Abstract
X-ray crystallography, molecular dynamics (MD) simulations and biochemistry were utilized to investigate the effect of introducing hydrophobic interactions in the 4-fold (N148L and Q151L) and B-pores (D34F) of Pseudomonas aeruginosa bacterioferritin B (BfrB) on BfrB function. The structures show only local structural perturbations and confirm the anticipated hydrophobic interactions. Surprisingly, structures obtained after soaking crystals in Fe2+-containing crystallization solution revealed that although iron loads into the ferroxidase centers of the mutants, the side chains of ferroxidase ligands E51 and H130 do not reorganize to bind the iron ions, as is seen in the wt BfrB structures. Similar experiments with a double mutant (C89S/K96C) prepared to introduce changes outside the pores show competent ferroxidase centers that function akin to those in wt BfrB. MD simulations comparing wt BfrB with the D34F and N148L mutants show that the mutants exhibit significantly reduced flexibility, and reveal a network of concerted motions linking ferroxidase centers and 4-fold and B-pores, which are important for imparting ferroxidase centers in BfrB with the required flexibility to function efficiently. In agreement, the efficiency of Fe2+ oxidation and uptake of the 4-fold and B-pore mutants in solution is significantly compromised relative to wt or C89S/K96C BfrB. Finally, our structures show a large number of previously unknown iron binding sites in the interior cavity and B-pores of BfrB, which reveal in unprecedented detail conduits followed by iron and phosphate ions across the BfrB shell, as well as paths in the interior cavity that may facilitate nucleation of the iron phosphate mineral.
Introduction
Iron, an essential nutrient for pathogenic bacteria, can also stimulate the formation of reactive oxygen species via the Haber Weiss cycle, in which free iron catalyzes the conversion of hydrogen peroxide and superoxide to the highly toxic hydroxyl radical (1, 2). Consequently, free levels of iron in bacteria are tightly regulated to ensure sufficiency for metabolic needs, while preventing iron-induced oxidative toxicity. To maintain iron homeostasis, pathogens must balance the need to obtain iron from their host with careful management of intracellular iron levels, which includes storage of iron reserves for subsequent utilization when the nutrient becomes scarce (2, 3). Bacteria have evolved two types of protein for storing iron, ferritin (Ftn) and bacterioferritin (Bfr); the latter is unique to bacteria (2, 4). The significance of bacterial Ftn and Bfr in the life cycle and virulence of pathogens is just beginning to emerge, as reflected in recent findings with Bfr and Ftn mutants of Mycobacterium tuberculosis, which are highly susceptible to antibiotics and unable to persist in mouse and guinea pig models of infection (5, 6). In the plant pathogen Erwinia chrysanthemi mutation of the bfr gene results in impaired iron utilization and growth defects (7).
The subunit architecture of eukaryotic Ftns, bacterial Ftns and Bfrs is highly conserved and consists of five α-helices (A-E) arranged in a four-helix bundle (A-D) and a short helix (E) that lies nearly perpendicular to the central axis of the bundle; helices B and C are connected by a long loop that traverses the length of the four-helix bundle. Although the structures of Bfrs are similar to those of Ftns in terms of the overall architecture of the subunits and how they assemble together into 24-mers, their amino acid sequences exhibit little homology (< 18%) (8, 9). In addition, Bfrs are unique in possessing intrinsic heme groups, which are bound at two-fold symmetric inter-subunit sites by coordinative interactions with methionine residues from adjacent subunits. (Figure 1A). Twenty four subunits and 12 hemes assemble into a spherical and hollow structure (Figure 1B) with an outer diameter of ∼120 Å and an inner diameter of ∼80 Å, where up to ∼3,500 iron atoms can be stored in the form of a Fe3+ mineral(10). Details pertaining the self-assembly and stability of the 24-mer bacterioferritin shells are beginning to emerge (11, 12). The formation of an iron core (iron uptake) requires binding of ferrous iron (Fe2+) to a ferroxidase catalytic site, where it is oxidized to the ferric (Fe3+) state (10, 13, 14), and then translocated to the interior cavity. Reincorporation of iron in metabolism requires accepting electrons to reduce Fe3+ in the internal cavity and releasing Fe2+ to the bacterial cytosol (15, 16). This dual function (iron uptake and iron release) enables an equilibrium that regulates the range of cytosolic Fe2+ concentrations that allow Fur (Fe uptake repressor) to perform a broad range of regulatory functions linking iron homeostasis to broader bacterial metabolism (17-19). Pseudomonas aeruginosa has two ferritin-like molecules, a bacterial ferritin (FtnA), and a bacterioferritin (BfrB) (20). The release of iron stored in BfrB involves reduction of the Fe3+ mineral, which requires that electrons originating in NADPH are shuttled to BfrB by way of an NADPH ferredoxin reductase (FPR) and a ferredoxin, termed bacterioferritin-associated ferredoxin (Bfd) (16). The X-ray crystal structure of the BfrB-Bfd complex revealed that residues at the complementary BfrB-Bfd interface are highly conserved in Bfr and Bfd sequences from a number of pathogenic bacteria, suggesting that the BfrB/Bfd interaction is important in the regulation of cytosolic iron concentrations in gram-negative pathogens (15).
Figure 1.
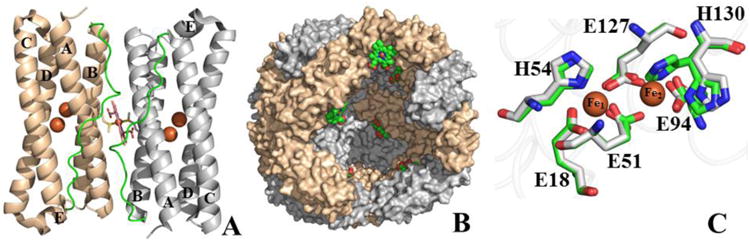
Structural organization of BfrB (PDB:3IS8). (A) Subunit dimer and associated heme; each subunit is composed of a four-helix bundle (A-D), a perpendicular short helix (E), and a loop connecting the B and C helices (green). The location of the two iron ions in the ferroxidase center of each subunit is shown as orange spheres. (B) Cross sectional view of the 24-mer assembly illustrating the internal cavity where mineral iron is stored; heme molecules are shown in sphere representation (green). (C) Overlay of ferroxidase center residues from the as-isolated (gray) and Fe-soaked (green) wt BfrB structures, illustrating the conformational rearrangements of E51 and H130 upon binding iron.
In Bfrs the ferroxidase catalytic centers are located in the middle of each subunit (Figure 1A and C). In the structure of as-isolated recombinant wt BfrB, the ferroxidase centers are devoid of iron, and although most of the ferroxidase residues are poised to bind iron, the side chain of H130 is rotated away; we termed this conformation of H130 “gate open” (gray in Figure 1C) (21). Soaking crystals of as-isolated wt BfrB in Fe2+ solution causes iron to load onto the ferroxidase center and the H130 side chain is seen in two conformations, one coordinated to Fe2 (“gate closed”) and the other in the “gate open” state (Figure 1C) (21). These structural observations, together with findings obtained from iron incorporation studies in solution, support the idea of a gating mechanism in BfrB, where a ferroxidase channel allows entry of Fe2+ from the protein exterior to the ferroxidase center, where a di-Fe2+ moiety is coordinated by the ferroxidase ligands, including H130. Oxidation to di-Fe3+ is thought to signal H130 to adopt the gate open conformation and allow Fe3+ translocation from the ferroxidase center to the internal cavity (21).
The structures of ferritin-like molecules suggest that in addition to the ferroxidase pores, the protein interior communicates with the exterior via 8 three-fold pores, 6 four-fold pores, and 24 B-pores (Figures 2A-B). The 3-fold pores assemble at the intersection of 3 subunits and in BfrB are formed by layers of alternating positively and negatively charged residues (Figure 2C) (21). The 4-fold pores, which assemble at the intersection of 4-subunits, are lined by the corresponding E-helices; their narrowest portion is composed of two layers made by the side chains of four N148 and four Q151 (Figure 2D). Bacterial Ftns and Bfrs also have B-pores, which are formed at the intersection of three subunits not aligned with an axis of symmetry. Four B-pores surround each 4-fold pore, such that there are 24 B-pores in the 24-mer. In BfrB the narrowest section of a B-pore is defined by D132 and T136 from one subunit, and D34 from a different subunit (Figure 2E). It has been noted that the size of B-pores may allow the traffic of Fe2+ across the Bfr shell (22, 23).
Figure 2.
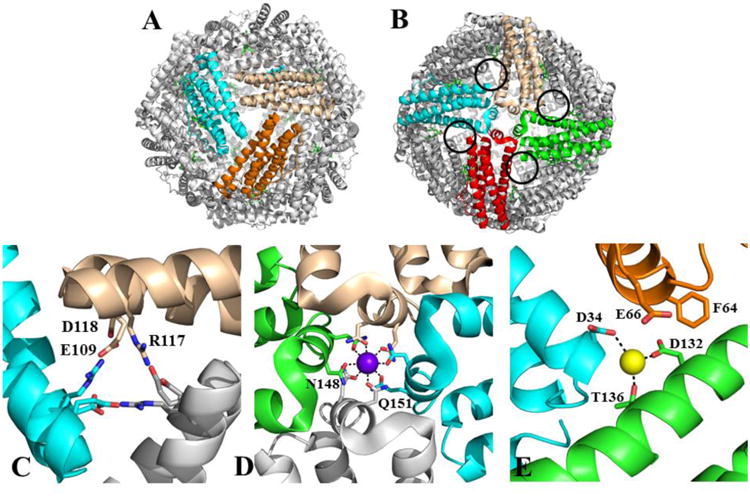
A 24-mer BfrB viewed (A) down one of the eight 3-fold pores, and (B) down one of the six 4fold pores; four B-pores surrounding a 4-fold pore are highlighted by black circles. (C) Zoomed-in view of a 3-fold pore illustrating the alternating arrangement of positively and negatively charged residues, (D) a 4-fold pore and associated potassium ion (purple sphere) coordinated by the side chains of N148 and Q151, and (E) a B-pore and associated sodium ion (yellow sphere) coordinated by D34, D132 and T136.
The low amino acid sequence conservation among Ftns, bacterial Ftns and Bfrs (8, 9) profoundly influence packing of the protein shell, charge distribution inside the pores, and possibly pore function: The 3-fold pores of eukaryotic Ftns are lined with negatively charged residues (24), which are thought to function as conduits for iron across the eukaryotic Ftn shell (25, 26). A similar function, however, is not obvious for the 3-fold pores in Bfrs, which have a layered structure of alternating positive and negative charge. The 4-fold pores of eukaryotic Ftn are lined with hydrophobic residues, but contain hydrophilic amino acids in BfrB (Figure 2D). In reported BfrB structures, a cation has been observed in the pores, which is best modeled as potassium (21) and sodium (15), whereas in two distinct structures of Azotobacter vinelandii Bfr the cation was modeled as a barium (27) or iron (23). Eukaryotic Ftns lack B-pores, whereas the 24 B-pores in BfrB are lined with hydrophilic and negatively charged residues (Figure 2E). Moreover, in molecular dynamics (MD) simulations carried out with wt BfrB, K+ ions were seen to traffic in and out of the BfrB shell via B- and ferroxidase pores, but not through the 3-fold or 4-fold pores (28). The MD simulations also suggest that movement of K+ through B-pores is enabled by periodic kinking/straightening, as well as folding/unfolding of the C-terminal half of helices D, and that these fluctuations are coupled to lateral oscillations of the short E-helices that constitute the 4-fold pores, such that the aggregate of the motions is a periodic expansion and contraction of the B-pores. More recently, elastic network models were used to predict extensive networks of highly correlated residues connecting ferroxidase centers with B- and 4-fold pores in Bfr and bacterial Ftn, leaving the 3-fold pores disconnected from the grid, while in contrast a network of correlated residues in eukaryotic Ftn connects ferroxidase centers and 3-fold pores (9). These predictions are not only in agreement with the putative distinct function of the different pores in Bfr and eukaryotic Ftn, but also support the idea that cooperative dynamics of the 24-mer architecture are pivotal for iron traffic and iron handling inside the ferritin cavity.
