Abstract
Purpose of review
To review recent developments in our understanding of endoplasmic reticulum (ER) aminopeptidase-1 (ERAP1) function in relation to its role in MHC class I peptide presentation and HLA class I-associated diseases.
Recent findings
ERAP1 polymorphisms exhibiting loss-of-function have been associated with protection from ankylosing spondylitis (AS). The aminopeptidase function of ERAP1 optimizes peptides for binding and presentation by MHC class I. Most studies have revealed reduced MHC class I expression in situations of reduced ERAP1 function. Under these circumstances the presented peptides are often N-terminally extended, and cell surface complexes are unstable and fall apart more readily. In contrast, peptides presented by HLA-B*27:05 when ERAP1 is silenced are frequently extended on the C-terminus. Recent work has emphasized the importance of assessing the function of allotypes encoded by ERAP1 haplotypes, rather than effects of single amino acid substitutions. The allotypes found in a series of AS patients were poorer at restoring HLA-B27 expression than allotypes found in unaffected controls, which may seem contrary to the genetic data linking loss-of-function to protection.
Summary
More work is needed to understand how ERAP1 variants associated with risk and protection influence the quality and quantity of peptides available for binding to HLA class I molecules in the ER. Moreover, we need to determine allele-specific effects of ERAP1 variants in the context of HLA-B*51 and HLA-Cw*6, which are associated with Behçet’s disease and psoriasis, respectively.
Keywords: ERAP1, HLA-B27, rheumatic disease, ankylosing spondylitis
Introduction
Endoplasmic reticulum (ER) aminopeptidase 1 (ERAP1) is an important component of the major histocompatibility complex (MHC) class I antigen processing and presentation pathway [1]. The discovery that ERAP1 polymorphisms are associated with the MHC (HLA) class I-linked diseases ankylosing spondylitis (AS) [2], psoriasis [3], and Behçet's disease (BD) [4], and that in each case there is evidence for gene-gene interaction (epistasis), has generated considerable interest in understanding how these variants affect the biology of HLA molecules.
Peptides derived from the cytosolic degradation of endogenous proteins are transported into the ER via peptide transporters (TAP1/TAP2). ER aminopeptidases 1 and 2 (ERAP1, ERAP2) trim peptides to optimize their length for binding to peptide-receptive major histocompatibility complex (MHC) class I molecules. To become peptide-receptive, MHC class I heavy chains (HCs) fold and associate with β2-microglobulin (β2m). To prevent expression of sub-optimally loaded or even ‘empty’ class I molecules, HC-β2m complexes are retained by tapasin as part of the peptide-loading complex until they are stabilized by peptides. Following peptide binding, MHC class I complexes are transported to the cell surface where they display peptides to CD8+ T cells and NK cells. In this pathway, peptides represent cargo that MHC class I molecules deliver to the cell surface, but also serve as ‘glue’ that holds HC-β2m complexes together. When ER peptide supply is greatly diminished, or in the absence of β2m, HCs misfold and are degraded in the ER-associated degradation (ERAD) pathway [5]. In contrast, in the absence of tapasin, MHC class I complexes escape the ER, but fall apart readily on the cell surface due to the presence of sub-optimal cargo.
In this issue Ombrello and colleagues review the genetic evidence implicating ERAP1 single nucleotide polymorphisms (SNPs) and haplotypes in disease [Ombrello M, et al., this issue]. The purpose of this review is to summarize and update what is known about the functional consequences of ERAP1 variation and loss-of-function.
ERAP1 Structure
ERAP1 belongs to the M1 zinc aminopeptidase family, so-named for the active site zinc-binding motif [6]. ERAP1 is located in a 200 kb region on chromosome 5q15 in humans along with endoplasmic reticulum aminopeptidase 2 (ERAP2) and leucyl/cystinyl aminopeptidase (LNPEP). ERAP2 and LNPEP are also associated with AS, psoriasis and/or inflammatory bowel disease, but at this point less is known about disease-associated variants. The ERAP2 gene is absent in rodents, and in mice the product of ERAP1 was originally abbreviated as ERAAP (ER aminopeptidase associated with antigen processing). LNPEP is also known as insulin-regulated aminopeptidase (IRAP), and it is involved in endosome-mediated cross-presentation of exogenous antigen by MHC class I in dendritic cells [7].
