Abstract
Atrial natriuretic peptide, B-type natriuretic peptide and C-type natriuretic peptide compose a family of three structurally related, but genetically distinct, signaling molecules that regulate the cardiovascular, skeletal, nervous, reproductive and other systems by activating transmembrane guanylyl cyclases and elevating intracellular cGMP concentrations. This review broadly discusses the general characteristics of natriuretic peptides and their cognate signaling receptors, then specifically discusses the tissue specific metabolism of natriuretic peptides and their degradation by neprilysin, insulin-degrading enzyme and natriuretic peptide receptor-C.
Keywords: ANF, cardiovascular disease, endochondrial ossification, receptor internalization, signaling, proteolysis, ANP, BNP, CNP, cGMP, NEP
General characteristics of natriuretic peptides and their receptors
All natriuretic peptides are synthesized as preprohormones that are processed to smaller mature forms containing an obligate C-terminal 17-amino acid disulfide ring structure. Please see previous articles in this series for more detailed reviews of natriuretic peptides and their receptors [1–3]. Atrial natriuretic peptide (ANP) is primarily stored as a propeptide in atrial granules and is secreted and cleaved to a 28-residue mature peptide as it enters the circulation in response to atrial stretch. A version of ANP called urodilatin containing four additional amino-terminal residues is primarily found in the kidney. B-type natriuretic peptide (BNP) is also in atrial granules, but is found at highest levels in ventricles from stressed hearts like those from congestive heart failure patients. BNP is not stored in granules in the ventricles. Instead, BNP is regulated at the transcriptional level. Plasma concentrations of ANP are several-fold higher than plasma concentrations of BNP in healthy humans [4, 5]. Both ANP and BNP concentrations are elevated in patients with severe heart failure, and in some cases, BNP levels exceed ANP levels [5–8]. Gene deletion experiments in mice indicate that ANP has broad systemic functions that lower blood pressure and cardiac preload, whereas BNP primarily prevents fibrosis in the heart [9, 10]. C-type natriuretic peptide (CNP) is found at low concentrations in the heart and is present at higher concentrations in chondrocytes where it stimulates long bone growth [11].
There are three known receptors for natriuretic peptides. Guanylyl cyclase-A (GC-A) is a particulate guanylyl cyclase that catalyzes the synthesis of cGMP upon binding by ANP or BNP (Fig. 1) [12]. It contains a large extracellular ligand-binding domain, single membrane-spanning region and a large intracellular region composed of kinase homology domain-regulatory, coiled-coil-dimerization and guanylyl cyclase-catalytic domains. Guanylyl cyclase-B (GC-B) is homologous to GC-A, but is activated by CNP. Most physiologic effects of natriuretic peptides are mediated by these two receptors. The best-characterized physiologic functions associated with the activation of GC-A are renal sodium and water excretion, vasorelaxation, antagonism of the renin-angiotensin-aldosterone system, and endothelial extravasation [13]. In contrast, gene deletion studies in mice and familial mutations in humans provide compelling data to indicate that the CNP/GC-B system mediates long bone growth [14–16].
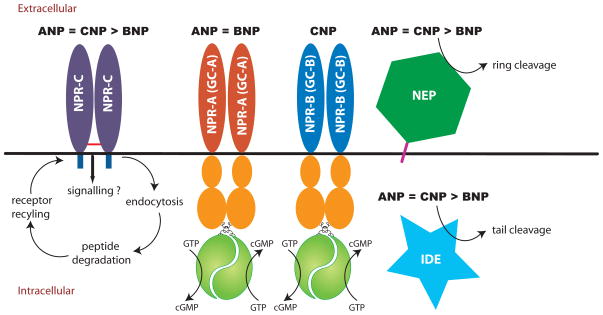
Figure 1.
Natriuretic peptides bind multiple cell surface proteins. Bolded black natriuretic peptide abbreviations indicate binding or substrate preference. NPR-C internalizes all three natriuretic peptides, which targets them for degradation by intracellular proteases. NEP is an extracellular metalloprotease that cleaves ANP and CNP at the C-F bond and breaks the ring. BNP is a much poorer substrate for NEP and is not cleaved at the conserved C-F bond. Insulin degrading enzyme (IDE) is depicted as a cytosolic enzyme but it has also been found in membrane preparations [64]. It initially cleaves ANP and BNP outside the ring.
All three natriuretic peptides also bind natriuretic peptide receptor-C (NPR-C). NPR-C is a disulfide-linked homodimer that is homologous to the extracellular domains of GC-A and GC-B but only contains 37 intracellular amino acids (Fig. 1). It may also have signaling functions, but the majority of physiologic data indicate that the primary role for NPR-C is to clear natriuretic peptides from the extracellular environment via a receptor mediated internalization and degradation process. NPR-C binds all three family members with similar affinities. The half-life of 125I-ANP in the circulation of NPR-C null mice is two-thirds longer than in wild-type mice, although total ANP concentrations were not reduced in the null animals [17]. Additionally, mice lacking functional NPR-C display mild hypotension, volume depletion and dilute urine associated with over-activation of GC-A and elongated long bones and kyphosis associated with over-activation of GC-B [17, 18]. In addition to receptor-mediated degradation, natriuretic peptides are also metabolized by extracellular proteases. Natriuretic peptide degradation is the focus of the remaining sections of this review.
Tissue-specific metabolism of natriuretic peptides
Natriuretic peptides are rapidly cleared from the body. Three mechanisms could formally contribute to this process: receptor-mediated degradation, degradation by extracellular proteases and secretion of the peptides into the body fluids like urine or bile. Since little evidence supports the latter process under normal conditions [19], it will not be further considered here.
The ability of individual organs to remove molecules from the circulation is described by the extraction ratio, which is calculated by subtracting the venous concentration from the arterial concentration and dividing this value by the arterial blood concentration of the molecule. This so called A/V difference quantifies how efficiently the organ removes or degrades the molecule in question. The extraction ratio for ANP varies from about 20 to 75%, but is generally around 35% for most organs [19]. To determine the net effect of the organ on whole body concentrations of the molecule, organ blood flow also must be taken into consideration. Thus, the extraction ratio is multiplied by organ plasma flow to generate the organ clearance rate, which is described in units of liters per minute.
Using these calculations, natriuretic peptide removal has been determined for several organs. However, caveats to the following discussion are that many of the human studies were conducted on sick patients, antibodies used to detect ANP were generated against peptides less than the full length 28 amino acid molecule and blood sampling sites varied between studies. Additionally, the vast majority of reports evaluated ANP but not BNP or CNP.