Despite the significant advances made toward understanding Bfr function, many important questions remain unanswered. For example, it is not yet clear how ions (positive and negative) enter or exit the interior cavity through the protein shell, or how these ions are “handled” in the protein interior. It is also not known how the dynamics of individual subunits and 24-mer assemblies contribute to the function of Bfr and the traffic of ions across the protein shell. In an attempt to provide answers to these questions, in this work we interrogated the effect that mutations in the 4-fold and B-pores may exert in the structure, iron uptake efficiency and dynamics of BfrB. N148 and Q151, the residues in helices E that line the 4-fold pores and normally coordinate K+ or Na+ (Figure 2D) were each replaced by leucine, and D34, one of the negatively charged residues that is part of the narrowest section of the B-pores, and coordinates a Na+ ion within the pore (Figure 2E), was replaced with phenylalanine. Our results show that the site-directed alterations not only exert a local effect on the targeted pores, but also influence the flexibility and reactivity of relatively distant ferroxidase centers. MD simulations carried out with the X-ray crystal structures of select mutants provide a platform for interpretation of the experimental observations in the context of cooperative motions, extending from 4-fold pores, via B-pores, into ferroxidase centers, which are required for efficient oxidation and mineralization of iron by the 24-mer bacterioferritin assembly. Structural studies of the BfrB mutants carried out after exposure of single crystals to Fe2+ solution provide experimental support for iron traffic through B-pores, and compilation of iron binding sites in the interior surface of the distinct mutants give unprecedented insight into the paths followed by iron and phosphate, from entry ports in the Bfr shell toward possible mineralization sites in the interior cavity.
Experimental Procedures
Site-Directed Mutagenesis and Protein Expression
The pET11a vector containing the bfrB gene (16) was mutated to D34F, N148L, Q151L and C89SK96C using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) using the manufacturer instructions. Primer pair sequences are provided in Supplementary Information. PCR products were digested using Dpn I and transformed into XL1-Blue Supercompetent cells (Agilent Technologies) for DNA amplification. Plasmid DNA was isolated using the QIAprep Spin Miniprep Kit (QIAGEN), and the sequences verified by SeqWright (Houston, TX). Recombinant DNA plasmids with the correct sequence were transformed to Escherichia coli ArcticExpress (DE3)RIL competent cells (Agilent Technologies) for subsequent protein expression. The protocols for protein expression, purification and reconstitution with heme have been described previously (16, 21).
Crystallization and Data Collection
All crystallization experiments were conducted in Compact 300 or CombiClover 500 (Rigaku Reagents) sitting drop vapor diffusion plates at 20 °C. Equal volumes of BfrB (10 mg/mL in 100 mM potassium phosphate buffer, pH 7.6 and 1 mM TCEP) and crystallization solutions were equilibrated against 75 μL reservoir volume. Within 1 day, red prismatic crystals were obtained from Wizard 2 (Rigaku Reagents) condition E2 (35% (v/v) 2-methyl-2,4-pentanediol, and 100 mM MES pH 6.5, 200 mM Li2SO4). Crystals were transferred to a fresh drop of crystallization solution, which served as the cryoprotectant, and stored in liquid nitrogen for data collection. Fe-soaked crystals were prepared by soaking native crystals for 10 minutes in 50 mM FeCl2 freshly dissolved in crystallization solution then frozen in liquid nitrogen. X-ray diffraction data were collected at the Advanced Photon Source beamline 17-ID (IMCA-CAT) using a Dectris Pilatus 6M pixel array detector.
Structure Solution and Refinement
Intensities were integrated using XDS (29) via the XDSAPP (30) interface or the Autoproc (31) software package and the Laue class analysis, and data scaling were performed with Aimless (32). All crystals were isomorphous (P212121, 24 molecules/asu) with the previously determined BfrB structure (PDB: 3IS7) (21). However, BfrB (C89S/K96C) crystals were found to belong to the Laue class 6/mmm (a=158.57 Å, c=152.38 Å) with the likely space group P6322 (4 molecules/asu). The Matthews coefficient (33) (Vm) suggested that there were likely four (Vm = 3.2 Å3/Da, 62% solvent) molecules in the asymmetric unit. This is similar to the Matthew's coefficient observed for the P212121 crystal form (3.0 Å3/Da, 59% solvent), indicating similar crystal packing in both crystals. Structure solution was conducted by molecular replacement using a single subunit from PDB:3IS7 as the search model with Phaser (34). All space groups with 622 point symmetry were tested in the molecular replacement searches. The top solution was found in the space group P6322 with four molecules in the asymmetric unit. A second crystal form of as-isolated BfrB (C89S/K96C) was obtained from crystals that had been incubating in the trays for over six months, which resulted in significantly different lattice constants of a=b=155.13 Å, c=155.18 Å. The biological assembly (24-mers) of C89S/K96C BfrB can be generated by application of the space group symmetry operators to the asymmetric units. Structure refinement and manual model building were conducted with Phenix (35) and Coot (36) respectively. Disordered side chains were truncated to the point where electron density could be observed. Structure validation was conducted with Molprobity (37). Coordinates and structure factors were deposited to the Worldwide Protein Data Bank and the accession codes are provided in Table 1.
Table 1.
Crystallographic Data for BfrB Structures.
| As isolated (N148L) |
Fe-soaked (N148L) |
As isolated (Q151L) |
Fe-soaked (Q151L) |
As-isolated (D34F) |
Fe-soaked (D34F) |
As-isolated (C89S/K96C) |
As-isolated-2 (C89S/K96C) |
Fe-soaked (C89S/K96C) |
|
|---|---|---|---|---|---|---|---|---|---|
| Data Collection | |||||||||
| Unit-cell parameters (Å, º) | a=125.73 | a=125.32 | a=125.67 | a=125.59 | a=125.68 | a=125.49 | a=b=158.57 | a=b=155.13 | a=b=153.99 |
| b=203.35 | b=203.29 | b=203.15 | b=203.22 | b=203.54 | b=203.40 | c=152.38 | c=155.18 | c=155.20 | |
| c=208.27 | c=207.18 | c=207.79 | c=207.40 | c=206.97 | c=207.25 | ||||
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P6322 | P6322 | P6322 |
| Resolution (Å)1 | 49.39-2.00 (2.03-2.00) | 49.36-1.95 (1.98-1.95) | 49.24-1.95 (1.98-1.95) | 49.35-2.25 (2.29-2.25) | 49.41-2.05 (2.09-2.05) | 49.39-2.20 (2.24-2.20) | 49.13-1.65 (1.68-1.65) | 48.27-1.80 (1.83-1.80) | 48.23-1.80 (1.83-1.80) |
| Wavelength (Å) | 1.74013 | 1.74013 | 1.00000 | 1.73964 | 1.00000 | 1.73769 | 1.00000 | 1.00000 | 1.73963 |
| Temperature (K) | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Observed reflections | 2,270,196 | 3,760,949 | 2,639,550 | 1,749,112 | 2,240,862 | 1,490,791 | 2,688,900 | 2,015,697 | 1,825,740 |
| Unique reflections | 356,578 | 373,157 | 384,285 | 250,626 | 328,340 | 257,692 | 134,788 | 101,666 | 99,837 |
| <I/σ(I)>1 | 10.8 (1.7) | 14.7 (2.2) | 10.8 (2.2) | 12.2 (2.4) | 13.1 (2.2) | 18.1 (3.4) | 12.3 (2.3) | 20.8 (2.3) | 22.3 (1.8) |
| Completeness (%)1 | 99.7 (94.1) | 97.7 (91.5) | 100 (100) | 100 (100) | 99.5 (98.9) | 96.3 (71.8) | 100 (99.9) | 100 (100) | 99.7 (96.5) |
| Multiplicity1 | 6.4 (4.7) | 10.1 (8.1) | 6.9 (6.4) | 7.0 (6.7) | 6.8 (6.6) | 5.8 (2.4) | 19.9 (20.3) | 19.8 (19.2) | 18.3 (10.2) |
| Rmerge (%)1, 2 | 12.1 (81.5) | 14.0 (92.9) | 14.4 (95.8) | 14.4 (96.7) | 11.7 (95.1) | 6.2 (25.9) | 16.8 (179.1) | 10.6 (160.0) | 12.1 (131.3) |
| Rmeas (%)1, 4 | 13.2 (91.6) | 14.8 (99.1) | 15.6 (103.3) | 15.2 (104.8) | 12.6 (103.1) | 7.4 (35.6) | 17.7 (188.2) | 10.9 (164.3) | 12.4 (138.3) |
| Rpim (%)1, 4 | 5.2 (40.8) | 4.6 (33.6) | 5.9 (40.9) | 5.7 (40.3) | 4.8 (39.7) | 2.4 (21.4) | 3.9 (41.2) | 2.4 (37.2) | 2.8 (42.7) |
| CC1/21, 5 | 0.996 (0.609) | 0.996 (0.712) | 0.996 (0.695) | 0.996 (0.680) | 0.998 (0.710) | 0.998 (0.885) | 0.997 (0.783) | 1.000 (0.818) | 0.999 (0.701) |
| Refinement | |||||||||
| Resolution (Å) | 49.39-2.00 | 49.36-1.95 | 49.34-1.95 | 49.35-2.25 | 41.06-2.05 | 48.39-2.20 | 38.10-1.65 | 37.27-1.80 | 48.23-1.80 |
| Reflections (working/test)6 | 654,121/34,746 | 688,869/36,492 | 364,760/19,327 | 461,135/24,449 | 311,642/16,559 | 485,422/24,416 | 127,956/6,770 | 96,523/5,074 | 180,676/9,520 |
| Rfactor / Rfree (%)3 | 16.9/21.1 | 15.5/19.3 | 15.6/18.8 | 15.4/19.2 | 15.3/19.2 | 14.9/18.7 | 14.5/16.2 | 15.3/17.8 | 14.7/17.2 |
| No. of atoms (Protein/Heme/K+/Iron/Sulfate/Water) | 30,466/1,032/6/0/0/2,588 | 30,665/1,032/6/126/40/3,005 | 30,252/5160/0/0/2,998 | 30,343/516/0/120/40/2,110 | 30,434/516/6/0/0/2,066 | 30,262/516/6/144/40/2,278 | 5,151/172/1/0/0/762 | 5,151/172/1/0/8/641 | 5,165/172/1/37/8/748 |
| Model Quality | |||||||||
| R.m.s deviations | |||||||||
| Bond lengths (Å) | 0.010 | 0.956 | 0.009 | 0.011 | 0.016 | 0.010 | 0.008 | 0.010 | 0.009 |
| Bond angles (º) | 0.975 | 0.017 | 1.008 | 1.023 | 1.265 | 0.985 | 1.107 | 0.996 | 1.013 |
| Average B-factor (Å2) | |||||||||
| All Atoms | 26.0 | 22.6 | 20.9 | 24.8 | 28.6 | 24.8 | 22.6 | 27.7 | 24.8 |
| Protein | 25.4 | 19.0 | 19.9 | 31.7 | 28.2 | 24.1 | 20.6 | 26.2 | 23.1 |
| Heme | 27.9 | 21.8 | 18.0 | 30.4 | 25.6 | 24.4 | 20.2 | 24.3 | 22.9 |
| K+ | 38.6 | 28.2 | - | - | 21.5 | 39.9 | 15.9 | 20.6 | 19.5 |
| Iron | - | 45.1 | - | 60.0 | - | 45.1 | - | - | 46.8 |
| Sulfate | - | 48.0 | - | 59.8 | - | 53.4 | - | 52.2 | 39.3 |
| Water | 32.9 | 20.0 | 29.5 | 39.3 | 35.7 | 31.2 | 34.0 | 38.8 | 35.4 |
| Coordinate error (maximum likelihood) (Å) | 0.24 | 0.19 | 0.17 | 0.22 | 0.20 | 0.20 | 0.15 | 0.17 | 0.18 |
| Ramachandran Plot | |||||||||
| Most favored (%) | 99.7 | 99.6 | 100 | 100 | 99.5 | 99.8 | 99.7 | 99.7 | 99.5 |
| Additionally allowed (%) | 0.3 | 0.4 | - | - | 0.5 | 0.2 | 0.3 | 0.3 | 0.5 |
| PDB Code | 4TO9 | 4TOA | 4TOB | 4TOC | 4TOD | 4TOE | 4TOF | 4TOG | 4TOH |
Values in parenthesis are for the highest resolution shell.