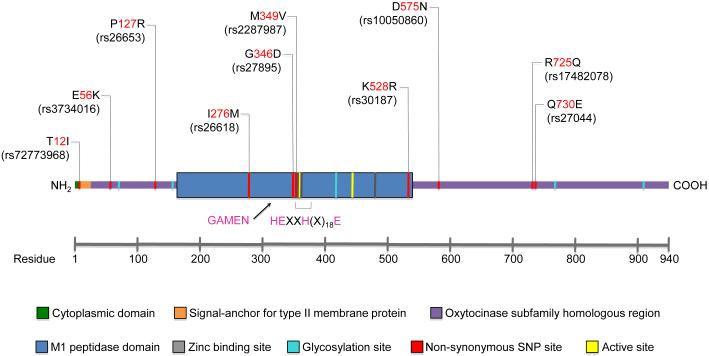
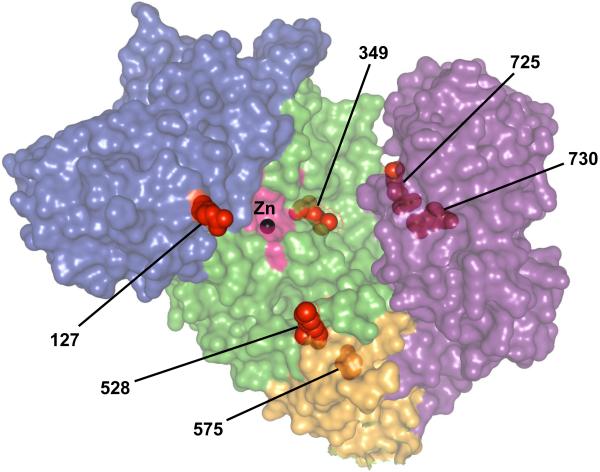
ERAP1 has four protein domains, with domain II having catalytic activity and carrying the GAMEN and Zn-binding HExxHx18E motifs [8] (Figures 1 & 2). Domain I docks on top of domain II, capping off the active site and providing binding sites for the N-terminus of peptide substrates. Domain III is a small β-sandwich domain, and domain IV extends away from the active site (Figure 2). Binding of long rather than short peptides induces a conformational change with reorientation of a key catalytic residue toward the active site, providing a mechanism to explain trimming of 9-16 amino acid peptides more efficiently than 8-mers. Six AS risk SNPs were mapped on this 3-D structure, revealing their location near putative substrate binding and regulatory sites (Figure 2). The K528R variant, although distant from the active site, alters aminopeptidase activity against many peptide substrates [9].
Figure 1. Schematic diagram of ERAP1.
The locations of common (>5% frequency) naturally occurring amino acid variants and their corresponding SNPs are shown. The rheumatic disease-associated variants (127, 349, 528, 575, 725, and 730) are shown in Figure 2.
Figure 2. Rheumatic disease-associated missense variants of ERAP1.
A surface map of ERAP1 demonstrates the locations of six rheumatic disease-associated amino acid substitutions of ERAP1 (shown as red spheres). The surface coloring indicates the domain structures (I = blue, II = green, III = orange, IV = purple) and identifies the enzyme’s catalytic site (pink), which contains the catalytic Zn2+ molecule (black sphere). This model was created using 3MDJ and PyMol software. (Courtesy of M. Ombrello)
ERAP1 Function: Effects on MHC Class I Expression
Using traditional loss-of-function approaches, effects of reducing or eliminating ERAP1 expression on MHC class I alleles have been examined in several systems. Small inhibitory RNA (siRNA)-mediated knockdown of ERAP1 in mouse cells caused a reduction in cell surface expression of MHC class I (H-2Kk and H-2Dl alleles) [10] with similar effects observed for H-2Kb and H-2Db in ERAP1 knockout (KO) mice [11]. MHC class I peptide loading, trafficking through the Golgi, and appearance on the cell surface was no different in ERAP1+/− and ERAP1−/− mouse cells, although ERAP1+/+ (wild type) cells were not compared [11]. However, the stability of cell surface complexes was significantly reduced with each allele falling apart more rapidly when assembled in the absence of ERAP1. This reflects the loading of sub-optimal peptides onto MHC class I when ERAP1 is lacking.
