| Waist | 67.2 ± 12.1 | 68.6 ± 12.4 | 74.1 ± 16.7 |
| Hip | 78.1 ± 11.2 | 79.6 ± 12.3 | 83.3 ± 14.3 |
| Thigh | 42.5 ± 7.3 | 43.7 ± 7.8 | 47.7 ± 9.5 |
| Calf | 30 ± 3.6 | 31.2 ± 5.1 | 32.6 ± 5.9 |
| Waist:hip ratio | 0.86 ± 0.05 | 0.86 ± 0.05 | 0.88 ± 0.06 |
| Skin fold thickness (mm) | |||
| Triceps | 20.8 ± 8.2 | 21.2 ± 11.1 | 24.2 ± 11.4 |
| Biceps | 10.2 ± 4.3 | 10.7 ± 6.6 | 12.5 ± 7.3 |
| Subscapular | 15.8 ± 8.9 | 15.6.0 ± 10.1 | 18.8 ± 10.6 |
| Suprailiac | 18.4 ± 8.9.0 | 16.8 ± 10.9.0 | 23.2 ± 12.1 |
| Sum of 4 skin folds | 56.6 ± 26.6 | 53.7 ± 33.6 | 66.2 ± 37.9 |
| DXA Lean Mass (kg) | 24.4 ± 6.4 | 24.6 ± 5.0 | 27.3 ± 10.4 |
| DXA Fat Mass (kg) | 14.3 ± 10.1 | 15.9 ± 10.0 | 19.6 ± 12.4 |
| DXA % Fat Mass | 32.5 ± 10.5 | 34.4 ± 12.5 | 36.5 ± 10.1 |
Mean ± SD.
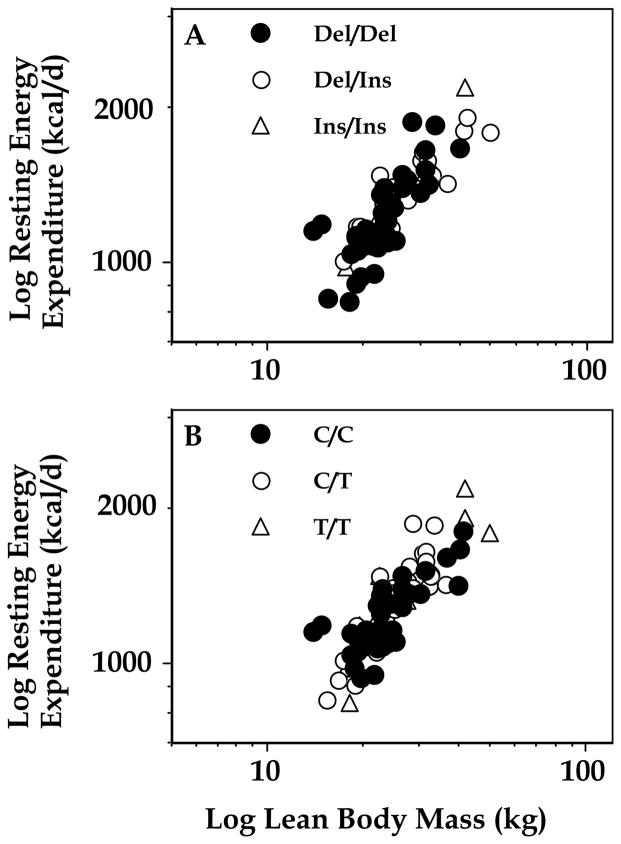
In analyses using lean body mass, body fat mass, and gender as covariates, resting energy expenditure, was not significantly different according to exon 4 or exon 8 genotype (Figure 3). Respiratory quotient was also not different in each group (for exon 8, del/del 0.87 ± 0.04; del/ins 0.86 ± 0.05; ins/ins 0.88 ± 0.05, p = NS, for exon 4, CC 0.85 ± 0.04; T/C 0.87 ± 0.04; T/T 0.86 ± 0.04, p = NS).
Figure 3.
Resting energy expenditure was not altered in children with different exon 8 or exon 4 polymorphisms. A: del/del, del/ins, and ins/ins subjects. B: C/C, T/C, and C/C subjects.