Rmerge = ΣhklΣi |Ii(hkl) - <I(hkl)>| /ΣhklΣi Ii(hkl), where Ii(hkl) is the intensity measured for the ith reflection and <I(hkl)> is the average intensity of all reflections with indices hkl.
Rfactor = Σhkl ‖Fobs (hkl) | - |Fcalc (hkl) ‖ / Σhkl |Fobs (hkl)|; Rfree is calculated in an identical manner using 5% of randomly selected reflections that were not included in the refinement.
Rmeas = redundancy-independent (multiplicity-weighted) Rmerge(32, 58). Rpim = precision-indicating (multiplicity-weighted) Rmerge(59, 60).
CC1/2 is the correlation coefficient of the mean intensities between two random half-sets of data (61, 62).
For the data sets obtained from iron soaked crystals or native data sets collected at the iron peak wavelength, the number of reflections used during refinement is greater than the number unique reflections reported for data scaling. This is due to the fact that Friedel pairs were kept separate during refinement and the anomalous scattering factors for the Fe2+ atoms were refined.
In order to assign the Fe sites for the Fe-soaked BfrB (C89S/K96C) structure, data were collected at a wavelength corresponding to the iron absorption edge (1.7396 Å). This structure was refined using data to 1.8 Å resolution. During subsequent data collection at the synchrotron, diffraction data were collected for another Fe-soaked BfrB (C89S/K96C) crystal and data were collected at wavelengths of 1.7398 Å and 1.7463 Å, which correspond to the absorption peak and low energy remote respectively. The diffraction data were a bit weaker for this sample and were scaled to 2.1 Å resolution. However, this allowed for the confirmation of the assigned Fe sites, particularly those that displayed weaker electron density and possibly lower occupancy. By computing phased anomalous difference maps the anomalous signal was compared at the peak/remote wavelengths. If Fe is indeed present at a particular site, the anomalous signal should disappear when using the low energy remote wavelength data. Conversely, an atom such as a K+ ion, which yields an appreciable anomalous signal at the Fe peak wavelength, would display an increased anomalous signal at the low energy remote wavelength. This second data set (peak and low energy remote) was included with the structure factors deposited to the PDB for the Fe-soaked C89S/K96C structure.
Effect of the mutations on iron oxidation and uptake in solution by wt and mutant BfrB
As-isolated BfrB (wt and mutants) contain only a very small amount of iron in their core (16). Reconstitution with an iron core was carried out as previously reported (16): A 10 mM solution of ferrous ammonium sulfate was prepared inside an anaerobic chamber (Coy Laboratories, Grass Lake, MI), placed in a container with a rubber septum, and removed from the chamber. Concentrated HCl was added to the ferrous ammonium sulfate solution (50 μL/100 mL) through the septum with the aid of a Hamilton microsyringe (final pH ≈ 2.0) and the resultant solution titrated into a stirred solution of 2 μM BfrB and 1.0 mM TCEP in 100 mM phosphate buffer, pH 7.6, in aliquots delivering 50 Fe2+ ions/BfrB. Fifteen minutes were allowed after the addition of each aliquot; upon the addition of the total iron load (500 Fe atoms/BfrB), the solutions were stirred overnight at 4 °C and then passed through a Sephadex G25M size exclusion column (GE Healthcare). The iron content of the samples, prior and after reconstitution with iron, was analyzed using a colorimetric ferrozine-based assay (38), as reported previously (16): 50 μL concentrated HCl was added to 50 μL mineralized BfrB and the mixture incubated for 15 min at room temperature prior to the addition of 50 μL ascorbic acid (25 mg/mL) and 250 μL saturated sodium acetate. The concentration of iron was determined using the absorbance at 562 nm (ε562nm = 27.9 mM-1cm-1) 15 min after the addition of ferrozine (5mg/mL).
Molecular dynamics simulations of BfrB D34F and N148L mutants
Each mutant BfrB (D34F and N148L) was first immersed in a pre-equilibrated cubic water box of 160 Å in all three dimensions. This box is the same size as that used in the Grand Canonical Monte Carlo/Brownian Dynamics simulations of wt BfrB described previously (28, 39). Both the box and the BfrB molecule were centered at the origin. The initial ion configurations were the same as those in the E2, E10 and E40 systems described previously (28); the free K+ ion numbers are 2,840 (E2), 1,042 (E10), and 714 (E40) and the numbers of HPO42- ions are 1,231 (E2), 332 (E10) and 168 (E40). In each of the D34F mutant systems, 24 K+ ions were added in the bulk solution to make the total charge of the systems neutral, and they were distributed with 2,000 Monte Carlo moves, which were either accepted or rejected based on Metropolis criteria. Water molecules within 2.4 Å from the ions and the BfrB heavy atoms were removed. The systems were then subjected to a 900-ps equilibration cycle with decreasing positional harmonic restraints on heavy atoms not including water oxygen. In the BfrB interior, the removed water molecules that were close to BfrB before the equilibration cycle were quickly replaced by water from the interior cavity of BfrB, creating small vacuum pockets. To overcome this problem, an additional sphere of pre-equilibrated water with 40 Å radius was added to the system on top of the other water molecules (also see (28)). Any of these newly added water molecules that were within 2.4 Å of either the BfrB heavy atoms or the other water molecules in the system were deleted. Each of the resultant systems was again equilibrated with diminishing positional-harmonic-restraint potentials on the heavy atoms for another 900 ps and then subjected to a 40-ns production.
All the simulations were carried out in NPT (constant particle number, pressure and temperature) ensembles using the NAMD2.9 (40) simulation package. The simulation inputs were obtained from the Quick MD Simulator module in CHARMM-GUI (41). The simulations were performed with CHARMM all-atom parameter set PARAM22 (42) including the dihedral cross-term corrections (CMAP) (43) and a modified TIP3P water model (44). The van der Waals interactions were smoothly switched off at 10-12 Å by a force switching function (45) and the electrostatic interactions were calculated using the particle-mesh Ewald method (46) with a mesh size of ∼1 Å for fast Fourier transformation, κ = 0.34 Å-1, and a sixth-order B-spline interpolation. The temperature (300 K) and pressure (1 atm) were kept constant during all the simulations by Langevin dynamics and the hybrid Nosé-Hoover Langevin piston method, respectively. The Langevin damping coefficient was set to 1 ps-1; the decay period and damping timescale were 50 fs, respectively.
Results
X-ray crystal structures
The structures of four BfrB mutants (N148L, Q151L, D34F and C89S/K96C) were determined using crystals grown from purified protein (as-isolated) and using crystals of pure protein after soaking in Fe2+ solution (Fe-soaked). Two mutants (N148L and Q151L) were prepared to investigate the effect of replacing the hydrophilic side chains that coordinate K+ in the 4-fold pores of BfrB (Figure 2D) with hydrophobic non-coordinative side chains. A third mutant (D34F) was prepared to study the effect of replacing a negatively charged residue, which was shown to bind a Na+ ion in the B-pores of BfrB (Figure 2E), with a hydrophobic side chain. As will be shown below, these mutations affect the flexibility of ferroxidase center ligands and the catalytic efficiency of Fe2+ oxidation by their respective ferroxidase centers. To show that mutation of residues outside of the pores has no effect on ferroxidase center function, we studied the surface mutant C89S/K96C, which had been prepared for a different investigation. Interestingly, the C89S/K96C mutant not only showed wt-like behavior of the ferroxidase center in crystallo and in solution, but also displayed all the iron binding sites observed in the structures of the different mutants reported here, and also revealed new ones. The unprecedented observation of six different types of iron binding sites (>200 iron ions not counting heme-iron) in one Bfr structure allows comparison of thermal factors for each type of iron binding site, and therefore provides insights into their relative occupancies.
The structures of all the mutants show strong conservation of the subunit architecture and 24-mer assemblies. Superposition (47) of subunit A from the as-isolated structure of each mutant reported here with subunit A of the previously determined wt BfrB structure (PDB: 3IS7) resulted in the following RMSD between Cα atoms for 153 residues aligned: N148L (0.15 Å), Q151L (0.15 Å), D34F (0.10 Å) and C89S/K96C (0.15 Å). Closer inspection shows that the mutations cause local structural perturbations in the corresponding 4-fold or B-pores relative to the wt structure, as expected. However, comparison of as-isolated and Fe-soaked structures for each of the mutants uncovers important differences: (i) With the exception of the C89S/K96C mutant, iron ions in the ferroxidase centers are not coordinated as in the wt protein, (ii) previously unobserved iron binding sites are present in the interior cavity, near the entrance to 3-fold, 4-fold and B-pores in all mutants, (iii) sulfate ions are found in the interior cavity at the entrance to 3-fold pores, where they are coordinated by iron ions, (iv) iron ions are observed in B-pores, and (v) the ubiquitous K+ ions observed in the 4-fold pores of wt and other mutant BfrB proteins are absent in the Q151L mutant. Pertinent details are presented below.
Four-fold pores
In wt BfrB the side chains of N148 and Q151, located in each of the two turns of helices E, point toward the pore interior, where they coordinate a K+ ion (Figures 2D and 3A). The structure of 4-fold pores in the as-isolated and Fe-soaked C89S/K96C mutant is nearly identical to that in wt BfrB, including the placement of the K+ ion (Figure 3B). As seen previously in the Fe-soaked structure of wt BfrB (21), in the interior cavity of Fe-soaked C89S/K96C four Fe ions (orange spheres) nest in the exterior wall of the 4-fold pore, where they are coordinated by H155 from one subunit (A) and H153 from a different subunit (B) (Figure 3B and Figure S1A of the Supplementary information). These sites were previously termed Fein to denote their location in the interior cavity. Herein, we will term these sites Fe4f-1 to indicate that they are the first Fe-sites observed near 4-fold pores. The C89S/K96C structure also revealed previously unobserved Fe sites located deeper in the interior cavity, along the perimeter of the entrance to 4-fold pores from the interior cavity, which we term Fe4f-2 (blue spheres). Although these Fe ions do not appear to be coordinated by a protein-provided ligand (Figure S1A), interactions with the side chains of one of the terminal residues (E157 or E158), for which there is no observable electron density, cannot be ruled out.
Figure 3.
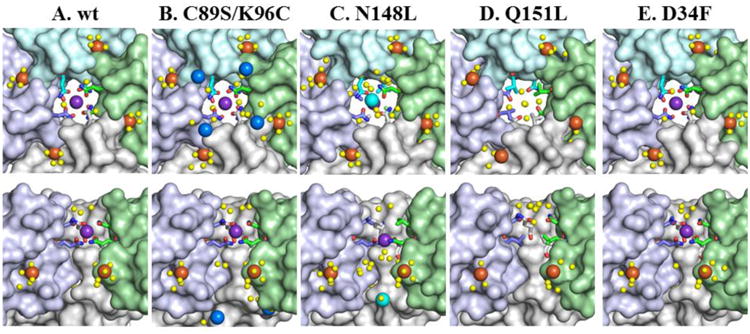
Zoomed in view of a 4-fold pore in wt and mutant BfrB molecules. The top row depicts views from the interior cavities, while the bottom row shows cross-sectional views with a subunit removed. Fe4f-1 iron ions are shown as orange spheres, Fe4f-2 iron ions as blue spheres, Fe4f-3 iron ions as cyan spheres, water molecules as yellow spheres, and potassium ions as purple spheres, nitrogen in blue and oxygen in red.
Diffraction data collected at a wavelength of 1.0000 Å did not yield anomalous signal for the K+ ions in the 4-fold pores. In contrast, diffraction data collected at low energy wavelength (∼1.74 Å, Fe-edge) resulted in appreciable anomalous difference density at the K+ sites, and naturally, at the iron and heme-iron sites. Notably, the anomalous-difference-map peak height increases at the K+ sites when computed using data collect at a low energy remote wavelength relative to the Fe-edge (Figure S2). This suggests that the assignment of K+ in the pore interior and Fe ions in the periphery of 4-fold pores in the interior cavity is the correct model.