With this information in mind, clearance of ANP can be generally characterized as relatively fast and on par with removal of other peptide hormones like vasopressin and angiotensin II. The reported half-life of ANP ranges from 0.5 to 4 minutes in mice, rats, rabbits, dogs and monkeys [20] and is about two minutes in normal human subjects [21, 22]. Most tissues remove ANP from the circulation, but some organs are more efficient at ANP extraction than others. Early human A/V studies indicated that about 30 to 50% of ANP is removed by the kidney, liver or lower limbs, while no extraction was observed across the lung [23, 24]. However, later reports in humans and dogs indicated that the lungs have a significant ANP extraction rate of between 19% and 24%. Importantly, lung clearance is the highest of all organs (269 ml/min) due to the high blood vessel surface area and perfusion rate of this tissue [25]. The difference between studies that observed significant verses no lung extraction appears to result from the use of different sampling sites [26]. The organ preference for ANP extraction is lung>liver>kidney [25].
Few studies have reported the clearance of BNP and CNP. Mukoyama and colleagues originally observed that the removal of BNP from the human circulation is composed of short and long half-life components of 3.9 and 20.7 minutes, respectively [5]. Other investigators reported a similarly long (22.6 min) half-life for BNP in humans [27]. Mukoyama and coworkers went on to report that BNP binds to human NPR-C 7% as tightly as it binds ANP and suggested that the increased half-life of BNP results from deceased removal by NPR-C-mediated internalization and degradation. A/V differences of BNP are less than those observed for ANP in humans, consistent with the longer half-life of BNP [28].
CNP has the shortest half-life (2.6 min.) of all the natriuretic peptides in humans [29] and a similarly low half-life (1.6 min.) in sheep [30]. When CNP was infused in sheep at rates of 1 or 10 pmol/kg/min, metabolic clearance rates of 3.1 and 2.5 l/min, respectively, were observed. Like ANP, CNP is removed in dogs by the lungs, kidney and vasculature of the lower body [31]. A recent study in humans reported positive CNP A/V gradients from the heart, head and neck and musculoskeletal system and negative gradients from renal, hepatic and pulmonary tissue, consistent with the former tissues secreting and the later tissues degrading CNP [32].
Receptor-mediated clearance of natriuretic peptides
NPR-C mediated ANP clearance was first demonstrated by Maack and colleagues in 1987 [33]. The key to these experiments was the development of C-ANF4–23, an ANP analog missing the complete carboxyl-terminal tail as well as five amino acids within the disulfide ring. This analog preferentially binds NPR-C over GC-A. Using ANF4–23 as a competing ligand the vast majority of ANP binding sites (> 90%) in the kidney and intact rat were attributed to NPR-C. Perfusion of relatively high concentrations of C-ANF4–23 (100 nM) into isolated kidneys did not stimulate glomerular filtration rate or sodium excretion, consistent with the inability of this peptide to activate GC-A at the infused concentrations. However, infusion of C-ANF4–23 into whole rats increased sodium excretion and decreased blood pressure in a manner that temporally correlated with elevations of full length ANP1–28. Infusions of full length ANP1–28 to levels observed during C-ANF4–23 infusions, yielded similar levels of sodium excretions and blood pressure reductions, consistent with C-ANF4–23 blocking NPR-C-mediated ANP degradation. C-ANF4–23 infusions also markedly decreased metabolic clearance, volume of distribution and appearance of radiolabeled hydrolytic products in anesthetized rats infused with 125I-ANP [34]. A separate study reported that C-ANF increased trichloroacetic acid-precipitable radiation from rats infused with 125I-ANP by seven-fold, consistent with NPR-C and/or other ANP binding molecules playing a predominant role in mediated ANP degradation [35].
It is worth noting that the evolution of a separate receptor to clear peptide signaling molecules from the cardiovascular system is relatively unique to the natriuretic peptide system because most other peptide signaling molecules like angiotensin II, endothelin or vasopressin are primarily degraded by extracellular proteases, whereas the vast majority of insulin is internalized by its cognate tyrosine kinase signaling receptor, not a separate non-tyrosine kinase receptor.
The cellular mechanics of NPR-C mediated natriuretic peptide internalization and degradation is similar to that of the receptors for low-density lipoprotein, asialoglycoprotein and hyaluronic acid. Like features include lysosomal ligand hydrolysis and recycling of the ligand-free receptor back to the plasma membrane. Internalization is speculated to occur through a clathrin-dependent mechanism but this has not been demonstrated. The effect of ligand binding on NPR-C internalization is disputed with one group indicating constitutive internalization and another group indicating downregulation [36, 37]. The internalization rate of NPR-C is about 5% per minute and is inhibited by hyperosmotic sucrose, low temperature and various agents that block lysosomal protein degradation [38]. Unlike GC-A that rapidly releases ANP after binding [39], dissociation of ANP from NPR-C is slower than the rate of receptor internalization, which ensures that the majority of bound ligand is delivered to the lysosome for degradation. NPR-C lacks known cytoplasmic internalization motifs like NPXY or YXXZ (where X is any amino acid) that are common to other receptors that are internalized via a clathrin-coated pit-dependent pathway. Mutation of individual intracellular amino acids only marginally reduces the rate of NPR-C internalization but removal of the complete 37 amino acid intracellular domain decreases internalization approximately 10 fold [38].
Proteolysis of natriuretic peptides
Natriuretic peptides are also degraded by extracellular proteases. Early studies indicated that rat and rabbit renal cortex brush border membranes but not basolateral membranes rapidly degrade human ANP [40, 41]. Subsequent reports indicated that the inactivating cleavage occurs between C7 and F8 (Fig. 2) and is inhibited by the metal chelating agents, 1,10 phenanthroline and EDTA [42, 43]. In pig microvillar membranes, ANP degradation was inhibited by phosphoramidon, an inhibitor of neprilysin (NEP) (EC 3.4.24.11), which is also known as neutral endopeptidase, enkephalinase, common acute lymphoblastic leukemia antigen and CD10 [44]. NEP was initially discovered in rabbit kidney brush border membranes as a metalloenzyme that degrades the insulin beta chain [45] and subsequently as an enkephalinase [46] and beta amyloid-degrading enzyme. NEP is a zinc-containing, membrane-bound, ectoenzyme that cleaves substrates on the amino side of hydrophobic residues [45] (Fig. 1). Stephenson and colleagues demonstrated that the HPLC elution pattern of ANP cleavage products from kidney membranes was similar to the products produced when ANP was degraded by purified NEP [47]. Subsequent studies indicated that ANP degrading activity in solubilized rat membranes copurifies with NEP and is blocked by specific NEP inhibitors [48].
