Abstract
Molecular computing based on enzymes or nucleic acids has attracted a great deal of attention due to the perspectives of controlling living systems in a way we control electronic computers. Enzyme-based computational systems can respond to a great variety of small molecule inputs. They have an advantage of signal amplification and highly specific recognition. DNA computing systems are most often controlled by oligonucleotide inputs/outputs and are capable of sophisticated computing, as well as controlling gene expressions. Here, we developed an interface that enables communication of otherwise incompatible nucleic acid and enzyme computational systems. The enzymatic system processes small molecules as inputs and produces NADH as an output. The NADH output triggers electrochemical release of an oligonucleotide, which is accepted by a DNA computational system as an input. This interface is universal since the enzymatic and DNA computing systems are independent of each other in composition and complexity.
Keywords: Molecular computation, NADH, deoxyribozyme, enzyme computation, electrochemistry
Modern silicon-based analog/digital computer technology has been one of the most successful and influential transformative developments in recent history. At the same time, natural biological molecules (e.g., nucleic acids and proteins) are organized in complex communicating networks responsible for growth of all living creatures through metabolism and reproduction. It was suggested that application of the well-developed computational approach to biological molecules may open new chapters in understanding biological signaling, neuron communication,[1] and cancer development,[2] as well as in improving diagnosis of infectious diseases and genetic disorders.[3] Indeed, biomolecular information processing has been an active research field[4] in the general framework of chemical[5] unconventional computing.[6] In this research area, DNA computing[4a-e] and enzyme-based computing[7] have received exceptional attention. DNA computing is believed to be a potential alternative to electronic computers[8] for some computational tasks, due to the advantage of massive parallel data processing,[9] a straightforward design of relatively complex circuits,[10] and affordability. Among the most obvious applications of DNA-based logic circuits is the analysis of genetic alterations that can be transformed into clinical testing of infectious and genetic diseases.[2,3] Despite advances in the development of in vitro selection, functional DNAs are still limited in the diversity and efficiency of catalytic reactions and are inferior to proteins in terms of affinity and diversity of ligands that DNA can recognize.[11] At the same time, enzymes are proven to be selective and sensitive receptors; they are known as the best catalysts, enabling rate enhancement up to 1017 fold in comparison with uncatalyzed reactions.[13] However, enzyme-based computing was experimentally limited to the systems mimicking operation of only few concatenated logic gates,[7] and the network complexity was restricted by enzymes cross-reactivity and noise build.[14] Combining enzyme and DNA computational systems in communicating enzyme-DNA (Enz/DNA) circuits may enable (i) highly selective recognition of a diverse spectrum of biological molecules or disease markers; (ii) catalytic signal amplification; (iii) massive parallel data processing and (iv) complex computational information processing for biologically generated signals. So far, mixed enzyme-DNA computational systems have been limited to those that involve enzymes directly acting on DNA, e.g., DNA polymerases, DNA ligases, endonuclease, etc.[15] However, DNA processing enzymes cannot detect such disease biomarkers as small biological molecules, sugars, proteins, etc. On the other hand, biocomputing systems based on general enzymes (not related to DNA) were successfully used for logic processing and binary sensing of various combinations of physiological biomarkers in the YES/NO format.[16] Therefore, a more universal interface for connecting an enzymatic output signal with DNA-processing circuits is needed. Here, we introduce such an Enz/DNA interface.
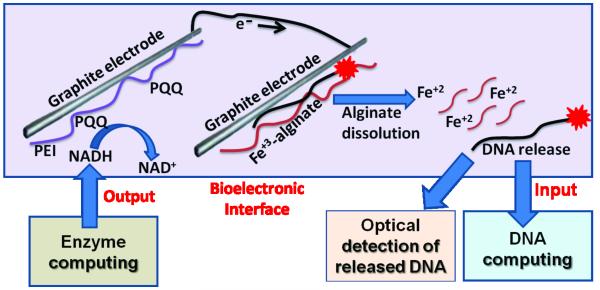
The interface recognizes NADH, which is produced as an output of an enzymatic system, and releases a DNA oligonucleotide, which can be processed by a downstream DNA computing system as an input (Figure 1). The interface was based on two modified electrodes (see experimental details in the SI). The first electrode communicating with the enzyme computing system (PQQ-electrode) was coated with adsorbed polyethyleneimine (PEI) and pyrroloquinoline quinone (PQQ) covalently attached to the PEI thin-film.[17] The immobilized PQQ served as a catalyst for electrochemical oxidation of NADH.[18] This process resulted in the formation of a negative potential of ca. −60 mV (vs. Ag|AgCl|KCl, 3 M, reference electrode; all other potentials are reported vs. this reference) and the corresponding current sufficient for reduction of Fe3+ as part of the Fe3+-cross-linked alginate film on the second connected electrode.[19] Note that Fe2+ cations are not capable of alginate cross-linking, and their formation results in the alginate thin-film dissolution and concomitant release of the entrapped molecules (Figure S1).