In the as-isolated N148L structure the conformation of L148 side chains is nearly identical to that of N148 in the wt protein, which results in a tight network of hydrophobic interactions in the pore interior of the mutant. Closer to the interior cavity, the Q151 side chains assume conformations identical to those observed in wt BfrB, and a K+ is present in the pore interior, but is coordinated only by the side chains of Q151 (Figure 3C). Consequently, K+ in the 4-fold pores of N148L BfrB is 1.7 Å closer to the interior cavity than equivalent K+ in the wt protein. As seen previously in the Fe-soaked structure of wt (21), and C89S/K96C BfrB, four Fe4f-1 ions (orange) nest in the exterior wall of the 4-fold pore inside the BfrB cavity (Figure 3C). The N148L structure also reveals a previously unobserved Fe ion at the entrance to 4-fold pores in the interior cavity, which is termed Fe4f-3 (cyan spheres). These Fe ions also do not appear to be coordinated by a protein-provided ligand (Figure S1B), although it is likely that the side chains of one of the terminal residues, for which there is no observable electron density (E157 or E158), provide the necessary stabilizing interactions.
In the structure of as-isolated and Fe-soaked Q151L BfrB the L151 side chains adopt the same conformation as the Q151 side chains in the wt protein, which creates a network of hydrophobic interactions that constrict the interior of 4-fold pores. Interestingly, the K+ ubiquitously found in Pa BfrB structures are absent in the 4-fold pores of the Q151L mutant, in both the as-isolated and Fe-soaked structures (Figure 3D). In the Fe-soaked structure, four Fe4f-1 ions are present in the exterior perimeter of the 4-fold pore, but unlike N148L, the Fe4f-3 ion is not observed. The presence of iron is confirmed by strong anomalous difference density observed in diffraction data sets collected at ∼1.74 Å (Figure S1C). It is also interesting that structured water is absent in the pores below the hydrophobic constriction formed by the L151 side chains in the as-isolated and Fe-soaked structures.
The D34F mutation does not have a structural effect on the 4-fold pores. The side chains of N148 and Q151 coordinate K+ (Figure 3E) in the as-isolated and Fe-soaked structures. Four Fe4f-1 ions are observed along the periphery of 4-fold pores in the interior cavity, in positions identical to those observed in wt BfrB. As with the previously discussed structures, anomalous difference density in data sets collected at ∼1.74 Å corroborate Fe ions at these sites (Figure S1D).
Three-fold pores
The 3-fold pores in the structures of the as-isolated N148L, Q151L, D34F and C89S/K96C mutants are almost identical to their counterparts in wt Bfr (Figure 4 top). The narrowest pore section is lined by side chains from three R117 and three K121, with E109 immediately above (closer to the cytosol) R117 and D118 immediately below (closer to the interior cavity) K121; the K121 side chains cannot be modeled entirely due to disorder in as-isolated BfrB. The innermost layer is wider and demarcated by the side chains of D122 and E125. Soaking crystals of wt BfrB in crystallization solution containing Fe2+ led to the observation of a sulfate ion in the 3-fold pores, located between the side chains of R117 and K121 (Figure 4A center) (21). Similar iron-soaking experiments carried out with crystals of all mutant proteins allowed observation of a sulfate ion in the innermost layer of each pore, where it is coordinated by three K121 side-chains (Figure 4B-E). Interestingly, in the mutant structures each sulfate ion also interacts with three Fe ions, termed Fe3f (avg. Fe-O distance 4.2 Å), which in turn are coordinated by the side chains of D118, D122 and E125. The side chains of D118 and E125 move from their conformation in the as-isolated structures to coordinate iron in the Fe-soaked structure. The presence of Fe at these sites is supported by anomalous difference density in data sets collected at ∼1.74 Å (Figure S3).
Figure 4.
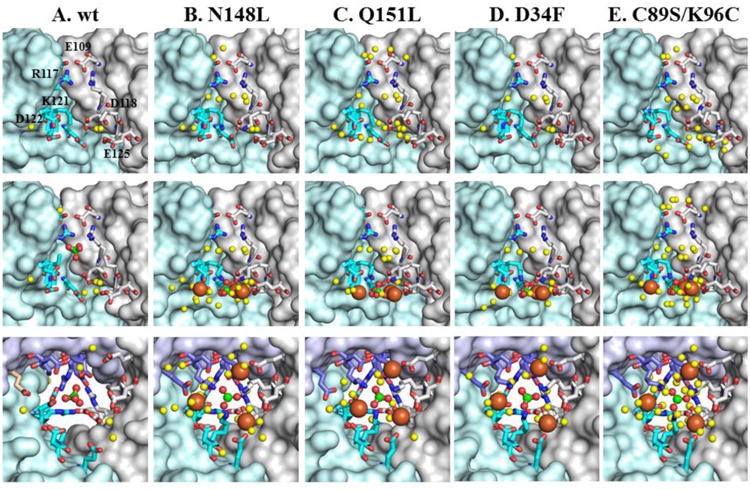
View of a 3-fold pore in wt BfrB and mutants. The top row shows cross sectional views of the 3-fold pores in the as-isolated structures. The middle row depicts an identical cross-section view of the 3-fold pores in the Fe-soaked structures, with Fe3f iron ions rendered as orange spheres, sulfate ions as green (S) and red (O) spheres, and structured water as yellow spheres. The bottom row depicts the 3-fold pores viewed from the interior cavity, to illustrate the network of interactions involving iron, sulfate and protein side chains.
It is interesting to note that sulfate is absent from the 3-fold pores in all the structures of as-isolated proteins, but is present in the corresponding Fe-soaked structures. An exception is the structure of as-isolated C89S/K96C BfrB obtained from a crystal that had been in the crystallization trays for over 6 months (hereafter as-isolated-2), which showed clear electron density consistent with sulfate ions in the 3-fold pores (Figure S4). In comparison, diffraction data from other as-isolated BfrB crystals was obtained within 20 days of crystal growth. Therefore, it may be reasonable to consider the as-isolated-2 crystal as being soaked for a prolonged period in high sulfate concentration (200 mM Li2SO4 from the crystallant), which causes sulfate to slowly occupy the 3-fold pores. As noted in the experimental section, the unit cell parameters differ significantly for the as-isolated and as-isolated-2 C89S/K96C structures. This appears to result in a “tightening up” of the lattice and formation of an intermolecular disulfide bond between C96 of subunit A and its counterpart related by a crystallographic 2-fold axis (Figure S8). It should be noted that soaking of as-isolated C89S/K96C crystals with iron also results in this lattice transformation (see Table 1). Comparison of the 3-fold pores before and after Fe-soak is illustrated in Figure 5: While in the as-isolated-2 structure sulfate nests between R117 and K121 in the middle of the pore, after Fe-soak sulfate is in the most interior pore layer, still coordinated by the side chains of K121, which likely “escort” the anion to its position at the exit of 3-fold pores, where it is seen interacting with iron ions.
Figure 5.
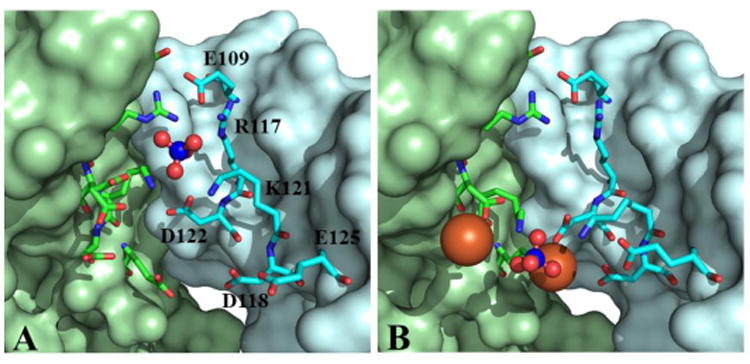
Comparison of 3-fold pores in (A) as-isolated-2 and (B) Fe-soaked C89S/K96C BfrB suggests a possible path for the ingress of sulfate to the interior cavity. Sulfate is shown as blue (sulfur) and red (oxygen) spheres, and Fe3f ions as orange spheres. Key side chains are shown in sticks with nitrogen in blue and oxygen in red.
In the context of anion traffic through 3-fold pores, it is also interesting to contrast the relatively low tendency of sulfate to populate 3-fold pores in as-isolated crystals of BfrB with its ubiquitous presence within the pores, or at the exit into the interior cavity, after crystals are soaked in Fe2+ solution. The comparisons suggest that 3-fold pores are conduits for anions, which are needed to compensate for the flux of positive charge that accompanies iron traffic. Note that the anions in 3-fold pores were modeled as sulfate because the crystallization conditions contain 200 mM Li2SO4. However, since the core mineral in Bfr and bacterial Ftn is known to be composed of iron and phosphate in nearly 1:1 ratio (48-50), our findings can be interpreted to suggest that phosphate traffics across Bfr through the 3-fold pores to counteract the charge flux created by the traffic of iron across the Bfr shell as iron enters or leaves the interior cavity. The bottom row in Figure 4 illustrates the arrangement of iron, phosphate and acidic side chains in the interior cavity at the exit of the 3-fold pores. This highly organized network of contacts encompassing electrostatic and coordinative interactions supports the idea that iron ions entering BfrB via different pores in the structure are directed toward the perimeter of 3-fold pores to interact with phosphate ions accessing the BfrB interior via 3-fold pores.
B-pores
The B-pore channels in wt BfrB are formed at the intersection of three subunits not aligned with an axis of symmetry, and are demarcated by the side chains of E66, D34, T136 and D132, arranged in a corkscrew leading from the outermost (E66) to the innermost (D132) layer (Figure 6A). The structure of B-pores in as-isolated and Fe-soaked N148L and Q151L BfrB are very similar to wt (Figure 6B-C), with the exception of the E66 side chain, which is rotated away and does not participate in the “corkscrew” of hydroxyl and carboxylate groups traversing the channel. In the B-pores of as-isolated D34F BfrB the E66 side chain is also rotated away from the corkscrew, and the side chain of F34 acts as a hydrophobic barrier that occludes the pores and disrupts the hydrophilicity of the corkscrew. Interestingly, the corresponding Fe-soaked structure shows an Fe ion (FeB-1) in the innermost layer of the pores, where it is coordinated by D132 (Figure 6D). Anomalous difference density in data sets collected at 1.74 Å support this conclusion (Figure S5A).
Figure 6.
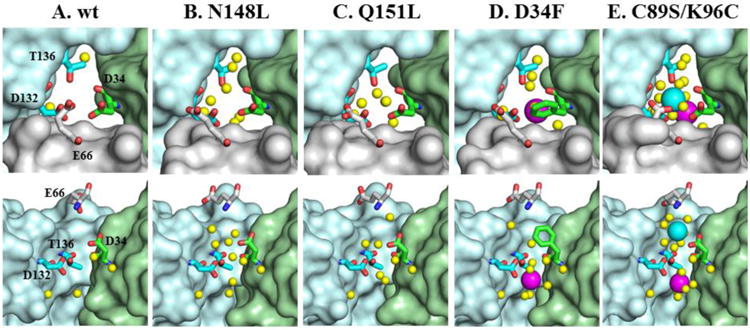
Zoomed-in view of B-fold pores in wt BfrB and mutants. The top row depicts the pores viewed from the protein exterior and the bottom row shows cross-sectional views with the wheat subunit omitted. FeB-1 iron ions are shown as magenta spheres, the FeB-2 iron ion is shown as a cyan sphere, water molecules as yellow spheres, nitrogen in blue and oxygen in red.
The corkscrew of hydroxyl and carboxylate residues seen in wt BfrB is preserved in the B-pores of as-isolated and Fe-soaked C89S/K96C. Remarkably, the Fe-soaked structure of the double mutant reveals two Fe ions aligned along the length of the B-pores (Figure 6E and Figure S5B). One is coordinated by D132 and is located near the interior cavity in a position nearly equivalent to that of the FeB-1 ion in the B-pores of Fe-soaked D34F BfrB. The second (FeB-2) is near the cytosolic surface, where it is coordinated by D34 and E66. These observations, which provide the first experimental evidence of Fe ions in B-pores, strongly support the idea derived from our MD simulations (51) that iron traffics across the Bfr shell using B-pores as conduits.