In contrast to the results in mouse cells, knocking down ERAP1 in HeLa cells increased the rate of assembly and cell surface expression of HLA class I, suggesting that when ERAP1 was present it was destroying more quality peptides than it was generating [12]. However, this result conflicts with another study using HeLa cells where ERAP1 knockdown resulted in a 10% decrease of HLA class I expression on cell surface [13]. These differences might be due to clonal variation in HeLa cells and loss of ERAP2 expression (Ken Rock, personal communication). This is of interest because ERAP1-ERAP2 interaction may enhance biological effects on peptides not explained by a simple additive model [13,14]. Moreover, there is a common variant in ERAP2 (rs2248374; allele frequency ~50%) that causes differential mRNA splicing which results in an in-frame premature stop codon. The variant mRNA undergoes nonsense-mediated decay and protein expression is lost [15]. The high frequency of the variant results in failure to express ERAP2 in about 25% of the population. Despite the fact that this ERAP2 variant (i.e. loss of expression) affects antigen presentation, MHC class I expression, and would be predicted to reduce ERAP1 function, the variant is not associated with AS [16]. It is worth noting that a risk haplotype of ERAP2 that is associated with increased mRNA and protein expression is associated with the HLA-A29-associated eye disease birdshot chorioretinopathy [17].
ERAP1 Function: Effects on Individual Peptides and Cross-Presentation
ERAP1 silencing can reduce the presentation of individual peptides to epitope-specific CD8+ T cells, consistent with the generation of many epitopes as extended peptides that require trimming in the ER for efficient MHC class I presentation [10,11,18]. However, the opposite has also been demonstrated, where the presence of ERAP1 reduces the supply of antigenic peptides and destroys certain epitopes that are generated more efficiently when ERAP1 is silenced [12]. The absence of ERAP1 in mice does not affect the development of CD8+ T cells, and the immune response to influenza virus and LCMV infection was not impaired, although specific epitopes were altered [19,20]. Cross-presentation of cell-associated antigens was impaired in ERAP1 KO mice, whereas soluble exogenous antigens were handled appropriately [19,20]. This is in contrast to LNPEP (IRAP), which is necessary for efficient cross-presentation of exogenous antigens [7].
ERAP1 Function: Specificity
ERAP1 has a preference for peptide substrates of 9-16 amino acids with large hydrophobic C-terminal residues, and demonstrates slower trimming when the C-terminus is charged [21]. Peptides of 8-9 residues are spared further trimming. In contrast, ERAP2, which does not exhibit the length preference seen with ERAP1, exhibits aminopeptidase activity that is independent of C-terminal residues [21]. There is also evidence that ERAP1 prefers substrates containing hydrophobic residues (e.g. Leu) on the N-terminal extension, whereas ERAP2 prefers basic residues (Arg, Lys) in this region. However factors other than the N-terminal residue also influence the efficiency of digestion. For example, in another study efficient trimming of an extension containing hydrophobic and basic residues required the joint action of ERAP1 and ERAP2 [13].
ERAP1 deficiency leads to increased recovery of N-terminally extended peptides from isolated MHC class I molecules [22]. However, a recent examination of HLA-B27-bound peptides presented in cell lines where ERAP1 was silenced revealed many C-terminally extended peptides. This paper presented additional evidence that binding of C-terminally extended peptides was a consequence of the A and B pockets of HLA-B27 [23**], which are major determinants of its peptide binding specificity with a predilection for basic residues (Arg/Lys at P1, and Arg P2). The B pocket also confers to HLA-B27 its tendency to misfold and dimerize [24]. Since ERAP1 is an aminopeptidase, the C-terminally extended peptides are unlikely to results from failed C-terminal trimming. Rather, as suggested by Chen et al., it is more likely that these peptides achieve stable binding via P2 and P1 residues and the A/B pockets of HLA-B27, and extend on their C-termini. This is of interest as it is conceivable that greater ERAP1 activity could destroy more epitopes than it creates for HLA-B27.