Figure 2.
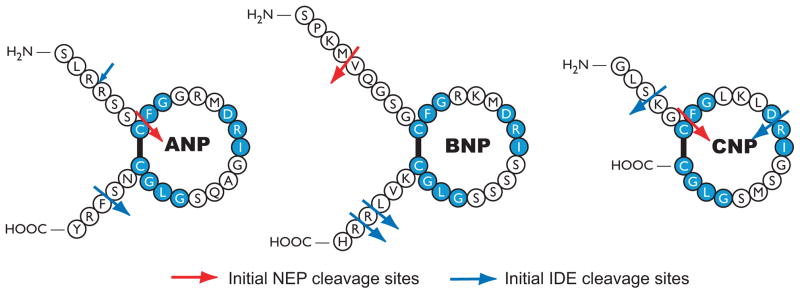
Initial NEP and IDE cleavage sites in human natriuretic peptides. NEP data are from studies by Kenny et al, Vanneste et al, Norman et al and Watanabe et al. [49, 50, 53, 55]. IDE cleavage sites are from studies performed by Muller et al using rat brain IDE and rat ANP, porcine BNP-26 and porcine CNP and Ralat et al using recombinant human IDE and human natriuretic peptides [68, 69]. Large blue arrows indicates primary initial IDE sites. The small blue arrow indicates a minor initial IDE site in ANP. Both groups observed IDE cleavage of CNP between D12 and R13 but only Ralat et al observed cleavage between S3 and K4.
Purified NEP binds and degrades natriuretic peptides similarly to other peptide hormones like angiotensin II [47]. Inhibition constants (Ki values) derived from blocking the degradation of the beta chain of insulin range from 2.5 μM for CNP to 172 μM for human BNP [49]. Seven ANP cleavage sites were identified (R4-S5, C7-F8, R11-M12, R14-I15, G16-A17, G20-L21 and S25-F26) but the initial attack occurs between C7 and F8, which breaks the ring and inactivates the peptide [47, 50]. Initial NEP cleavage sites for ANP, BNP and CNP are shown in Figure 2. Interestingly, a frameshift mutant of ANP containing 12 additional C-terminal amino acids is resistant to NEP degradation and is elevated in patients with familial atrial fibrillation [51, 52]. NEP also efficiently cleaves CNP at multiple sites (C6-F7, G8-L9, K10-L11, R13-I14, S16-M17 and G19-L20), and like ANP, the initial cleavage site is between the conserved C and F residues [49, 53]. The ring structures of both ANP and CNP are essential for hydrolysis because reduction and alkylation of the peptides greatly decreases degradation [53].
In contrast to ANP or CNP, which have one or zero amino acid differences between human and rodent forms of the peptides, BNP varies greatly between species [54]. For instance rat BNP is 45-residues and human BNP is 32-residues with 16 differences within the common 32-residue core structure. Studies using purified enzymes indicated that BNP is a poorer substrate for human or porcine NEP compared to ANP or CNP [49, 53]. NEP cleaves human BNP at M5-V6 and R17-I18 but not at the conserved C10-F11 bond [53, 55]. Kenny and colleagues found that cleavage at M5-V6 precedes cleavage at R17-I18 (Fig. 2)[49]. Urodilatin is a four residue amino-extended form of ANP that is also a poorer substrate for NEP than ANP [56]. These data suggest that the additional terminal residues in ANP, BNP and urodilatin reduce access of NEP to the primary C10-F11 cleavage site [53]. Consistent with the idea of ANP being a better substrate for NEP, phosphoramidon dramatically increased ANP-dependent, but not BNP-dependent cGMP elevations, in mouse kidney slices [57]. The degradation preference of porcine NEP for human natriuretic peptides is CNP ≥ ANP > urodilation ≫ BNP [49, 53, 56]. The kcat/Km values for human ANP, BNP and CNP are 5.1 M−1s−1, 0.5 M−1s−1 and 7.8 M−1s−1, respectively [53]. We recently demonstrated that NEP-dependent degradation of BNP is species specific. Although NEP accounts for most of the BNP degrading activity in rat kidney membranes, NEP inhibitors failed to block BNP degradation by human kidney membranes, which suggest that NEP is not a significant regulator of BNP concentrations in the human kidney [58].
Oral NEP inhibitors elevate natriuretic peptide concentrations in humans and animals models and increase sodium excretion during heart failure, consistent with NEP or another enzyme that is blocked by NEP inhibitors contributing to normal natriuretic peptide degradation [56, 59–62]. Natriuretic peptide levels have not been reported in mice lacking NEP, but these mice show no obvious signs of increased natriuretic peptide receptor activation, consistent with other degradation pathways compensating for the loss of NEP activity in this species [63].
ANP is also cleaved by insulin degrading enzyme (IDE), a zinc metalloprotease that is found in both cytoplasmic and membrane fractions and has diverse substrate specificity (Fig. 1) [44, 64]. Initial studies revealed that conditioned medium from smooth muscle and endothelial cells contained an EDTA- and EGTA-inhibited proteolytic activity that cleaves the bond between S25 and F26 of ANP [65, 66]. Cross-linking studies by Muller and colleagues revealed that 125I-ANP binds with high affinity (Kd = 60 nM) to a cytosolic 112 kDa protein from rat olfactory bulb homogenates [67]. Based on competition with insulin for ANP degradation and partial amino acid sequence of the 112 kilodalton protein, IDE was suggested to be an ANP degrading enzyme [67]. Additional studies demonstrated that ANP binding to IDE was blocked by full length ANP but not by an ANP variant lacking the last three C-terminal residues or amino-terminally truncated porcine BNP-26.
Proteolysis of rat ANP, porcine BNP-26 and CNP with purified IDE revealed that ANP is the preferred substrate [68]. The half-life for degradation of ANP by purified IDE was approximately one third that for BNP or CNP. HPLC purification and mass spectrometry analysis indicated that ANP was sequentially cleaved four times by IDE whereas BNP and CNP were cleaved three and two times, respectively. ANP was initially cleaved at the S25-F26 bond (Fig. 2), and then in successive order and at much slower rates, the R3-R4, D13-R14 and C7-F8 bonds were hydrolyzed. Longer incubations with IDE resulted in the initial cleavage of BNP at the R24-R25 bond followed by cleavage at the G6-R7 and D10-R11 bonds. In contrast to initial cleavages outside the disulfide ring, CNP was initially cleaved between D12 and R13 (Fig. 2).