The ferroxidase sites
In all the BfrB structures obtained from as-isolated protein, the ferroxidase centers are devoid of iron. Soaking crystals of as-isolated protein in crystallization solution containing iron invariably leads to iron incorporation in the ferroxidase center. In the structures of wt and C89S/K96C BfrB, iron binding at the ferroxidase center is accompanied by a conformation change of the H130 side chain from gate open to gate closed state (Figures 1C and 7A). In contrast, although iron is observed in the ferroxidase centers of D34F, N148L and Q151L BfrB, H130 remains in its gate open conformation, and E51 does not rotate to coordinate and bridge Fe1 and Fe2 (Figure 7B-D). The presence of Fe ions in the ferroxidase centers of Fe-soaked structures is supported by anomalous difference density peaks using diffraction data sets collected at ∼1.74 Å (Figures 7 and S6). The distance between ferroxidase iron ions in all structures is 4.1 Å, which suggests di-Fe2+ sites (52).
Figure 7.
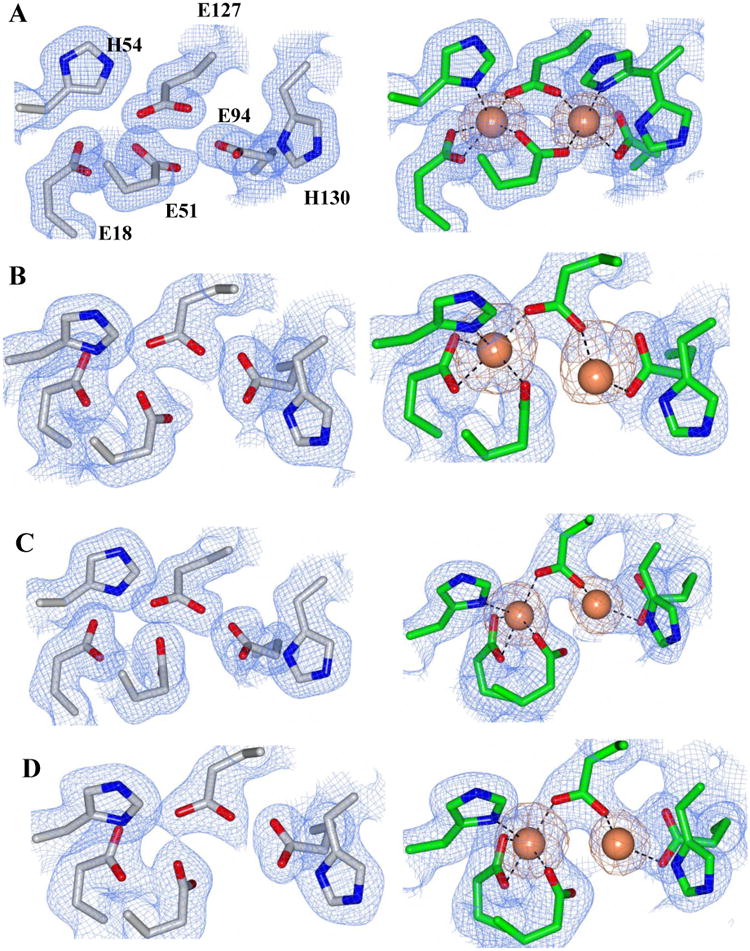
2Fo-Fc electron density map of the ferroxidase ligands (blue mesh) contoured at 1σ and phased anomalous difference map of the ferroxidase center iron atoms (orange mesh) contoured at 3σ for the following BfrB mutant structures: A) C89S/K96C, B) N148L C) Q151L and D) D34F. Ferroxidase centers from as isolated proteins (devoid of iron) are on the left (gray sticks), and ferroxidase centers from iron-soaked structures are on the right (green sticks). Iron atoms are drawn as orange spheres and contacts to the ferroxidase center ligands are represented as dashed lines.
The Fe-soaked structure of C89S/K96C BfrB also shows an iron site on the inner surface of the protein, directly below each of the ferroxidase centers (Feb-fc), where it is coordinated by H46 (Figure 8 and Figure S7), and close to D50 (∼4.5 Å). The Feb-fc site is equivalent to the iron site observed upon soaking crystals of E. coli Bfr in iron-containing solution of crystallant, which was shown to be important in the process of iron mineral growth (53). In the asymmetric unit cell of Fe-soaked C89S/K96C BfrB (subunits A, B, C and D), the side chains of H46 and H130 in subunit A (grey in Figure 8) are observed in two conformations: In one conformation ferroxidase ligand H130 is coordinated to Fe2 (gate closed) and H46 is coordinated to Feb-fc, whereas in their alternate conformations both side chains are rotated away from their respective iron ions. In subunits B, C and D, the H46 and H130 side chains are seen only in their iron bound conformations. The observations in subunit A permit a glimpse into the possible cooperative process of ferroxidase iron translocation, in which the transition of H130 from gate closed to gate open is accompanied by a change in conformation of H46, which serves to coordinate incoming Fe2 and usher it to the Feb-fc site. The model is also consistent with the Feb-fc site serving as an iron depot for the distribution of iron entering via the ferroxidase centers to nucleation and mineralization sites.
Figure 8.
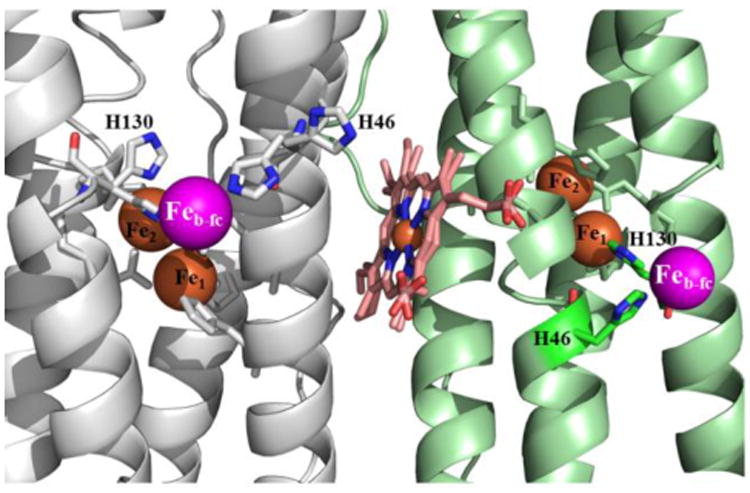
The iron ions below the ferroxidase centers, Feb-fc (magenta spheres) in the Fe-soaked structure of C89S/K96C BfrB are coordinated by H46. In subunit A (grey) H46 adopts two conformations, one of which coordinates Feb-fc; H130 also adopts two conformations, one (gate closed) coordinates Fe2 (orange), while the second (gate open) is thought to enable its translocation into the interior cavity, aided by H46. In the remainder subunits of the asymmetric unit (B, C and D), H46 and H130 are observed only coordinated to Feb-fc, and Fe2, respectively. The heme is shown in two orientations, which are made evident by the relatively high resolution (1.80 Å) of the structure.
Effect of the mutations on iron oxidation and uptake function
The X-ray crystal structures show that H130 in the ferroxidase centers of D34F, N148L, and Q151L BfrB does not rotate to coordinate Fe2, and that of E51 does not bridge Fe1 and Fe2 (see Figures 7 and S6). These observations suggest that mutations in the 4-fold and B-pores adversely affect the function of relatively distant ferroxidase centers. To investigate this idea, we carried out experiments in solution to compare the efficiency of Fe2+ oxidation and Fe3+ storage exhibited by each mutant relative to wt BfrB. This was carried out by challenging each of the bacterioferritins with 500 Fe2+/BfrB, added in aliquots delivering 50 Fe2+/ BfrB, as described in Experimental Methods. Because Fe2+ autoxidizes rapidly in aqueous solution at pH above 6.5, and even faster in the presence of phosphate (54), the addition of Fe2+ to aerobic solutions of BfrB in phosphate buffer, pH 7.6, results in two competing reactions: (i) oxidation of Fe2+ at the ferroxidase centers of BfrB and storage of the resultant Fe3+ in the interior cavity (Fe3+ is not captured by Bfr or Ftn), and (ii) autoxidation of Fe2+ in solution, followed by hydrolysis of Fe3+ into an insoluble ferric colloid. The results show that wt and C89S/K96C BfrB oxidize and capture iron with >85% efficiency (420 ± 20 Fe2+/BfrB, and 430 ± 22 Fe2+/BfrB, respectively), indicating that the dominant reaction is the oxidation of Fe2+ by BfrB. In contrast, the amounts of Fe2+ oxidized and captured by the mutants are significantly lower: D34F BfrB oxidized and captured only 110 ± 30 Fe2+/BfrB (24%), Q151L captured 320 ± 30 (64%) and the N148L mutant 300 ± 20 Fe2+/BfrB (60%). These observations, which show that oxidation of Fe2+ by the BfrB mutants does not compete effectively with Fe2+ autoxidation, indicate that mutation of important residues at 4-fold pores (N148L, Q151L) and B-pores (D34F) compromises the function of the relatively distant ferroxidase centers. Consequently, the inability of H130 and E51 side chains to rotate and bind iron at their corresponding ferroxidase centers in crystallo manifests in solution in the decreased ferroxidase activity exhibited by the 4-fold and B-pore mutants. No attempts were made to determine rates of Fe2+ oxidation by BfrB because the only spectroscopic handle to follow the oxidation of Fe2+ by BfrB is a broad absorption near 300 nm, which is caused by the formation of Fe3+-O- moieties. Since the ferroxidase activity of the BfrB mutants is compromised, the competing Fe2+ autoxidation contributes significantly to the changes in absorbance at 300 nm, and therefore precludes meaningful analysis of the progress curves in the context of Fe2+ oxidation by BfrB.
Molecular dynamics simulations
In an attempt to explain the nature of the disorganization of the ferroxidase centers in the Fe-soaked structures of the D34F and N148L mutants, we investigated their dynamic properties relative to wt BfrB. The per-residue backbone root mean square fluctuation (RMSF) plots in the top row of Figure 9 illustrate the relative fluctuations in a BfrB subunit. In wt BfrB (Figure 9A) the flexibility increases with increasing K2HPO4 concentrations (from system E40 to E2). In contrast, the flexibility of the mutants is significantly dampened and minimally affected by the same K2HPO4 concentrations (Figure 9B and C); the only regions exhibiting a strong tendency to fluctuate are the long loops connecting helices B and C and the N- and C-termini. This is in stark contrast with wt BfrB, where in addition to the BC loops and the N- and C-termini, the C-terminal half of helices D exhibit a strong tendency to fluctuate, followed by the N-terminal half of helix C (Figure 9 middle). A minimal assembly of six subunits containing a 3-fold pore (green star), a 4-fold pore (blue star), and a B-pore (red star), show that the regions with the highest fluctuations in a subunit of wt BfrB pack to form B- and 4-fold pores (Figure 9 bottom). In contrast, these regions are far less flexible in D34F and N148L BfrB (Figure 9B-C and supporting information movies S1-S3).
Figure 9.
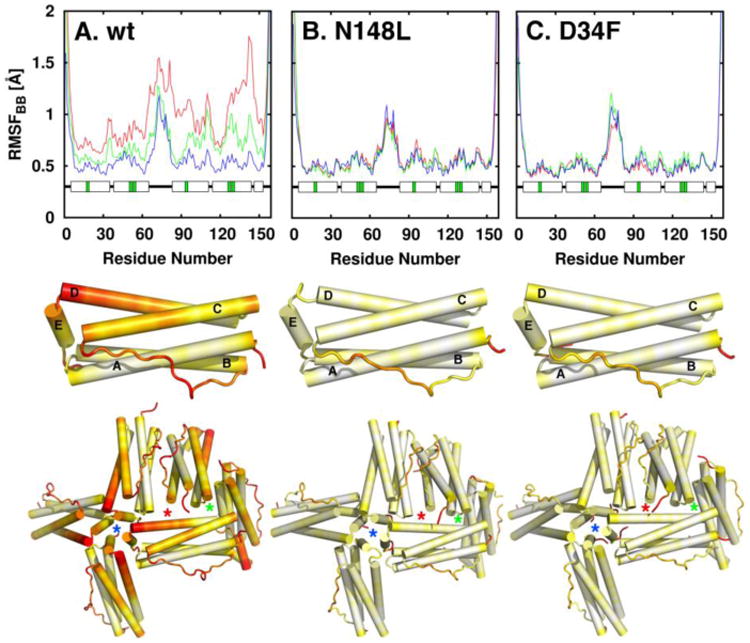
Flexibility in the wt and mutant BfrB molecules. The top row shows plots of per-residue backbone RMSF in system E2 (red), E10 (green), and E40 (blue). The middle row depicts the per-residue backbone RMSF (systems E2) mapped onto a BfrB subunit, and the bottom row shows per-residue backbone RMSF mapped on six subunits of the 24-mer assembly to illustrate relative flexibility at the 4-fold (blue stars), 3-fold (green stars) and B-pores (red stars). Flexibility increases in the color scale from white to red.