Figure 1.
Enzyme computing system produces NADH as an output, which is oxidized on an electrode and reduces Fe3+ to Fe2+ on another electrode. This leads to dissolution of Fe3+-cross-linked alginate polymer and release of an entrapped DNA output. The DNA output can be then used as an input by a DNA computing system.
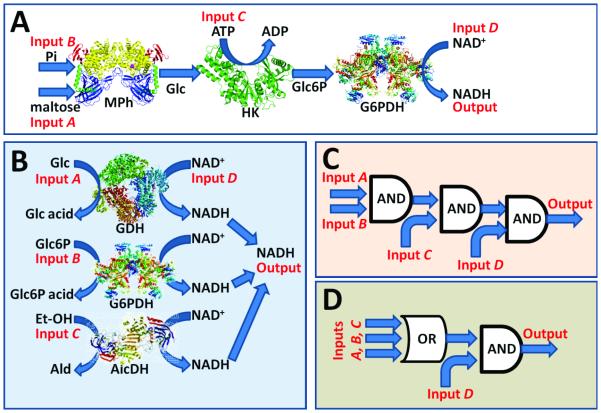
In this study we took advantage of two enzyme systems, either of which produced NADH (Figure 2). For binary operation of the enzyme systems, digital input 0 was defined as the absence of the corresponding substrate, whereas digital input 1 was defined as experimentally optimized concentrations of the substrates. The first system (Figure 2A) operated as a cascade of reactions catalyzed by three enzymes – maltose phosphorylase, hexokinase and glucose-6-phosphate dehydrogenase. It mimicked three concatenated Boolean AND logic gates (Figure 2C), and the high output signal (production of NADH) was observed only in the presence of all four input substrates (see legend to Figure 2). The second system (Figure 2B) operated as a 3-input OR gate connected to an AND gate (Figure 2D). For this system, the NADH production was activated in the presence of any of substrate inputs A, B, C with the mandatory presence of NAD+ (input D). Enzymatically formed NADH reacted with the PQQ-electrode producing a negative potential and re-oxidized to the NAD+ state (Figure 1).
Figure 2.
Two enzyme systems used in this study and their corresponding logic schemes. A) A cascade of three AND gates made of maltose phosphorylase (MPh; E.C. 2.4.1.8), hexokinase (HK; E.C. 2.7.1.1), and glucose-6-phosphate dehydrogenase (G6PDH; E.C. 1.1.1.49). The biocatalytic reaction of MPh was activated in the presence of maltose (input A) and inorganic phosphate (Pi, input B) resulting in glucose (Glc) and glucose-1-phosphate byproduct formation. In the next reaction step catalyzed by HK Glc is converted to glucose-6-phosphate (Glc6P) in the presence of ATP (input C). Finally, Glc6P reduces NAD+ (Input D) to NADH in the process biocatalyzed by G6PDH. Overall, the NADH production is only possible in the presence of all 4 input signals activating the enzyme-based system. B) A combination of three parallel reactions biocatalyzed by three NAD+-dependent enzymes: glucose dehydrogenase (GDH; E.C. 1.1.1.47), G6PDH and alcohol dehydrogenase (AlcDH; E.C. 1.1.1.1). Each biocatalytic reaction was activated by the corresponding substrate: Glc (Input A), Glc6P (Input B) and ethanol (Et-OH), (Input C). The NAD+ cofactor (input D) was needed for all reactions, thus none of them could proceed in the absence of NAD+. C-D) The logic schemes corresponding to the biocatalytic cascades are shown in A and B, respectively.
