Abstract
The identification of amino acid residues in proteins involved in binding small molecule ligands is an important step for their functional characterization, as the function of a protein often depends on specific interactions with other molecules. The accuracy of computational methods aiming to predict such binding residues was evaluated within the “Function prediction (prediction of binding sites, FN)” category of the Critical Assessment of protein Structure Prediction (CASP) experiment. In the last edition of the experiment (CASP10), 17 research groups participated in this category, and their predictions were evaluated on 13 prediction targets containing biologically relevant ligands. The results of this experiment indicate that several methods achieved an overall good performance, showing the usefulness of such methods in predicting ligand binding residues. As in previous years, methods based on a homology transfer approach were dominating. In comparison to CASP9, a larger fraction of the top predictors are automated servers. However, due to the small number of targets and the characteristics of the prediction format, the differences observed among the first ten methods were not statistically significant and it was also not possible to analyze differences in accuracy for different ligand types or overall structure prediction difficulty. To overcome these limitations and to allow for a more detailed evaluation, in future editions of CASP prediction methods in the FN category will no longer be evaluated on the “normal” CASP targets, but assessed continuously by CAMEO (Continuous Automated Model Evaluation) based on weekly pre-released sequences from the PDB.
Keywords: protein function, protein structure, evaluation, assessment, binding site, active site, cofactor, ligand, CASP
Introduction
Proteins interact with a broad range of molecules to perform their function. While the majority of these interactions are unspecific and transient (e.g. with water molecules, ions and other solutes in the cell), others are very specific and essential for the function of the protein. Specific interactions can be stable, e.g. oligomeric proteins, or transient e.g. in signaling networks or motor proteins. Binding partners of a protein are not limited to other proteins, but can include the whole range of other molecule types. Typical examples include complexes of enzymes with substrates and co-factors, receptors and ligands, antibodies and epitopes, transcription factors and cognate DNA, protein-RNA assemblies such as the ribosome, or ligands in a protein structure with a structural role. For the characterization of a new protein, information about ligands, cofactors and binding sites often provides crucial hints about its function. However, when determining the structure of a protein experimentally, ligands with medium to low binding affinities are often lost during the purification procedure, and the resulting structures often do not contain ligands. Additionally, in many cases neither the 3-dimensional structure of the protein itself is known, nor the location and identity of possible ligands. To overcome these limitations, computational methods were established to predict from a protein’s sequence its 3-dimensional structure and possible ligand binding residues. Several computational approaches for predicting ligand binding sites have been developed, which differ with respect to the information they are based on: (1) only the sequence of the protein; (2) its structural properties; (3) both sequence and structure; (4) homologue proteins.1-12
The aim of the Critical Assessment of protein Structure Prediction (CASP) experiment is to assess the current state of the art of such methods, and to highlight bottlenecks and opportunities for further development. The accuracy of predictions of three-dimensional structures is assessed as part of the template-based (TBM) and free-modeling (FM) categories,13-16 which includes the accuracy of binding site coordinates.17,18 Function prediction (FN) was introduced as a new category in CASP6.19 In later editions, the definition of function predictions was specified as prediction of residues involved in binding relevant ligands.20-22 Here, we describe the results of the assessment in the category “prediction of binding sites (function prediction, FN)” of the CASP10 experiment.
Materials and Methods
Prediction format
As in previous CASP experiments, the format for the predictions of binding residues for a given target protein consisted of a list of the amino-acid positions that were predicted to be in contact with a biologically relevant ligand. The CASP format did not include a confidence score, so that residues are classified in a binary way, either binding or not binding to any ligand. Predictors could optionally propose the name and the category of the compounds that could bind to these residues. One consequence of the prediction format is that it is not possible to correctly assess over-predictions; neither in case a target did not include any biologically relevant ligand, nor if a prediction indicated a binding site for a different ligand elsewhere in the target.
Prediction targets
All CASP10 target sequences were sent out as prediction targets in the FN category.23 For the assessment, a subset of the target structures (coordinates available as of 2013-08-05) were selected, which contained at least one biologically relevant ligand. To define which ligands were considered as “biologically relevant”, we used information coming from scientific literature, Swiss-Prot24 annotations, sequence conservation of functionally important residues, and information from homologous structures. For the purpose of the assessment, covalently bound ligands in the reference structure were handled the same way as non-covalently bound ones. In case of oligomeric assemblies, the “biological assembly units” as defined by the authors were used as reference target structures.
Binding site definition
A binding site was defined by all protein residues in the target structure having at least one (non-hydrogen) atom within a certain distance (di,j) to biologically relevant ligand atoms:
where di,j is the distance between a residue atom i and a ligand atom j, ri and rj are the Van der Waals radii of the involved atoms, while c is a tolerance distance of 0.5 Å. In case the biological assembly of the experimental target structure represents a homo-oligomeric protein, or in case of NMR ensembles, residues were included in the binding site definition if they fulfilled the distance criterion in at least half of the reference chains. The binding site definitions used for the assessment are shown in Table II. Analysis of ligand binding sites was implemented using OpenStructure (version 1.4).25,26
Table 2.
Definition of ligand binding residues. A residue in the target structure was defined as binding if it had at least one heavy atom of a biologically relevant ligand within 0.5 Å distance of the sum of the Van der Waals radii of the involved atoms.
