Abstract
Cellulose is a fundamentally important component of cell walls of higher plants. It provides a scaffold that allows the development and growth of the plant to occur in an ordered fashion. Cellulose also provides mechanical strength, which is crucial for both normal development and to enable the plant to withstand both abiotic and biotic stresses. We quantified the cellulose concentration in the culm of 288 two – rowed and 288 six – rowed spring type barley accessions that were part of the USDA funded barley Coordinated Agricultural Project (CAP) program in the USA. When the population structure of these accessions was analysed we identified six distinct populations, four of which we considered to be comprised of a sufficient number of accessions to be suitable for genome-wide association studies (GWAS). These lines had been genotyped with 3072 SNPs so we combined the trait and genetic data to carry out GWAS. The analysis allowed us to identify regions of the genome containing significant associations between molecular markers and cellulose concentration data, including one region cross-validated in multiple populations. To identify candidate genes we assembled the gene content of these regions and used these to query a comprehensive RNA-seq based gene expression atlas. This provided us with gene annotations and associated expression data across multiple tissues, which allowed us to formulate a supported list of candidate genes that regulate cellulose biosynthesis. Several regions identified by our analysis contain genes that are co-expressed with CELLULOSE SYNTHASE A (HvCesA) across a range of tissues and developmental stages. These genes are involved in both primary and secondary cell wall development. In addition, genes that have been previously linked with cellulose synthesis by biochemical methods, such as HvCOBRA, a gene of unknown function, were also associated with cellulose levels in the association panel. Our analyses provide new insights into the genes that contribute to cellulose content in cereal culms and to a greater understanding of the interactions between them.
Background
Cellulose is crucially important to plant cell walls for several reasons. Through its presence in the primary cell wall, cellulose provides a strong yet flexible scaffold allowing growth of cells, while maintaining a defined structure and three-dimensional shape for the cell. In the secondary cell wall, which is produced once cells have stopped enlarging, cellulose provides the mechanical strength necessary to support the plant against the forces of gravity and to help it withstand biotic and abiotic stresses. For example, cellulose content per unit length of the culm has been shown to be a major determinant of straw strength [1]. A weaker culm makes a plant more prone to lodging, which leads to reduced grain yields. In secondary cell walls, cellulose and non-cellulosic wall polysaccharides are often embedded in an amorphous lignin matrix, together forming an abundant but relatively intractable bio-composite (lignocellulose) that has great potential as a raw material in second generation biofuel production. Understanding more about the genes regulating cellulose content will therefore contribute to several key global challenges related to crop yields and renewable bioenergy.
Cellulose is a long unbranched polysaccharide comprised of a chain of glucosyl residues joined by glycosidic linkages between the first and fourth carbon atoms of a series of glucosyl monomers, (1→4)- β-glucan. A sturdy and strong scaffold is constructed by multiple cellulosic chains aligning to form a crystalline microfibril comprised of between 18 and 36 glucan chains [2,3,4]. A single microfibril is believed to be synthesised by a complex of particle rosettes located at the plasma membrane [5,6,7].
Previous studies have identified the CELLULOSE SYNTHASE A (CesA) genes as being crucial in the synthesis of cellulose. The first CesA genes to be identified were GsCesA1 and GsCesA2 in cotton [8]. In barley eight CesA genes have been described, six of which form two groups of closely co-expressed genes [9]. Thus, HvCesA1, HvCesA2, and HvCesA6 comprise group one, and group two is composed of HvCesA4, HvCesA7, and HvCesA8, with HvCesA3 and HvCesA5 falling into neither cluster. Group one has been implicated in primary cell wall development, whereas group two has a clear role in secondary cell wall construction [9], based on the types and developmental stages of the tissues in which they are transcribed. This means that to some degree the expression patterns of these two groups of genes differ both spatially and temporally. A recent study by Zhang et al [10] illustrated this at a very localised and detailed scale by comparing expression patterns of the maize orthologs of these genes along an elongating internode at stage V16 (when the collar of the 16th leaf is visible) [11]. When expression levels of genes from group one and two are compared across sections of the internode it is only in section 1, the meristematic zone, that group one genes (ZmCesA2, ZmCesA5, ZmCesA7, and ZmCesA8) have a higher level of expression than group two genes (ZmCesA10, ZmCesA11, and ZmCesA13). This corresponds with increasing cellulose content from low levels in section 1 (20% by weight), to section 10, the upper maturation zone where cellulose content reaches its maximum of 42% by weight.
Mutation studies of genes involved in both primary and secondary cell wall development have also helped to characterise and typify these processes. Mutations in genes such as PROCUSTE1 (PRC1), which encodes AtCesA6 and RSW1 (AtCesA1), result in lower levels of cellulose, coupled with gaps between cells and abnormally shaped cells due to restricted cell elongation [12]. Mechanical strength for the plant overall is provided primarily by secondary cell walls. The fragile stem locus described by Burton et al.[13], fs2, is the result of mutations in HvCesA4. Two independent fs2 mutants cause reductions of crystalline cellulose by 40% and 60% when compared with their respective wild type parental lines. As variation in cellulose content can be characteristic of both primary and/or secondary wall development, we may expect to identify genes involved in either or both of these processes by carrying out a GWAS for this trait.
In addition to genes responsible for the synthesis of cellulose, several genes have been shown to be involved in cellulose breakdown. These include members of several glycoside hydrolase families [14]. There is also emerging evidence that some genes that encode polysaccharide hydrolases from these families are actually involved in the synthesis of cellulose, especially with regards to plant development. The GH9 family includes endo-(1,4)-β-glucanases, also referred to as cellulases [15]. One member of this gene family, known as KORRIGAN (KOR) (HvCel1 in barley), has been studied comprehensively to demonstrate its role in cellulose synthesis. In Arabidopsis, a reduction in crystalline cellulose coupled with an increase in the amount of pectin and non-crystalline cellulose has been observed in mutant and T-DNA insertion lines of kor. These lines also exhibit reduced cell elongation, have a dwarf phenotype and reduced wall separation between cells [16–21].
Here we describe the results of a GWAS carried out to identify regions of the barley genome associated with variation in cellulose concentration of the culm. Although such an approach has been applied to understanding regions within the genome associated with cellulose content in Miscanthus [22] and Populus [23], we are not aware of any other GWAS for this trait on other members of the Poaceae, an economically important family of plants that includes grasses and cereals. We quantified this trait in several sets of spring barley germplasm, chosen to reflect both 2-rowed and 6-rowed breeding germplasm, as well as malting and feed type barley, currently the two main uses of this crop. The germplasm samples used in our analysis are part of the USDA-funded barley CAP project and therefore represent accessions that are currently or have been included in contemporary breeding programs. We identified significant associations in regions containing glycosyl hydrolases, and others that overlap with the map positions of two other fragile stem loci, fs1 and fs3. Several of the regions contained genes known to be highly co-expressed with HvCesA genes involved in both primary and secondary cell wall development, and we show here that they are co-expressed across an extended range of barley tissues, providing independent support for their role in determining final culm cellulose concentration.
