Abstract
Automated methods to detect new information in clinical notes may be valuable for navigating and using information in these documents for patient care. Statistical language models were evaluated as a means to quantify new information over longitudinal clinical notes for a given patient. The new information proportion (NIP) in target notes decreased logarithmically with increasing numbers of previous notes to create the language model. For a given patient, the amount of new information had cyclic patterns. Higher NIP scores correlated with notes having more new information often with clinically significant events, and lower NIP scores indicated notes with less new information. Our analysis also revealed “copying and pasting” to be widely used in generating clinical notes by copying information from the most recent historical clinical notes forward. These methods can potentially aid clinicians in finding notes with more clinically relevant new information and in reviewing notes more purposefully which may increase the efficiency of clinicians in delivering patient care.
Keywords: Electronic Health Records, Natural Language Processing, Text Mining, Information Storage and Retrieval
Introduction
Electronic health record (EHR) systems are becoming increasingly ubiquitous and vital for health care delivery due to several key advantages over paper-based health records, including information accessibility, sharing, and integration. The widespread adoption of EHR systems has resulted in a rapid increase of patient information in the form of electronic narratives or clinical notes. In EHR systems, a clinician can create a note by using information from previous notes, which often results in redundant information between multiple notes [1].
Redundant information may decrease health care efficiency and have a negative impact on patient safety. Reviewing and synthesizing clinical notes is a fundamental and vital patient care task for clinicians. Redundant information increases the volume of text in clinical notes and de-emphasizes important new information, thus placing an additional cognitive load on clinicians who must function in a time-constrained clinical environment. A mixture of important new information and less important redundant information (especially outdated and no longer accurate information) in clinical notes may interfere with decision-making [2]. Moreover, replicated erroneous information in clinical notes has been demonstrated to create medical errors and be a patient safety issue [1]. It has also been reported that redundant information in notes results in decreased use of clinical notes by clinicians [3]. Healthcare organizations are increasingly recognizing the issues caused by redundant information in electronic notes and are asking for solutions [4].
Patient records containing collections of unstructured or semistructured clinical notes (clinical texts) are prevalent for both documentation and communication between clinicians and for billing. While structured information (i.e., lab tests and vital signs) may be easy to aggregate and compute, this information can be difficult for clinicians to interpret, as contextual information is often lost. Clinical texts can provide this context, providing clinicians with nuanced and detailed information to interpret, treat, and diagnose patients more easily. A number of researchers have focused on information extraction from clinical notes with natural language processing (NLP) techniques, creating computational tools to analyze and extract information from clinical texts. Statistical language models, which were used for speech recognition, machine translation, and information retrieval, are one such NLP technique.
Synthesizing multiple clinical notes, especially with redundant information, can be challenging. Thus, there is an immediate need for computer-based tools designed to assist clinicians with identifying and synthesizing clinical notes by identifying redundant information or its counterpart, new information, within clinical notes. Recent studies in the area have focused more on automatic summarization techniques to generate a separate short document containing only key information [5]. However, such summarization loses detailed contextual information and makes it difficult for clinicians to comprehensively understand and synthesize clinical notes. Tools that can accurately identify and visualize new information within electronic notes in EHR systems without losing crucial contextual information are desirable.
Some studies have reported that redundant information is ubiquitous in inpatient [6] and outpatient [7] clinical notes by quantifying redundancy. We previously developed a linguistic model to identify relevant new information and visualized this information through the use of highlighting [8]. We also found for a small set of outpatient notes that redundancy increased over time and appeared to have a cyclic pattern of scores with dips relating to significant clinical new events. The visualization of new information as a feature of clinical note user interfaces has also been demonstrated in a prototype to save time in reviewing notes with a set of controlled patient scenarios and to improve navigation of these notes [9]. Information navigation using statistical language models, however, has not been well investigated.