DISCUSSION
We found a significant association between an allelic variant in the 3’ UTR of UCP2 exon 8 and measures of obesity in this small sample of children. Subjects with the del/ins genotype had a significantly greater BMI than those with del/del. Body fat measures, determined either by anthropometry or by DXA, were also greater in those having the del/ins genotype. Although too few ins/ins subjects were identified to find statistically significant increases in any measured variable, ins/ins results were generally similar to del/ins. To our knowledge, these findings are the first linking any gene locus relevant for energy expenditure to body weight regulation in childhood. By contrast, as others have reported in adults and children (17, 23, 43) we found no significant associations between UCP2 exon 4 allelic variation and body mass index or body composition. Thus it appears that the exon 4 Ala to Val variant at amino acid 55 is of little or no importance for child or adult body weight regulation. Although there was some concordance between the exon 8 and exon 4 variants (Table 3), the C/T genotype was fairly evenly divided between those with Del/Del (19% of subjects) and those with Del/Ins (29% of subjects), perhaps explaining why the C/T genotype was not associated with childhood measures of body adiposity.
Some previous studies in adults have found body weight or energy expenditure linkage to the UCP2/UCP3 locus in distinct subpopulations (17, 20). We found an association of UCP2 genotype with BMI and body fat mass in all three racial subgroups examined. These findings suggest that a gene either within or close to the UCP2/UCP3 locus, may be an important determinant of body weight in children.
At present, little is known about the effect of the exon 8 insertion variant on UCP2 function. The ins/del polymorphism has been hypothesized to influence UCP2 protein levels by altering mRNA concentrations or translation rates. In a study of muscle biopsy specimens from Pima Indians, UCP2 expression was not significantly different in 10 del/del and 12 del/ins adults (17). However, animal and human data each suggest that adipose tissue UCP2 mRNA expression may be more tightly associated with body weight (44–49). In humans, BMI is correlated only with adipose tissue UCP2 mRNA expression but not with skeletal muscle UCP2 mRNA (50). Thus it is possible that adipose tissue UCP2 mRNA expression might differ according to the ins/del polymorphism even if muscle mRNA does not. Finally, the degree to which mRNA expression reflects UCP2 protein levels may vary with genotype. UCP2 mRNA and protein levels in both adipose tissue and skeletal muscle of individuals with all three possible genotypes should be examined to test the hypothesis that the UCP2 ins/del polymorphism influences UCP2 levels, and functional studies of the ins/del variants should be undertaken.
Previous studies that have examined the UCP2 exon 8 insertion variant in adults have reached different conclusions with regard to the importance of this polymorphism for body weight and energy expenditure. In a study of adult French Canadians, Otabe et al found no difference in ins/del gene frequency between morbidly obese and non-obese subjects, and no relationships between UCP2 ins/del genotype and either body composition or resting energy expenditure (23). In adult Pima Indians, Walder et al found that exon 8 ins/del heterozygotes had higher metabolic rates than either del/del or ins/ins homozygotes, and that the ins/del genotype was associated with a lower BMI in subjects over age 45 y (17). One study in Caucasian children found allele frequencies of exon 8 or exon 4 polymorphisms were not significantly altered in a subgroup of 25 children characterized by low REE (43). Population differences in age, race, and possibly diet may underlie these disparate results. It remains unknown whether the exon 8 variant studied is etiologic for our finding of a greater BMI in children with del/ins genotype, or merely in linkage disequilibrium with another nearby important genetic variant, such as a mutation in UCP3. The lack of differences in REE or respiratory quotient found in our cohorts with differing UCP2 genotypes may be evidence for this possibility.
Unlike other studies that have found associations between the UCP2/UCP3 locus and energy expenditure or respiratory quotient (17, 18, 20), we found no such difference. Since this UCP2 insertion has been estimated to contributes less than 5 ± 11% of the variance in adult sleeping metabolic rate (17), it may be necessary to study considerably larger sample sizes to detect changes in energy expenditure or respiratory quotient related to this UCP2 variant in childhood, where the variability of measurements is often higher than that observed in adults.