Material and Methods
Germplasm
Germplasm from six US spring breeding programs, each represented by 96 accessions (S1 Table) was grown and phenotyped for cellulose content. The germplasm has been described previously by Hamblin et al. [24] and Zhou et al [25]. Here, a combination of both 2- and six- rowed genotypes from the Montana (MT) and North Dakota (N2), Minnesota (MN), North Dakota (N6) Washington (WA) and Utah (UT) programs were included. A list of the genotypes used is given in S3 Table. All lines were genotyped with barley oligonucleotide pool assays (BOPA) 1 and 2 yielding data for 3,072 SNP loci in total per individual [24, 26]. The BOPA assays were designed using Expressed Sequence Tags (ESTs), therefore all of these SNP loci reside in protein coding regions of the genome. Genotypic data can be accessed at http://www.hordeumtoolbox.org/.
Phenotyping
The barley accessions were grown according to standard agronomic practices at the Minnesota Agricultural Experiment Station on the St. Paul campus of the University of Minnesota in 2008. Plots consisted of 30 cm long rows sown with a four-row planter with the centre two rows being the test entries and the two outside rows the border plots. Two grams of seed was used for each plot with 30 cm spacing between entries within a row and between rows. The experiment was conducted using a completely randomized design. When plants reached maturity, (hard dough stage, and leaf tissue all senescent), all lines were harvested within the same 3 day interval at the end of the season and tissue was sampled from one internode (the first below the peduncle). Internode tissue from at least three culms was bulked and ground per line. Crystalline cellulose was determined by the acetic acid/nitric acid method [27], with modifications as described in [28] and is presented in Tables 1 and 2.
Table 1. Cellulose quantities by breeding program and subpopulation in barley culms.
| Abbreviation | Breeding program | sample size | mean cellulose concentration | range of cellulose concentration |
|---|---|---|---|---|
| WA | Washington | 94 | 0.394 | 0.222–0.464 |
| MT | Montana | 96 | 0.433 | 0.340–0.497 |
| N2 | North Dakota | 84 | 0.456 | 0.264–0.648 |
| UT | Utah | 96 | 0.409 | 0.292–0.498 |
| N6 | North Dakota | 94 | 0.361 | 0.035–0.477 |
| MN | Minnesota | 96 | 0.402 | 0.179–0.459 |
| All | 560 | 0.408 | 0.035–0.648 |
Sample size, mean cellulose concentration, and range of cellulose concentration in mg cellulose / mg dry weight.
Populations are based on Barley CAP breeding programs.
Table 2. Cellulose quantities by subpopulation defined by STRUCTURE in barley culms.
| Subpopulation | sample size | mean cellulose concentration | range of cellulose concentration |
|---|---|---|---|
| 1 | 18 | 0.418 | 0.364–0.468 |
| 2 | 84 | 0.356 | 0.035–0.427 |
| 3 | 87 | 0.401 | 0.264–0.648 |
| 4 | 98 | 0.401 | 0.179–0.459 |
| 5 | 183 | 0.414 | 0.222–0.597 |
| 6 | 58 | 0.405 | 0.292–0.498 |
| All | 528 | 0.399 | 0.035–0.648 |
Sample size, mean cellulose concentration, and range of cellulose concentration in mg cellulose / mg dry weight.
Populations are based on subpopulations lines were assigned to after STRUCTURE analysis.
Genetic analysis
GenALEx 6.5 [29] was used to construct a genetic distance matrix and carry out a principal coordinates analysis (PCoA) to identify subgroups within the data set based on differences in allele frequency. We then visualised the first 3 principal coordinates using CurlyWhirly (http://bioinf.hutton.ac.uk/curlywhirly). The Bayesian clustering method STRUCTURE [30] provides Fst values by population, allowing quantification of differentiation between subgroups. Because our PCoA analysis grouped the data into between 5 and 9 subgroups, and Evanno et al., [31] suggest testing a range of values starting at 1 or 2, up to and including the true number of populations plus 3, we used 2 to 12 as our priors (K values) for STRUCTURE [30]. The admixture model was selected with a burn in length of 10 x 103 and Markov Chain Monte Carlo (MCMC) of 10 x 103 using 20 reps per value of K. The results file was uploaded into Structure Harvester v0.6 [32] to estimate the most likely number of populations within the dataset (ΔK) [31]. This corresponds to the K value with the highest ΔK. Linkage disequilibrium (LD) was calculated using Haploview [33] for both the entire dataset, and for the four subpopulations with more than 88 lines.
In total 2,842 markers that had less than 10% missing data and minimum allele frequency (MAF) >5% were included in the GWAS analysis in Genstat v.14 (VSN International). A null and kinship analysis, using a kinship matrix also generated in GenStat was performed. This analysis was carried out using the subpopulations identified as described in the previous section separately. A stringent false discovery rate (FDR) < 5% was calculated using the qvalue package [34] in R version 3.1.1 (R core team 2014) to provide adjusted p values. This set the significance threshold to (-log10 p ≥ 3.0) for the adjusted p≤0.05, and (-log10 p ≥ 5.69) for the adjusted p ≤0.001. Significant SNPs (−log10 p ≥ 3.0) positioned within 5 cM were considered to be linked to the same QTL, with the most significant SNP chosen as representing the QTL. Package stats in R Statistical Computing Environment (R Development Core Team 2014) was used to calculate the measure of association between the marker and the phenotype, the cellulose content. As the data was not expected to follow bivariate normal distribution, the squared rank-based correlation coefficient was calculated based on Kendall’s tau statistic.
Nomenclature for identified QTLs broadly follows that described in [35] and OWB-DGGT (http://wheat.pw.usda.gov/ggpages/maps/OWB/), using Cel as an abbreviation for cellulose content.
Regions that contain significant associations with cellulose concentration were anchored using the barley sequence assembly and map positions determined for all markers that had a significant association with the trait using three genetic maps, a 9K i-Select map [36], the Barley Genome Zipper [37], the POPSEQ map [38] and one physical map, i.e. the barley genome assembly [39]. However in Table 3 we just provide the name of the marker with the highest LOD score for each region that showed a significant association with cellulose content. These genetic and physical maps were used in combination with http://floresta.eead.csic.es/barleymap/ to provide annotations for genes within the intervals identified by the association analysis. When querying http://floresta.eead.csic.es/barleymap/, we extended the interval by 2.5 cM in both directions from markers that flanked our QTL (i.e. a total of 5cM) to take account of map order uncertainty and LD.