Information navigation of electronic notes is essential for physicians reviewing a complex patient (with a long series of longitudinal clinical notes with historical medical information). The ability to highlight new and relevant information in clinical notes provides clinicians with the ability to navigate notes more purposefully. Moreover, there is limited investigation into the sources of redundant information within a specific clinical note, which can be important in understanding the behaviors of clinicians in generating new clinical notes, as well as inform the development of future tools for new information identification. The aim of this study is to describe an automated method to quantify new information and navigate to notes with new information, and to investigate possible new information (or its inverse - redundant information) patterns for individual patient records. As a secondary aim, we also sought to understand “copy and paste” behaviors and to provide a potential method to navigate notes.
Methods
Data Collection
EHR notes were retrieved from University of Minnesota Medical Center affiliated Fairview Health Services. For this study, we selected patients with multiple co-morbidities, allowing for relatively large numbers of longitudinal records in the outpatient clinic setting. These notes were extracted in text format from the Epic™ EHR system 1 during a six-year period (06/2005 to 06/2011). To simplify the study, we limited the notes to office visit notes (Fig. 1, see part B). Each note was indexed based on chronological order (e.g., note A1 indicates the 1st note of patient A). Institutional review board approval was obtained and informed consent waived for this minimal risk study.
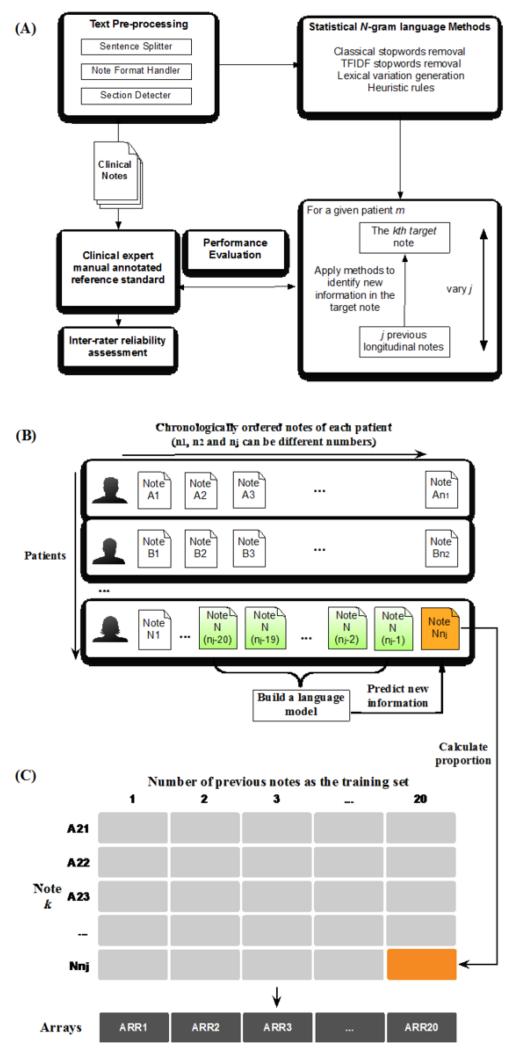
Figure 1.
(A) statistical language model development; (B) longitudinal data set; (C) score matrix of new information proportion (NIP). Build a language model (A) to calculate the NIP of note k (B) and generate the corresponding cell in the matrix (C).
Manually reviewed annotation as gold standard
Two medical interns (physicians aged 26 and 30) were asked to identify new information within each document (starting from the second document) based on all the preceding documents chronologically for each patient record using their clinical judgment. Each medical expert annotated five patient records with one record overlapping with both. Annotation of new information in clinical notes was implemented by using the General Architecture for Text Engineering (GATE) 2. GATE allows for the annotation of text and XML outputs through a graphical user interface, with a customized annotation schema.
To achieve a high-quality gold standard, we first asked the physicians to annotate one sample note (based on historical notes) and then compare and discuss the annotations with each other to reach a consensus on annotation standards for new information. Each physician later manually annotated another 10 notes based on the same historical notes to measure agreement. Cohen’s Kappa statistic and percent agreement [10] were used to assess inter-rater reliability at a sentence or statement level.
Overall, longitudinal outpatient clinical notes from 15 patients were selected for annotation. To better evaluate our method, raters annotated the same last 3 notes as the target notes compared to historical notes of each patient’s note set, but used different numbers of previous notes as the reference clinical history (e.g., one used the previous 5 notes, the other used the previous 10 notes). Overall, each medical intern annotated 45 notes. Twenty of them were used for training and developing the system and another twenty-five for evaluation. Performance of automated methods was then compared to the reference standard and measured for accuracy, precision, recall, and F-measure at a sentence or statement level.