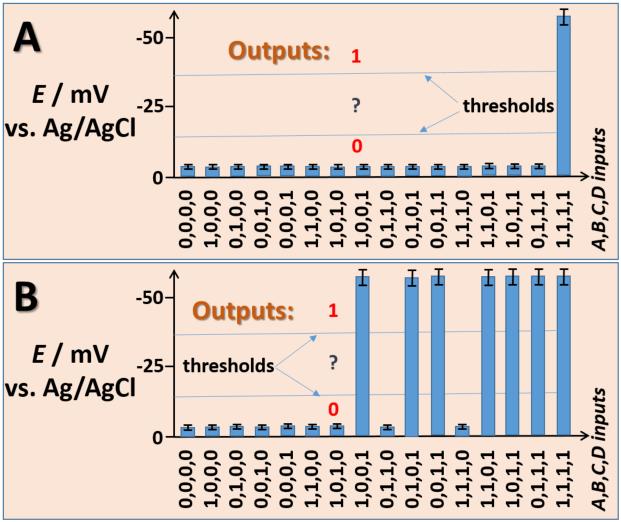
Figure 3 demonstrates the correct digital behavior of the enzymatic logic gate systems interacting with the PQQ-electrode. A negative potential of ca. −60 mV (digital 1) was achieved when NADH was produced by either of the enzyme logic systems. Otherwise, the potential less negative than −10 mV (digital 0) was measured (Figure 3). For the enzyme logic system mimicking three concatenated AND gates (Figure 2A,C), logic output 1 (ca. −60 mV) was measured only in the presence of all reacting input species (input combination 1,1,1,1). All other input combinations (15 different variants) resulted in the electrode potential less negative than −10 mV, digital 0 (Figure 3A). Alternatively, operation of the enzyme system mimicking a 3-input OR gate followed by an AND gate (Figure 2 B,D) resulted in output 1 (ca. −60 mV) generated in the following input combinations (A,B,C,D): 0,0,1,1; 0,1,0,1; 1,0,0,1; 0,1,1,1; 1,0,1,1; 1,1,0,1; 1,1,1,1, while all other input combinations resulted in output signal 0 (Fig. 3B).
Figure 3.
A-B) Electric potentials generated on the PQQ-modified electrode interfaced with the biocatalytic systems shown in Figures 2A and 2B, respectively, when different combinations of input signals were applied. The bars show the potential values achieved after 30 min of exposing the PQQ-modified electrode to the enzyme systems. The data are average of three independent experiments. The potential produced on the PQQ-modified electrode has a logarithmic dependence on the NADH concentration (according to the Nernst equation), thus resulting in very small variations of the measured potentials. Threshold lines separate logic output 0, undefined area and logic output 1.
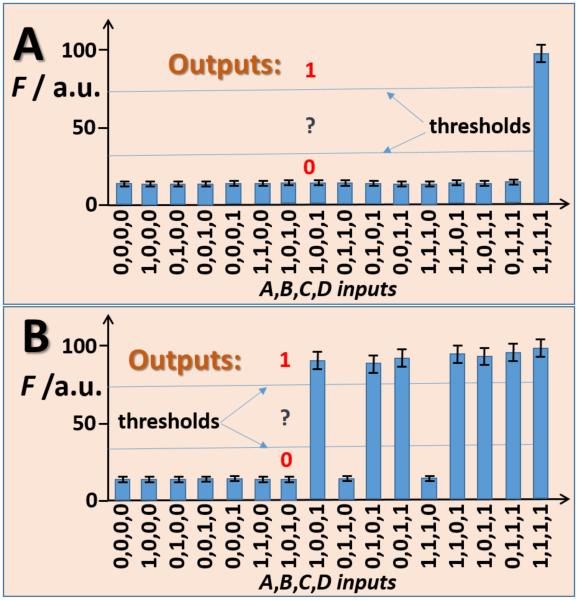
To enable transfer of the output signal produced by the enzyme computing systems (Figure 3) into a DNA input signal, the PQQ-electrode was connected to another electrode, which was coated with Fe3+-alginate film entrapping a fluorescently labeled DNA oligonucleotide output OP1 (FITC-5’-TGC AGA CGT TGA AGG ATC CTC). Generation of the negative potential on the PQQ-electrode resulted in subsequent reduction of Fe3+ into Fe2+ on the Fe3+-alginate-coated electrode. It triggered the alginate film dissolution and OP1 release. It was observed that when the potential of ca. −60 mV (digital 1 output of the enzymatic computing systems) was applied to the second electrode, the alginate film was substantially degraded (Figure S3). At the same time, no visible changes in the film structure were observed at the potential of ca. −5 mV (digital 0 output of the enzyme computing systems), on the same experimental time-scale (data not shown). Fluorescent signal of the solution containing released OP1 was measured in the presence of different combinations of enzymatic system inputs (Figure 4). As expected, high fluorescence (digital 1) was registered upon the electrochemically stimulated release of OP1 in the presence of NADH. When no NADH was produced, the fluorescent signal remained low (digital 0). The concentration of the released (digital 1) and leaking (digital 0) OP1 was reaching ca. 5 nM and 0.8 nM, respectively, after 30 min. This result shows significant discrimination between the leakage and stimulated release of OP1 entrapped in the Enz/DNA interface. It should be noted that there is perfect correlation between the output signals produced by the enzymatic systems in the form of the potentials (Figure 3) with the fluorescence of released OP1 (Figure 4). In other words, the enzyme-generated output was consistently converted into OP1, which served as input for DNA computing as detailed below.