A recent and exciting report by Ralat and colleagues suggests that IDE plays multiple roles in modulating the signaling response to natriuretic peptides [69]. Like Muller et al. they found that human IDE purified from E. coli binds ANP five times tighter (IC50 = 40 nM) than insulin. They also determined that human versions of ANP and CNP were much better IDE substrates than BNP, having Kcat values of 10 s−1, 20 s−1 and 0.2 s−1, respectively.
Studies involving siRNA knockdown of IDE in 293 cells stably expressing GC-A or GC-B revealed novel effects of IDE on receptor activation. Reduced IDE expression enhanced stimulation of GC-A and GC-B by ANP and CNP, respectively, consistent with IDE-dependent degradation and inactivation of ANP and CNP. In contrast, reduced IDE expression was correlated with decreased activation of GC-A by BNP, consistent with IDE producing a superactive BNP variant. Incubation of these peptides with purified ICE increased and decreased activation of GC-A by BNP and ANP, respectively. Surprisingly, IDE exposure decreased CNP activation of GC-B, but increased cross-activation of GC-B by ANP and BNP. These in vitro data are consistent with IDE modulating natriuretic peptide potency and receptor preference.
Ralat and colleagues also determined major and minor cleavage sites of the natriuretic peptides (Fig. 2). Like Muller et al., they found that the major cleavage site of ANP was at the S25-F26 bond, but they also observed a small amount of cleavage products from breaking the R3-R4 bond after incubating the peptide with IDE for 1 sec. For this reason, they proposed a “biased stochastic” as opposed to “sequential” cleavage model. Longer incubations resulted in near complete breakdown of ANP. Major cleavage sites for BNP were between L29-R30 and R30-R31, whereas major sites for CNP were between S3-K4 and at the F12-R13. The general observation was that cleavage occurs first at the tails for peptides that have N- and C-terminal extensions, but within the disulfide loop in peptides lacking extensions. As with NEP, the C-terminally extended frameshift mutant of ANP was a poor substrate for IDE. Interestingly, when IDE was incubated with mutant ANP and either wild type ANP or CNP, the mutant peptide was preferentially and efficiently degraded. Finally, mice lacking IDE exhibit increased levels of amyloid beta protein and insulin, but natriuretic peptide concentrations in these animals have not been reported [70].
As described above, BNP is a poor substrate for NEP and IDE, which suggest that another protease is responsible for its cleavage. Consistent with this notion, Pankow and colleagues reported that a 32 amino acid version of the normal 45-residue mouse BNP molecule is degraded by the multimeric renal metalloprotease, meprin A [71]. Initial data indicated that BNP, but not ANP, is degraded similarly in wild type and NEP “knockout” mice, consistent with a NEP-independent proteolytic event. HPLC purification and mass spectrometric identification determined that the initial meprin cleavage site is at the H6-I7 bond. Interestingly, the resulting BNP8–32 product retains the ability to activate GC-A in cell culture but has reduced renal activating activity in dogs [71, 72]. The H6-I7 sequence of the truncated mouse BNP used by Pankow and colleagues corresponds to Q6-G7 in human BNP. Hence, the meprin cleavage site is not conserved in human BNP and is not shown in Fig. 2. Protease inhibitor screening indicated that compounds known to inhibit meprin A (EDTA or actinonin), completely blocked BNP degradation in kidney membranes from the NEP deficient animals. Purified mouse meprin A efficiently degraded mouse BNP1-32, rat BNP and porcine BNP, but not CNP. Importantly, BNP1–32 degradation was severely blunted in kidney membranes from mice lacking meprin A and cleavage of BNP1–32 with meprin increased susceptibility of the peptide to ring cleavage by NEP. Thus, it was suggested that meprin A cleavage of BNP1–32 facilitates subsequent cleavage and inactivation of BNP7–32 by NEP [71]. However, the meprin cleavage site is not conserved in human BNP, and we found that meprin and NEP cleave rat BNP but do not cleave human BNP when measured in their respective kidney membrane preparations [58]. Interestingly, the serine protease inhibitor leupeptin was the most effective inhibitor of human BNP degradation, but the specific protease inhibited by leupeptin has not been identified.
Relative contribution of NPR-C and NEP to natriuretic peptide degradation
The relative contribution of NPR-C and NEP to ANP degradation has been investigated in a number of animal systems using various NPR-C blocking peptides and NEP inhibitors. However, an assumption of these studies is that the NPR-C blocking peptides do not inhibit the proteases that degrade natriuretic peptides, which has not been tested to my knowledge.
Under normal conditions, infusion of NPR-C blocking peptides has a slightly greater or an equal effect on circulating ANP concentrations and associated physiologic functions as various NEP inhibitors [60, 73–75]. However, in all cases examined, maximum ANP concentrations require inhibition of both degradation pathways. During pathological or pharmacological scenarios where natriuretic peptide concentrations are elevated and NPR-C may be saturated, NEP plays a more significant role in ANP degradation [76]. Both NPR-C and NEP pathways contribute to the degradation of BNP and CNP as well, although the exact contribution of each pathway to BNP concentrations is unclear [5, 31, 59, 77]. In dogs, total metabolic clearance rate of infused CNP was significantly reduced by infusion of C-ANF4–23 or a NEP inhibitor [31, 59]. NEP inhibition reduced CNP clearance by the kidney but not the lung, suggesting that NEP significantly contributes to CNP degradation in some but not all tissues.
Conclusion and perspectives
A vast amount of data has been published regarding the metabolism and degradation of natriuretic peptides. From these reports it is clear that NPR-C is a specific natriuretic peptide degrading receptor and that NEP and IDE are general proteases that degrade natriuretic and other peptides. However, recent reports of increased half-lives of natriuretic peptides associated with disease as well as improved clinical benefits of proteolytic resistant natriuretic peptides suggests that natriuretic peptide degradation may be more important than previously appreciated [51, 52, 78, 79].
Several important questions remain regarding natriuretic peptide degradation. Specifically, what role does IDE play in regulating in vivo natriuretic peptide concentrations, and does C-terminal cleavage by IDE produces natriuretic peptide variants with unique binding and activation characteristics? A specific IDE inhibitor would be extremely useful in illuminating the physiologic significance of IDE in natriuretic peptide signaling. Other important questions are what is the sequence of the BNP derivative produced by incubation with IDE, and what is the identity of the leupeptin-sensitive protease that degrades human BNP? Regarding receptor-dependent peptide clearance, several questions involving the molecular nature of NPR-C internalization have yet to be answered. Although this receptor clearly internalizes and degrades natriuretic peptides, the molecular transport system and adaptor protein partners required for this process are not known. Finally, it remains to be seen whether ramifications of basic science research on natriuretic peptide degradation will find its way into the clinic. Can new versions of nesiritide (recombinant human BNP), which is approved for the treatment of acute decompensated congestive heart failure, be engineered that are resistant to degradation or that have more desirable therapeutic profiles? For instance, can peptides be engineered that are degraded slowly in the kidney and rapidly in the vasculature? Only time will tell whether basic information on natriuretic peptide metabolism will translate into better therapeutic options for patients.