Also, a 5th year resident physician (JTL) manually reviewed two randomly selected patient records. Without seeing our results, the physician first reviewed the most current history (11-20th notes of a given patient) and then the target notes (21-38th). Any new information found in the target notes not in the previous 10 notes was noted. The physician then also reviewed another 10 historical notes prior (1-10th), and marked if there was any additional new information within notes 21-38th not recognized with review of the earlier notes. We then compared this with the automatically computed redundancy results.
New information pattern analysis
We first used the automated method to identify new information and quantify the new information proportion (NIP) in each note. The principle method used for this study is based on the updated n-gram statistical language models reported previously [8]. In short, we used a bigram language model with classic stopword removal3, term frequency-inverse document frequency (TF-IDF) stopword removal, application of lexical variation generation (LVG) [11], and the adjustment of the model through several heuristic rules (Fig. 1A).
The developed computational model was used to identify new information in all notes (starting from the 21st note) based on the previous n (=1, 2 … 20) longitudinal clinical notes. We then obtained the matrix of NIP (number of sentences with new information/number of all sentences per note) with the dimension of 2,918 × 20. 2,918 is the number of all notes having at least 20 historical notes for the whole 100 patient corpus. For example, the orange cell in column 20 and row Nnj (Fig. 1C) represents the NIP contained in the note Nnj (Fig. 1B) calculated based on the previous 20 clinical notes. Thus, each row represents each note, and columns are the corresponding numbers of previous notes used in the language model to predict new information in that target note. We used this matrix to investigate the impact of the number of previous clinical notes in the model on the NIP scores. Twenty arithmetic means were obtained by averaging NIP scores in each array, and correlated with the previous note numbers to find the relationship.
We clustered notes of the same patient as a group (e.g., longitudinal notes of the patient A: A21, A22 … A38) and averaged notes sharing the same note index from different patients (e.g., 21st notes from all patients: A21, B21 … N21) to get representative NIP scores based on all patient notes. We then plotted NIP scores to investigate the overall patterns of how new information changed over time.
Results
Annotation evaluation and model performance
The two raters showed good agreement on the task of identifying new information on the overlapped annotation. Cohen’s Kappa coefficient of two annotators for the overlap clinical documents was 0.80 and percent agreement was 97% on new information identification at the sentence/statement level. We compared the results generated by automated results with the refined reference standard. The accuracy, precision, recall, and F-measure are 0.83, 0.72, 0.71, 0.72, respectively.
Changes in the amount of new information
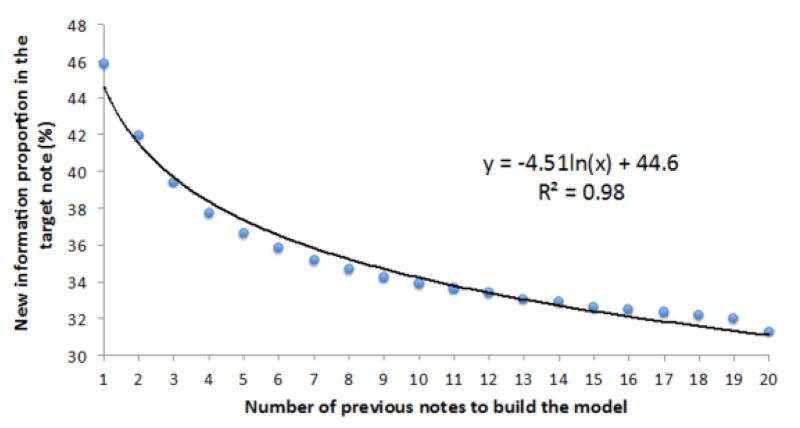
After averaging all 2,918 NIP scores for the array (Fig. 1C), we obtained 20 arithmetic mean NIP scores. The means were then plotted with the number of previous notes and fitted with a logarithm function (Fig. 2).