Effect of ERAP1 Polymorphisms on Aminopeptidase Activity and Expression
There are multiple missense SNPs in ERAP1 that result in amino acid (AA) substitutions. Those with >5% frequency in the population are as follows: T12I, E56K, P127R, I276M, G346D, M349V, K528R, D575N, R725Q, and Q730E, where the first letter reflects the ancestral AA, followed by the position in the protein, and then the non-ancestral AA (see Table 1 in Ombrello M et al., this issue) and Figure 1. Genome wide association studies (GWAS) identified risk associated with K528 and Q730, while 575N, 349V and 725Q are protective [2,25,26], with follow-up fine mapping studies identifying/confirming 127R, M349, K528, D575, R725, and Q730 as being associated with risk [26]. These studies are summarized in detail in the paper by Ombrello et al. in this issue. To understand how these missense polymorphisms affect ERAP1 function many have been generated by site-directed mutagenesis and studied in vitro and/or in cells in tissue culture. Changing K528 to 528R (K528R), R725Q, and Q730E individually, revealed a reduction in activity for N-terminally extended peptide precursors. However, the effect was dependent on individual peptide substrates as well as the peptide concentration [14]. In general, loss-of-function variants either have little effect on cell surface expression or cause a small reduction of canonical forms of HLA-B27, consistent with reduced editing and the studies discussed above where ERAP1 expression led to an overall reduction in MHC class I expression [14]. ERAP1 K528R reduced its ability to produce a 9-mer peptide from a 13-mer precursor [9], and caused a reduction in the presentation of an HLA-B27-restricted epitope to CD8+ T cells in ERAP1-deficient mouse embryonic fibroblasts [23**].
Table 1.
ERAP1 Haplotypes and Allotypes
ERAP1 variants exist as haplotypes encoding allotypes [27**,28]. In a study of the peptides bound to HLA-B*27:04 (peptidome) using 4 cell lines with different ERAP1 alleles, the presence of more loss-of-function (protective) SNPs resulted in a greater abundance of long peptides and reduced thermostability of cell surface complexes [29]. These results are consistent with reduced optimization by protective variants. Another study of ERAP1 haplotypes from 20 individuals showed that R725Q/Q730E and K528R/R725Q exhibited generalized reduced trimming and epitope generation from precursor peptides. However, for individual substrates there were examples where variants exhibited normal, hypofunctional, or hyperfunctional aminopeptidase activity [28].
A more recent study sequenced full-length cDNAs generated from ERAP1 mRNAs expressed in 17 AS patients and 19 controls, and then analyzed the function of the ERAP1 allotypes after re-expression in ERAP1-deficient cells [27**]. Thirteen allotypes were present in various combinations in the 36 individuals. Strikingly, there was complete segregation of allotype combinations (pairs) found in AS patients versus controls. Allotype combinations from AS patients exhibited a reduction in generation of a specific epitope presented by the mouse class I molecule, H-2Kb, and 5 out of 6 allotype pairs associated with AS were unable to restore H-2Db and H-2Kb expression. In contrast, allotype pairs from controls restored MHC class I on the cell surface. When effects of ERAP1 allotypes on HLA-B*27:05 were examined, similar results were obtained, with a failure of disease-associated allotypes to increase HLA-B*27:05 expression. These data provide evidence that loss-of-function ERAP1 allotypes may confer risk, which seems inconsistent with the genetic data discussed above. Surprisingly, the 528R and 730E variants, which are protective SNPs in the GWAS studies, were overrepresented in AS cases from this study. The 528R variant was present in 82.4% of allotypes (28/34 alleles from 17 patients) in AS vs. 44.7% (17/38 alleles from 19 controls) of controls, with 730E present in 50% (17/34) of cases but only 34.2% (13/38) controls. Thus the data indicating that these variants confer loss-of-function relative to other variants are consistent, whereas the genetics are inconsistent. From a genetic standpoint this study had a very small sample size, and thus must be interpreted with caution. Similarly, studies examining functional effects of single AA variants must be interpreted with caution in terms of disease-relatedness given the existence of functionally distinct allotypes.