Acknowledgments
I thank the members of my laboratory for helpful comments on this manuscript. National Institutes of Health grant NIHR21HL093402 and the American Heart Association grant 0950053G supported this work.
Abbreviations
- ANP
atrial natriuretic peptide
- BNP
B-type natriuretic peptide
- CNP
C-type natriuretic peptide
- IDE
insulin degrading enzyme
- NEP
neprilysin
- GC-A
natriuretic peptide receptor-A
- GC-B
natriuretic peptide receptor-B
- NPR-C
natriuretic peptide receptor-C
References
- 1.Kishimoto I, Tokudome T, Nakao K, Kangawa K. The cardiovascular significance of the natriuretic peptide system. FEBS J. 2011 doi: 10.1111/j.1742-4658.2011.08116.x. [DOI] [PubMed] [Google Scholar]
- 2.Misono K, Philo J, Tsutomu A, Ogata C, Qiu Y, Ogawa H, Young S. Structure, signaling mechanism and regulation of natriuretic peptide receptor-guanylate cyclase. FEBS J. 2011 doi: 10.1111/j.1742-4658.2011.08083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pandey K. Latest Perspectives and Paradigms on the Functional Genomics of Guanylyl Cyclase/Natriuretic Peptide Receptor-A. FEBS J. 2011 doi: 10.1111/j.1742-4658.2011.08081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckley MG, Sethi D, Markandu ND, Sagnella GA, Singer DR, MacGregor GA. Plasma concentration and comparisons of brain natriuretic peptide and atrial natriuretic peptide in normal subjects, cardiac transplant recipients and patients with dialysis-independent or dialysis-dependent chronic renal failure. Clin Sci (Lond) 1992;83:437–444. doi: 10.1042/cs0830437. [DOI] [PubMed] [Google Scholar]
- 5.Mukoyama M, Nakao K, Hosoda K, Suga S, Saito Y, Ogawa Y, Shirakami G, Jougasaki M, Obata K, Yasue H, et al. Brain natriuretic peptide as a novel cardiac hormone in humans. Evidence for an exquisite dual natriuretic peptide system, atrial natriuretic peptide and brain natriuretic peptide. J Clin Invest. 1991;87:1402–1412. doi: 10.1172/JCI115146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnett JC, Jr, Kao PC, Hu DC, Heser DW, Heublein D, Granger JP, Opgenorth TJ, Reeder GS. Atrial natriuretic peptide elevation in congestive heart failure in the human. Science. 1986;231:1145–1147. doi: 10.1126/science.2935937. [DOI] [PubMed] [Google Scholar]
- 7.Mukoyama M, Nakao K, Saito Y, Ogawa Y, Hosoda K, Suga S, Shirakami G, Jougasaki M, Imura H. Increased human brain natriuretic peptide in congestive heart failure. N Engl J Med. 1990;323:757–758. doi: 10.1056/NEJM199009133231114. [DOI] [PubMed] [Google Scholar]
- 8.Yandle TG, Richards AM, Gilbert A, Fisher S, Holmes S, Espiner EA. Assay of brain natriuretic peptide (BNP) in human plasma: evidence for high molecular weight BNP as a major plasm component in heart failure. J Clin Endocrinol Metab. 1993;76:832–838. doi: 10.1210/jcem.76.4.8473392. [DOI] [PubMed] [Google Scholar]
- 9.John SW, Krege JH, Oliver PM, Hagaman JR, Hodgin JB, Pang SC, Flynn TG, Smithies O. Genetic decreases in atrial natriuretic peptide and salt-sensitive hypertension [published erratum appears in Science 1995 Mar 24;267(5205):1753] Science. 1995;267:679–681. doi: 10.1126/science.7839143. [DOI] [PubMed] [Google Scholar]
- 10.Tamura N, Ogawa Y, Chusho H, Nakamura K, Nakao K, Suda M, Kasahara M, Hashimoto R, Katsuura G, Mukoyama M, et al. Cardiac fibrosis in mice lacking brain natriuretic peptide. Proc Natl Acad Sci U S A. 2000;97:4239–4244. doi: 10.1073/pnas.070371497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chusho H, Tamura N, Ogawa Y, Yasoda A, Suda M, Miyazawa T, Nakamura K, Nakao K, Kurihara T, Komatsu Y, et al. Dwarfism and early death in mice lacking C-type natriuretic peptide. Proc Natl Acad Sci U S A. 2001;98:4016–4021. doi: 10.1073/pnas.071389098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potter LR. Regulation and therapeutic targeting of peptide-activated receptor guanylyl cyclases. Pharmacology and Therapeutics. 2011 doi: 10.1016/j.pharmthera.2010.12.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Potter LR, Abbey-Hosch S, Dickey DM. Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocr Rev. 2006;27:47–72. doi: 10.1210/er.2005-0014. [DOI] [PubMed] [Google Scholar]
- 14.Bartels CF, Bukulmez H, Padayatti P, Rhee DK, van Ravenswaaij-Arts C, Pauli RM, Mundlos S, Chitayat D, Shih LY, Al-Gazali LI, et al. Mutations in the transmembrane natriuretic peptide receptor NPR-B impair skeletal growth and cause acromesomelic dysplasia, type Maroteaux. Am J Hum Genet. 2004;75:27–34. doi: 10.1086/422013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura N, Doolittle LK, Hammer RE, Shelton JM, Richardson JA, Garbers DL. Critical roles of the guanylyl cyclase B receptor in endochondral ossification and development of female reproductive organs. Proc Natl Acad Sci U S A. 2004;101:17300–17305. doi: 10.1073/pnas.0407894101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsuji T, Kunieda T. A loss-of-function mutation in natriuretic peptide receptor 2 (Npr2) gene is responsible for disproportionate dwarfism in cn/cn mouse. J Biol Chem. 2005;280:14288–14292. doi: 10.1074/jbc.C500024200. [DOI] [PubMed] [Google Scholar]