The structure of D34F BfrB shows that the F34 side chain partially occludes the B-pores and also packs against the L63 side chain. The MD simulations add insight by providing evidence for dampened flexibility in the B pores of the mutant relative to wt BfrB: Figures 10A and 11A depict regions surrounding the B-pores during the MD simulations of wt BfrB, illustrating an unfolded state of the C-terminal section of helix D (cyan), which undergoes frequent folded/unfolded transitions. Such transitions are likely linked to the conformational exchange of D34 and F64 side chains observed in wt BfrB. Figures 10C and 11C, respectively, show the distributions of rotameric states (χ1, χ2) for the D34 and F64 side chains in wt BfrB. The dynamic conformational exchange exhibited by these side chains in wt BfrB contributes significantly to the breathing motions of the B-pores and facilitates the translocation of K+ across the B-pores (Table 2). In contrast, analyses of B-pores in the MD trajectories of D34F and N148L BfrB show a low-propensity for folding/unfolding transitions in the C-termini of helices D (Figures 10B and 11B), as well as dampened breathing motions of the B-pores (movies S1-S3). It is likely that the dampened motions in the B-pore of the D34F mutant stem from the hydrophobic packing between the F34 and L63 side chains (Figure 10B), which limits the conformational freedom of the F34 (Figure 10D) and F64 (Figure 10B) side chains to only one rotamer. In the N148L mutation, hydrophobic packing of L148 side chains inside the 4-fold pores likely stabilizes the interactions of the four E-helices that form a 4-fold pore (Figure 11B). As a result the C-terminal folding/unfolding transitions of helices D are nearly absent, as is the conformational freedom of the F64 side chains (Figure 11D). Thus, the packing between F34 and L63 in the B-pores of D34F, or the hydrophobic packing of L148 side chains in N148L, which stabilize interactions of the four E-helices that from a 4-fold pore, largely dampen the folding/unfolding transitions of the C-terminal portions of helices D and the breathing motions of the B-pores. This has the effect of reducing the permeability of B-pores to K+ ions that cross the BfrB shell using these channels (Table 2).
Figure 10.
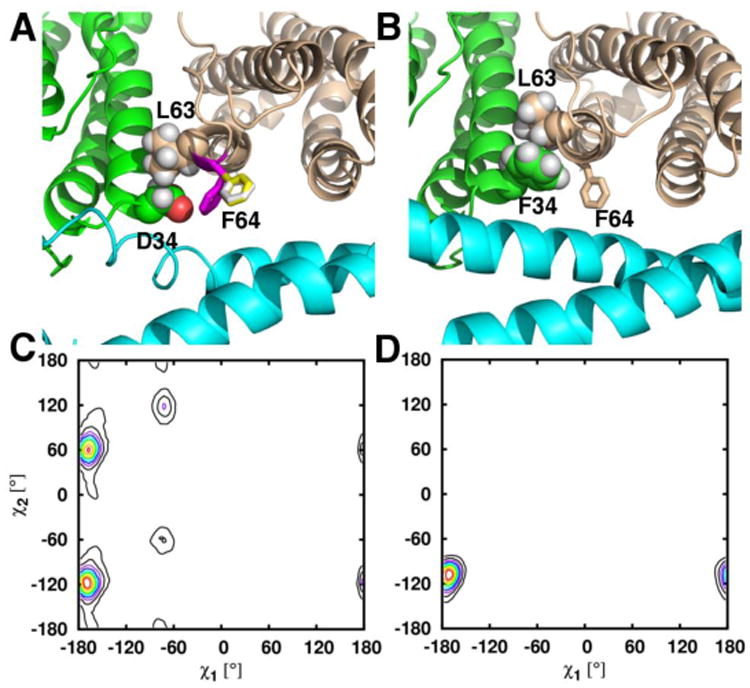
Close up view of a B-pore taken during the MD simulations of (A) wt and (B) D34F BfrB. In wt BfrB, the three rotameric states of the F64 side chain in wt BfrB are depicted by white, yellow and magenta sticks in (A), and the three rotameric states of D34 are indicated in the plot shown in (C). The rotameric exchange of D34 and F64 contributes to the breathing motions of the B-pores as well as ion traffic across B-pores in wt BfrB. In the D34F strcture packing of F34 against L63 (spheres) likely contributes to the lower flexibility of B-pores in the mutant, which is manifested in only one conformational rotamer of the F34 side chain (D), and only one conformation of the F64 side chain, wheat sticks in (B).
Figure 11.
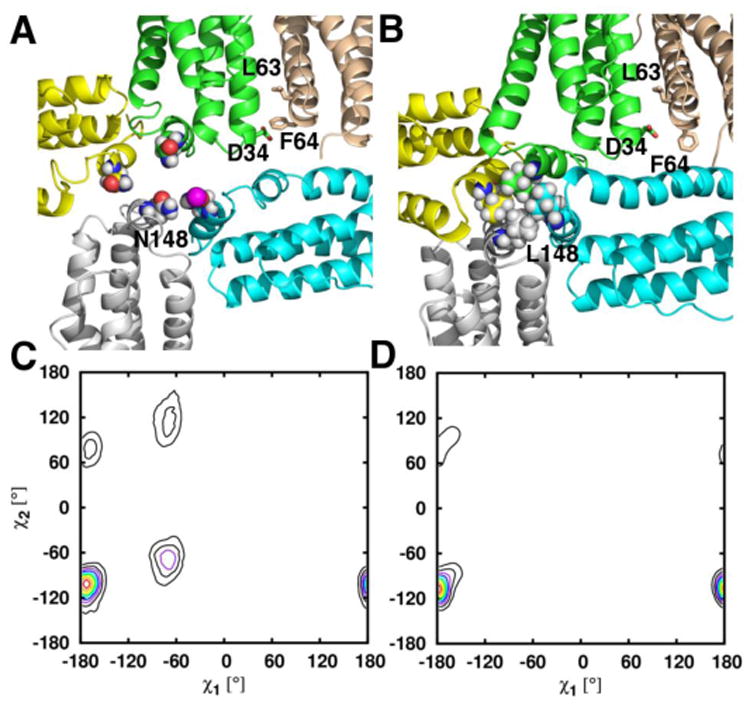
Snapshot obtained from MD simulations illustrating the packing of residues 148 in the 4-fold-pores of (A) wt and (B) N148L BfrB. The comparison of torsion angles sampled by F64 in the wt (C) and N148L (D) simulations are indicative of the dampened B-pore breathing motions observed in the N148L MD simulation relative to those observed in the wt BfrB simulation. Side chains of residue 148 in the 4-fold pores are rendered in spheres, whereas the side chains of D34, L63, and F64 in the B-pores are shown in stick representation.
Table 2. Number of K+ ions escaped from the BfrB shell during the simulations.
| E2 | E10 | E40 | |
|---|---|---|---|
| WT | 63 | 63 | 0 |
| N148L | 5 | 14 | 4 |
| D34F | 30 | 22 | 8 |
Structural perturbations in the 4-fold and B-pores affect the dynamics of distant ferroxidase centers
In the MD simulations of wt BfrB, the conformational agility of the H130 side chains is evident in the average density of H130 side chain conformations sampled during the simulations (Figure 12A), which is located halfway between the gate-open (green) and gate-closed (cyan) conformations observed in the as-isolated and Fe-soaked crystal structures. In contrast, in the MD simulations of the D34F and N148L mutants the flexibility of helix D is lower, and the average (conformation) density of H130 side chains matches the gate-open conformation (Figures 12B-C). These observations, which suggest low conformational freedom of the H130 side chain in the mutants, are in excellent agreement with the findings from the as-isolated and Fe-soaked structures, where only the gate open conformation is observed (Figure 7). Consequently, our results strongly suggest that flexibility in the C-terminal half of helix D in wt BfrB functions to couple breathing motions in 4-fold and B-pores with ferroxidase center ligands. It is likely that these networked motions facilitate efficient rearrangement of H130 side chains, thus enabling the gating function of this residue in Fe2+ capture and oxidation at the ferroxidase centers, and subsequent translocation of Fe3+ to the internal cavity for mineralization.
Figure 12.
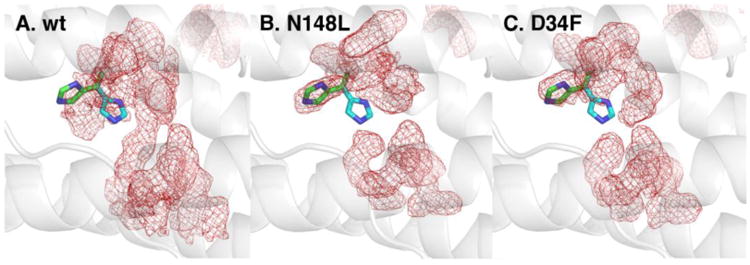
The average heavy atom density of side chain conformations sampled by ferroxidase center residues during the MD simulations is shown as red mesh for wt (A), N148L (B), and D34F BfrB (C). The H130 side chains in the “gate-open” (green) and “gate-closed” (cyan) conformations are shown in sticks. The view is from the exterior surface.
Discussion
A network of molecular fluctuations connecting 4-fold pores, B-pores and ferroxidase centers are crucial to BfrB function
Bacterioferritins serve as dynamic regulators of cytosolic iron concentrations by performing two functions: (i) iron storage, which is carried out by capturing Fe2+ followed by its oxidation to Fe3+ and storage as a ferric-phosphate mineral, and (ii) iron release, which requires reduction of the Fe3+ mineral, followed by Fe2+ mobilization from the interior cavity. The 24-mer spherical assemblies (Ftn and Ftn-like molecules) are typically referred to as protein cages, where iron can be stored. Although a protein cage may suggest a relatively rigid shell that protects the ferric mineral from the reducing potential of the cytosol, recent evidence suggests that the Bfr shell is highly dynamic. Application of a network-weaving algorithm coupled to normal mode analysis revealed long-range communication between ferroxidase centers and functionally related pores in Ftn-like molecules, which is mediated by a network of highly correlated residues, encompassing approximately 40% of residues in each subunit (9). In the case of eukaryotic Ftn (M Ftn), the network connects ferroxidase centers with 3-fold pores, an observation that is in agreement with the idea that the 3-fold pores of eukaryotic Ftn are conduits for iron traffic (25, 26). In comparison, the network of correlated residues in bacterial Ftn and Bfr, connect ferroxidase centers with 4-fold and B-pores. MD simulations carried out with BfrB showed that K+ ions move across the protein shell through B-pores, in a process that is largely aided by breathing motions driven by the folding/unfolding of the C-terminal half of the D-helices, which are coupled to lateral displacements of the E-helices that constitute the 4-fold pores (28). The lateral displacements of E-helices typically move two E-helices closer to one another, while the other two are separated further, thus narrowing the 4-fold pores. Although in the MD simulations the 4-fold pores do not permeate K+ ions, the lateral displacement of the E-helices, which is driven by the folding/unfolding transitions of the D-helices, enable periodic open/close transitions of the B-pores and facilitate K+ traffic. Hence, results obtained from coarse grain methods (normal mode analysis) and atomistic MD simulations suggest a model in which the Bfr function is intimately related to extended networks of coordinated motions that affect a large portion of the protein shell.