Figure 4.
A-B) Fluorescence signal corresponding to the dye-labelled oligonucleotide OP1 released from the alginate thin-film when the PQQ-modified electrode was interfaced with the enzymatic logic gate systems shown in Figures 2A and 2B, respectively, when different combinations of input signals were applied. The bars show the fluorescence measured after 30 min of exposing the electrodes to the enzyme systems. The fluorescence is represented by normalized arbitrary values. The data are average of three independent experiments. Threshold lines separate logic output 0, undefined area and logic output 1.
Oligonucleotide OP1 released by the interface was recognized by a 3-input deoxyribozyme AND gate (3iAND) (Figure 5). Deoxyribozyme logic gates controlled by DNA oligonucleotide inputs are most well-developed DNA logic constructs up to date.[20-22] Indeed, such gates can be assembled in automaton that plays tic-tac-toe game with human,[20b] they can be organized in multi-layer computational cascades[21] and a molecular calculator with 7-segment digital display.[22] The design of 3iAND takes advantage of the concept of split (binary) deoxyribozyme sensors[23] and consists of two DNA strands folded in the stem-loop structures (3iANDa and 3iANDb in Figure 5A). The strands are dissociated in the absence of input oligonucleotides. However, hybridization of two oligonucleotide inputs to the loop fragments opens the hairpins and the third input bridges 3iANDa and 3iANDb, which results in the formation of a catalytic core (Figure 5B). The deoxyribozyme cleaves a fluorophore- and quencher-labeled substrate (F substrate in Figure 5), thus producing high fluorescence. In this study, OP1, an output of the Enz/DNA interface, was used as a bridging input for 3iAND. In experiments with 3iAND gates, OP1 without fluorescent label was used. Two other inputs (I1 and I2) mimicked the sequences of microRNAs shown to be promising molecular markers of human cancers.[24]
Figure 5.
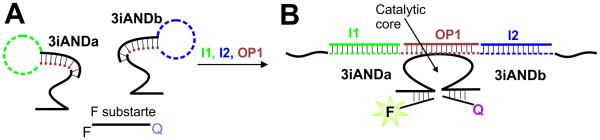
Principal scheme of a three-input deoxyribozyme AND gate (3iAND). A) Strands 3iANDa and 3iANDb of the gate are dissociated in the absence of inputs. Dashed lines indicate the input-recognition fragments of the strands. B) Catalytic Dz complex formed in the presence of all three inputs (I1, OP1, and I2). The Dz catalytic core cleaves the fluorophore- and quencher-labelled F substrate and increases fluorescent signal. Note that OP1 is not fluorescently labelled.
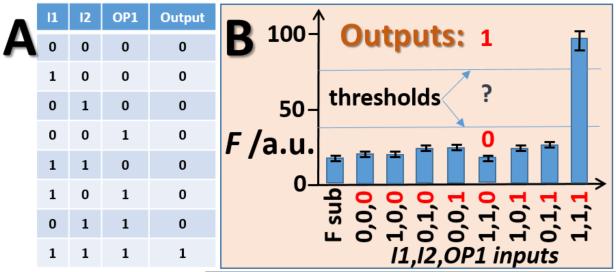
According to the truth table (Figure 6A), 3iAND produces high fluorescence output (digital 1) only in the presence of all 3 inputs. In our experiments, the OP1 input was produced in situ by the stimulated release (digital 1) or leakage (digital 0) from the alginate-modified electrode and its concentration was set by the system as a function of logic operation of the enzyme systems. Two other inputs, I1 and I2, were either used in concentration of 10 nM or absent for digital 1 and 0, respectively.
Figure 6.
Digital performance of the 3iAND gate. A) Truth table; B) Fluorescent response of 3iAND in the presence of all possible DNA input combinations. The concentrations of the inputs were as follows. For low inputs (digital 0): I1, 0 nM; I2, 0 nM; OP1 was produced in situ with the concentration set by the system corresponding to output 0 (when substrate inputs A,B,C,D for the enzyme systems were 0,0,0,0). For high inputs (digital 1): I1, 10 nM; I2, 10 nM; OP1 was produced in situ with the concentration set by the system corresponding to output 1 (when inputs A,B,C,D for the enzyme systems were 1,1,1,1). Digital values for OP1 are shown in red. F sub bar, a control sample containing only fluorescent substrate (F substrate, see Figure 5). The bars show the fluorescence measured after 30 min of exposing the electrode to the enzyme systems. The fluorescence is represented by normalized arbitrary values. The data are average of three independent experiments. Threshold lines separate logic output 0, undefined area and logic output 1.