- 17.Matsukawa N, Grzesik WJ, Takahashi N, Pandey KN, Pang S, Yamauchi M, Smithies O. The natriuretic peptide clearance receptor locally modulates the physiological effects of the natriuretic peptide system. Proc Natl Acad Sci U S A. 1999;96:7403–7408. doi: 10.1073/pnas.96.13.7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaubert J, Jaubert F, Martin N, Washburn LL, Lee BK, Eicher EM, Guenet JL. Three new allelic mouse mutations that cause skeletal overgrowth involve the natriuretic peptide receptor C gene (Npr3) Proc Natl Acad Sci U S A. 1999;96:10278–10283. doi: 10.1073/pnas.96.18.10278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerbes AL, Vollmar AM. Degradation and clearance of atrial natriuretic factors (ANF) Life Sci. 1990;47:1173–1180. doi: 10.1016/0024-3205(90)90208-9. [DOI] [PubMed] [Google Scholar]
- 20.Ruskoaho H. Atrial natriuretic peptide: synthesis, release, and metabolism. Pharmacol Rev. 1992;44:479–602. [PubMed] [Google Scholar]
- 21.Nakao K, Sugawara A, Morii N, Sakamoto M, Yamada T, Itoh H, Shiono S, Saito Y, Nishimura K, Ban T, et al. The pharmacokinetics of alpha-human atrial natriuretic polypeptide in healthy subjects. Eur J Clin Pharmacol. 1986;31:101–103. doi: 10.1007/BF00870995. [DOI] [PubMed] [Google Scholar]
- 22.Yandle TG, Richards AM, Nicholls MG, Cuneo R, Espiner EA, Livesey JH. Metabolic clearance rate and plasma half life of alpha-human atrial natriuretic peptide in man. Life Sci. 1986;38:1827–1833. doi: 10.1016/0024-3205(86)90137-2. [DOI] [PubMed] [Google Scholar]
- 23.Crozier IG, Nicholls MG, Ikram H, Espiner EA, Yandle TG, Jans S. Atrial natriuretic peptide in humans. Production and clearance by various tissues. Hypertension. 1986;8:II11–15. doi: 10.1161/01.hyp.8.6_pt_2.ii11. [DOI] [PubMed] [Google Scholar]
- 24.Schutten HJ, Henriksen JH, Warberg J. Organ extraction of atrial natriuretic peptide (ANP) in man. Significance of sampling site. Clin Physiol. 1987;7:125–132. doi: 10.1111/j.1475-097x.1987.tb00154.x. [DOI] [PubMed] [Google Scholar]
- 25.Hollister AS, Rodeheffer RJ, White FJ, Potts JR, Imada T, Inagami T. Clearance of atrial natriuretic factor by lung, liver, and kidney in human subjects and the dog. J Clin Invest. 1989;83:623–628. doi: 10.1172/JCI113926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bates ER, McGillem MJ, Mancini GB, Grekin RJ. Pulmonary extraction of immunoreactive atrial natriuretic factor in dogs. Am J Cardiol. 1989;63:372–373. doi: 10.1016/0002-9149(89)90353-6. [DOI] [PubMed] [Google Scholar]
- 27.Holmes SJ, Espiner EA, Richards AM, Yandle TG, Frampton C. Renal, endocrine, and hemodynamic effects of human brain natriuretic peptide in normal man. J Clin Endocrinol Metab. 1993;76:91–96. doi: 10.1210/jcem.76.1.8380606. [DOI] [PubMed] [Google Scholar]
- 28.Richards AM, Crozier IG, Yandle TG, Espiner EA, Ikram H, Nicholls MG. Brain natriuretic factor: regional plasma concentrations and correlations with haemodynamic state in cardiac disease. Br Heart J. 1993;69:414–417. doi: 10.1136/hrt.69.5.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunt PJ, Richards AM, Espiner EA, Nicholls MG, Yandle TG. Bioactivity and metabolism of C-type natriuretic peptide in normal man. J Clin Endocrinol Metab. 1994;78:1428–1435. doi: 10.1210/jcem.78.6.8200946. [DOI] [PubMed] [Google Scholar]
- 30.Charles CJ, Espiner EA, Richards AM, Nicholls MG, Yandle TG. Biological actions and pharmacokinetics of C-type natriuretic peptide in conscious sheep. Am J Physiol. 1995;268:R201–207. doi: 10.1152/ajpregu.1995.268.1.R201. [DOI] [PubMed] [Google Scholar]
- 31.Brandt RR, Heublein DM, Aarhus LL, Lewicki JA, Burnett JC., Jr Role of natriuretic peptide clearance receptor in in vivo control of C-type natriuretic peptide. Am J Physiol. 1995;269:H326–331. doi: 10.1152/ajpheart.1995.269.1.H326. [DOI] [PubMed] [Google Scholar]
- 32.Palmer SC, Prickett TC, Espiner EA, Yandle TG, Richards AM. Regional release and clearance of C-type natriuretic peptides in the human circulation and relation to cardiac function. Hypertension. 2009;54:612–618. doi: 10.1161/HYPERTENSIONAHA.109.135608. [DOI] [PubMed] [Google Scholar]
- 33.Maack T, Suzuki M, Almeida FA, Nussenzveig D, Scarborough RM, McEnroe GA, Lewicki JA. Physiological role of silent receptors of atrial natriuretic factor. Science. 1987;238:675–678. doi: 10.1126/science.2823385. [DOI] [PubMed] [Google Scholar]
- 34.Almeida FA, Suzuki M, Scarborough RM, Lewicki JA, Maack T. Clearance function of type C receptors of atrial natriuretic factor in rats. Am J Physiol. 1989;256:R469–475. doi: 10.1152/ajpregu.1989.256.2.R469. [DOI] [PubMed] [Google Scholar]
- 35.Chiu PJ, Tetzloff G, Romano MT, Foster CJ, Sybertz EJ. Influence of C-ANF receptor and neutral endopeptidase on pharmacokinetics of ANF in rats. Am J Physiol. 1991;260:R208–216. doi: 10.1152/ajpregu.1991.260.1.R208. [DOI] [PubMed] [Google Scholar]
- 36.Nussenzveig DR, Lewicki JA, Maack T. Cellular mechanisms of the clearance function of type C receptors of atrial natriuretic factor. J Biol Chem. 1990;265:20952–20958. [PubMed] [Google Scholar]
- 37.Rathinavelu A, Isom GE. Differential internalization and processing of atrial-natriuretic-factor B and C receptor in PC12 cells. Biochem J. 1991;276:493–497. doi: 10.1042/bj2760493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen D, Koh GY, Nikonova LN, Porter JG, Maack T. Molecular determinants of the clearance function of type C receptors of natriuretic peptides. J Biol Chem. 1996;271:9863–9869. doi: 10.1074/jbc.271.16.9863. [DOI] [PubMed] [Google Scholar]
- 39.Koh GY, Nussenzveig DR, Okolicany J, Price DA, Maack T. Dynamics of atrial natriuretic factor-guanylate cyclase receptors and receptor-ligand complexes in cultured glomerular mesangial and renomedullary interstitial cells. J Biol Chem. 1992;267:11987–11994. [PubMed] [Google Scholar]