| Target | Binding site (residue numbers) |
|---|---|
| T0652 | 74, 79, 80, 99, 100, 101, 102, 103, 104, 165, 180, 182, 183 |
| T0657 | 121, 132, 133, 143 |
| T0659 | 43, 48, 63 |
| T0675 | 21, 24, 37, 42, 49, 52, 65, 70 |
| T0686 | 28, 30, 103 |
| T0696 | 18, 69, 104 |
| T0697 | 91, 150, 151, 152, 190, 243, 245, 247, 272, 274, 301, 303, 304, 351 |
| T0706 | 25, 27, 99, 101, 129, 130 |
| T0720 | 32, 34, 35, 62, 99, 113, 114, 115, 182, 188, 191, 194, 197, 200 |
| T0721 | 10, 12, 13, 14, 33, 34, 35, 36, 37, 38, 39, 42, 45, 46, 60, 78, 79, 80, 109, 110, 111, 114, 126, 136, 235, 237, 268, 269, 277, 278, 281 |
| T0726 | 273, 277, 307 |
| T0737 | 37, 40, 41, 42, 44, 45, 49, 78, 83, 114, 117, 118, 120, 121, 123, 124, 128, 130, 135, 138, 174, 237 |
| T0744 | 22, 23, 24, 26, 58, 61, 120, 121, 122, 124, 196, 214, 216, 270, 271, 272, 273, 314, 316 |
Binding site prediction evaluation
According to the binding site definition in the experimental reference structure, predicted binding residues were classified as True Positives (TP: correctly predicted binding site residue), True Negatives (TN: correctly predicted non-binding residue), False Negatives (FN: incorrectly not predicted binding site residue), False Positives (FP: incorrectly predicted non-binding residue). As in the previous CASP assessment22 the evaluation of the quality of the binding site predictions was performed using the Matthews Correlation Coefficient (MCC):
MCC is a useful measure when the two classes (in our case binding and non-binding residues) are of very different sizes. MCC ranges from +1 (perfect prediction) to −1 (inverse prediction), where a MCC of 0 corresponds to random prediction. Raw scores and confusion matrices for all groups and all targets are provided in Table S1, S4 and Figure S1. Finally, the MCCs were standardized by calculating their Z-scores to allow the combination of scores for targets of different difficulty:
where MCCP,T is the MCC of the predictor P for target T, is the mean MCC for the target T by all predictors P and σT is the standard deviation of the MCCs for the target T by all predictors P. The final ranking of the methods was based on the average value of the MCC across all targets. Cumulative confusion matrices are provided in supplementary data. For completeness, we also assessed the accuracy of the predictions using the distance based method BDT27 (see Table S3).
Statistical significance and robustness of the ranking
To measure the statistical significance of the assessment results, we applied two-tailed Student’s paired t-test and Wilcoxon signed-rank test on the MCCs values for each target’s predictions. Both tests were performed using the R statistical package (version 2.11.1).28 The robustness of the ranking based on MCC values was assessed by 100 rounds of random sampling using 70% of the targets.
Results and discussion
Prediction targets
Although the CASP10 experiment had in total about 100 prediction targets, only very few of them had ligands which were classified as biologically relevant.23,29 In total, we identified 13 targets with biologically relevant ligands, as listed in Table I and II. In 8 targets, metal ions (Zn2+, Mg2+, Mn2+, Na+) are present, one contained an iron-sulfur cluster (SF4), one bound an Adenine-Mono-Phosphate (AMP), one had a reduced Flavin mononucleotide (FNR), two had Flavin-adenine-dinucleotide (FAD) ligands, and one had LPP (N’-pyridoxyl-lysin-5′-monophosphate) covalently bound at the dimer interface. It is worth emphasizing that the presence or absence of a ligand in a target structure depends on the experimental conditions, i.e. the same binding site can be occupied by a ligand under one condition, and be empty or occupied by a different ligand under different conditions. Therefore, target structures without bound ligands can therefore not be considered as reference in the assessment. This issue is especially pronounced in prediction targets solved by high-throughput methods, where the experimental conditions often do not contain the biologically relevant ligands or cofactors. As a consequence, the number of targets that bind a relevant compound and that can be used for further prediction assessment in CASP10 is quite small. The following paragraph provides a short overview of the assessed targets:
Table I. Targets with biologically relevant ligands used in the FN prediction assessment.
| Target | PDB ID | Ligand ID | Type | Interface |
|---|---|---|---|---|
| T0652 | 4HG0 | AMP | Non-metal | No |
| T0657 | 2LUL | ZN | Metal | No |
| T0659 | 4ESN | ZN (2) | Metal | No |
| T0675 | 2LV2 | ZN (2) | Metal | No |
| T0686 | 4HQO | MG | Metal | No |
| T0696 | n.a | NA | Metal | No |
| T0697 | n.a | LLP (2) | Non-metal | A-A |
| T0706 | n.a | MG (2) | Metal | A-A |
| T0720 | 4IC1 | MN(10)/SF4(10) | Metal | No |
| T0721 | 4FK1 | FAD (2) | Non-metal | No |
| T0726 | 4FGM | ZN | Metal | No |
| T0737 | 3TD7 | FAD | Non-metal | No |
| T0744 | 2YMV | FNR | Non-metal | No |
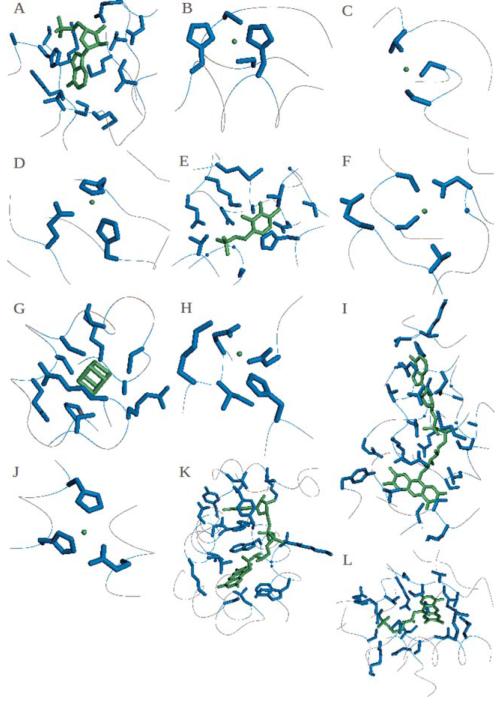
Target T0652 (PDB: 4HG0)
The magnesium and cobalt efflux protein CorC contains two CBS (cystathionine-beta-synthase) domains, which bind an Adenosine monophosphate (AMP) (Figure 1A), next to a transporter associated domain (CorC_HlyC) at the C terminus of the protein. CBS is a small intracellular module, mostly found in two or four copies next to a wide range of protein domains in bacteria, archaea, and eukaryotes.21,30 Pairs of CBS domains can bind adenosyl groups such as AMP, ATP or SAM, thus they could regulate the activity of the attached domains13 and they may act as sensors of intracellular metabolites.14 The CorC_HlyC transporter associated domain is found in a family of proteins of unknown function with CBS domain and also in CorC involved in Magnesium and Cobalt efflux; it is hypothesized that it could modulate the transport of ion substrates.