The full logic network includes 6 independent logic inputs: 4 inputs (A, B, C, D) in the enzyme part and 2 inputs in the DNA part (I1 and I2), while OP1 is not an independent input. Therefore, the full truth table includes 26 = 64 variants of logic input combinations. Figure 6 shows a simplified representation of the logic process considering only the DNA logic part. Logic value 0 and 1 for the intermediate output/input OP1 can be realized with various combinations of the enzyme-inputs A, B, C, D. For simplicity and for minimizing number of experiments we used A, B, C, D enzyme inputs in combinations 0,0,0,0 and 1,1,1,1 for realizing the OP1 digital values 0 and 1, respectively. This simplification is justified by very small signal variations of for all combinations of the A, B, C, D inputs generating either by output 0 or 1 (Figure 4). In other words, the leakage of OP1 and the release of OP1 are almost the same regardless of the input combinations.
The correct digital response of 3iAND was registered at all possible DNA input combinations (Figure 6B). Importantly, the high output signal (last bar in Figure 6B) could be statistically distinguished from the low output (about 4-fold fluorescence increase, see also Figure S3 for raw fluorescent data). The fluorescent results were supported by the analysis of the samples by gel electrophoresis (Figure S4). The data proves the expected digital response of 3iAND and the possiblity of using an electrode–released oligonucleotide for transfering the signal from enzymatic to DNA computational systems.
This study demonstrates the possibility to design an interface that enables communication between enzymatic and DNA-based computing systems. The whole system includes two individual logic sub-systems (enzyme-based and DNA-based) connected electrically to allow the output signal produced by the enzyme logic gates operate as the input signal for the DNA logic gates. The system operated in two distinct steps, first the enzyme-logic process and then the DNA logic process (see the step-by-step process description in the Supporting Information). To the best of our knowledge, the system reported here is the first Enz/DNA interface that connects a non-DNA processing enzyme computation system with DNA logic gates. We call this interface ‘universal’ because it is compatible with a variety of both enzymatic and DNA molecular logic circuits. NADH communicating between the enzyme system and the interface electrode allows great versatility for the selection of enzymes participating in the biocomputing process, since NADH is produced in a broad variety of reactions. In addtion, it is possible to replace NADH with other reducing molecules (e.g., glucose).[25] The deoxyribozyme gate-based computational systems are also known to show great versatility and complexity.[20-22] The limitations of the interface are the following. (i) The enzyme-based computing system must produce NADH or other reductive species as an output. (ii) The DNA-based computing system must accept nM-range concentration of oligonucleotide as an input. However, the amount of the released DNA could be increased if larger electrodes or thicker alginate films are used for the DNA entrapment and release. (iii) In its current design, the signal can be transferred in only one direction: from the enzyme to DNA system. (iv) Only one kind of DNA (or a set of DNA sequences) can be released per an electrode pair. More DNA outputs could be released in the controlled way, if a multi-electrode array is applied. Despite the limitations, the reported Enz/DNA system can find some important practical applications. Indeed, the enzymatic and DNA-based computing systems used in this study proved to be relevant to diagnosis of human diseases,[3,26] as well as to very complex information processing.[20-22] The reported data represnt ouputs after the system came to the saturation (the end of the process) similarly to most other studies in the field.[5f-h,7] It would be interesting to study the time-dependence of output production. The time-dependent outputs were studied experimentally and modelled theoretically for some multi-step biocatalytic reactions applied for logic operations.[27] Also time-dependent dissolution of alginate thin-film and concomitant release of loaded substances were reported recently.[28] The present system includes a number of processes with complex kinetics (biocatalytic cascades, electric potential formation, reductive dissolution of alginate, OP1 release and finally DNA reactions). Study of the combination of these time-dependent processes and their kinetics could become the subject of subsequent invesigation. It should be also noted that the alginate film dissolution and subsequent DNA release could be achieved in none-electrochemical systems using direct chemical communication with enzyme logic systems. Similar release processes have been studied using enzyme systems producing citrate as the final output.[29] However, in this case the choice of potentially useful enzymes is very limited, and EnzDNA interface is not as general as the one reported in this study.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (R15AI10388001A1) and NSF CCF (24066076) to DMK and NSF CBET (1403208) to EK.