Figure 2.
Scatter plot and fitted line of new information proportion with the numbers of the previous notes.
New information patterns
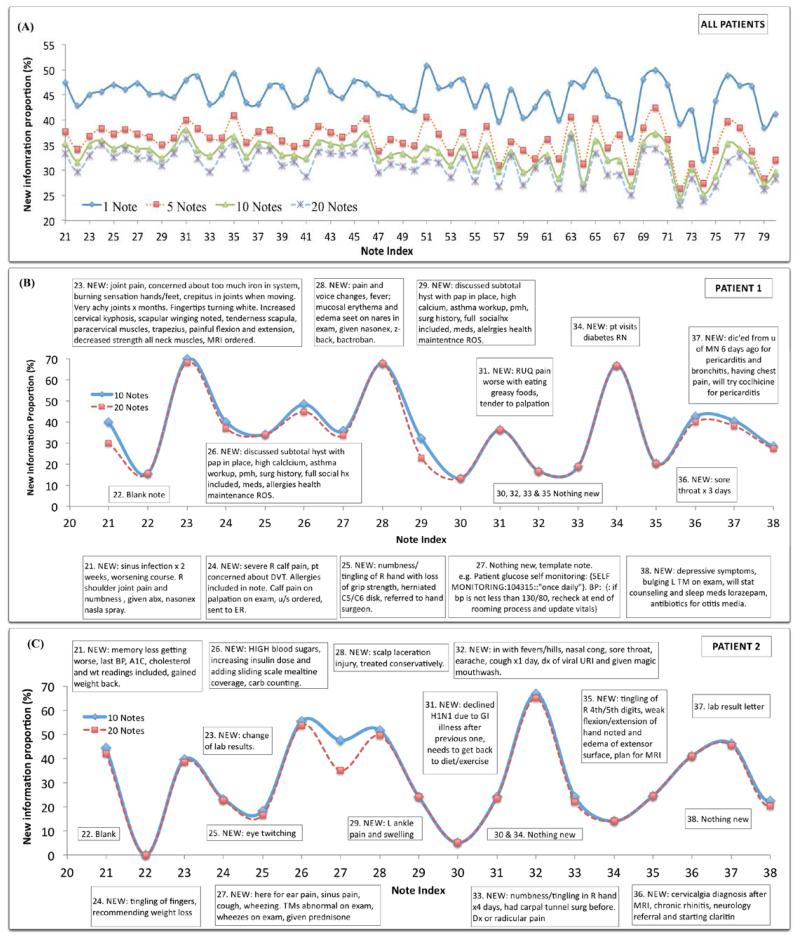
Figure 3A showed patterns of NIP scores in longitudinal clinical notes with the consideration of all patient notes. The four cyclic patterns indicated similar shapes, although the four curves had different NIP values. When the number of previous notes changed from 1 to 5, NIP dropped significantly; whereas the change was more gradual from note 5 to note 10 and even less from note 10 to note 20.
Figure 3.
(A) Overall pattern of new information proportions based on the averaged scores over patients; (B) & (C) New information proportions of longitudinal notes based on the previous 10 and 20 notes from two individual patients. New information contents shown in the boxes were annotated by the expert and compared with the previous 10 notes.
Figure 3B & 3C show longitudinal clinical notes of two patients with new information manually identified by JTL (in boxes). Solid lines show the NIP based on the previous 10 notes, and dotted lines show new information based on the previous 20 notes. Longitudinal clinical notes from both patients appeared to have a cyclic pattern, characterized by alternating periods of peaks (larger NIP) and troughs (smaller NIP). Based on the NIP scores of two patient notes, larger NIP scores (usually larger than 20%) correlated to notes with more new information content and a smaller NIP scores (usually less than 20%) corresponded to notes without a significant amount of new information. One exception is that the high NIP in note #27, patient 1 (Fig. 3B) did not contain new patient clinical history but instead had newly information of note template, including “glucose self monitoring: SELF MONITORING:104315::‘once daily’”. Also, note #25, patient 2 (Fig. 3C) had a relatively lower NIP score but was judged to have new information of clinical significance (eye twitching).