In this work we tested the model with mutants designed to decrease pore flexibility: (i) The side chains of N148 and Q151, which coordinate a K+ ion in the 4-fold pores of wt BfrB (Figure 2), were replaced with Leu to install hydrophobic interactions, which were expected to affect the lateral displacement of the E-helices. (ii) The D34F mutation was carried out with the expectation of occluding the B-pores and installing hydrophobic packing that would reduce B-pore flexibility. Structural determinations reveal local structural perturbations that confirm the installment of hydrophobic packing in the 4-fold and B-pores of the mutants. A surprising finding is the significant loss of Fe2+ oxidation (ferroxidase) efficiency exhibited by the mutants, particularly the D34F mutant, which oxidizes and stores only 20% of the iron presented. Remarkably, the Fe-soaked structures of D34F, N148L and Q151L show that although iron loads onto the ferroxidase center, the diiron center is not coordinated by a full complement of ferroxidase ligands (Figure 7); E51 does not bridge Fe1 and Fe2, and the H130 side chain remains in its gate open conformation. Taken together, the observations indicate that mutations in the 4-fold and B-pores reduce the conformational agility of the ferroxidase center, thus reducing its catalytic efficiency. In this context, it is noteworthy that mutations outside the pores (C89S/K96C) do not have an effect on the efficiency of iron oxidation and storage, or in the conformational fitness of ferroxidase ligands, as evaluated by the conformational competency of E51 and H130 in crystallo (Figure 7A), and the unaffected efficiency of iron oxidation and uptake in solution exhibited by the double mutant.
We suspected that the reduced fitness of the ferroxidase centers in the 4-fold and B-pore mutants may be related to changes in the dynamic properties of the pores resulting from the hydrophobic interactions installed by the mutations. Consequently, in search of additional insight, we turned to MD simulations. As previously revealed by our simulations of wt BfrB, folding/unfolding transitions of the C-terminal half or helices-D drive the dynamic behavior of 4-fold and B-pores. Given that H130 is near the center of helix D, we asked if the folding/unfolding transitions of the helix affect the conformational flexibility of H130 side chains. The MD trajectory shows that in wt BfrB the average position of the H130 side chain is midway between the gate open and gate closed conformations (Figure 12A), a finding that is in agreement with the gating function of the H130 side chain in the binding of Fe2+ at ferroxidase centers, oxidation to a di-Fe3+ moiety, and subsequent entry of Fe3+ to the interior cavity (14, 21). In contrast, the MD trajectories obtained with the D34F and N148L mutants reveal that helices D exhibit low propensity to undergo folding/unfolding transitions, which results in dampening of motions at the 4-fold and B-pores relative to wt BfrB. Analysis of the conformational freedom of H130 in the mutants showed that the average position of the H130 side chains in the simulations is aligned with the gate-open conformation (Figures 12B-C) observed in the crystal structures. Consequently, it is reasonable to conclude that in BfrB the 4-fold pores, their surrounding B-pores, and the ferroxidase centers are connected by networked fluctuations, and that these motions enable the gating properties of the ferroxidase center and the ion permeation properties of the pores.
Iron traffic paths in BfrB
In previous work we showed that soaking crystals of wt BfrB in crystallization solution containing Fe2+ prior to X-ray diffraction reveal iron bound at the ferroxidase center, and iron bound at the periphery of 4-fold pores in the interior cavity, now termed Fe4f-1 (Figure 3A). Similar experiments conducted with each of the BfrB mutants allowed us to discover many additional iron binding sites in the interior surface of BfrB, and remarkably, within the B-pores. Visualizing all the iron sites simultaneously provides unprecedented insight into conduits used by iron as it traverses the BfrB shell, as well as the routes it may take on the surface of the interior cavity on its way to mineralization or cavity exit sites. It is noteworthy that all Fe sites except Fe4f-3 are observed in the structure C89S/K96C BfrB, which allows comparison of the relative thermal factors for the experimentally observed sites (Table 3), and Figure 13 depicts these iron sites as spheres, color-coded by B-factor range. Since atomic occupancies and B-factors are highly correlated parameters (55), Fe ions were refined with occupancy factors of 1.0 and the isotropic B-factors were examined in an effort to distinguish fully occupied and partially occupied/transient ions. Note that iron sites ubiquitously found in all Fe-soaked structures (Fe1, Fe2, and Fe4f-1) have the lowest B-factors (red), which are comparable in magnitude to the protein and heme thermal factors. Together the observations suggest that iron at these sites is relatively stable and nearly 100% occupied. Iron in the B-pores (FeB-1/FeB-2) and iron near the entrance to 3-fold pores (Fe3f) exhibit B-factors nearly twice as large (orange), which suggests these sites as more transient. Finally, the Feb-fc and Fe4f-2 sites (yellow) show thermal factors four- to five-fold larger than the protein B-factors, which suggests that these sites are only transiently occupied. To aid visualization of the full-complement of iron sites observed in this study, in Figure 13 we also include Fe4f-3 (orange), the iron site observed immediately above the entrance to 4-fold pores in Fe-soaked N148L BfrB (see Figure 3-C). The iron ions in Figure 13 are superposed on the structure of the Fe-soaked D34 mutant, which was selected because it is one of the structures where the H130 side chains remain in the gate open position even after iron loads onto the ferroxidase center. With the H130 side chain in the gate open position, it is possible to observe the ferroxidase iron ions Fe1 and Fe2 (red) within ferroxidase centers, and how the gate open conformation of H130 may enable translocation of Fe2 into the interior cavity, where it binds to H46 at site Feb-fc (yellow). This is most evident in the ferroxidase centers located at the extreme left or top sections of Figure 13. Residues lining the B-pores are colored in cyan and their inspection illustrates how B-pores may function as conduits for iron traffic across the Bfr shell, with two iron ions (FeB-1, FeB-2, orange) aligned along the length of the channels (see also Figure 6E). Residues delineating the 4-fold pores are colored in magenta. The 4-fold pore at the center of the figure illustrates the three layers of iron binding sites located around the external perimeter of the pore inside the BfrB cavity (see also Figure 3): (1) Four Fe4f-1 ions (red) surround the largest diameter of the pore, where the pore wall meets the interior surface. (2) Four Fe4f-2 ions (yellow) surround the narrowest perimeter of the pore, where the pore wall meets the interior cavity. (3) One Fe4f-3 ion (orange) is located immediately above the entrance to the pore from the interior cavity. Remarkably, iron ions are not observed inside 4-fold pores, which in our structures always bind K+, with the exception of the pores in the Q151L structure. Residues lining the 3-fold pores are blue. Note how each of the sulfate ions at the entrance to each of the pores from the interior cavity (green and red sticks) is surrounded by three Fe3f ions (orange; see also Figure 4).
Table 3. Thermal factors of iron ions in the Fe soak C89S/K96C BfrB structure.
| Iron site | Location | B-factor range (Å2) | Color in Figure 12 |
|---|---|---|---|
| Feheme | heme | 26-28 | red |
| Fe4f-1 | 4-fold pore | 18-25 | red |
| Fe4f-2 | 4-fold pore | 81-105 | yellow |
| Fe3f | 3-fold pore | 37-40 | orange |
| FeB1, FeB2 | B pore | 45-63 | orange |
| Fe1, Fe2 | fc1 | 18-30 | red |
| Feb-fc | below fc | 56-81 | yellow |
fc: ferroxidase center
Figure 13.

Transverse view of the BfrB interior cavity depicting all iron sites observed in the crystal structure of Fe-soaked C89S/K96C BfrB, color coded according to the corresponding B-factor ranges listed in Table 3; red (Fe1, Fe2, Fe4f-1, Feheme), orange (Fe3f, FeB-1, FeB-2) and yellow (Fe4f-2, Feb-fc). The Fe4f-3 iron, shown immediately on top of the 4-fold pore in the center of the figure is from the Fe-soaked N148L structure, rendered in orange to reflect the range of thermal factors exhibited by these iron ions) (31-54 Å2 relative to those in Table 3. Residues lining 4-fold pores are in magenta, residues lining 3fold pores are in blue and those lining B-pores are in cyan. Sulfate ions are in green and red sticks and heme molecules are rendered as wheat sticks.
The picture that emerges from this analysis suggests that iron ions can enter the BfrB cavity via ferroxidase centers or through B-pores. Iron processed through ferroxidase centers will likely enter the cavity as Fe3+, which may then be transported by coordinating residues on the interior surface to mineralization sites, or toward 3-fold pores where it binds phosphate ions accessing the interior cavity via 3-fold pores, prior to mineralization. Iron entering through B-pores is likely in the ferrous oxidation state. Given the large number of B-pores (24 in total) on the Bfr structure, Fe2+ accessing the interior cavity via B-pores would be in relative close proximity to a ferroxidase center, where it can be oxidized by electron exchange with Fe3+ bound to the ferroxidase sites, or, at the growing Fe3+ mineral, as has been proposed for eukaryotic Ftn (56) and E. coli Bfr (57). It is noteworthy that in spite of the cumulative large number of iron binding sites we have been able to observe, we have not yet found evidence of iron ions inside the 4-fold pores of BfrB. Instead, the structures reveal several iron ions bound to the external perimeter of these pores inside the BfrB cavity, which could have reached those sites after accessing the interior cavity via B-pores. Hence, although we cannot rule out the possibility that iron ions traverse BfrB using 4-fold pores, it is tempting to speculate that the 4-fold pores in BfrB function mainly as binding sites for Fe2+ in the interior cavity, prior to oxidation at ferroxidase centers, or as nucleation sites for incorporation of iron ions into a growing Fe3+ mineral. The data also suggest that iron and phosphate combine near the 3-fold pores, which seem to be the channels used by the anion to traverse the BfrB shell.
In conclusion, our approach, which combined biochemistry with X-ray crystallography and MD simulations, provides unprecedented insight into Bfr function. Our findings demonstrate that a vast network of concerted motions connecting 4-fold and B-pores with ferroxidase centers is crucial for efficient ferroxidase activity and for permeation of ions across the Bfr cavity. Our results also reveal a large number of iron binding sites in the interior cavity, which aid in the traffic, oxidation and mineralization of the iron, and that the 3-fold pores are the likely conduits of phosphate, which is necessary to form the ferric phosphate mineral core in Bfr.
Supplementary Material
Acknowledgments
The assistance of Dr. Saroja Weeratunga in the preparation of N148L and Q151L mutants of BfrB is acknowledged. Use of the University of Kansas Protein Structure Laboratory was supported by grants from the National Center for Research Resources (5P20RR017708-10), and the National Institute of General Medical Sciences (8 P20 GM103420-10) from the National Institutes of Health. Use of the IMCA-CAT beamline 17ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with the Hauptman-Woodward Medical Research Institute. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Footnotes
This study was supported by a grant from the National Science Foundation (M.R., MCB-1158469), funds from the University of Kansas Strategic Initiative – Center for Antimicrobial Discovery and Development (M.R.), a grant from the National Institutes of Health (W.I. R01-GM092950), and a grant from XSEDE (W.I., MCB070009).
Coordinates and crystallographic structure factors for the distinct BfrB structures have been deposited in the protein data bank under accession codes listed in Table 1.