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- [1].a) Qian L, Winfree E, Bruck J. Nature. 2011;475:368–372. doi: 10.1038/nature10262. [DOI] [PubMed] [Google Scholar]; b) Genot AJ, Fujii T, Rondelez Y. J. R. Soc. Interface. 2013;10:20130212. doi: 10.1098/rsif.2013.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Goldental A, Guberman S, Vardi R, Kanter I. Front. Comput. Neurosci. 2014;8:52. doi: 10.3389/fncom.2014.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].a) Wang D, Fu Y, Yan J, Zhao B, Dai B, Chao J, Liu H, He D, Zhang Y, Fan C, Song S. Anal. Chem. 2014;86:1932–1936. doi: 10.1021/ac403661z. [DOI] [PubMed] [Google Scholar]; b) Gil B, Kahan-Hanum M, Skirtenko N, Adar R, Shapiro E. Nano Lett. 2011;11:2989–2996. doi: 10.1021/nl2015872. [DOI] [PubMed] [Google Scholar]; c) Hemphill J, Deiters A. J. Am. Chem. Soc. 2013;135:10512–10518. doi: 10.1021/ja404350s. [DOI] [PubMed] [Google Scholar]
- [3].a) Konry T, Walt DR. J. Am. Chem. Soc. 2009;131:13232–1323. doi: 10.1021/ja905125b. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cornett EM, Campbell EA, Gulenay G, Peterson E, Bhaskar N, Kolpashchikov DM. Angew. Chem. Int. Ed. Engl. 2012;51:9075–9077. doi: 10.1002/anie.201203708. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2012;124:9209–9211. [Google Scholar]; c) Gerasimova YV, Kolpashchikov DM. Chem. Commun. 2015;51:870–8722. doi: 10.1039/c4cc08241a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Pei R, Matamoros E. Chem. Commun. 2015;51:870–872. [Google Scholar]; Liu M, Stefanovic D, Stojanovic MN. Nature Nanotechnol. 2010;5:773–777. doi: 10.1038/nnano.2010.194. [DOI] [PubMed] [Google Scholar]; b) Stojanovic MN, Stefanovic D, Rudchenko S. Acc. Chem. Res. 2014;47:1845–1852. doi: 10.1021/ar5000538. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Stojanovic MN, Stefanovic D. J. Comput. Theor. Nanosci. 2011;8:434–440. [Google Scholar]; d) Benenson Y. Nature Rev. Genetics. 2012;13:455–468. doi: 10.1038/nrg3197. [DOI] [PubMed] [Google Scholar]; e) Kahan M, Gil B, Adar R, Shapiro E. Physica D. 2008;237:1165–1172. [Google Scholar]; f) Privman V. Nature Nanotechnol. 2010;5:767–768. doi: 10.1038/nnano.2010.221. [DOI] [PubMed] [Google Scholar]; g) Katz E, editor. Biomolecular Computing – From Logic Systems to Smart Sensors and Actuators. Willey-VCH; Weinheim, Germany: 2012. [Google Scholar]
- [5].a) De Silva AP, Uchiyama S, Vance TP, Wannalerse B. Coord. Chem. Rev. 2007;251:1623–1632. [Google Scholar]; b) Szacilowski K. Chem. Rev. 2008;108:3481–3548. doi: 10.1021/cr068403q. [DOI] [PubMed] [Google Scholar]; c) Credi A. Angew. Chem. Int. Ed. 2007;46:5472–5475. doi: 10.1002/anie.200700879. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2007;119:5568–5572. [Google Scholar]; d) Privman V. Israel J. Chem. 2011;51:118–131. [Google Scholar]; e) Andreasson J, Pischel U. Chem. Soc. Rev. 2010;39:174–188. doi: 10.1039/b820280j. [DOI] [PubMed] [Google Scholar]; f) Katz E, editor. Molecular and Supramolecular Information Processing: From Molecular Switches to Logic Systems. Wiley-VCH; Weinheim, Germany: 2012. [Google Scholar]; g) Szacilowski K. Infochemistry. Wiley, Chichester; 2012. [Google Scholar]; h) de Silva AP. Molecular Logic-Based Computation. Royal Society of Chemistry; Cambridge: 2013. [Google Scholar]
- [6].a) Calude CS, Costa JF, Dershowitz N, Freire E, Rozenberg G, editors. Lecture Notes in Computer Science. Vol. 5715. Springer; Berlin: 2009. Unconventional Computation. [Google Scholar]; b) Adamatzky A, De Lacy Costello B, Bull L, Stepney S, Teuscher C, editors. Unconventional Computing. Luniver Press; Frome, UK: 2007. [Google Scholar]