Table 3. Regions of the barley genome identified by GWAS as having significant associations with cellulose content.
| Pop 3 | Pop 5 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (2-row) | (2-row) | 3rd | |||||||||||||||
| Ch | Peak | Marker | i-select cM | POPSEQ | IBSC | Zipper MxBk | -log 10 (pvalue) | R2 | -log 10 (pvalue) | R2 | Candidate annotation | Barley Gene id (MLOC) | cM (IBSC) | Morex Contig | internode FPKM | ||
| 1H | QCel1H1 | 11_10516 | 65.5 | 59.1 | 61 | 60.8 | 0.8 | 0 | 3.6 | * | 0.04 | ? | n/a | n/a | n/a | n/a | |
| QCel1H2 | 11_11037 | 84.7 | n/a | n/a | n/a | 0 | 0.01 | 6.1 | *** | 0.1 | HvGT1 | MLOC_66574 | 72 | contig_51769 | 611.2 | ||
| 5H | QCel5H1 | 11_20097 | 100.3 | 80.3 | 80.3 | n/a | 1 | 0.04 | 5.7 | *** | 0.08 | HvGH1 | MLOC_66740 | 83.5 | contig_52038 | 20.7 | |
| HvChitinase | AK249826 | 80.4 | n/a | n/a | |||||||||||||
| QCel5H2 | 11_20134 | 106.2 | n/a | 92.4 | n/a | 0.1 | 0 | 3.9 | * | 0.05 | HvGH1 | AK252806 | 88.2 | contig_230478 | 5.6 | ||
| HvCel1 | AK373680 | 97.3 | contig_137207 | 0.1 | |||||||||||||
| 11_10094 | 122.4 | 95.5 | n/a | n/a | 0 | 0.01 | 3.8 | * | 0.07 | HvGT2 | MLOC_1996 | 97.7 | contig_117772 | 20.9 | |||
| QCel5H3 | 11_20487 | 134.6 | n/a | n/a | n/a | 2.3 | 0.03 | 4.1 | * | 0.06 | EXO67725 (GH3) | MLOC_67725 | 122.4 | contig_53512 | 3 | ||
| EXO75585 (GH3) | MLOC_75585 | 122.4 | contig_68278 | 0.2 | |||||||||||||
| GH3 | MLOC_56792 | 126.1 | contig_41156 | 0 | |||||||||||||
| HvCobra1 (GH1) | MLOC_55526 | 125.8 | contig_40045 | 482.2 | |||||||||||||
| QCel5H4 | 12_30502 | 0 | 169.4 | n/a | n/a | 0 | 0 | 5.9 | *** | 0.08 | Sugar transferase | MLOC_11345 | 166.67 | contig_1560890 | 71.5 | ||
| 6H | QCel6H1 | 11_10427 | 34.4 | 38 | n/a | 37.4 | 0.1 | 0 | 4.2 | * | 0.06 | HvCesA9 | AK367031 | 37.4 | contig_39637 | 777.1 | |
| QCel6H2 | 11_10239 | 112.3 | 105.1 | 104.8 | n/a | 0 | 0 | 3.6 | * | 0.05 | ? | n/a | n/a | n/a | n/a | ||
| 7H | QCel7H1 | 11_10069 | 83.4 | 75.2 | n/a | 74.4 | 4.7 | * | 0.04 | 0 | 0 | HvCslF6 | MLOC_57200 | 72.5 | contig_41513 | 244.1 | |
| HvNAM | MLOC_19933 | 73.2 | contig_1587220 | 7.3 |
This table includes just pop 3 and 5 as these were the two subpopulatons in which significant association with culm cellulose content were identified. Significant (−log10 p ≥ 3) marker-trait associations which pass the False Discovery Rate (FDR) adjusted p value threshold of ≤ 0.05 are indicated by *,
≤ 0.01 are indicated by **,
and ≤ 0.001 are indicated by ***
R-squared values for each marker are also included. The marker name and location for the SNP with the highest lod score is provided for each QTL. Germplasm included in the analysis was a subset of 2-rowed and 6- rowed spring accessions from the Barley CAP project. Subpopulations, referred to here as “Pop”, are those determined by STRUCTURE analysis based on degree of shared genetic ancestry. Barley Gene ID (MLOC), Morex Contig, 3rd internode FPKM—fragments per kilobase of exon per million fragments mapped from [39] are provided for each candidate gene identified under each peak where information is available. Four different versions of the barley map were used to provide information for each locus: 9K i-Select [35], Barley Genome Zipper [37], POPSEQ [38] and the IBSC [39]. Question marks indicate where no obvious candidate was identified for particular regions based on the available annotations.
For those genes that we considered to be candidates for influencing cellulose content in the culm we referred to the barley genome assembly [39] to extract information on gene expression in the third internode of the culm, a similar tissue to that assayed in the current experiment.
For those genes that we suspected to be the most likely candidates for influencing cellulose content we compared expression patterns across a range of tissues using data from the IBSC [39], and our unpublished data. The developmental stages and tissues are embryo (germinating), root (10cm seedlings), shoot (10cm seedlings), inflorescence (0.5cm), inflorescence (1–1.5cm), tillers (3rd internode), grain (5 days post anthesis (DPA)), grain (15 DPA), etiolated (10 day seedlings), lemma (42 days after planting (DAP)), lodicule (42 DAP), palea (42 DAP), epidermis (28 DAP), rachis (35 DAP), root (4 week seedling), and senescing leaf (2 months). The gene expression data are available as normalised transcript abundance in fragments per kilobase of exon per million fragments mapped (FPKM). Details regarding how the reads were mapped, total number of reads per stage and how FPKM was calculated can be found in IBSC 2012 [39]. For groups of genes we identified as being coexpressed we retrieved the rice orthologs and expression levels in 16 tissues from http://rice.plantbiology.msu.edu/. The genetic position of these associations was compared to that of introgressions containing mutant alleles of fragile stem (fs) loci, namely fs1 on chromosome 5H, fs2 on 1H, and fS3 on 7H, [40] as these have been shown to have decreased cellulose content in the culm.
Results
Population structure and linkage disequilibrium
The first axis of the PCoA explained 58% of variation within this dataset, clearly separating lines based on row type (Fig 1). Axis 2 and 3 explained a further 15.6% and 11.5% respectively. The PCoA identified between 5 and 9 putative subpopulations. Structure analysis showed that there was clearly admixture in some subpopulations, and ΔK indicated that the most likely number of subpopulations within the dataset was 6 (Fig 1, S1 Fig). Setting the shared ancestry threshold, Q, to 0.6 allocated individual lines into the populations summarised in S2 Table, with more detail provided in S3 Table. Lines that did not meet these criteria were excluded from further analysis. Intra chromosomal LD analysis revealed that the average extent of LD across all chromosomes varied between subpopulations when they were analysed separately (pop2 = 21.2cM, pop 3 = 10.8cM, pop 4 = 7.5cM, and pop5 = 13.2 cM). The average extent of LD was much smaller when all lines were analysed together (5.5cM).