We chose to compare normal weight children who have overweight parents with children who were already overweight before adolescence, because we hypothesized that the genetic determinants of pediatric- and adult-onset obesity would be different. We believe the present study validates this approach. It can be anticipated that other genetic factors involved in pediatric weight regulation can be uncovered through such comparisons.
In conclusion, we have shown that a genetic variant of UCP2 appears to be associated with childhood-onset obesity. These findings suggest that the UCP2/UCP3 genetic locus may be involved in determining childhood body weight.
Acknowledgments
JAY is supported by the ORMH, NIH and NIH HD-00641. CHW and ALD are supported by NIH DK-52581 and HL-35773.
Footnotes
J. Yanovski and N. Sebring are Commissioned Officers in the USPHS
References
- 1.Stunkard AJ, Harris JR, Pedersen NL, McClearn GE. The body-mass index of twins who have been reared apart. N Engl J Med. 1990;322(21):1483–7. doi: 10.1056/NEJM199005243222102. [DOI] [PubMed] [Google Scholar]
- 2.Fabsitz RR, Sholinsky P, Carmelli D. Genetic influences on adult weight gain and maximum body mass index in male twins. Am J Epidemiol. 1994;140(8):711–20. doi: 10.1093/oxfordjournals.aje.a117319. [DOI] [PubMed] [Google Scholar]
- 3.Chagnon YC, Peruse L, Bouchard D. Familial aggregation of obesity, candidate genes and quantitative trait loci. Curr Opin Lipidol. 1997;8:205–211. doi: 10.1097/00041433-199708000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Borecki IB, Blangero J, Rice T, Perusse L, Bouchard C, Rao DC. Evidence for at least two major loci influencing human fatness. Am J Hum Genet. 1998;63(3):831–8. doi: 10.1086/302006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klingenberg M. Mechanism and evolution of the uncoupling protein of brown adipose tissue. Trends Biochem Sci. 1990;15(3):108–12. doi: 10.1016/0968-0004(90)90194-g. [DOI] [PubMed] [Google Scholar]
- 6.Fumeron F, Durack-Bown I, Betoulle D, et al. Polymorphisms of uncoupling protein (UCP) and beta 3 adrenoreceptor genes in obese people submitted to a low calorie diet. Int J Obes Relat Metab Disord. 1996;20(12):1051–4. [PubMed] [Google Scholar]
- 7.Oppert JM, Vohl MC, Chagnon M, et al. DNA polymorphism in the uncoupling protein (UCP) gene and human body fat. Int J Obes Relat Metab Disord. 1994;18(8):526–31. [PubMed] [Google Scholar]
- 8.Clement K, Ruiz J, Cassard-Doulcier AM, et al. Additive effect of A-->G (-3826) variant of the uncoupling protein gene and the Trp64Arg mutation of the beta 3-adrenergic receptor gene on weight gain in morbid obesity. Int J Obes Relat Metab Disord. 1996;20(12):1062–6. [PubMed] [Google Scholar]
- 9.Gagnon J, Lago F, Chagnon YC, et al. DNA polymorphism in the uncoupling protein 1 (UCP1) gene has no effect on obesity related phenotypes in the Swedish Obese Subjects cohorts. Int J Obes Relat Metab Disord. 1998;22(6):500–5. doi: 10.1038/sj.ijo.0800613. [DOI] [PubMed] [Google Scholar]
- 10.Urhammer SA, Fridberg M, Sorensen TI, et al. Studies of genetic variability of the uncoupling protein 1 gene in Caucasian subjects with juvenile-onset obesity. J Clin Endocrinol Metab. 1997;82(12):4069–74. doi: 10.1210/jcem.82.12.4414. [DOI] [PubMed] [Google Scholar]