- [7].Katz E, Privman V. Chem. Soc. Rev. 2010;39:1835–1857. doi: 10.1039/b806038j. [DOI] [PubMed] [Google Scholar]
- [8].a) Benenson Y. Mol. Biosyst. 2009;5:675–685. doi: 10.1039/b902484k. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Stojanovic MN. Isr. J. Chem. 2011;51:99–105. [Google Scholar]
- [9].Adleman LM. Science. 1994;266:1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- [10].Santini CC, Bath J, Turberfield AJ, Tyrrell AM. Int. J. Mol. Sci. 2012;13:5125–5137. doi: 10.3390/ijms13045125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].a) Schlosser K, Li Y. Chem. Biol. 2009;16:311–322. doi: 10.1016/j.chembiol.2009.01.008. [DOI] [PubMed] [Google Scholar]; b) Toh SY, Citartan M, Gopinath SC, Tang TH. Biosens. Bioelectron. 2014;64C:392–403. doi: 10.1016/j.bios.2014.09.026. [DOI] [PubMed] [Google Scholar]
- [13].Radzicka A, Wolfenden R. Science. 1995;267:90–931. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- [14].a) Privman V, Zavalov O, Halámková L, Moseley F, Halámek J, Katz E. J. Phys. Chem., B. 2013;117:14928–14939. doi: 10.1021/jp408973g. [DOI] [PubMed] [Google Scholar]; b) Privman V, Arugula MA, Halámek J, Pita M, Katz E. J. Phys. Chem., B. 2009;113:5301–5310. doi: 10.1021/jp810743w. [DOI] [PubMed] [Google Scholar]; c) Privman V, Strack G, Solenov D, Pita M, Katz E. J. Phys. Chem. B. 2008;112:11777–11784. doi: 10.1021/jp802673q. [DOI] [PubMed] [Google Scholar]
- [15].a) Benenson Y, Paz-Elizur T, Adar R, Keinan E, Livneh Z, Shapiro E. Nature. 2001;414:430–434. doi: 10.1038/35106533. [DOI] [PubMed] [Google Scholar]; b) Kolpashchikov DM, Stojanovic MN. J. Am. Chem. Soc. 2005;127:11348–11351. doi: 10.1021/ja051362f. [DOI] [PubMed] [Google Scholar]; c) Han D, Zhu Z, Wu C, Peng L, Zhou L, Gulbakan B, Zhu G, Williams KR, Tan W. J. Am. Chem. Soc. 2012;134:20797–20804. doi: 10.1021/ja310428s. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Beyer S, Simmel FC. Nucleic Acids Res. 2006;34:1581–1587. doi: 10.1093/nar/gkl075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].a) Katz E, Wang J, Privman M, Halámek J. Anal. Chem. 2012;84:5463–5469. doi: 10.1021/ac3007076. [DOI] [PubMed] [Google Scholar]; b) Wang J, Katz E. Isr. J. Chem. 2011;51:141–150. [Google Scholar]; c) Halámková L, Halámek J, Bocharova V, Wolf S, Mulier KE, Beilman G, Wang J, Katz E. Analyst. 2012;137:1768–1770. doi: 10.1039/c2an00014h. [DOI] [PubMed] [Google Scholar]; d) Halámek J, Bocharova V, Chinnapareddy S, Windmiller JR, Strack G, Chuang M-C, Zhou J, Santhosh P, Ramirez GV, Arugula MA, Wang J, Katz E. Molec. Biosys. 2010;6:2554–2560. doi: 10.1039/c0mb00153h. [DOI] [PubMed] [Google Scholar]
- [17].Moore ANJ, Katz E, Willner I. Electroanalysis. 1996;8:1092–1094. [Google Scholar]
- [18].Katz E, Lötzbeyer T, Schlereth DD, Schuhmann W, Schmidt H-L. J. Electroanal. Chem. 1994;373:189–200. [Google Scholar]
- [19].a) Mailloux S, Halámek J, Katz E. Analyst. 2014;139:982–986. doi: 10.1039/c3an02162a. [DOI] [PubMed] [Google Scholar]; b) Mailloux S, Guz N, Zakharchenko A, Minko S, Katz E. J. Phys. Chem. B. 2014;118:6775–6784. doi: 10.1021/jp504057u. [DOI] [PubMed] [Google Scholar]; c) Mailloux S, Guz N, Gamella Carballo M, Pingarrón JM, Katz E. Anal. Bioanal. Chem. 406. 2014:4825–4829. doi: 10.1007/s00216-014-7936-z. [DOI] [PubMed] [Google Scholar]; d) Jin Z, Güven G, Bocharova V, Halámek J, Tokarev I, Minko S, Melman A, Mandler D, Katz E. ACS Appl. Mater. Interfaces. 2012;4:466–475. doi: 10.1021/am201578m. [DOI] [PubMed] [Google Scholar]