Fig 1. Genotypic data and population structure analysis.
(A) Principal coordinates analysis of all lines phenotyped, colour coded by breeding program. (B) STRUCTURE bar plot for K = 6 based on bOPA 1&2 genotyping data for spring barley lines ordered by breeding programs, but colour coded by K value. Please note colours in A. are independent to those in B. Breeding program 1 = Washington (WA), 2 = Montana (MT), 3 = 2- row North Dakota (N2), 4 = Utah (UT), 5 = 6-row North Dakota (N6), and 6 = Minnesota (MN). Colours represent subpopulation defined by shared genetic ancestry. Q value represents proportion of ancestry to a given subpopulation. (C) Output from Structure Harvester showing K as calculated based on ΔK method, in this case K = 6. L(K) represents the likelihood distribution of K, and L”(K) represents the second order rate of change from L(K).
Cellulose concentration
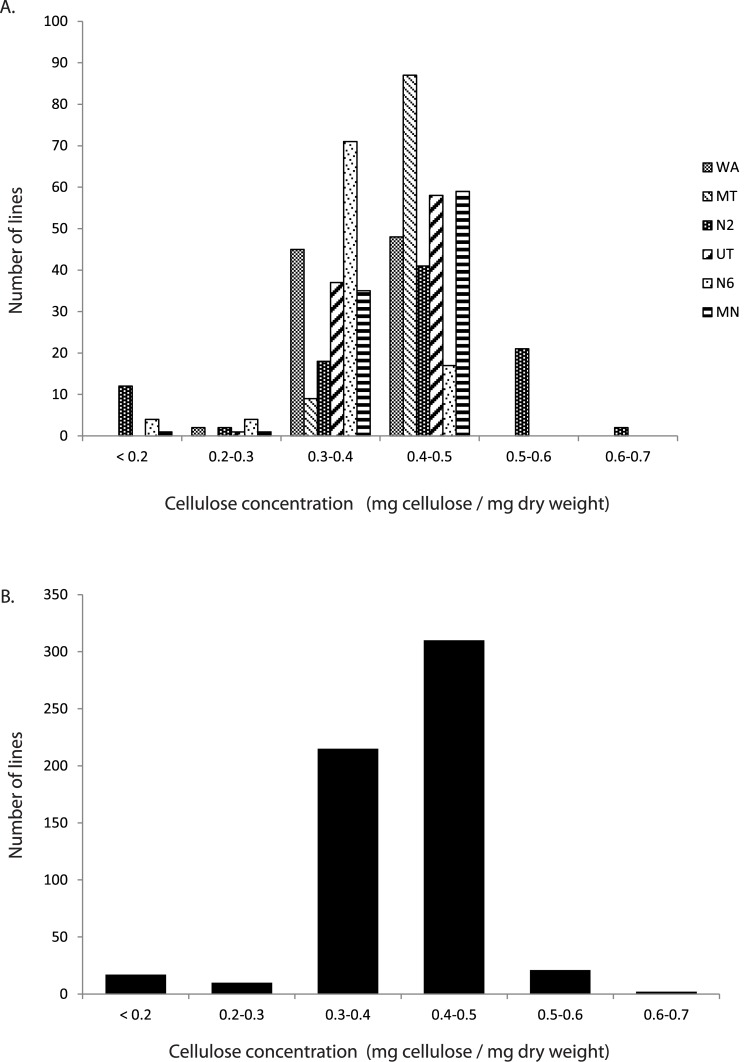
For all subpopulations most lines (between 61.5–100%) had between 0.3 and 0.5 mg cellulose /mg of dry weight. The average concentration of cellulose differed significantly among breeding programs [F (4) = 3.58, p = 0.007], ranging from 0.39 mg cellulose /mg of dry weight (MN) to 0.43 mg cellulose /mg of dry weight (MT) (Fig 2). Sample size, mean cellulose concentration, range of (cellulose concentration) values, and heritability (of cellulose concentration), within each breeding program and subpopulation as well as across all lines can be found in Table 1.
Fig 2. Phenotypic data used to carry out a genome wide association study (GWAS).
(A) Mean culm cellulose content for 2-row and 6 row spring barley accessions by breeding program. (B) Mean culm cellulose content for all lines to illustrate the distribution of this trait in the barley CAP programs included in this analysis.
GWAS
GWAS on each of the four largest subpopulations individually (subpopulations 2, 3, 4 and 5), detected nine regions containing significant associations (using a FDR corrected adjusted p value of −log10 p ≥3.0 with adjusted p≤0.05) with cellulose concentration (Table 3, S4 Table, Fig 3). Significant associations were detected in the two two-rowed subpopulations (subpopulations 3 and 5) but not in the two six-rowed subpopulations (subpopulations 2 and 4). The largest subpopulation, subpopulation 5, was the germplasm in which we detected most associations, and several had adjusted p values of p≤0.001 (−log10 p ≥5.69) (Table 3,S4 Table). A further significant association was identified in subpopulation 3. GWAS was not carried out on populations one and six due to the small number of individuals in each. When considering the PCoA (Fig 1) it is quite clear that the two rowed subpopulations, subpopulations 3 (dominated by N2), and 5 (mostly consisting of WA and MT) are almost as distinct from each other as they are from the six rowed subpopulations 2 and 4.
Fig 3. Manhattan plots of culm cellulose content (mg cellulose / mg dry weight) genome wide association scans (GWAS) using the Kinship relationship model.
The-log10 (p-values) from a genome-wide scan are plotted against the position on each of the 7 barley chromosomes. Manhattan plots are displayed for those populations which had associations that passed the significance threshold set by FDR p > 0.05 (-log10P >3.0), i.e. populations 3 and 5. Manhattan plots of the null models are provided for comparison. (A) Population 3 Kinship model. (B) Population 5 Kinship model. (C) Population 3 Null model. (D) Population 5 Null model.
We focused our attention on these 9 loci, in particular those regions for which annotations of closely associated genes implied a possible functional link to cellulose content. Regional gene content and gene annotations were derived from the barley, Brachypodium, and rice genome assemblies (Table 3) using the resources described in the material and methods. This allowed us to predict putative candidate genes for 7 of the 9 associations. The candidates fell within three groups dictated by function; primary cell wall cellulose synthesis, secondary cell wall cellulose synthesis, and glycoside hydrolases. We then referred to deep RNA-seq expression data across multiple tissues and developmental stages to explore patterns of expression for each of the candidate genes [39]. Although cellulose content in the current study was assayed using the first internode, a dataset from the 3rd internode of the culm was included in the IBSC study. We used these data to help prioritise an individual candidate gene’s likely contribution to cellulose content. We did not strictly interpret lack of expression as ruling a gene out of having a role in cellulose content; however, we did assume that those expressed in the target tissue are more likely to be important for the trait.