Figure 1. Binding sites and ligands of the assessed targets.
Biologically relevant ligands are coloured in green and the residues included in the binding site are coloured in blue. Targets and ligands included here: (A) T0652: AMP, (B) T0675: ZN, (C) T0686: MG, (D) T0696: NA, (E) T0697: LLP, (F) T0706: MG, (G-H) T0720: SF4 and MN, (I) T0721: FAD, (J) T0726: ZN, (K) T0737: FAD, (L) T0744: FNR. Targets T0657 and T0659 are in Figure 7.
Target T0657 (PDB: 2LUL)
The Tyrosine-protein kinase Tec is composed by a PH (Pleckstrin homology) domain and a BTK (Btk-type zinc finger) domain that binds a Zn2+ cation (Figure 7A). The first occurs in many proteins involved in intracellular signaling or as part of the cytoskeleton,15 such beta/gamma subunits of heterotrimeric G proteins.16 This domain has specificities for different membrane phosphoinositides phosphorylated at different sites within the inositol ring, so the function of PH-containing proteins is modulated by enzymes that de-phosphorylate such rings. PH recruits proteins to different cellular compartments or it allows them to be involved in signal transduction pathways. The structure of this domain consists of two perpendicular anti-parallel beta sheets followed by an amphiphatic helix; the loop between the beta strands has a very variable length. The BTK domain contains a conserved zinc-binding motif of one histidine and three cysteine residues, it is very close to the PH domain and it consists in a long loop held together by a zinc ion. Target T0659 (PDB: 4ESN). There are no sequence annotations on this target, a homo-dimer that binds a Zn2+ ion in both chains at the same position (Figure 7B). A DELTA-BLAST31 search revealed a conserved domain of unknown function homologous to Listeria innocua Lin0431, a protein similar to the N-Utilization Substance G (NusG) N terminal (NGN) insert (domain II, DII). Lin0431 has a similar structure and charged surface distribution to Aquifex aeolicus NusG DII, indicating a possible role in transcription or translation regulating functions.
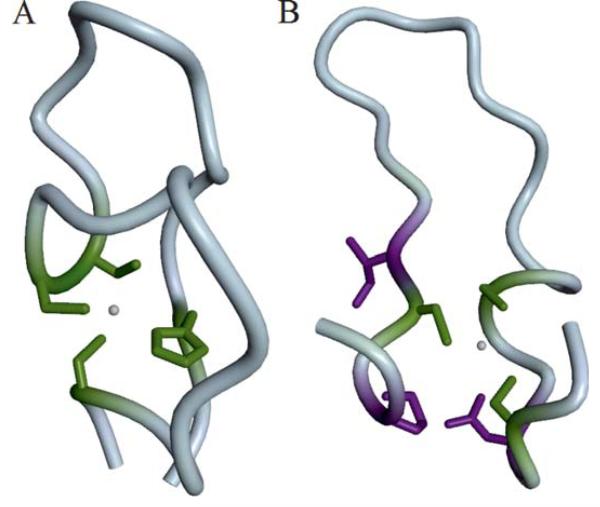
Figure 7. Binding site prediction examples.
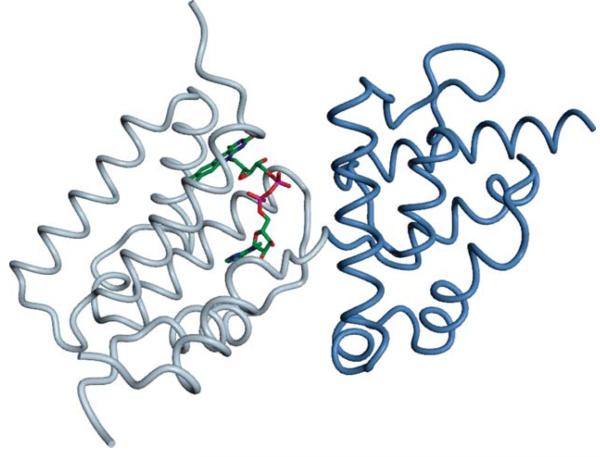
Residues are colored as “correctly predicted” (true positive, green) and “wrongly predicted” (false positive, violet). (A) The Zn2+ binding site in a Btk-type zinc finger in the PH domain from the Tyrosine-protein kinase Tec (T0657) is formed by His 121, Cys 132, Cys 133, and Cys 143. Coloring according to prediction by group “Binding_Kihara” (FN231). (B) Structure of a hypothetical protein T0659 with a Zn2+ ion bound by three conserved Cysteine residues (Cys 43,48,63). Coloring according to predictions by group “SP-ALIGN” (FN326).
Target T0675 (PDB: 2LV2)
The insulinoma-associated protein 1 contains two Zinc finger domains (Figure 1B), which are stable structural motifs that bind DNA, RNA, protein or lipid substrates.17,23,25,29,32 Some types of this domain use Zinc, others use Iron or form salt-bridges to create the correct fold, which often does not change conformation upon binding the target. Zinc fingers are usually found in groups and they have different binding specificities depending on their amino acid sequence and on the overall structure of the protein containing them. The domains in this target are of the C2H2 type, where two conserved cysteines and histidines coordinate a zinc ion inside of two short beta strands followed by an alpha helix; this “finger” binds the major groove of the DNA.