- 11.Hamann A, Tafel J, Busing B, et al. Analysis of the uncoupling protein-1 (UCP1) gene in obese and lean subjects: identification of four amino acid variants. Int J Obes Relat Metab Disord. 1998;22(9):939–41. doi: 10.1038/sj.ijo.0800725. [DOI] [PubMed] [Google Scholar]
- 12.Fleury C, Neverova M, Collins S, et al. Uncoupling protein-2: a novel gene linked to obesity and hyperinsulinemia. Nat Genet. 1997;15(3):269–72. doi: 10.1038/ng0397-269. [DOI] [PubMed] [Google Scholar]
- 13.Gong DW, He Y, Karas M, Reitman M. Uncoupling protein-3 is a mediator of thermogenesis regulated by thyroid hormone, beta3-adrenergic agonists, and leptin. J Biol Chem. 1997;272(39):24129–32. doi: 10.1074/jbc.272.39.24129. [DOI] [PubMed] [Google Scholar]
- 14.Solanes G, Vidal-Puig A, Grujic D, Flier JS, Lowell BB. The human uncoupling protein-3 gene. Genomic structure, chromosomal localization, and genetic basis for short and long form transcripts. J Biol Chem. 1997;272(41):25433–6. doi: 10.1074/jbc.272.41.25433. [DOI] [PubMed] [Google Scholar]
- 15.Vidal-Puig A, Solanes G, Grujic D, Flier JS, Lowell BB. UCP3: an uncoupling protein homologue expressed preferentially and abundantly in skeletal muscle and brown adipose tissue. Biochem Biophys Res Commun. 1997;235(1):79–82. doi: 10.1006/bbrc.1997.6740. [DOI] [PubMed] [Google Scholar]
- 16.Boss O, Giacobino JP, Muzzin P, et al. Genomic structure of uncoupling protein-3 (UCP3) and its assignment to chromosome 11q13. Genomics. 1998;47(3):425–6. doi: 10.1006/geno.1997.5135. [DOI] [PubMed] [Google Scholar]
- 17.Walder K, Norman RA, Hanson RL, et al. Association between uncoupling protein polymorphisms (UCP2-UCP3) and energy metabolism/obesity in Pima Indians. Hum Mol Genet. 1998;7(9):1431–5. doi: 10.1093/hmg/7.9.1431. [DOI] [PubMed] [Google Scholar]
- 18.Bouchard C, Perusse L, Chagnon YC, Warden C, Ricquier D. Linkage between markers in the vicinity of the uncoupling protein 2 gene and resting metabolic rate in humans. Hum Mol Genet. 1997;6(11):1887–9. doi: 10.1093/hmg/6.11.1887. [DOI] [PubMed] [Google Scholar]
- 19.Barbe P, Millet L, Larrouy D, et al. Uncoupling protein-2 messenger ribonucleic acid expression during very- low-calorie diet in obese premenopausal women. J Clin Endocrinol Metab. 1998;83(7):2450–3. doi: 10.1210/jcem.83.7.4962. [DOI] [PubMed] [Google Scholar]
- 20.Argyropoulos G, Brown AM, Willi SM, et al. Effects of mutations in the human uncoupling protein 3 gene on the respiratory quotient and fat oxidation in severe obesity and type 2 diabetes. J Clin Invest. 1998;102(7):1345–51. doi: 10.1172/JCI4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urhammer SA, Dalgaard LT, Sorensen TI, et al. Mutational analysis of the coding region of the uncoupling protein 2 gene in obese NIDDM patients: impact of a common amino acid polymorphism on juvenile and maturity onset forms of obesity and insulin resistance. Diabetologia. 1997;40(10):1227–30. doi: 10.1007/s001250050811. [DOI] [PubMed] [Google Scholar]
- 22.Urhammer SA, Dalgaard LT, Sorensen TI, et al. Organisation of the coding exons and mutational screening of the uncoupling protein 3 gene in subjects with juvenile-onset obesity. Diabetologia. 1998;41(2):241–4. doi: 10.1007/s001250050897. [DOI] [PubMed] [Google Scholar]