ERAP1 Expression
ERAP1 haplotypes associated with AS have been shown to result in increased ERAP1 mRNA expression in monocyte-derived dendritic cells (MD-DC) from both AS patients and healthy controls. Furthermore, ERAP1 transcript and protein levels correlated with a haplotype risk score in B-lymphoblastoid cell lines (B-LCL). Enzymatic activity of ERAP1 against fluorescent substrates was increased for AS-predisposing haplotypes in both MD-DC and B-LCL [30*]. To the extent that increased expression of ERAP1 would result in greater overall aminopeptidase activity in the ER, these data are consistent with increased activity being associated with risk.
Effect of ERAP1 on the Immunobiology of HLA-B27
HLA-B27 has a tendency to misfold and can exist on the cell surface as a disulfide-linked homodimer, or as a monomer free of β2m. Other MHC class I alleles can lose peptide/β2m and exist as monomers, whereas misfolding and dimerization is very uncommon if not unique to HLA-B27. The question of whether ERAP1 variants affect the relative amount of free heavy chain was investigated using monocytes from AS patients. Comparing C/C and C/G at the rs27044 SNP (C encodes Q730, while G specifies 730E) with G/G revealed increased MHC class I free heavy chain with CC and CG [31]. Since the SNP encoding 730E is considered protective, this would be inconsistent with free cell surface heavy chains contributing to disease. However, this study did not identify whether HLA-B27 was contributing to the cell surface free heavy chains. Interestingly, this study also examined effects of ERAP1 knockdown (loss-of-function) and found increased antibody staining for unfolded (or pre-folded) HLA-B27 heavy chains inside permeabilized cells, as well as increased cell surface staining with an antibody that recognizes HLA-B27 complexed with longer peptides. The effect of ERAP1 knockdown on HLA-B27 accumulation was only seen with AS-associated subtypes B*27:04 and B*27:05, and not with B*27:09 and B*27:06, which are weakly or not associated with disease, respectively [31]. No published studies have examined whether ERAP1 knockdown affects the formation of disulfide-linked HLA-B27 dimers. In our own experiments ERAP1 knockdown mimicking loss-of-function “protective” effects results in greater expression of HLA-B27 with long peptides (recognized by the MARB4 antibody) and greater accumulation of disulfide linked dimers on the cell surface (submitted). This result is consistent with the increased tendency of suboptimal complexes to fall apart, which is a precursor to cell surface dimerization. If indeed loss-of-function is protective, then this result would suggest that ERAP1 effects on homodimer formation do not correlate with disease susceptibility.
In mice expressing a mouse-human hybrid form of HLA-B27 or -B7 (where the peptide binding regions are fused to mouse class I α3 domains), ERAP1-deficiency reduced the expression of cell surface HLA-B27 much more than HLA-B7, consistent with there being allelic differences in the effects of ERAP1 [32*]. In these mice, ERAP1 loss-of-function caused either a reduction or enhancement in peptide presentation by HLA-B27 to CD8+ cytotoxic T cells (CTL) depending on the peptide studied. Interestingly, the CTL response to the HLA-B27-restricted immunodominant influenza nucleoprotein (NP)383-391 epitope is reduced when ERAP1 is absent, whereas the CTL response to the HLA-B7-restricted NP418-426 was not affected [32].
Other Biological Effects of ERAP1
ERAP1 can be secreted by macrophages when cells are activated by LPS and IFNγ, resulting in enhanced phagocytic activity. These observations were extended to include other Toll-like receptor agonists (TLR1/2, TLR2/6, and TLR9), and shown to be dependent on autocrine IFNβ and TNFα through increases in cytosolic Ca2+ and calmodulin activation [33**,34]. Interestingly, ERAP1 contributes to increased nitric oxide (NO) synthesis as well [35*]. Thus, in addition to aminopeptidase activity that influences the availability of optimally sized peptides for MHC class I, ERAP1 also plays a direct role in inflammation. Whether this activity is affected by polymorphisms is not known.