Target T0686 (PDB: 4HQO)
The sporozoite surface protein 2 is the ectodomain of a thrombospondin repeat anonymous protein (TRAP), a mediator in the infection of mosquito and vertebrate cells and in the gliding motility of sporozoites, which is an important target of pre-erythrocytic malaria vaccines. TRAP passes through the plasma membrane and is attached to the actin cytoskeleton by aldolase.25 This structure has a Von Willebrand factor type A (VWA) domain binding a Mg2+ ion (Figure 1C) which is additionally coordinated by three water molecules, and a Thrombospondins (TSP) domain. The first is found in various plasma proteins, e.g. complement factors or integrins, and is often involved in protein complexes which participate in various biological process (for example signal transduction, cell adhesion, pattern formation and migration);26 it contains a metal ion site at the surface that could represent a general metal ion-dependent adhesion site (MIDAS) for binding protein ligands.33 This site binds magnesium in the I-domain of Integrins CD11b33 and Manganese in CD11a34 by slightly different coordination of the same conserved residues.34 TSP is a multimeric multidomain glycoprotein functioning in the extracellular matrix and it regulates cell interactions.
Target T0696 (PDB: n.a.)
A DELTA-BLAST search relates this target with a conserved domain superfamily called “Glyoxalase/fosfomycin resistance/dioxygenase domain”, which is found in a variety of structurally related, but functionally diverse metallo-proteins, including glyoxalase I, type I extradiol dioxygenases and some antibiotic resistance proteins. They use different metal cations for their catalytic activity (for example Fe2+, Mn2+, Zn2+, Ni2+, or Mg2+). In this target the binding site is occupied by a Na+ (Figure 1D), which substitutes one of mentioned metal ions.
Target T0697 (PDB: n.a.)
belongs to the pyridoxal phosphate (PLP)-dependent decarboxylase family (EC number 4.1.1) group 2, which includes glutamate, histidine, tyrosine, and aromatic-L-amino-acid decarboxylases. This family is involved in the biosynthesis of amino acids, their derived metabolites, amino sugars and in the synthesis or catabolism of neurotransmitters. The PLP cofactor (Figure 1E) forms a Schiff base with a conserved lysine in the active site, which is temporarily displaced by the substrate; the resulting aldimine is the common central intermediate for PLP-catalyzed reactions.35
Target T0706 (PDB: n.a.)
A DELTA-BLAST search indicates that the target belongs to the Von Willebrand factor type A domain family, which contains a metal ion-dependent adhesion site (MIDAS) for binding protein ligands (for details, see also target T0686). In this target, a Mg2+ ion is bound to the adhesion site (Figure 1F).
Target T0720 (PDB: 4IC1)
The CRISPR-associated exonuclease Cas4 (EC=3.1.−.−) protein is involved in the mobile genetic elements immunity of the CRISPR (clustered regularly interspaced short palindromic repeat) system in most bacteria and archea.36 Short DNA sequences from viruses, the “spacers”, are flanked by CRISPR repeats in the host genome and transcribed into CRISPR RNAs (crRNAs), which are used by Cas (CRISPR-associated) proteins to recognize and degrade viral cognate sequences. This target in particular belongs to the Cas4 family of proteins, which resembles the RecB family37 and contains a cysteine-rich motif similar to the AddB family.38 It is a 5′ ssDNA metal-dependent (magnesium or manganese) exonuclease that needs an iron-sulfur cluster for structural stability (Figure 1G, 1H).39
Target T0721 (PDB: 4FK1)
The putative Thioredoxin reductase TrxB contains a FAD-dependent pyridine nucleotide-disulphide oxidoreductase domain with a FAD bound (Figure 1I).
Target T0726 (PDB: 4FGM)
contains an M61 glycyl aminopeptidase and a PDZ domain. Metalloproteases containing the first domain bind a divalent cation, through His, Glu, Asp or Lys amino acids, that activates the water molecule; usually a Zinc ion is bound by three residues which often can be described by an HEXXH motif (X can be any amino acid).40 The target binds a Zn2+ ion with the motif’s Histidines and a different Glutamate (Figure 1J). The second domain is found in eukaryotes41 and it binds the target protein by extending its beta-sheet with a strand from the partner C-terminus, so acting as a bridge between transmembrane proteins and the cytoskeleton in signaling pathways.42
Target T0737 (PDB: 3TD7)
is the probable FAD-linked sulfhydryl oxidase R596 from the Acanthamoeba polyphaga mimivirus. Its sequence contains a ERV/ALR sulphydryl oxidase domain which catalyzes disulphide bond formations. This module has a CXXC motif next to a FAD cofactor (in Figure 1K) which is used to transfer electrons from the thiol substrates to the (non-thiol) acceptor. A structure with bound FAD (PDB code: 3GWN) was available at the time of prediction for this target.