- 40.Hori R, Inui K, Saito H, Matsukawa Y, Okumura K, Nakao K, Morii N, Imura H. Specific receptors for atrial natriuretic polypeptide on basolateral membranes isolated from rat renal cortex. Biochem Biophys Res Commun. 1985;129:773–779. doi: 10.1016/0006-291x(85)91959-x. [DOI] [PubMed] [Google Scholar]
- 41.Olins GM, Spear KL, Siegel NR, Reinhard EJ, Zurcher-Neely HA. Atrial peptide inactivation by rabbit-kidney brush-border membranes. Eur J Biochem. 1987;170:431–434. doi: 10.1111/j.1432-1033.1987.tb13717.x. [DOI] [PubMed] [Google Scholar]
- 42.Koehn JA, Norman JA, Jones BN, LeSueur L, Sakane Y, Ghai RD. Degradation of atrial natriuretic factor by kidney cortex membranes. Isolation and characterization of the primary proteolytic product. J Biol Chem. 1987;262:11623–11627. [PubMed] [Google Scholar]
- 43.Olins GM, Spear KL, Siegel NR, Zurcher-Neely HA. Inactivation of atrial natriuretic factor by the renal brush border. Biochim Biophys Acta. 1987;901:97–100. doi: 10.1016/0005-2736(87)90260-4. [DOI] [PubMed] [Google Scholar]
- 44.Malito E, Hulse RE, Tang WJ. Amyloid beta-degrading cryptidases: insulin degrading enzyme, presequence peptidase, and neprilysin. Cell Mol Life Sci. 2008;65:2574–2585. doi: 10.1007/s00018-008-8112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kerr MA, Kenny AJ. The purification and specificity of a neutral endopeptidase from rabbit kidney brush border. Biochem J. 1974;137:477–488. doi: 10.1042/bj1370477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malfroy B, Swerts JP, Guyon A, Roques BP, Schwartz JC. High-affinity enkephalin-degrading peptidase in brain is increased after morphine. Nature. 1978;276:523–526. doi: 10.1038/276523a0. [DOI] [PubMed] [Google Scholar]
- 47.Stephenson SL, Kenny AJ. The hydrolysis of alpha-human atrial natriuretic peptide by pig kidney microvillar membranes is initiated by endopeptidase-24.11. Biochem J. 1987;243:183–187. doi: 10.1042/bj2430183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sonnenberg JL, Sakane Y, Jeng AY, Koehn JA, Ansell JA, Wennogle LP, Ghai RD. Identification of protease 3.4.24.11 as the major atrial natriuretic factor degrading enzyme in the rat kidney. Peptides. 1988;9:173–180. doi: 10.1016/0196-9781(88)90024-1. [DOI] [PubMed] [Google Scholar]
- 49.Kenny AJ, Bourne A, Ingram J. Hydrolysis of human and pig brain natriuretic peptides, urodilatin, C-type natriuretic peptide and some C-receptor ligands by endopeptidase-24.11. Biochem J. 1993;291 ( Pt 1):83–88. doi: 10.1042/bj2910083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanneste Y, Michel A, Dimaline R, Najdovski T, Deschodt-Lanckman M. Hydrolysis of alpha-human atrial natriuretic peptide in vitro by human kidney membranes and purified endopeptidase-24.11. Evidence for a novel cleavage site. Biochem J. 1988;254:531–537. doi: 10.1042/bj2540531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dickey DM, Yoder AR, Potter LR. A familial mutation renders atrial natriuretic Peptide resistant to proteolytic degradation. J Biol Chem. 2009;284:19196–19202. doi: 10.1074/jbc.M109.010777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hodgson-Zingman DM, Karst ML, Zingman LV, Heublein DM, Darbar D, Herron KJ, Ballew JD, de Andrade M, Burnett JC, Jr, Olson TM. Atrial natriuretic peptide frameshift mutation in familial atrial fibrillation. N Engl J Med. 2008;359:158–165. doi: 10.1056/NEJMoa0706300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe Y, Nakajima K, Shimamori Y, Fujimoto Y. Comparison of the hydrolysis of the three types of natriuretic peptides by human kidney neutral endopeptidase 24.11. Biochem Mol Med. 1997;61:47–51. doi: 10.1006/bmme.1997.2584. [DOI] [PubMed] [Google Scholar]
- 54.Potter LR, Yoder AR, Flora DR, Antos LK, Dickey DM. Natriuretic peptides: their structures, receptors, physiologic functions and therapeutic applications. Handb Exp Pharmacol. 2009:341–366. doi: 10.1007/978-3-540-68964-5_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Norman JA, Little D, Bolgar M, Di Donato G. Degradation of brain natriuretic peptide by neutral endopeptidase: species specific sites of proteolysis determined by mass spectrometry. Biochem Biophys Res Commun. 1991;175:22–30. doi: 10.1016/s0006-291x(05)81194-5. [DOI] [PubMed] [Google Scholar]
- 56.Abassi ZA, Golomb E, Agbaria R, Roller PP, Tate J, Keiser HR. Hydrolysis of iodine labelled urodilatin and ANP by recombinant neutral endopeptidase EC. 3.4.24.11. Br J Pharmacol. 1994;113:204–208. doi: 10.1111/j.1476-5381.1994.tb16194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kishimoto I, Hamra FK, Garbers DL. Apparent B-type natriuretic peptide selectivity in the kidney due to differential processing. Can J Physiol Pharmacol. 2001;79:715–722. [PubMed] [Google Scholar]
- 58.Dickey DM, Potter LR. Human B-type natriuretic peptide is not degraded by meprin A. Biochem Pharmacol. 2010;80:1007–1011. doi: 10.1016/j.bcp.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brandt RR, Mattingly MT, Clavell AL, Barclay PL, Burnett JC., Jr Neutral endopeptidase regulates C-type natriuretic peptide metabolism but does not potentiate its bioactivity in vivo. Hypertension. 1997;30:184–190. doi: 10.1161/01.hyp.30.2.184. [DOI] [PubMed] [Google Scholar]