Conclusion
ERAP1 missense SNPs that have functional significance are associated with several immune-mediated inflammatory diseases that have strong HLA class I associations, including AS, psoriasis, and BD. ERAP1 SNPs exist as haplotypes that encode functionally different proteins, or allotypes. SNPs that are associated with protection from AS confer reduced aminopeptidase activity to ERAP1 and are associated with lower expression levels, and thus can be mimicked at lest in part by knocking down or eliminating expression of ERAP1. Loss of ERAP1 function reduces peptide trimming in the ER and results in the presentation of longer peptides on MHC class I molecules. For HLA-B27, the peptides can be extended either on the N- or C-terminus, in contrast to other alleles that exhibit predominantly N-terminally extended epitopes. In an extensive analysis of allotypes and allotype pairs derived from HLA-B27+ individuals with AS, there was consistent evidence for reduced ERAP1 function in AS patients compared to unaffected controls, with 528R and 730E protective variants being overrepresented in AS patients. This study is provocative, but needs further genetic confirmation. Our understanding of how ERAP1 affects the biology of HLA-B51 (BD), HLA-Cw6 (psoriasis), and even HLA-B27 (AS) is incomplete. It will be essential to identify and study differences between key risk/protective haplotypes and how they affect the biology of different HLA alleles
Key Points.
ERAP1 trims peptides longer than 8-9 amino acids at their N-terminus, optimizing ligands for binding to MHC class I molecules.
Common genetic variants of ERAP1 that are associated with protection from ankylosing spondylitis (AS) confer reduced aminopeptidase activity against many substrates.
ERAP1 haplotypes encode protein allotypes that exhibit functional differences.
ERAP1 allotypes found in AS are poor at restoring mouse MHC class I and HLA-B*27:05 expression in ERAP1-deficient cells, suggesting they are hypofunctional relative to allotypes found in unaffected controls.
Additional studies of naturally occurring allotypes and their effects on HLA class I alleles involved in susceptibility to AS, psoriasis and Behcet’s disease are needed to better understand how these HLA molecules contribute to disease.
Acknowledgements
We thank Sohee Hong and Joseph Gonzalez for contributing to Figure 1, Michael Ombrello for providing Figure 2, and Michael Ombrello and Elaine Remmers for helpful discussions.
Financial Support and Sponsorship
This work was supported by the Intramural Research Program of the National Institute of Arthritis Musculoskeletal and Skin Diseases (Z01 AR041184) of the National Institutes of Health, Bethesda, MD, USA
Footnotes
Conflict of Interest
None
References
- 1.Shastri N, Nagarajan N, Lind KC, et al. Monitoring peptide processing for MHC class I molecules in the endoplasmic reticulum. Curr Opin Immunol. 2014;26:123–127. doi: 10.1016/j.coi.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wellcome Trust Case Control C, Australo-Anglo-American Spondylitis C. Burton PR, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Genetic Analysis of Psoriasis C, the Wellcome Trust Case Control C. Strange A, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirino Y, Bertsias G, Ishigatsubo Y, et al. Genome-wide association analysis identifies new susceptibility loci for Behcet's disease and epistasis between HLA-B*51 and ERAP1. Nat Genet. 2013;45:202–207. doi: 10.1038/ng.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes EA, Hammond C, Cresswell P. Misfolded major histocompatibility complex class I heavy chains are translocated into the cytoplasm and degraded by the proteasome. Proc Natl Acad Sci U S A. 1997;94:1896–1901. doi: 10.1073/pnas.94.5.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hattori A, Tsujimoto M. Endoplasmic reticulum aminopeptidases: biochemistry, physiology and pathology. J Biochem. 2013;154:219–228. doi: 10.1093/jb/mvt066. [DOI] [PubMed] [Google Scholar]