Target T0744 (PDB: 2YMV)
is a homologue of Mycobacterium tuberculosis Acg (Rv2032) in the reduced form from Mycobacterium smegmatis. The proteins in the Acg family are monomers that resemble the nitroreductase homodimer fold, with a single flavin mononucleotide binding site (Figure 1L) closed by a lid, instead of two open binding sites as in homodimeric nitroreductases. The structure and the lack of reduction by NADPH suggest that this proteins has lost the nitroreductase function and instead they may act as inhibitor of another nitroreductase by storing the flavin cofactor during the dormancy state of the bacteria.43
Overall performance
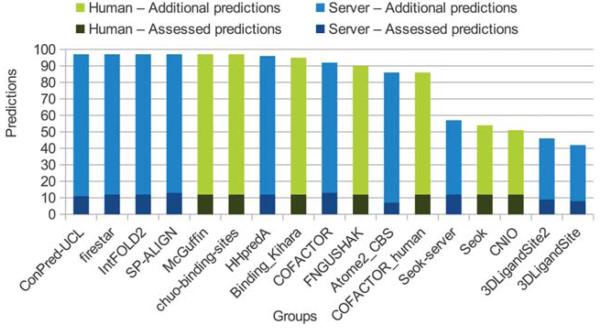
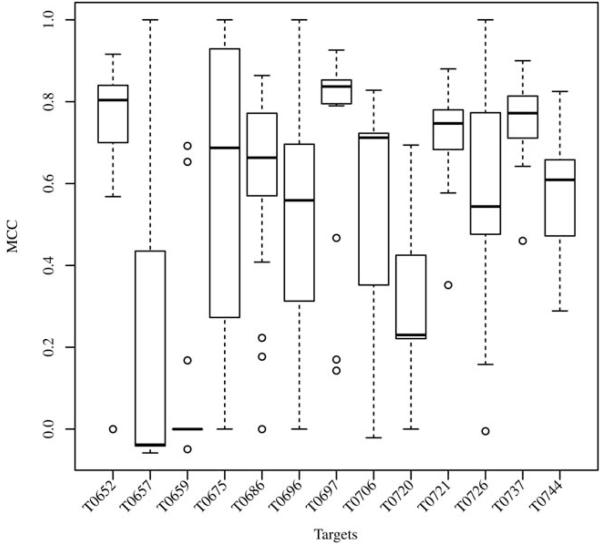
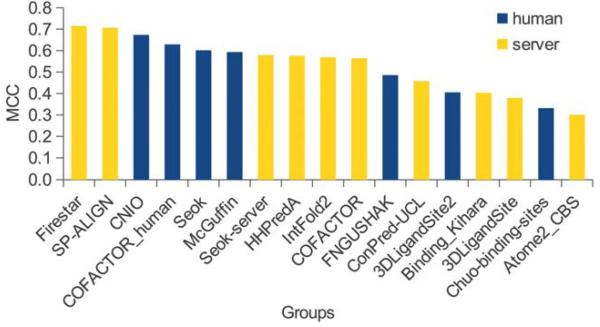
As in previous years, the evaluation of the binding site prediction accuracy was based on the Matthew Correlation Coefficient. A total of 1′817 submissions by 19 groups for the FN category were received by the Prediction Center. In CASP10 only 13 target proteins contained relevant ligands, i.e. only a small sub-set of all submissions could be used for the assessment (Figure 2). Of the 17 groupsa in the assessment, most of them submitted predictions for all 13 targets. Missing predictions were assigned a MCC score of zero, corresponding to a random prediction. Figure 3 shows a box plot representing the MCC distributions for each target, which gives a first estimate of the prediction difficulty. On most targets the predictors achieved on average a good performance around an MCC of 0.6, except in three cases, where in two (T0657 and T0659) the median scores were around zero and in one (T0720) was around 0.2. For comparison of the method’s overall performance, groups were ranked according to the average value of their MCCs normalized on all prediction targets (Figure 4; supplementary Table S1). Within the first ten groups, there were more “servers” at CASP10 than in CASP9, six instead of two, with an average MCC of 0.62. Their performance was indistinguishable from the “human” predictors, which is an improvement with respect to the results obtained in CASP9. The main differences between “human” and “server” methods is that the former could access human-only readable data (e.g. literature or databases) to identify relevant ligands, and have access to the pool of 3D structure predictions by servers due to the late submission deadline. While there was only a difference of 0.15 between the top ten groups based on average MCC, group FN119 (Firestar10) and FN326 (SP-ALIGN11) achieved the best scores of 0.715 and 0.707 respectively. These two methods had an overall different behavior: Firestar was one of the two predictors, together with HHPredA, with the highest number of top scores; it had the best MCC in three targets (T0696, T0726, T0744) while SP-ALIGN only for T0659, which was the most difficult target.
Figure 2. Number of predictions per group.
Since only a small number of all CASP10 targets contained relevant ligands, only a few predictions could be used for the assessment (dark blue and dark green), while the majority of the predictions could not be evaluated (blue and green).
Figure 3. Target difficulty.
Distribution of the predictor’s MCC for each target shown as Box plot (1st quartile, median and 3rd quartile), indicating the difficulty in the prediction of the various binding sites.
Figure 4. Groups ranking by MCC.
The predictors are ranked in decreasing order by the average value of the MCC, calculated on all the evaluated targets. Human predictors are shown in blue and server predictors in yellow.
We also evaluated the predictor performance based on the distance based BDT measure (supplementary table S3), which gave a ranking very similar to the MCC averages that was within deviations expected from the robustness test described below. To better understand to which extend these methods were useful in practice, we compared their performance with a baseline predictor that inferred the target’s binding sites using the first ten templates with ligands found by DELTA-BLAST and collecting all the residues in contact with them. The resulting average MCC was 0.339, which is only half of the performance obtained by the top predictors, and only two methods in the experiment performed worse than this baseline. This result indicates that most of the methods assessed in CASP10 give advantages in the ligand binding site prediction compared to a naïve homology search approach and could positively support the characterization of a protein’s function.
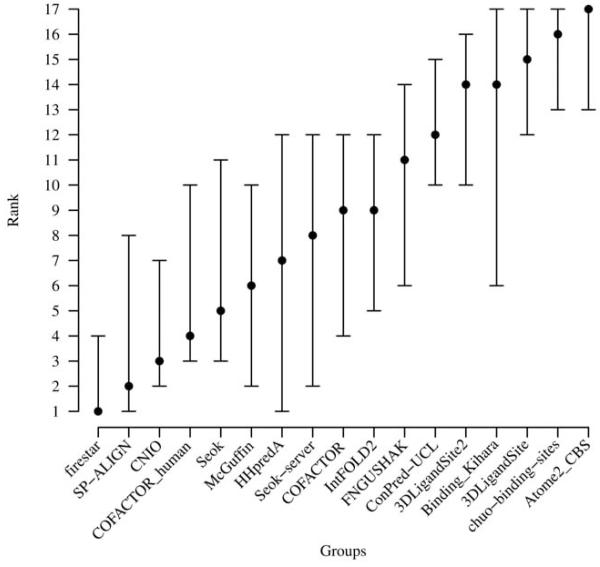
Assessment robustness
Since the number of prediction targets was extremely small, we assessed the robustness of the ranking by calculating MCC distributions with 100 cycles of random sampling using 70% of the targets (Figure 5). Although, the median values confirm the order of the top groups ranked by MCC, the rank spread is rather large and fluctuations by 10 positions are not unusual, i.e. the ranking is strongly influenced by the composition of the data set and does therefore not necessarily correctly reflect the differences in prediction accuracy of the individual methods. When calculating the statistical significance of the overall ranking by applying Student’s T-test (data not shown) and Wilcoxon signed-rank test (Table S2), the results indicated that the ranking was not robust and the differences between the top ten groups were not statistically significant. Both results are not surprising, considering the fact that the assessment had to be based on a very small number of target structures.