Glycosyl transferases (GT) and associated genes
Several regions identified in our analysis included members of GT gene families [14] (QCel1H2, QCel5H2, QCel6H1, and QCel7H1), such as the cellulose synthase (HvCesA) genes of the GT2 family of glycosyl transferases [41, 42] that have been shown to influence cellulose content. Supportive of the GWAS approach was the identification of a region containing HvCesA9 (QCel6H1), a member of the GT2 family that shows very high levels of transcript abundance in the 3rd internode (777.1 FPKM) and has high sequence similarity to HvCesA6, which itself is associated with primary cell wall cellulose synthesis [9]. Another member of the GT2 family, a gene with a known function in (1,3;1,4)-β-glucan synthesis during primary cell wall development, CELLULOSE SYTHASE LIKE F6 (HvCslF6) [43], is located within the region under QCel7H1 on chromosome 7H. This gene is represented by MLOC_ 572000 and shows high levels of transcript abundance in the culm tissue (244.1 FPKM). Further support for the validity of our approach was provided by associations that collocated with two of the three fragile stem loci known in barley: fs1 on chromosome 5H (QCel5H1) and fs3 on chromosome 7H (QCel7H1) [40]. Burton et al., [13] demonstrated that the fs2 brittle stem mutant on chromosome 1H was attributed to the insertion of a retroelement in the first intron of the CesA4 gene. Another gene annotation within the introgression on chromosome 7H containing fs1 is MLOC_19933, which is known to be a transcription factor master switch of secondary wall development. This gene is an ortholog of AtVND7 which activates transcription of several MYB transcript factors, including MYB46, that regulate secondary cell wall development in Arabidopsis [44].
Glycoside hydrolases
Genes encoding glycosyl hydrolase enzymes (GH) were found in three of the nine regions associated with cellulose levels (QCel5H1, QCel5H2, and QCel5H3), including members of families GH1, GH3, GH9 and GH19 [14]. These glycosyl hydrolase families have been previously shown to hydrolyse the glycosidic linkages between the β-linked glucosyl residues in cellulose or cellodextrins, with the exception of the GH19 family, which includes chitinases and lysozymes [14]. Some members of the GH1 family, which include β-glucosidases and the Cobra-like enzymes, have been discussed above. One of the candidate gene annotations for QCel5H2 was HvCel2, which is a member of hydrolase family GH9 and is closely related to HvCel1, also known as KOR [15]. This GH family includes endo-β-glucanases, which have been implicated in cellulose synthesis [2], and cellobiohydrolases, which are important in the complete depolymerisation of cellulose to glucose [14]. However, it must be noted that the GH9 family also includes endo-xylanases and xyloglucanases [14].
Co-expression across various tissues and developmental stages
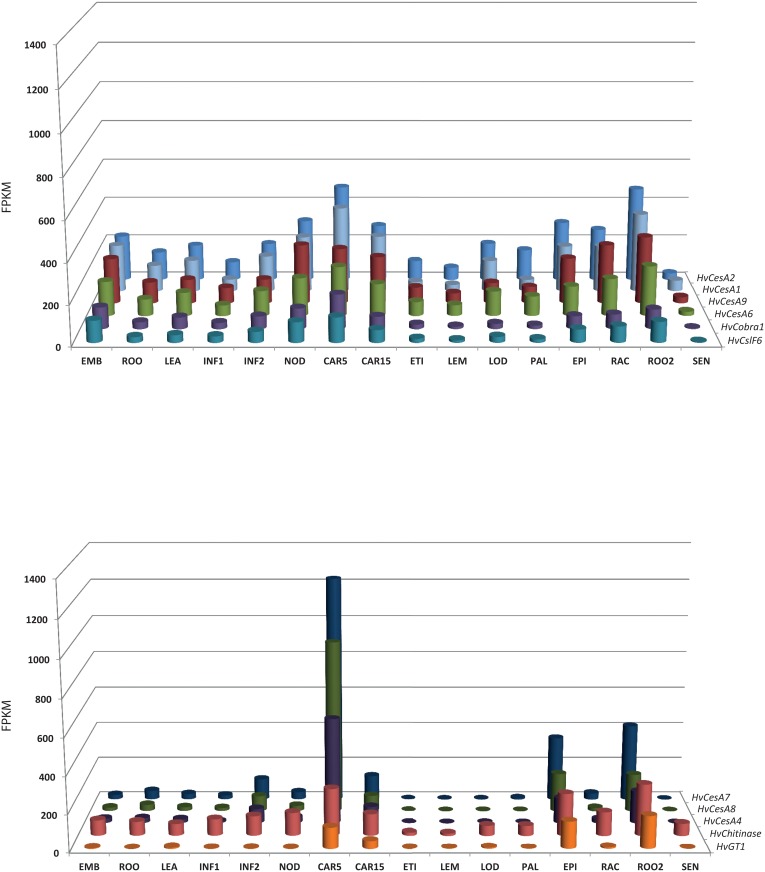
We were interested to see if any of the candidate genes from the GWAS were co-expressed, as this could provide further evidence for their being responsible for the association peaks identified. This is particularly relevant for understanding variation in cellulose content, because the synthesis of cellulose is likely to involve a number of co-factors and interacting proteins. The co-expression analysis allowed us to prioritise genes on our list of potential candidates in Table 3. We extended the number of tissues and developmental stages for which we compared candidate gene expression to sixteen (our unpublished data). When we considered the data for the 14 candidate genes from the nine regions highlighted in Table 3, we observed that five could be classified into two groups of co-expressed genes, groups 1 and 2, which are strongly co-expressed based on pairwise regression analysis between each gene across these 16 tissues with members of the CesA gene family (Fig 4). The output from the regression analysis can be found in S5 Table. Group 1 genes comprised HvCobra1, HvCslF6, and HvCesA9 (a close ortholog /paralog of HvCesA6), and formed a co-expression complex across the 16 different tissues with HvCesA1, HvCesA2, and HvCesA6, genes previously implicated in primary cell wall cellulose biosynthesis [9]. Group 2 genes, HvChitinase and HvUDP-glucosyltransferase (HvGT), were co-expressed with genes important for secondary cell wall development, namely HvCesA4, HvCesA7, and HvCesA8. Where possible we compared the coexpression of the rice orthologs of these genes, however there was no ortholog of HvCslF9 or HvChitinase1. Across both species we observed that genes in group 1 showed a higher and more significant degree of coexpression than those in group 2, with the expression of the candidate genes in barley showing a higher correlation to the known primary and secondary cell wall genes than their rice orthologs (S5 Table).
Fig 4. Co-expression of two groups of genes (HvCesA9, HvCobra1, HvCslF6, and HvChitinase, HvGT1) identified by GWAS as putatively linked to culm cellulose content with HvCesA genes known to be involved in primary and secondary cell wall development.