Comparing solid lines (10 notes) to dotted lines (20 notes), we found that most NIP scores did not decrease much, with some keeping the same score and a few notes having significantly lower scores. For example, note #21, patient 1 (Fig. 3B) had a lower score when using longer patient history, which correlated to an old sinus infection found in the note #8. Also in patient 1, note #29 (Fig. 3B) contained new information based on the previous 10 notes (i.e., note #19-28), including symptoms, surgical history, and social history, which were also found in the note #18, thus the NIP dropping compared with all the pervious 20 notes (i.e., note #9-18). In patient 2 record, Fig. 3C, we also found a similar change for note #27, where new information of symptoms was found in note #13.
Discussion
This study focuses on a relatively understudied topic in clinical informatics: methods to detect new information and to improve note navigation in longitudinal notes. Preliminary studies have shown that improved information navigation with notes may be a promising feature in future EHR document interface design [9]. There is a need for both back-end algorithms design to facilitate this, as well as improved front-end user interfaces with these capabilities within EHRs.
To build such an information navigation system, annotation is a vital step but also a challenging task that we faced in our previous work [8] and in this study. We found that although inter-rater agreement was high, one rater annotated more carefully. Another issue was that the exact boundary of redundant versus new information was not well defined. To obtain a high-quality gold standard, which can help develop more accurate methods, we established good baseline communication between annotators before performing actual annotation for our method development. For example, one rater only annotated the piece of new information (lab values), but another rater still marked the corresponding information such as the title, date of the lab. Enhanced communications between annotators along with clearer guidelines improved the gold standard’s quality.
Changing the number of previous clinical notes in longitudinal patient records changes the size of training data for the language model. For a given note, as the model includes a longer history before the target note, it includes more clinical history about the patient and the model can recognize relatively more redundant information if information in the target note was included earlier in the patient’s history. If new information continues to decrease when the model “sees” more historical notes, it indicates several possibilities about a patient’s history including 1) that the history may contain information in the target note copied from earlier notes, 2) that clinicians may express events similarly and that there is a balance between what is old or new and that some events that repeat may actually be new (i.e., a repeat flu infection one year later), or 3) that by adding large amounts of notes to the model, it at some point may contain too much noise to detect new events.
We used our method to estimate NIP and averaged all NIP scores of 2,918 notes from 100 patients. NIP scores decreased as the number of previous notes increased (Fig. 2), indicating that physicians either copy information from as far as 20 previous notes or use similar forms of expression to describe similar events. Interestingly, the trend almost perfectly (R2 = 0.98) fits a logarithmic function. The decrease of new information logarithmically appears related to the length of history. Based on the trend generated from all patient notes, approximately 55% of information in the current note was redundant compared to the immediate previous note. In other words, 55% of redundant information may have been copied and pasted from or present in the previous along with the current note for other reasons. Approximately an additional 11% of information in the current note was propagated from previous 2-10 notes; and another additional 4% from of information from the previous 11-20 notes. These numbers can be different for individual patient’s records, but this overall trend indicates the boundaries of the source of redundant information.
Chronically diseased patients come to the office quite often, thus generating as many as 90 longitudinal notes (Fig. 3A) over the study period. The cyclic pattern indicated that the collection of longitudinal notes contains both important notes with new information and less important notes with mostly redundant information. This finding was consistent with our previous finding using global alignment methods [7]. In that study, we randomly chose a set of 10 notes from three patients and used up to 10 previous notes to quantify redundancy scores. We saw similar cyclic patterns and an overall uncharacterized trend of increasing redundancy.
Here, we further investigated new information (and its counterpart - redundancy) patterns of notes for individual patients, comparing our scores with human judgment. There was high correlation between calculated NIP scores and clinically significant events. Higher NIP scores correlated to more new information in the notes and lower NIP scores indicated less new information. We observed that 20% may be a suitable threshold value only based on the NIP scores of the two patient notes (Fig. 3B&C). However, further work is necessary to calibrate our measure threshold to distinguish notes with more new information from those with less. We also observed that unexpectedly high NIP scores can be seen with the introduction of templates absent in historical notes. One prerequisite condition for an accurate statistical language model is that the training set should be representative of the test set. This finding also provides us with a challenge that potentially could be addressed by incorporating EHR document templates into our model.