Figure 5. Groups ranking robustness.
Methods were ranked using the median value of the MCC distributions after 100 cycles of random sampling using 70% of the targets. Bars indicate best, median and worst ranking for each group.
Top predictors’ methods are based on homology transfer
Let’s take a closer look at the groups ranked highest by MCC: Firestar (FN119), SP-ALIGN (FN326), CNIO (FN475) and Cofactor_human (FN208); the first two were registered to CASP10 as “server”, while the last two as “human” predictor groups. Firestar10 bases its predictions on homology transfer of functionally important residues, found by local evolutionary sequence conservation; SP-ALIGN, an update to FINDSITE,11 is a threading based method to detect ligand binding sites by the employment of remote template identification and superimposition, structure-pocket alignment and binding site clustering guided by the template ligands; CNIO combines predictions from Firestar and 3DLigandSite,12 which clusters superimposed ligands from homologous structures to identify the binding residues; Cofactor_human requires human assistance to validate the binding residues found by the Cofactor algorithm,32 which employs a local superimposition of conserved residues taken from the target’s templates. The common theme among these methods is that they are all based on the analysis of the ligands bound to homologous structures. Firestar and CNIO use FireDB44, Cofactor employs structures from BioLip 45, while SP-ALIGN uses an ad-hoc template library. As a consequence, the performance of these methods is tied to the availability of annotated protein structures and the ability of finding homologue templates. Nevertheless, it has to be noted that the protocol to transfer the information on binding residues is different among those methods.
In recent years, homology based methods for structure prediction have started to reach a substantial coverage for proteins of interest: today some form of structural information – either experimental or computational – is available for the majority of amino acids encoded by common model organism genomes.33 For almost all known protein-protein interactions for which the individual components are structurally characterized, structures of complexes can be identified in the PDB which can be used for template-based prediction approaches.34-36 The overall good performances of methods such as Firestar and SP-ALIGN in CASP10, and their ability to identify ligand binding sites in different families of proteins in the absence of close homologue targets indicates that the field of ligand binding site prediction shows a similar trend.
It should be noted that in previous editions of CASP, almost all FN targets were classified as “template based modeling” (TBM) and only very few as “free modeling” (FM). In this round of CASP10, none of the relevant ligand binding sites were located in FM targets. Although target T0737 is classified as “free modeling”, the part of the protein to which the ligand FAD is bound has experimental structure information (Figure 6). This directly implies that the CASP assessment is mainly suitable to evaluate methods based on homology transfer to predict binding residues, but unable to measure the performance on harder targets, for which template structure information is not a useful source of information.
Figure 6. FAD binding site in target T0737.
Residues 176-292 (D1, blue) have been classified as “free modeling”. However, the N-terminal domain (grey) where the ligand FAD is bound, is covered by experimental structures. (Image generated with OpenStructure).
Prediction examples
Two targets, T0657 and T0659, appeared to be most challenging as predictors obtained on average the lowest scores. The first (PDB: 2LUL) was a solution NMR structure of the PH domain from the “Tyrosine-protein kinase Tec”, bound to a Zn2+ ion in a Btk-type zinc finger (Figure 7A). On a first view, this appears to be a simple template-based modeling target, since at least one template with the correct ion bound (e.g. PDB:1B55) is easily detectable with BLAST. However, the median MCC achieved for this target was −0.05, where the best predictor (“Binding_Kihara”, FN231) achieved an MMC of 1 (supplementary table S1). Other predictors achieved a lower MCC of about 0.3, mainly because they predicted more binding residues than were present in the reference structure, some of which have been assigned to other ligands than Zinc as indicated in the comments field. This example illustrates one of the limitations of the current binary prediction format.
The second target, T0659 (PDB: 4ESN), was a crystal structure of a hypothetical protein that bound a Zn2+ ion by three conserved Cysteines (Figure 7B). The median MCC was zero, while the best score, an MCC of 0.69 (supplementary table S1), was obtained for a prediction by SP-ALIGN (FN326), which is shown in Figure 7B. Easily detectable homologous structures of this protein did not contain any ligand, which explains the overall weak performance on this target. Interestingly, SP-ALIGN predicted an iron ion bound at this position; potentially, this could be due to the employment of its threading based method that detected a remote homologue bound to that ion.
Conclusions
Predicting a protein’s binding site is an important step towards understanding its function, and has implications for gene product characterization, drug design and enzyme engineering. The 13 targets evaluated in the assessment include proteins with interesting functions. For example T0686, which contains a metal ion-dependent adhesion site (MIDAS) which mediates the invasion of vertebrate cells by malaria Sporozoites; or T0720 – a CRISPR-associated (Cas) protein involved in the genetic mobile elements defense and it contains a catalytic magnesium ion plus a structural Iron-Sufur cluster. As in previous years, homology transfer approaches, in which the target binding residues are inferred from homologous proteins, have scored best with an average MCC of 0.71.
As in previous rounds of the CASP experiment, only a very limited number of targets with biologically relevant ligands (13 out of 97 targets) were available. Consequently, the assessment did not lead to a stable ranking of the participating methods, and it was not possible to differentiate methods by their performance on different types of targets or ligands. Another limitation originates from the current binary prediction format (“binding” or “not binding”), which does not include any information on the type of compounds or a level of confidence for the prediction. For a more detailed discussion, see assessment of ligand binding predictions in CASP9.22
During the CASP10 predictors meeting in Gaeta, it was recognized that the current procedure is not appropriate to assess the state of the art in ligand binding site predictions, and therefore does not stimulate the development of new approaches. In order to overcome these limitations, the following improvements should be implemented: a) Binary predictions should be replaced by predicting continuous probability values. b) The prediction format should include the specification of ligand type / ligand identity. c) The number of prediction targets, specifically those without trivial templates, needs to be increased substantially.