- 7.Saveanu L, Carroll O, Weimershaus M, et al. IRAP identifies an endosomal compartment required for MHC class I cross-presentation. Science. 2009;325:213–217. doi: 10.1126/science.1172845. [DOI] [PubMed] [Google Scholar]
- 8.Tsujimoto M, Hattori A. The oxytocinase subfamily of M1 aminopeptidases. Biochim Biophys Acta. 2005;1751:9–18. doi: 10.1016/j.bbapap.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 9.Kochan G, Krojer T, Harvey D, et al. Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc Natl Acad Sci U S A. 2011;108:7745–7750. doi: 10.1073/pnas.1101262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serwold T, Gonzalez F, Kim J, et al. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. 2002;419:480–483. doi: 10.1038/nature01074. [DOI] [PubMed] [Google Scholar]
- 11.Hammer GE, Gonzalez F, Champsaur M, et al. The aminopeptidase ERAAP shapes the peptide repertoire displayed by major histocompatibility complex class I molecules. Nat Immunol. 2006;7:103–112. doi: 10.1038/ni1286. [DOI] [PubMed] [Google Scholar]
- 12.York IA, Chang SC, Saric T, et al. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8-9 residues. Nat Immunol. 2002;3:1177–1184. doi: 10.1038/ni860. [DOI] [PubMed] [Google Scholar]
- 13.Saveanu L, Carroll O, Lindo V, et al. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat Immunol. 2005;6:689–697. doi: 10.1038/ni1208. [DOI] [PubMed] [Google Scholar]
- 14.Evnouchidou I, Kamal RP, Seregin SS, et al. Cutting Edge: Coding single nucleotide polymorphisms of endoplasmic reticulum aminopeptidase 1 can affect antigenic peptide generation in vitro by influencing basic enzymatic properties of the enzyme. J Immunol. 2011;186:1909–1913. doi: 10.4049/jimmunol.1003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andres AM, Dennis MY, Kretzschmar WW, et al. Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet. 2010;6:e1001157. doi: 10.1371/journal.pgen.1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harvey D, Pointon JJ, Karaderi T, et al. A common functional variant of endoplasmic reticulum aminopeptidase 2 (ERAP2) that reduces major histocompatibility complex class I expression is not associated with ankylosing spondylitis. Rheumatology (Oxford) 2011;50:1720–1721. doi: 10.1093/rheumatology/ker199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuiper JJ, Van Setten J, Ripke S, et al. A genome-wide association study identifies a functional ERAP2 haplotype associated with birdshot chorioretinopathy. Hum Mol Genet. 2014;23:6081–6087. doi: 10.1093/hmg/ddu307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.York IA, Brehm MA, Zendzian S, et al. Endoplasmic reticulum aminopeptidase 1 (ERAP1) trims MHC class I-presented peptides in vivo and plays an important role in immunodominance. Proc Natl Acad Sci U S A. 2006;103:9202–9207. doi: 10.1073/pnas.0603095103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan J, Parekh VV, Mendez-Fernandez Y, et al. In vivo role of ER-associated peptidase activity in tailoring peptides for presentation by MHC class Ia and class Ib molecules. J Exp Med. 2006;203:647–659. doi: 10.1084/jem.20052271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Firat E, Saveanu L, Aichele P, et al. The role of endoplasmic reticulum-associated aminopeptidase 1 in immunity to infection and in cross-presentation. J Immunol. 2007;178:2241–2248. doi: 10.4049/jimmunol.178.4.2241. [DOI] [PubMed] [Google Scholar]
- 21.Chang SC, Momburg F, Bhutani N, et al. The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a "molecular ruler" mechanism. Proc Natl Acad Sci U S A. 2005;102:17107–17112. doi: 10.1073/pnas.0500721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanchard N, Kanaseki T, Escobar H, et al. Endoplasmic reticulum aminopeptidase associated with antigen processing defines the composition and structure of MHC class I peptide repertoire in normal and virus-infected cells. J Immunol. 2010;184:3033–3042. doi: 10.4049/jimmunol.0903712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **23.Chen L, Fischer R, Peng Y, et al. Critical role of endoplasmic reticulum aminopeptidase 1 in determining the length and sequence of peptides bound and presented by HLA-B27. Arthritis Rheumatol. 2014;66:284–294. doi: 10.1002/art.38249. This paper demonstrates that ERAP1 silencing results in the presentation of longer peptides by HLA-B27, with many more C-terminally extended epitopes compared to those with N-terminal extensions. [DOI] [PubMed] [Google Scholar]