- 60.Charles CJ, Espiner EA, Nicholls MG, Richards AM, Yandle TG, Protter A, Kosoglou T. Clearance receptors and endopeptidase 24.11: equal role in natriuretic peptide metabolism in conscious sheep. Am J Physiol. 1996;271:R373–380. doi: 10.1152/ajpregu.1996.271.2.R373. [DOI] [PubMed] [Google Scholar]
- 61.Margulies KB, Barclay PL, Burnett JC., Jr The role of neutral endopeptidase in dogs with evolving congestive heart failure. Circulation. 1995;91:2036–2042. doi: 10.1161/01.cir.91.7.2036. [DOI] [PubMed] [Google Scholar]
- 62.Wegner M, Stasch JP, Hirth-Dietrich C, Dressel J, Voges KP, Kazda S. Interaction of a neutral endopeptidase inhibitor with an ANP-C receptor ligand in anesthetized dogs. Clin Exp Hypertens. 1995;17:861–876. doi: 10.3109/10641969509033640. [DOI] [PubMed] [Google Scholar]
- 63.Lu B, Gerard NP, Kolakowski LF, Jr, Bozza M, Zurakowski D, Finco O, Carroll MC, Gerard C. Neutral endopeptidase modulation of septic shock. J Exp Med. 1995;181:2271–2275. doi: 10.1084/jem.181.6.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Duckworth WC, Bennett RG, Hamel FG. Insulin degradation: progress and potential. Endocr Rev. 1998;19:608–624. doi: 10.1210/edrv.19.5.0349. [DOI] [PubMed] [Google Scholar]
- 65.Johnson GR, Arik L, Foster CJ. Metabolism of 125I-atrial natriuretic factor by vascular smooth muscle cells. Evidence for a peptidase that specifically removes the COOH-terminal tripeptide. J Biol Chem. 1989;264:11637–11642. [PubMed] [Google Scholar]
- 66.Johnson GR, Foster CJ. Partial characterization of a metalloendopeptidase activity produced by cultured endothelial cells that removes the COOH-terminal tripeptide from 125I-atrial natriuretic factor. Biochem Biophys Res Commun. 1990;167:110–116. doi: 10.1016/0006-291x(90)91737-d. [DOI] [PubMed] [Google Scholar]
- 67.Muller D, Baumeister H, Buck F, Richter D. Atrial natriuretic peptide (ANP) is a high-affinity substrate for rat insulin-degrading enzyme. Eur J Biochem. 1991;202:285–292. doi: 10.1111/j.1432-1033.1991.tb16374.x. [DOI] [PubMed] [Google Scholar]
- 68.Muller D, Schulze C, Baumeister H, Buck F, Richter D. Rat insulin-degrading enzyme: cleavage pattern of the natriuretic peptide hormones ANP, BNP, and CNP revealed by HPLC and mass spectrometry. Biochemistry. 1992;31:11138–11143. doi: 10.1021/bi00160a026. [DOI] [PubMed] [Google Scholar]
- 69.Ralat LA, Guo Q, Ren M, Funke T, Dickey DM, Potter LR, Tang WJ. Insulin-degrading enzyme modulates the natriuretic peptide- mediated signaling response. J Biol Chem. 2010 doi: 10.1074/jbc.M110.173252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Farris W, Mansourian S, Chang Y, Lindsley L, Eckman EA, Frosch MP, Eckman CB, Tanzi RE, Selkoe DJ, Guenette S. Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc Natl Acad Sci U S A. 2003;100:4162–4167. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pankow K, Wang Y, Gembardt F, Krause E, Sun X, Krause G, Schultheiss HP, Siems WE, Walther T. Successive action of meprin A and neprilysin catabolizes B-type natriuretic peptide. Circ Res. 2007;101:875–882. doi: 10.1161/CIRCRESAHA.107.153585. [DOI] [PubMed] [Google Scholar]
- 72.Boerrigter G, Costello-Boerrigter LC, Harty GJ, Huntley BK, Cataliotti A, Lapp H, Burnett JC., Jr B-type natriuretic peptide 8–32, which is produced from mature BNP 1–32 by the metalloprotease meprin A, has reduced bioactivity. Am J Physiol Regul Integr Comp Physiol. 2009;296:R1744–1750. doi: 10.1152/ajpregu.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koepke JP, Tyler LD, Trapani AJ, Bovy PR, Spear KL, Olins GM, Blaine EH. Interaction of non-guanylate cyclase-linked atriopeptin receptor ligand and endopeptidase inhibitor in conscious rats. J Pharmacol Exp Ther. 1989;249:172–176. [PubMed] [Google Scholar]
- 74.Kukkonen P, Vuolteenaho O, Ruskoaho H. Basal and volume expansion-stimulated plasma atrial natriuretic peptide concentrations and hemodynamics in conscious rats: effects of SCH 39.370, an endopeptidase inhibitor, and C-ANF-(4–23), a clearance receptor ligand. Endocrinology. 1992;130:755–765. doi: 10.1210/endo.130.2.1531129. [DOI] [PubMed] [Google Scholar]
- 75.Okolicany J, McEnroe GA, Koh GY, Lewicki JA, Maack T. Clearance receptor and neutral endopeptidase-mediated metabolism of atrial natriuretic factor. Am J Physiol. 1992;263:F546–553. doi: 10.1152/ajprenal.1992.263.3.F546. [DOI] [PubMed] [Google Scholar]
- 76.Hashimoto Y, Nakao K, Hama N, Imura H, Mori S, Yamaguchi M, Yasuhara M, Hori R. Clearance mechanisms of atrial and brain natriuretic peptides in rats. Pharm Res. 1994;11:60–64. doi: 10.1023/a:1018941626731. [DOI] [PubMed] [Google Scholar]
- 77.Smith MW, Espiner EA, Yandle TG, Charles CJ, Richards AM. Delayed metabolism of human brain natriuretic peptide reflects resistance to neutral endopeptidase. J Endocrinol. 2000;167:239–246. doi: 10.1677/joe.0.1670239. [DOI] [PubMed] [Google Scholar]
- 78.Lee CY, Chen HH, Lisy O, Swan S, Cannon C, Lieu HD, Burnett JC., Jr Pharmacodynamics of a novel designer natriuretic peptide, CD-NP, in a first-in-human clinical trial in healthy subjects. J Clin Pharmacol. 2009;49:668–673. doi: 10.1177/0091270009336233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lisy O, Huntley BK, McCormick DJ, Kurlansky PA, Burnett JC., Jr Design, synthesis, and actions of a novel chimeric natriuretic peptide: CD-NP. J Am Coll Cardiol. 2008;52:60–68. doi: 10.1016/j.jacc.2008.02.077. [DOI] [PMC free article] [PubMed] [Google Scholar]