Based on these considerations, prediction methods in the FN category in future editions of CASP will no longer be evaluated based on the regular set of CASP target proteins. Instead, ligand binding site prediction servers will be evaluated continuously using an automated system called Continuous Automated Model Evaluation (CAMEO, http://www.cameo3d.org/), which is based on weekly pre-released sequences from the PDB. Continuous evaluation allows developers to constantly monitor the performance of new developments. Thanks to the larger number of targets, continuous evaluation also provides statistically robust assessment of ligand binding site predictions and allows for a more detailed assessment of methods, e.g. by ligand type or target difficulty. We hope that these new developments will stimulate new methods and approaches in this important area of structural Bioinformatics.
Supplementary Material
Acknowledgements
We would like to thank the previous CASP assessors Tobias Schmidt and Florian Kiefer for fruitful discussions and their contributions to the assessment methods for the ligand binding category. We are grateful to Konstantin Arnold (SIB Swiss Institute of Bioinformatics and Biozentrum University of Basel) for professional systems support. Computational resources were provided by the [BC]2 Basel Computational Biology Center. The development of CAMEO was supported by the SIB Swiss Institute of Bioinformatics and the National Institute of General Medical Sciences as a sub-grant with Rutgers University, under Prime Agreement Award Number U01 GM093324-01.
Abbreviations
- MCC
Matthews Correlation Coefficient
- TBM
Template based modeling category in CASP
- FM
Free-modeling category in CASP
Footnotes
Predictions by two groups were excluded from the assessment by the CASP organizers.
References
- 1.Pupko T, Bell RE, Mayrose I, Glaser F, Ben-Tal N. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18(Suppl 1):S71–77. doi: 10.1093/bioinformatics/18.suppl_1.s71. [DOI] [PubMed] [Google Scholar]
- 2.Capra JA, Singh M. Predicting functionally important residues from sequence conservation. Bioinformatics. 2007;23(15):1875–1882. doi: 10.1093/bioinformatics/btm270. [DOI] [PubMed] [Google Scholar]
- 3.Fischer JD, Mayer CE, Soding J. Prediction of protein functional residues from sequence by probability density estimation. Bioinformatics. 2008;24(5):613–620. doi: 10.1093/bioinformatics/btm626. [DOI] [PubMed] [Google Scholar]
- 4.Laurie AT, Jackson RM. Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics. 2005;21(9):1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- 5.Binkowski TA, Joachimiak A. Protein functional surfaces: global shape matching and local spatial alignments of ligand binding sites. BMC Struct Biol. 2008;8:45. doi: 10.1186/1472-6807-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghersi D, Sanchez R. EasyMIFS and SiteHound: a toolkit for the identification of ligand-binding sites in protein structures. Bioinformatics. 2009;25(23):3185–3186. doi: 10.1093/bioinformatics/btp562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang B, Schroeder M. LIGSITEcsc: predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct Biol. 2006;6:19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glaser F, Morris RJ, Najmanovich RJ, Laskowski RA, Thornton JM. A method for localizing ligand binding pockets in protein structures. Proteins. 2006;62(2):479–488. doi: 10.1002/prot.20769. [DOI] [PubMed] [Google Scholar]
- 9.Capra JA, Laskowski RA, Thornton JM, Singh M, Funkhouser TA. Predicting protein ligand binding sites by combining evolutionary sequence conservation and 3D structure. PLoS Comput Biol. 2009;5(12):e1000585. doi: 10.1371/journal.pcbi.1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez G, Valencia A, Tress ML. firestar--prediction of functionally important residues using structural templates and alignment reliability. Nucleic Acids Res. 2007;35(Web Server issue):W573–577. doi: 10.1093/nar/gkm297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brylinski M, Skolnick J. A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation. Proc Natl Acad Sci U S A. 2008;105(1):129–134. doi: 10.1073/pnas.0707684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wass MN, Kelley LA, Sternberg MJ. 3 DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res. 2010;38(Web Server issue):W469–473. doi: 10.1093/nar/gkq406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mariani V, Kiefer F, Schmidt T, Haas J, Schwede T. Assessment of template based protein structure predictions in CASP9. Proteins. 2011;79(Suppl 10):37–58. doi: 10.1002/prot.23177. [DOI] [PubMed] [Google Scholar]
- 14.Kinch L, Yong Shi S, Cong Q, Cheng H, Liao Y, Grishin NV. CASP9 assessment of free modeling target predictions. Proteins. 2011;79(Suppl 10):59–73. doi: 10.1002/prot.23181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao B, Huang YJ, Aramini JM, Montelione GT. Assessment of template based protein structure predictions in CASP10. Proteins. 2013 doi: 10.1002/prot.24488. This Volume(This Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tai CH, Bai H, Taylor TJ, Lee BK. Assessment of template free modeling in CASP10 and ROLL. Proteins. 2013 doi: 10.1002/prot.24470. This volume(This issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kopp J, Bordoli L, Battey JN, Kiefer F, Schwede T. Assessment of CASP7 predictions for template-based modeling targets. Proteins. 2007;69(Suppl 8):38–56. doi: 10.1002/prot.21753. [DOI] [PubMed] [Google Scholar]
- 18.Battey JN, Kopp J, Bordoli L, Read RJ, Clarke ND, Schwede T. Automated server predictions in CASP7. Proteins. 2007;69(Suppl 8):68–82. doi: 10.1002/prot.21761. [DOI] [PubMed] [Google Scholar]
- 19.Soro S, Tramontano A. The prediction of protein function at CASP6. Proteins. 2005;61(Suppl 7):201–213. doi: 10.1002/prot.20738. [DOI] [PubMed] [Google Scholar]