- 24.Colbert RA, DeLay ML, Klenk EI, et al. From HLA-B27 to spondyloarthritis: a journey through the ER. Immunol Rev. 2010;233:181–202. doi: 10.1111/j.0105-2896.2009.00865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans DM, Spencer CC, Pointon JJ, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011;43:761–767. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harvey D, Pointon JJ, Evans DM, et al. Investigating the genetic association between ERAP1 and ankylosing spondylitis. Hum Mol Genet. 2009;18:4204–4212. doi: 10.1093/hmg/ddp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **27.Reeves E, Colebatch-Bourn A, Elliott T, et al. Functionally distinct ERAP1 allotype combinations distinguish individuals with Ankylosing Spondylitis. Proc Natl Acad Sci U S A. 2014;111:17594–17599. doi: 10.1073/pnas.1408882111. This is the first paper to compare the function of ERAP1 allotypes and allotype pairs found in AS patients with those from unaffected controls, with evidence that AS-associated-allotypes are less able to restore class I expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reeves E, Edwards CJ, Elliott T, et al. Naturally occurring ERAP1 haplotypes encode functionally distinct alleles with fine substrate specificity. J Immunol. 2013;191:35–43. doi: 10.4049/jimmunol.1300598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Medel N, Sanz-Bravo A, Barnea E, et al. The origin of proteasome-inhibitor resistant HLA class I peptidomes: a study with HLA-A*68:01. Mol Cell Proteomics. 2012;11:M111011486. doi: 10.1074/mcp.M111.011486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *30.Costantino F, Talpin A, Evnouchidou I, et al. ERAP1 gene expression is influenced by non-synonymous polymorphisms associated with predisposition to spondyloarthritis. Arthritis Rheumatol. 2015 doi: 10.1002/art.39072. This is the first report of ERAP1 SNPs being associated with differential expression. [DOI] [PubMed] [Google Scholar]
- 31.Haroon N, Tsui FW, Uchanska-Ziegler B, et al. Endoplasmic reticulum aminopeptidase 1 (ERAP1) exhibits functionally significant interaction with HLA-B27 and relates to subtype specificity in ankylosing spondylitis. Ann Rheum Dis. 2012;71:589–595. doi: 10.1136/annrheumdis-2011-200347. [DOI] [PubMed] [Google Scholar]
- *32.Akram A, Lin A, Gracey E, et al. HLA-B27, but not HLA-B7, immunodominance to influenza is ERAP dependent. J Immunol. 2014;192:5520–5528. doi: 10.4049/jimmunol.1400343. This paper demonstrates differential effects of ERAP1 on the production of immunodominant epitope for HLA-B27 and HLA-B7. [DOI] [PubMed] [Google Scholar]
- **33.Goto Y, Ogawa K, Nakamura TJ, et al. TLR-mediated secretion of endoplasmic reticulum aminopeptidase 1 from macrophages. J Immunol. 2014;192:4443–4452. doi: 10.4049/jimmunol.1300935. This paper shows ERAP secretion from macrophages is promoted by the activation of certain TLRs. [DOI] [PubMed] [Google Scholar]
- 34.Goto Y, Ogawa K, Hattori A, et al. Secretion of endoplasmic reticulum aminopeptidase 1 is involved in the activation of macrophages induced by lipopolysaccharide and interferon-gamma. J Biol Chem. 2011;286:21906–21914. doi: 10.1074/jbc.M111.239111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *35.Goto Y, Ogawa K, Nakamura TJ, et al. Substrate-dependent nitric oxide synthesis by secreted endoplasmic reticulum aminopeptidase 1 in macrophages. J Biochem. 2015 doi: 10.1093/jb/mvv001. This paper demonstrates effects of secreted ERAP1 on nitric oxide synthesis in macrophages. [DOI] [PubMed] [Google Scholar]