Transcript abundance across a range of tissues shown in fragment per kilobase of exon per million fragments mapped (FPKM) for group 1, primary cell wall including HvCesA1, HvCesA2, and HvCesA6 (A) for reference, and group 2, secondary cell wall including HvCesA4, HvCesA7 and HvCesA8 for reference (B). Abbreviations for tissues/ developmental stages as follows; EMB = Embryo tissues (germinating), ROO = Root (10cm seedlings), LEA = Shoot (10cm seedlings), INF1 = Inflorescence (0.5cm), INF2 = Inflorescence (1–1.5cm), NOD = Tillers (3rd internode), CAR5 = Grain (5 Days Post Anthesis—DPA), CAR15 = Grain (15 DPA), ETI = Etiolated (10 day seedlings), LEM = Lemma (6 weeks Days After Planting—DAP), LOD = Lodicule (42 DAP), PAL = Palea (42 DAP), EPI = Epidermis (28 DAP), RAC = Rachis (35 DAP), ROO2 = Root (28 DAP seedling), SEN = Senescing leaf (63 DAP).
Discussion
The barley CAP program, from which the plant material used in the current study was sourced, is based upon a well-studied set of accessions. Several papers describe the population structure within this germplasm [24,25,45], and more recently a GWAS for one quality trait, grain protein content, and five agronomic traits, plant height, heading date, percent kernel plump, grain test weight, and yield, has been published [45]. While we used only a subset of the germplasm from the CAP project in our analysis, our population structure analysis broadly agrees with the groups identified by others [24,25,45]. From the germplasm we sampled here, the program STRUCTURE resolved six sub-populations and due to sample size constraints we used only four of these separately in our analyses.
Using GWAS, we identified associations between cellulose content and nine regions of the barley genome in three subpopulations of 2-rowed barleys. Analysis of subpopulations 2 and 4, which are comprised of six-rowed barley lines, failed to identify any significant associations with cellulose content using FDR correction. This was likely due to limitations of our GWAS in terms of numbers of lines and numbers of SNPs used. This also indicates a relatively narrow gene pool, with the North Dakota 6 rowed program (N6) which constitutes subpopulation 2 having the narrowest genetic diversity [25]. Consequently greater population size, and, or marker density would likely provide higher confidence in our genetic results. However, despite the relatively low genetic resolution, by using the barley genome assembly [39] we were able to anchor the regions in which associations were detected, and screen gene annotations within them for putative candidate genes that could possibly be related to cellulose content. This provided us with a list of genes, and we subsequently used a gene expression atlas to further investigate their candidature based on tissue specific expression and patterns of co-expression with members of the CesA gene family and other genes known to be involved in cellulose synthesis.
While several GWAS studies concerning cellulose content have been carried out to date [23,46–48], none to our knowledge have specifically focused on barley. However, it has been the focus of QTL mapping studies taking a biparental mapping population approach [49–54]. These studies allowed us to compare the associations identified in our current work with the work of previous authors, and gauge how many of our associations are supported by the literature, and how many appeared novel/specific to the germplasm we have surveyed. Previous studies did identify QTLs collocating with either members of gene families highlighted in our current analysis, and/or genes that we observed were co-expressed with our candidate genes. Two QTL studies in Eucalyptus nitens [48,49] consisting of 420 and 296 individuals, respectively, identified a QTL in a region of the genome which includes EniCOBL4, an ortholog of AtCOBL4, which is in the same gene family as the two COBRA genes identified in our analysis. In poplar, the ortholog of one of the CesA genes that is implicated in secondary cell wall synthesis and co-expressed with genes identified in our analysis, PtiCesA7-B, was also found to be associated with cellulose content in a GWAS [23]. Collocation of a QTL for cellulose content and hemicellulose content in sorghum was reported in two bi-parental mapping populations [53,54]. Moreover, a recent GWAS for (1,3:1,4)-β-glucan content in the CAP populations [55] identified a QTL for this trait which maps to the same cM position as the most significant marker defining QCel7H1 reported in the current study.
Leveraging the barley genome assembly and published (and unpublished) RNA-seq expression data [39], we were able to highlight genes potentially contributing to variation in cellulose content based on annotation and subsequently by comparing their expression patterns to the CesA genes that are central to cellulose synthesis. This identified a group of co-expressed genes linked to primary cell wall cellulose synthesis and a second group involved in secondary cell wall development. For the primary cell wall group, there is also evidence from the literature that the genes highlighted by our analysis are co-expressed, and potentially involved in a regulatory complex [9]. HvCobra1, a likely ortholog of ZmBk2L3, has been shown in maize to be co-expressed with primary cell wall CesA genes [56]. The HvCslF6 gene is down-regulated by a small interfering RNA antisense transcript of HvCesA6, illustrating the link between several of these co-expressed genes [57]. The current work provides independent evidence of the co-expression of these genes, and their possible involvement in enzyme-protein complexes.
The secondary cell wall association group includes HvCesA4, HvCesA7, HvCesA8, HvChitinase, and HvGT1 (MLOC_66574) (Table 3). The Arabidopsis homolog of HvChitinase, AtPom1, has previously been shown to be co-expressed with genes involved in primary cell wall development in seedlings and mature roots, rosette leaves, various floral tissues, and siliques, and has been described as having a role in cellulose microfibril assembly [58]. However, while we observed similar patterns of expression to the primary cell wall CesA genes in equivalent tissues, a survey of the 16 tissues available from the barley genome assembly [39] we revealed a strong trend of co-expression with the secondary cell wall CesA genes. This does not disprove the hypothesis of Sanchez-Rodriguez et al., [58], that chitinases such as AtPom1 bind to cellulose microfibrils during assembly to regulate this process, but instead suggests a role in both processes, depending on the tissue. It must be noted that plants and their cell walls do not contain chitin and that the chitinases have long been identified as pathogenesis-related proteins that are involved in defence against fungal pathogens [59]. Finally, a putative role for the HvGT1, a UDP-glucosyltransferase, in secondary cell wall synthesis has yet to be proposed, however the enzyme is has been annotated as a 3-O-glucosyltransferase and a UDP-glucosyl transferase [14] and may be involved in the provision of the UDP-glucose donor substrate for cellulose biosynthesis.