- 20.Lopez G, Rojas A, Tress M, Valencia A. Assessment of predictions submitted for the CASP7 function prediction category. Proteins. 2007;69(Suppl 8):165–174. doi: 10.1002/prot.21651. [DOI] [PubMed] [Google Scholar]
- 21.Lopez G, Ezkurdia I, Tress ML. Assessment of ligand binding residue predictions in CASP8. Proteins. 2009;77(Suppl 9):138–146. doi: 10.1002/prot.22557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt T, Haas J, Gallo Cassarino T, Schwede T. Assessment of ligand-binding residue predictions in CASP9. Proteins. 2011;79(Suppl 10):126–136. doi: 10.1002/prot.23174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor TJ, Tai CH, Huang YJ, Block J, Bai H, Kryshtafovych A, Montelione GT, Lee BK. Definition and Classification of Evaluation Units for CASP10. Proteins. 2013 doi: 10.1002/prot.24434. This volume(This issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magrane M, Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011;2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biasini M, Mariani V, Haas J, Scheuber S, Schenk AD, Schwede T, Philippsen A. OpenStructure: a flexible software framework for computational structural biology. Bioinformatics. 2010;26(20):2626–2628. doi: 10.1093/bioinformatics/btq481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biasini M, Schmidt T, Bienert S, Mariani V, Studer G, Haas J, Johner N, Schenk AD, Philippsen A, Schwede T. OpenStructure: an integrated software framework for computational structural biology. Acta Crystallographica Section D. 2013;69(5):701–709. doi: 10.1107/S0907444913007051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roche DB, Tetchner SJ, McGuffin LJ. The binding site distance test score: a robust method for the assessment of predicted protein binding sites. Bioinformatics. 2010;26(22):2920–2921. doi: 10.1093/bioinformatics/btq543. [DOI] [PubMed] [Google Scholar]
- 28.R Development Core Team . R: A language and environment for statistical computing. 2.11.1. R Foundation for Statistical Computing; Vienna, Austria: 2011. [Google Scholar]
- 29.Kryshtafovych A, Moult J, Bales P, Bazan F, Biasini M, Burgin A, Cochran F, Das R, Fass D, Garcia-Doval C, Herzberg O, Lorimer D, Luecke HH, Ma X, Nelson D, van Raaij M, Rohwer F, Segall A, Seguritan V, Zeth K, Schwede T. Challenging the state-of-the-art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10. Proteins. 2013 doi: 10.1002/prot.24489. This volume(This issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ignoul S, Eggermont J. CBS domains: structure, function, and pathology in human proteins. Am J Physiol Cell Physiol. 2005;289(6):C1369–1378. doi: 10.1152/ajpcell.00282.2005. [DOI] [PubMed] [Google Scholar]
- 31.Boratyn GM, Schaffer AA, Agarwala R, Altschul SF, Lipman DJ, Madden TL. Domain enhanced lookup time accelerated BLAST. Biol Direct. 2012;7:12. doi: 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roy A. Protein-ligand binding sites prediction using COFACTOR; The EMBO Conference: Critical assessment for protein structure prediction (CASP10); Gaeta, Italy. 2012.pp. 57–58. [Google Scholar]
- 33.Schwede T. Protein Modeling: What Happened to the Protein Structure Gap? Structure (London, England: 1993) 2013;21(9):1531–1540. doi: 10.1016/j.str.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kundrotas PJ, Zhu Z, Janin J, Vakser IA. Templates are available to model nearly all complexes of structurally characterized proteins. Proc Natl Acad Sci U S A. 2012;109(24):9438–9441. doi: 10.1073/pnas.1200678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein A, Mosca R, Aloy P. Three-dimensional modeling of protein interactions and complexes is going ‘omics. Curr Opin Struct Biol. 2011;21(2):200–208. doi: 10.1016/j.sbi.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 36.Xu Q, Dunbrack RL., Jr. The protein common interface database (ProtCID)--a comprehensive database of interactions of homologous proteins in multiple crystal forms. Nucleic Acids Res. 2011;39(Database issue):D761–770. doi: 10.1093/nar/gkq1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43(6):1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 38.Yeeles JT, Cammack R, Dillingham MS. An iron-sulfur cluster is essential for the binding of broken DNA by AddAB-type helicase-nucleases. J Biol Chem. 2009;284(12):7746–7755. doi: 10.1074/jbc.M808526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Kasciukovic T, White MF. The CRISPR associated protein Cas4 Is a 5′ to 3′ DNA exonuclease with an iron-sulfur cluster. PLoS One. 2012;7(10):e47232. doi: 10.1371/journal.pone.0047232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rawlings ND, Barrett AJ. Evolutionary families of metallopeptidases. Methods Enzymol. 1995;248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- 41.Ponting CP. Evidence for PDZ domains in bacteria, yeast, and plants. Protein Sci. 1997;6(2):464–468. doi: 10.1002/pro.5560060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ranganathan R, Ross EM. PDZ domain proteins: scaffolds for signaling complexes. Curr Biol. 1997;7(12):R770–773. doi: 10.1016/s0960-9822(06)00401-5. [DOI] [PubMed] [Google Scholar]
- 43.Chauviac FX, Bommer M, Yan J, Parkin G, Daviter T, Lowden P, Raven EL, Thalassinos K, Keep NH. Crystal structure of reduced MsAcg, a putative nitroreductase from Mycobacterium smegmatis and a close homologue of Mycobacterium tuberculosis Acg. J Biol Chem. 2012;287(53):44372–44383. doi: 10.1074/jbc.M112.406264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopez G, Valencia A, Tress M. FireDB--a database of functionally important residues from proteins of known structure. Nucleic Acids Res. 2007;35(Database issue):D219–223. doi: 10.1093/nar/gkl897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J, Roy A, Zhang Y. BioLiP: a semi-manually curated database for biologically relevant ligand-protein interactions. Nucleic Acids Res. 2013;41(Database issue):D1096–1103. doi: 10.1093/nar/gks966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.