This analysis also helped to identify the original notes that served as the sources of redundant information found in target notes. A significant drop indicated that that the target note contained a lot of replicated information. Some notes were found to have negligible score changes between 11 and 20 notes in the model, due to the existence of few additional pieces of additional significant information in the previous 11-20 notes.
Future research includes development of more robust automated methods for identifying new information as well as the development of a user interface and navigation tools to assist clinicians with identification of new information and efficient utilization of clinical notes. Summarizing or providing key words of new information may be another approach for providing clinicians better tools for improved note navigate in EHRs at the point of care. We plan to perform usability testing with clinicians of these methods.
Conclusion
We investigated the use of statistical language models for information navigation with patient longitudinal clinical notes. New information in longitudinal notes had a cyclic pattern and an overall logarithmic relationship with the length of historical notes used to create the language model. Physicians tended to copy information from the most current notes. The analysis can also help find source notes of redundant information of a given note. Language models may be used as a potential information navigation tool for clinical notes.
Acknowledgments
This research was supported by the American Surgical Association Foundation Fellowship (GM), UMN Institute for Health Informatics Seed Grant (GM & SP), National Library of Medicine (#R01LM009623-01) (SP) and the UMN Graduate School Doctoral Dissertation Fellowship (RZ). The authors thank Fairview Health Services for support of this research.
Footnotes
References
- [1].Markel A. Copy and paste of electronic health records: a modern medical illness. Am J Med. 2010;123(5):9. doi: 10.1016/j.amjmed.2009.10.012. [DOI] [PubMed] [Google Scholar]
- [2].Patel VL, Kaufman DR, Arocha JF. Emerging paradigms of cognition in medical decision-making. J Biomed Inform. 2002;35(1):52–75. doi: 10.1016/s1532-0464(02)00009-6. [DOI] [PubMed] [Google Scholar]
- [3].Hripcsak G, Vawdrey DK, Fred MR, Bostwick SB. Use of electronic clinical documentation: time spent and team interactions. J Am Med Inform Assn. 2011;18(2):112–7. doi: 10.1136/jamia.2010.008441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hammond KW, Helbig ST, Benson CC, Brathwaite-Sketoe BM. Are electronic medical records trustworthy? Observations on copying, pasting and duplication. AMIA Annu Symp Proc. 2003:269–73. [PMC free article] [PubMed] [Google Scholar]
- [5].Van Vleck TT, Stein DM, Stetson PD, Johnson SB. Assessing data relevance for automated generation of a clinical summary. AMIA Annu Symp Proc. 2007:761–5. [PMC free article] [PubMed] [Google Scholar]
- [6].Wrenn JO, Stein DM, Bakken S, Stetson PD. Quantifying clinical narrative redundancy in an electronic health record. J Am Med Inform Assoc. 2010;17(1):49–53. doi: 10.1197/jamia.M3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhang R, Pakhomov S, MaInnes BT, Melton GB. Evaluating Measures of Redundancy in Clinical Texts. AMIA Annu Symp Proc. 2011:1612–20. [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang R, Pakhomov S, Melton GB. Automated Identification of Relevant New Information in Clinical Narrative. IHI’12 ACM Interna Health Inform Sym Proc. 2012:837–41. [Google Scholar]
- [9].Farri O, Rahman A, Monsen KA, Zhang R, Pakhomov S, Pieczkiewicz DS, Speedie SM, Melton GB. Impact of a prototype visualization tool for new information in EHR clinical documents. Appl Clin Inform. 2012;3(4):404–18. doi: 10.4338/ACI-2012-05-RA-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hunt RJ. Percent agreement, Pearson’s correlation, and kappa as measures of inter-examiner reliability. J Dent Res. 1986;65(2):128–30. doi: 10.1177/00220345860650020701. [DOI] [PubMed] [Google Scholar]
- [11].McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies. Proc Annu Symp Compt Appl Med Care. 1994:253–9. [PMC free article] [PubMed] [Google Scholar]