Two regions associated with cellulose content, QCel6H1 and QCel7H1, include members of glycosyl transferase family 2, namely HvCesA9 on chromosome 6H and HvCslF6 on chromosome 7H. In addition, QCel5H3 on chromosome 5H contained genes annotated as COBRA—LIKE (COBL). There is evidence from mutant studies that the COBRA gene family influences cellulose content based on a reduction in cellulose content in cobl mutants. Schindelman et al.[60], described COB mutants as having reduced cellulose content in the elongation zone of the root where the orientation of cell expansion increased in a radial direction compared to the wild type (that exhibited longitudinal expansion in this zone). Based on work using the cob-1 Arabidopsis mutant, COB was subsequently implicated in the deposition of cellulose microfibrils in the primary cell wall of developing roots [61]. MLOC_55526, the likely ortholog of AtCOB [57], falls within the interval identified on chromosome 5H, which is significantly associated with cellulose content. This gene is also expressed highly in the 3rd internode of the culm (482.2 FPKM) and is co-expressed with three members of the CesA family, ZmCesA1, ZmCesA2, and ZmCes6. MLOC_11345.4 is the best blastn hit for ZmBk2L3, a maize gene previously shown to be co-expressed with the CesA genes. Bk2 is another gene that encodes a member of the COBRA LIKE family, a likely ortholog of AtCobL4, and shows high levels of correlated co-expression with AtCesA10, AtCesA11, and AtCesA12, genes involved in secondary cell wall development. Plants with the mutant allele of Bk2 show a reduction in cellulose content in the culm and this is thought to be due to a fault in the cellulose synthase complex. Ultimately, this not only reduces the cellulose content but leads to a reduction in mechanical strength of the culm [62]. The mechanism by which a COBRA co-expressed with the secondary cell wall associated CesA genes influences cellulose crystallization has recently been described [63]. The NH2- terminal region of the protein encoded by Brittle Culm1 (BC1), a COBRA-LIKE gene, contains a carbohydrate binding module that interacts with crystalline cellulose and determines the size of microfibril crystallite size, regulating cellulose assembly [63]. To our knowledge the equivalent mechanism for the COBRA-LIKE genes associated with primary cell wall CesA genes is yet to be described.
We identified additional genes in regions associated with cellulose content that are known to have an impact on secondary cell wall development in barley. QCel5H1 in population 5 on chromosome 5H co-locates with the introgression of fs1 [40]. Similarly, a region identified as being associated with cellulose content on chromosome 7H, QCel7H1 in population 3, overlaps with the introgression known to contain fs3 [40]. Both fs1 and fs3 mutant accessions have reduced mechanical strength in their culms; a phenotype commonly linked to mutations in genes involved in secondary cell wall synthesis. Burton et al. [13] identified GH17 genes as being differentially transcribed in the maturation zone of internodes of stems from a wild type barley line (Kobinkatagi (J066)) and M382, a near isogenic fs3 mutant line. The same authors [13] also showed that the fragile stem phenotype of fs2, not identified in the current study, was due to a retroelement in the HvCesA4 gene, thereby illustrating the link between the fragile stem loci and key genes known to determine cellulose content.
Three of these nine regions contain members of the glycoside hydrolase families GH1, GH3, GH9, and GH19, which have previously been implicated in cellulose synthesis and degradation [64]. Several glycoside hydrolase (GH) families are capable of hydrolysing (1,4)-β-glycosidic linkages in cellulose and other polysaccharides, together with oligosaccharide products generated by endo-hydrolase activity [14]. Therefore, (1,4)-β-glucan endohydrolases (cellulases) of the GH9 families can hydrolyse cellulose chains, although their activity on crystalline cellulose is generally low. Certain β-glucosidases and β-glucan exohydrolases of the GH1 and GH3 families hydrolyse oligosaccharides derived from cellulose by endo-hydrolase action. An association identified on chromosome 6H contained a GH9 gene, which includes the endo-(1,4)-β-glucanases (cellulases)[64]. By constitutively expressing a poplar cellulase gene (PaPopCel1) in Arabidopsis thaliana, Park et al., [65] concluded that this group of enzymes trims noncrystalline regions of a cellulose molecule, releasing the trapped xyloglucan from the cellulose microfibrils. Cosgrove [2] also highlighted the potential role of the cellulase gene KOR in the maintenance of normal development of cellulose microtubules in terms of proper crystallisation into microfibrils. QCel5H1 on chromosome 5H contained an ortholog of AtCTL1, which is a GH19 (HvChitinase) and has been implicated in cellulose synthesis and assembly [59, 66] in both the primary and secondary cell walls [67].
We carried out our GWAS using subpopulations identified by rigorous population genetics analysis as described in [68]. We did also consider whether the Q +K method in Tassel version 5.2.6 [69]would be suitable for this germplasm set using a trait, row type, which is both genetically well characterised and simple compared to cellulose content. Row type is represented by equal numbers of two rowed and six rowed accessions in our germplasm, and the key genes influencing this trait are either cloned i.e. HvVrs1 on 2H [70], HvInt.c on 4H [71], HvVrs4 on 3H [72], or have been delimited to a small interval i.e. HvVrs2 on 5H and HvVrs3 on 1H [40]. We know from Ramsay et al., [71] that taking a population of two and six rowed lines can detect row type genes when a kinship matrix is applied. However even if we used a relaxed adjusted p- value after FDR of p<0.05 no significant associations were identified (S2 Fig, S6 Table). The strongest association with row type was identified using marker 12_30880 on 7H, with a—log10 (p) = 3.42, and (non-significant) adjusted p value = 0.53. We also observed very different patterns of LD between subpopulations and with different regions showed LD depending on subpopulation. A much faster rate of LD decay was observed when the dataset was analysed as whole compared to when subpopulations were analysed separately, concurrent with Mohammadi et al. [73]. Taken together with the clear divisions within this germplasm evident from population structure analysis we decided that analysing all lines together might “dilute” any potential population specific marker trait associations.
Conclusion
In summary, the GWAS analysis revealed a number of candidate genes that are likely to influence cellulose synthesis in barley. Participation of the products of these genes in cellulose biosynthesis has, in many cases, been validated in previous QTL analyses of biparental mapping populations and through biochemical, co-expression and other approaches that link these genes to the cellulose synthesis process. Of particular importance is the possibility that one of the products of these candidate genes interacts directly with the CESA enzyme to form an active complex. Despite the best attempts of many groups around the world over several decades, expression of CesA genes in cell-free systems has rarely produced credible levels of cellulose synthesis in vitro. A possible explanation for this has been presented by Morgan et al. [74] who showed that the cellulose synthase enzyme (designated BcsA) in the bacterium Rhodobacter was inactive in the absence of a second protein (BcsB) and solved the three-dimensional structure of the two co-crystallised proteins of the active complex. If higher plant CESA enzymes also require a second partner to form an active cellulose synthase complex, one of the candidate genes identified in the present GWAS analysis might encode the ‘missing’ partner protein. This possibility can now be explored through further validation of the role of these candidate genes using transgenesis and directed mutagenesis approaches, co-expression in heterologous expression systems and through in vitro assays.
Supporting Information
Breeding program 1 = Washington (WA), 2 = Montana (MT), 3 = 2- row North Dakota (N2), 4 = Utah (UT), 5 = 6-row North Dakota (N6), and 6 = Minnesota (MN). Q value represents proportion of ancestry to a given subpopulation.
(EPS)
(EPS)
* = included in this study.
(DOCX)
The probability that an individual belongs to a population, Q = 0.6, and the number of subpopulations within the dataset, ΔK = 6. Numbers in brackets representing the number of lines from each breeding program assigned into subpopulation.