- [20].a) Stojanovic MN, Mitchell TE, Stefanovic D. J. Am. Chem. Soc. 2002;124:3555–3561. doi: 10.1021/ja016756v. [DOI] [PubMed] [Google Scholar]; b) Stojanovic MN, Stefanovic D. Nature Biotechnol. 2003;21:1069–1074. doi: 10.1038/nbt862. [DOI] [PubMed] [Google Scholar]; c) Macdonald J, Li Y, Sutovic M, Lederman H, Pendri K, Lu W, Andrews BL, Stefanovic D, Stojanovic MN. Nano Lett. 2006;6:2598–2603. doi: 10.1021/nl0620684. [DOI] [PubMed] [Google Scholar]
- [21].a) Stojanovic MN, Semova S, Kolpashchikov D, Macdonald J, Morgan C, Stefanovic D. J. Am. Chem. Soc. 2005;127:6914–6915. doi: 10.1021/ja043003a. [DOI] [PubMed] [Google Scholar]; b) Yashin R, Rudchenko S, Stojanovic MN. J. Am. Chem. Soc. 2007;129:15581–15584. doi: 10.1021/ja074335t. [DOI] [PubMed] [Google Scholar]; c) Brown CW, 3rd, Lakin MR, Horwitz EK, Fanning ML, West HE, Stefanovic D, Graves SW. Angew. Chem. Int. Ed. Engl. 2014;53:7183–7187. doi: 10.1002/anie.201402691. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2014;126:7311–7315. [Google Scholar]; d) Brown CW, 3rd, Lakin MR, Stefanovic D, Graves SW. Chembiochem. 2014;15:950–954. doi: 10.1002/cbic.201400047. [DOI] [PubMed] [Google Scholar]
- [22].Poje JE, Kastratovic T, Macdonald AR, Guillermo AC, Troetti SE, Jabado OJ, Fanning ML, Stefanovic D, Macdonald J. Angew. Chem. Int. Ed. Engl. 2014;53:9222–9225. doi: 10.1002/anie.201402698. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014;126:9376–9379. [Google Scholar]
- [23].Kolpashchikov DM. Chembiochem. 2007;8:2039–2042. doi: 10.1002/cbic.200700384. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gerasimova YV, Cornett E, Kolpashchikov DM. Chembiochem. 2010;11:811–817. doi: 10.1002/cbic.201000006. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Mokany E, Bone SM, Young PE, Doan TB, Todd AV. J. Am. Chem. Soc. 2010;132:1051–1059. doi: 10.1021/ja9076777. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Gerasimova YV, Kolpashchikov DM. Chem. Biol. 2010;104:106. doi: 10.1016/j.chembiol.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu Z, Sall A, Yang D. Int. J. Mol. Sci. 2008;9:978–999. doi: 10.3390/ijms9060978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mailloux S, Halámek J, Halámková L, Tokarev A, Minko S, Katz E. Chem. Commun. 2013;49:4755–4757. doi: 10.1039/c3cc42027b. [DOI] [PubMed] [Google Scholar]
- [26].a) Gerasimova YV, Cornett EM, Edwards E, Su X, Rohde KH, Kolpashchikov DM. ChemBioChem. 2013;14:2087–2090. doi: 10.1002/cbic.201300471. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gerasimova YV, Kolpashchikov DM. Angew. Chem. Int. Ed. Engl. 2013;52:10586–10588. doi: 10.1002/anie.201303919. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2013;125:10780–10782. [Google Scholar]
- [27].a) Privman V, Domanskyi S, Mailloux S, Holade Y, Katz E. J. Phys. Chem. B. 2014;118:12435–12443. doi: 10.1021/jp508224y. [DOI] [PubMed] [Google Scholar]; b) Halámek J, Bocharova V, Arugula MA, Strack G, Privman V, Katz E. J. Phys. Chem. B. 2011;115:9838–9845. doi: 10.1021/jp2041372. [DOI] [PubMed] [Google Scholar]
- [28].Gamella M, Guz N, Mailloux S, Pingarrón JM, Katz E. ACS Appl. Mater. Interfaces. 2014;6:13349–13354. doi: 10.1021/am504561d. [DOI] [PubMed] [Google Scholar]
- [29].Mailloux S, Zavalov O, Guz N, Katz E, Bocharova V. Biomaterials Science. 2014;2:184–191. doi: 10.1039/c3bm60197h. [DOI] [PubMed] [Google Scholar]; Bocharova V, Zavalov O, MacVittie K, Arugula MA, Guz NV, Dokukin ME, Halámek J, Sokolov I, Privman V, Katz E. J. Mater. Chem. 2012;22:19709–19717. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.