- 23.Otabe S, Clement K, Rich N, et al. Mutation screening of the human UCP 2 gene in normoglycemic and NIDDM morbidly obese patients: lack of association between new UCP 2 polymorphisms and obesity in French Caucasians. Diabetes. 1998;47(5):840–2. doi: 10.2337/diabetes.47.5.840. [DOI] [PubMed] [Google Scholar]
- 24.Hanson RL, Ehm MG, Pettitt DJ, et al. An autosomal genomic scan for loci linked to type II diabetes mellitus and body-mass index in pima indians. Am J Hum Genet. 1998;63(4):1124–32. doi: 10.1086/302061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pratley RE, Thompson DB, Prochazka M, et al. An autosomal genomic scan for loci linked to prediabetic phenotypes in Pima Indians. J Clin Invest. 1998;101(8):1757–64. doi: 10.1172/JCI1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norman RA, Tataranni PA, Pratley R, et al. Autosomal genomic scan for loci linked to obesity and energy metabolism in Pima Indians. Am J Hum Genet. 1998;62(3):659–68. doi: 10.1086/301758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kubota T, Mori H, Tamori Y, et al. Molecular screening of uncoupling protein 2 gene in patients with noninsulin-dependent diabetes mellitus or obesity. J Clin Endocrinol Metab. 1998;83(8):2800–4. doi: 10.1210/jcem.83.8.4994. [DOI] [PubMed] [Google Scholar]
- 28.Klannemark M, Orho M, Groop L. No relationship between identified variants in the uncoupling protein 2 gene and energy expenditure. Eur J Endocrinol. 1998;139(2):217–23. doi: 10.1530/eje.0.1390217. [DOI] [PubMed] [Google Scholar]
- 29.Whitaker RC, Wright JA, Pepe MS, Seidel KD, Dietz WH. Predicting obesity in young adulthood from childhood and parental obesity. N Engl J Med. 1997;337(13):869–73. doi: 10.1056/NEJM199709253371301. [DOI] [PubMed] [Google Scholar]
- 30.Wurmser H, Laessle R, Jacob K, et al. Resting metabolic rate in preadolescent girls at high risk of obesity. Int J Obes Relat Metab Disord. 1998;22(8):793–9. doi: 10.1038/sj.ijo.0800662. [DOI] [PubMed] [Google Scholar]
- 31.Must A, Dallal GE, Dietz WH. Reference data for obesity: 85th and 95th percentiles of body mass index (wt/ht2) and triceps skinfold thickness. Am J Clin Nutr. 1991;54(5):773. doi: 10.1093/ajcn/53.4.839. [DOI] [PubMed] [Google Scholar]
- 32.Anonymous. Clinical guidelines on the identification, evaluation, and treatment of overweight and obesity in adults. Obes Res. 1998;6(Suppl 2):51S–209S. [PubMed] [Google Scholar]
- 33.Lohman TG, Roche AF, Martorell R. Anthropometric standardization reference manual. Champaign IL: Human Kinetics Publishers, Inc; 1988. [Google Scholar]
- 34.Gruelich WW, Pyle SI. Radiographic atlas of skeletal development of the hand and wrist. 2. Stanford, CA: Stanford University Press; 1959. [Google Scholar]
- 35.Yanovski SZ, Reynolds JC, Boyle A, Yanovski JA. Resting metabolic rate in Caucasian and African American girls. Obesity Research. 1997;5:321–325. doi: 10.1002/j.1550-8528.1997.tb00558.x. [DOI] [PubMed] [Google Scholar]
- 36.Weir JB. New methods for calculating metabolic rate with special reference to protein metabolism. J Physiol. 1949;109:1–9. doi: 10.1113/jphysiol.1949.sp004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yanovski JA, Yanovski SZ, Cutler G, Jr, Chrousos GP, Filmer KM. Differences in the hypothalamic-pituitary-adrenal axis of black girls and white girls. J Pediatr. 1996;129(1):130–5. doi: 10.1016/s0022-3476(96)70199-3. [DOI] [PubMed] [Google Scholar]