Supporting Information Available: Phased anomalous difference maps of the different iron sites in observed in the structures of Fe-soaked D34F, N148L, Q151L and C89S/K96C BfrB are shown in Figures S1-S3 and S5-S7. 2Fo-Fc maps for sulfate ions in the 3-fold pores of Fe-soaked N148L, Q151L and C89S/K96C BfrB are shown in Figure S3, and 2Fo-Fc maps of sulfate ions in the 3-fold pores of as-isolated-2 C89S/K96C BfrB are depicted in Figure S4. Anomalous peak heights and B-factors for iron atoms in the Fe-soaked structure of C89S/K96C structure are listed in Table S1, and anomalous peak heights and B-factor ranges for iron atoms in the structures of Fe-soaked D34F, N148L, and Q151L BfrB are listed in Table S2. Sequence of primer pairs used to introduce the sited directed mutations. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Touati D. Iron and oxidative stress in bacteria. Archives of biochemistry and biophysics. 2000;373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- 2.Andrews S, Norton I, Salunkhe AS, Goodluck H, Aly WS, Mourad-Agha H, Cornelis P. Control of iron metabolism in bacteria. Metal ions in life sciences. 2013;12:203–239. doi: 10.1007/978-94-007-5561-1_7. [DOI] [PubMed] [Google Scholar]
- 3.Andrews SC. Iron Storage in Bacteria. Advan Microbial Physiol. 1998;40:281–351. doi: 10.1016/s0065-2911(08)60134-4. [DOI] [PubMed] [Google Scholar]
- 4.Andrews SC, Robinson AK, Rodríguez-Quiñones F. Bacterial Iron Homeostasis. FEMS Microbiology Reviews. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 5.Pandey R, Rodriguez GM. A ferritin mutant of Mycobacterium tuberculosis is highly susceptible to killing by antibiotics and is unable to establish a chronic infection in mice. Infection and immunity. 2012;80:3650–3659. doi: 10.1128/IAI.00229-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy PV, Puri RV, Khera A, Tyagi AK. Iron storage proteins are essential for the survival and pathogenesis of Mycobacterium tuberculosis in THP-1 macrophages and the guinea pig model of infection. Journal of bacteriology. 2012;194:567–575. doi: 10.1128/JB.05553-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Expert D, Boughammoura A, Franza T. Sidreophore-Controlled Iron Assimilation in the Enterobacterium Erwinia chrysanthemi. J Biol Chem. 2008;283:36564–36572. doi: 10.1074/jbc.M807749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrews SC. The Ferritin-Like Superfamily: Evolution of the Biological Iron Storeman from a Rubrerythrin-Like Ancestor. Biochim et Biophys Acta. 2010;1800:691–705. doi: 10.1016/j.bbagen.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Ruvinsky AM, Vakser IA, Rivera M. Local packing modulates diversity of iron pathways and cooperative behavior in eukaryotic and prokaryotic ferritins. The Journal of chemical physics. 2014;140:115104. doi: 10.1063/1.4868229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewin A, Moore GR, Le Brun NE. Formation of Protein-Coated Iron Minerals. Dalton Trans. 2005:3597–3610. doi: 10.1039/b506071k. [DOI] [PubMed] [Google Scholar]
- 11.Ardejani MS, Li NX, Orner BP. Stabilization of a protein nanocage through the plugging of a protein-protein interfacial water pocket. Biochemistry. 2011;50:4029–4037. doi: 10.1021/bi200207w. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Raudah S, Teo H, Teo GW, Fan R, Sun X, Orner BP. Alanine-shaving mutagenesis to determine key interfacial residues governing the assembly of a nano-cage maxi-ferritin. The Journal of biological chemistry. 2010;285:12078–12086. doi: 10.1074/jbc.M109.092445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Brun NE, Crow A, Murphy MEP, Mauk AG, Moore GR. Iron Core Mineralization in Prokaryotic Ferritins. Biochim et Biophys Acta. 2010;1800:732–744. doi: 10.1016/j.bbagen.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Rivera M. Bacterioferritin: Structure Function and Protein-Protein Interactions. In: Kadish KK, Smith KM, Guilard R, editors. Handbook of Porphyrin Science. 2014. pp. 136–179. [Google Scholar]
- 15.Yao H, Wang Y, Lovell S, Kumar R, Ruvinsky AM, Battaile KP, Vakser IA, Rivera M. The Structure of the BfrB-Bfd Complex Reveals Protein-Protein Interactions Enabling Iron Release from Bacterioferritin. Journal of the American Chemical Society. 2012;134:13470–13481. doi: 10.1021/ja305180n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weeratunga S, Gee CE, Lovell S, Zeng Y, Woodin CL, Rivera M. Binding of Pseudomonas aeruginosa Apobacterioferritin-Associated Ferredoxin to Bacterioferritin B Promotes Heme Mediation of Electron Delivery and Mobilization of Core Mineral Iron. Biochemistry. 2009;48:7420–7431. doi: 10.1021/bi900561a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasil ML. How We Learnt About Iron Acquisition in Pseudomonas aeruginosa: A Series of Very Fortunate Events. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine. 2007;20:587–601. doi: 10.1007/s10534-006-9067-2. [DOI] [PubMed] [Google Scholar]
- 18.Oglesby-Sherrouse AG, Murphy ER. Iron-responsive bacterial small RNAs: variations on a theme. Metallomics : integrated biometal science. 2013;5:276–286. doi: 10.1039/c3mt20224k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masse E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. Journal of bacteriology. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao H, Jepkorir G, Lovell S, Nama PV, Weeratunga SK, Battaille KP, Rivera M. Two Disctinct Ferritin-Like Molecules in P. aeruginosa: The Product of the bfrA Gene is a Bacterial Ferritin (FtnA) not a bacterioferritin (Bfr) Biochemistry. 2011;50:5236–5248. doi: 10.1021/bi2004119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weeratunga S, Lovell S, Yao H, Battaile KP, Fischer CJ, Gee CE, Rivera M. Structural Studies of Bacterioferritin B (BfrB) from Pseudomonas aeruginosa Suggest a Gating Mechanism for Iron Uptake via the Ferroxidase Center. Biochemistry. 2010;49:1160–1175. doi: 10.1021/bi9015204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macedo S, Romão CV, Mitchell E, Matias PM, Liu MY, Xavier AV, LeGall J, Teixeira M, Lindley P, Carrondo MA. The Nature of the Di-Iron Site in the Bacterioferritin from Desulfovibrio desulfuricans. Nat Struct Biol. 2003;10:285–290. doi: 10.1038/nsb909. [DOI] [PubMed] [Google Scholar]
- 23.Swartz L, Kuchinskas M, Li H, Poulos TL, Lanzilotta WN. Redox-Dependent Structural Changes in the Azotobacter vinelandii Bacterioferritin: New Insights into the Ferroxidase and Iron Transport Mechanism. Biochemistry. 2006;45:4421–4428. doi: 10.1021/bi060146w. [DOI] [PubMed] [Google Scholar]
- 24.Hempstead PD, Yewdall SJ, Fernie AR, Lawson DM, Artymiuk PJ, Rice DW, Ford GC, Harrison PM. Comparison of the Three-Dimensional Structures of Recombinant Human H and Horse L Ferritins at High Resolution. J Mol Biol. 1997;268:424–448. doi: 10.1006/jmbi.1997.0970. [DOI] [PubMed] [Google Scholar]
- 25.Tosha T, Ng HL, Bhattasali O, Alber T, Theil EC. Moving Metal Ions Through Ferritin-Protein Nanocages from Three-Fold Pores to Catalytic sites. J Am Chem Soc. 2010;132:14562–14569. doi: 10.1021/ja105583d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertini I, Lalli D, Mangani S, Pozzi C, Rosa C, Theil EC, Turano P. Structural insights into the ferroxidase site of ferritins from higher eukaryotes. Journal of the American Chemical Society. 2012;134:6169–6176. doi: 10.1021/ja210084n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu HL, Zhou HN, Xing WM, Zhao JF, Li SX, Huang JF, Bi RC. 2.6 Å Resolution Crystal Structure of the Bacterioferritin from Azotobacter vinelandii. FEBS letters. 2004;573:93–98. doi: 10.1016/j.febslet.2004.07.054. [DOI] [PubMed] [Google Scholar]
- 28.Rui H, Rivera M, Im W. Protein dynamics and ion traffic in bacterioferritin. Biochemistry. 2012;51:9900–9910. doi: 10.1021/bi3013388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kabsch W. Automatic indexing of rotation diffraction patterns. Journal of Applied Crystallography. 1988;21:67–72. [Google Scholar]
- 30.Krug M, Weiss MS, Heinemann U, Mueller U. XDSAPP: a graphical user interface for the convenient processing of diffraction data using XDS. Journal of Applied Crystallography. 2012;45:568–572. [Google Scholar]
- 31.Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, Bricogne G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 34.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ringeling PL, Davy SL, Monkara FA, Hunt C, Dickson DPE, McEwan AG, Moore GR. Characterization of Bacterioferritin and Formation of Non-Haem Iron Particles in Intact Cells. Eur J Biochem. 1994;223:847–855. doi: 10.1111/j.1432-1033.1994.tb19061.x. [DOI] [PubMed] [Google Scholar]
- 39.Lee KI, Jo S, Rui H, Egwolf B, Roux B, Pastor RW, Im W. Web interface for Brownian dynamics simulation of ion transport and its applications to beta-barrel pores. Journal of computational chemistry. 2012;33:331–339. doi: 10.1002/jcc.21952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. Journal of computational chemistry. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. Journal of computational chemistry. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 42.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkewicz-Kuczera J, Yin D, Karplus J. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 43.Mackerell AD, Jr, Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. Journal of computational chemistry. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 44.Durell SR, Brooks B, Arieh BN. Solvent-Induced Forces Between Two Hydrophilic Groups. J Phys Chem. 1994;98:2198–2202. [Google Scholar]
- 45.Steinbach PJ, Brooks B. New Spherical-Cutoff Methods for Long-Range Forces in Macromolecular Simulation. J Comput Chem. 1994;15:667–683. [Google Scholar]
- 46.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A Smooth Particle Mesh Ewald Method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 47.Krissinel E, Henrick K. Secondary Structure Matching (SSM), A New tool for Fast Protein Structure Alignment in Three Dimensions. Acta Cryst. 2004;D60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 48.Mann S, Bannister J, Williams RJP. Structure and Composition of Ferritin Cores Isolated from Human Spleen, Limpet (Patella vulgata) Hemolymph and Bacterial (Pseudomonas aeruginosa) Cells. J Mol Biol. 1986;188:225–232. doi: 10.1016/0022-2836(86)90307-4. [DOI] [PubMed] [Google Scholar]
- 49.Mann S, Williams JM, Treffry A, Harrison PM. Reconstituted and Native Iron-Cores of Bacterioferritin and Ferritin. J Mol Biol. 1987;198:405–416. doi: 10.1016/0022-2836(87)90290-7. [DOI] [PubMed] [Google Scholar]
- 50.Watt GD, Frankel RB, Jacobs D, Huang H. Fe2+ and Phosphate Interactions in Bacterial Ferritin from Azotobacter vinelandii. Biochemistry. 1992;31:5672–5679. doi: 10.1021/bi00139a035. [DOI] [PubMed] [Google Scholar]
- 51.Rui H, Lee KI, Pastor RW, Im W. Molecular dynamics studies of ion permeation in VDAC. Biophysical journal. 2011;100:602–610. doi: 10.1016/j.bpj.2010.12.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kurtz DM. Stuctural Similarity and Functional Diversity of Diiron-Oxo Proteins. J Biol Inorg Chem. 1997;2:159–167. [Google Scholar]
- 53.Crow A, Lawson TL, Lewin A, Moore GR, Le Brun NE. Structural Basis for Iron Mineralization by Bacterioferritin. J Am Chem Soc. 2009;131:6808–6813. doi: 10.1021/ja8093444. [DOI] [PubMed] [Google Scholar]
- 54.Welch KD, Davis TZ, Aust SD. Iron autoxidation and free radical generation: effects of buffers, ligands, and chelators. Archives of biochemistry and biophysics. 2002;397:360–369. doi: 10.1006/abbi.2001.2694. [DOI] [PubMed] [Google Scholar]
- 55.Tronrud DE. Introduction to Macromolecular Refinement. Acta crystallographica Section D, Biological crystallography. 2004;60:2156–2168. doi: 10.1107/S090744490402356X. [DOI] [PubMed] [Google Scholar]
- 56.Zhao G, Bou-Abdallah F, Arosio P, Levi S, Janus-Chandler C, Chasteen ND. Multiple Pathways for Mineral Core Formation in Mammalian Apoferritin The Role of Hydrogen Peroxide. Biochemistry. 2003;42:3142–3150. doi: 10.1021/bi027357v. [DOI] [PubMed] [Google Scholar]
- 57.Bradley JM, Moore GR, Le Brun NE. Mechanisms of iron mineralization in ferritins: one size does not fit all. Journal of biological inorganic chemistry : JBIC : a publication of the Society of Biological Inorganic Chemistry. 2014;19:775–785. doi: 10.1007/s00775-014-1136-3. [DOI] [PubMed] [Google Scholar]
- 58.Evans P. Scaling and assessment of data quality. Acta crystallographica Section D, Biological crystallography. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 59.Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- 60.Weiss MS. Global indicators of X-ray data quality. Journal of Applied Crystallography. 2001;34:130–135. [Google Scholar]
- 61.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Evans P. Biochemistry Resolving some old problems in protein crystallography. Science. 2012;336:986–987. doi: 10.1126/science.1222162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