- 38.Oerter KE, Manasco P, Barnes KM, Jones J, Hill S, Cutler G., Jr Adult height in precocious puberty after long-term treatment with deslorelin. J Clin Endocrinol Metab. 1991;73(6):1235–40. doi: 10.1210/jcem-73-6-1235. [DOI] [PubMed] [Google Scholar]
- 39.Shinoki T, Suehiro T, Ikeda Y, et al. Mutation of the Uncoupling Protein 2 Gene in Patients with Non-insulin Dependent Diabetes Mellitus. Metabolism. doi: 10.1016/s0026-0495(99)90054-9. In Press. [DOI] [PubMed] [Google Scholar]
- 40.Winter JS, Faiman C, Reyes FI, Hobson WC. Gonadotrophins and steroid hormones in the blood and urine of prepubertal girls and other primates. Clin Endocrinol Metab. 1978;7(3):513–30. doi: 10.1016/s0300-595x(78)80007-3. [DOI] [PubMed] [Google Scholar]
- 41.Cutler G, Jr, Glenn M, Bush M, Hodgen GD, Graham CE, Loriaux DL. Adrenarche: a survey of rodents, domestic animals, and primates. Endocrinology. 1978;103(6):2112–8. doi: 10.1210/endo-103-6-2112. [DOI] [PubMed] [Google Scholar]
- 42.Korth-Schutz S, Levine LS, New MI. Dehydroepiandrosterone sulfate (DS) levels, a rapid test for abnormal adrenal androgen secretion. J Clin Endocrinol Metab. 1976;42(6):1005–13. doi: 10.1210/jcem-42-6-1005. [DOI] [PubMed] [Google Scholar]
- 43.Tu N, Chen H, Winnikes U, et al. Structural organization and mutational analysis of the human uncoupling protein-2 (hUCP2) gene. Life Sciences. 1998;64(3):PL41–PL50. doi: 10.1016/s0024-3205(98)00555-4. [DOI] [PubMed] [Google Scholar]
- 44.Boss O, Samec S, Dulloo A, Seydoux J, Muzzin P, Giacobino JP. Tissue-dependent upregulation of rat uncoupling protein-2 expression in response to fasting or cold. FEBS Lett. 1997;412(1):111–4. doi: 10.1016/s0014-5793(97)00755-2. [DOI] [PubMed] [Google Scholar]
- 45.Boyer BB, Barnes BM, Lowell BB, Grujic D. Differential regulation of uncoupling protein gene homologues in multiple tissues of hibernating ground squirrels. Am J Physiol. 1998;275(4 Pt 2):R1232–8. doi: 10.1152/ajpregu.1998.275.4.R1232. [DOI] [PubMed] [Google Scholar]
- 46.Oberkofler H, Liu YM, Esterbauer H, Hell E, Krempler F, Patsch W. Uncoupling protein-2 gene: reduced mRNA expression in intraperitoneal adipose tissue of obese humans. Diabetologia. 1998;41(8):940–6. doi: 10.1007/s001250051011. [DOI] [PubMed] [Google Scholar]
- 47.Samec S, Seydoux J, Dulloo AG. Role of UCP homologues in skeletal muscles and brown adipose tissue: mediators of thermogenesis or regulators of lipids as fuel substrate? Faseb J. 1998;12(9):715–24. doi: 10.1096/fasebj.12.9.715. [DOI] [PubMed] [Google Scholar]
- 48.Hidaka S, Kakuma T, Yoshimatsu H, Yasunaga S, Kurokawa M, Sakata T. Molecular cloning of rat uncoupling protein 2 cDNA and its expression in genetically obese Zucker fatty (fa/fa) rats. Biochim Biophys Acta. 1998;1389(3):178–86. doi: 10.1016/s0005-2760(97)00188-4. [DOI] [PubMed] [Google Scholar]
- 49.Matsuda J, Hosoda K, Itoh H, et al. Cloning of rat uncoupling protein-3 and uncoupling protein-2 cDNAs: their gene expression in rats fed high-fat diet. FEBS Lett. 1997;418(1–2):200–4. doi: 10.1016/s0014-5793(97)01381-1. [DOI] [PubMed] [Google Scholar]
- 50.Millet L, Vidal H, Andreelli F, et al. Increased uncoupling protein-2 and -3 mRNA expression during fasting in obese and lean humans. J Clin Invest. 1997;100(11):2665–70. doi: 10.1172/JCI119811. [DOI] [PMC free article] [PubMed] [Google Scholar]