(DOCX)
(DOCX)
Marker positions are as described in Comadran et al [36].
(DOCX)
Regression analysis of the rice orthologs of these genes is included for comparison.
(DOCX)
Marker positions are as described in Comadran et al [36].
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the USDA National Institute of Food and Agriculture Barley Coordinated Agricultural Project (#2006-55606-16722) DuPont Agricultural Biotechnology, the Australian Research Council and the BBSRC responsive mode scheme Grant number BB/J014869/1 and the Scottish Government Research Program Work Package 5.2. During the period in which the research was undertaken, JAR and KSD were affiliated with the DuPont Agricultural Biotechnology division at the commercial company DuPont Pioneer. DuPont Pioneer provided support in the form of salaries for authors JAR and KSD, and reviewed and approved the manuscript for publication. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1. Appenzeller L, Doblin M, Barreiro R, Wang HY, Niu XM, Kollipara K, et al. Cellulose synthesis in maize: isolation and expression analysis of the cellulose synthase (CesA) gene family. Cellulose 2004, 14 (3–4):287–299. [Google Scholar]
- 2. Cosgrove DJ. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol 2005, 6 (11):850–861. [DOI] [PubMed] [Google Scholar]
- 3. Carpita NC. Update on mechanisms of plant cell wall biosynthesis: how plants make cellulose and other (1→4)-β-d-Glycans. Plant Physiol 2011, 14 (1):171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thomas LH, Forsyth VT, Sturcova A, Kennedy CJ, May RP, Altaner CM, et al. Structure of cellulose microfibrils in primary cell walls from collenchyma. Plant Physiol 2013, 161:465–476. 10.1104/pp.112.206359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Giddings TH, Brower DL, Staehelin LA. Visualization of particle complexes in the plasma membrane of Micrasterias denticulata associated with the formation of cellulose fibrils in primary and secondary cell walls. J. Cell Biol. 1980, 84 (2):327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mueller SC, Brown RM. Evidence for an intramembrane component associated with a cellulose microfibril-synthesizing complex in higher plants. J. Cell Biol. 1980, 84(2): 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doblin MS, Kurek I, Jacob-Wilk D, Delmer DP. Cellulose biosynthesis in plants: from genes to rosettes. Plant Cell Physiol 2002, 14(12):1407–1420 [DOI] [PubMed] [Google Scholar]
- 8. Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. U.S.A. 1996, 93(22): 12637–12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burton RA, Shirley NJ, King BJ, Harvey AJ, Fincher GB. The CesA Gene Family of Barley. Quantitative Analysis of Transcripts Reveals Two Groups of Co-Expressed Genes. Plant Physiol 2004, 134(1):224–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang Q, Cheetamun R, Dhugga KS, Rafalski JA, Tingey SV, Shirley NJ, et al. Spatial gradients in cell wall composition and transcriptional profiles along elongating maize internodes. BMC Plant Biol. 2014, 14(1): 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McWilliams DA, Berglund DR, Endres GJ. (1999) Corn growth and management quick guide. In vol. A-1173 North Dakota: North Dakota State University. [Google Scholar]
- 12. Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, et al. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell, 2000, 12(12):2409–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burton RA, Ma G, Baumann U, Harvey AJ, Shirley NJ, Taylor J, et al. A customized gene expression microarray reveals that the brittle stem phenotype fs2 of barley is attributable to a retroelement in the HvCesA4 cellulose synthase gene. Plant Physiol 2010, 14(4):1716–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 2009, 37:D233–D238. 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Buchanan M, Burton RA, Dhugga KS, Rafalski AJ, Tingey SV, Shirley NJ, Fincher GB. Endo-(1, 4)-β-Glucanase gene families in the grasses: temporal and spatial. Co-transcription of orthologous genes. BMC Plant Biol. 2012, 12(1): 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou HL, He SJ, Cao YR, Chen T, Du BX, Chu CC,et al. OsGLU1, A putative membrane-bound Endo-1, 4-ß-d-glucanase from rice, affects plant internode elongation. Plant Mol. Biol. 2006, 60(1):137–151. [DOI] [PubMed] [Google Scholar]
- 17. His I, Driouich A, Nicol F, Jauneau A, Höfte H. Altered pectin composition in primary cell walls of korrigan, a dwarf mutant of Arabidopsis deficient in a membrane-bound endo-1, 4-β-glucanase. Planta 2001, 212 (3): 348–358. [DOI] [PubMed] [Google Scholar]
- 18. Lane DR, Wiedemeier A, Peng L, Höfte H, Vernhettes S, Desprez T,et al. Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1, 4-β-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol.2001, 126(1): 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nicol F, HisI, Jauneau A, Vernhettes S, Canut H, Höfte H. A plasma membrane‐bound putative endo‐1, 4‐β‐d‐glucanase is required for normal wall assembly and cell elongation in Arabidopsis. The EMBO Journal 1998, 17(19): 5563–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sato S, Kato T, Kakegawa K, Ishii T, Liu YG, Awano T,et al. Role of the putative membrane-bound endo-1, 4-β-glucanase KORRIGAN in cell elongation and cellulose synthesis in Arabidopsis thaliana. Plant Cell Physiol. 2001, 42 (3):251–263. [DOI] [PubMed] [Google Scholar]
- 21. Szyjanowicz PM, McKinnon I, Taylor NG, Gardiner J, Jarvis MC, Turner SR. The irregular xylem 2 mutant is an allele of korrigan that affects the secondary cell wall of Arabidopsis thaliana. Plant J. 2004, 37(5):730–740. [DOI] [PubMed] [Google Scholar]
- 22. Slavov GT, Nipper R, Robson P, Farrar K, Allison GG, Bosch M, et al. Genome‐wide association studies and prediction of 17 traits related to phenology, biomass and cell wall composition in the energy grass Miscanthus sinensis. New Phytol. 2013, 201(4):1227–39 10.1111/nph.12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Porth I, Klapšte J, Skyba O, Hannemann J, McKown AD, Guy RD, et al. (2013). Genome‐wide association mapping for wood characteristics in Populus identifies an array of candidate single nucleotide polymorphisms. New Phytol. 2013, 200(3): 710–726. 10.1111/nph.12422 [DOI] [PubMed] [Google Scholar]
- 24. Hamblin MT, Close TJ, Bhat PR, Chao S, Kling JG, Abraham KJ, et al. Population structure and linkage disequilibrium in US barley germplasm: Implications for association mapping. Crop Sci. 2010, 50(2): 556–566. [Google Scholar]
- 25. Zhou H, Muehlbauer G, Steffenson B. Population structure and linkage disequilibrium in elite barley breeding germplasm from the United States. J. Zhejiang Univ. Sci. 2012, 13(6):438–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Close TJ, Bhat PR, Lonardi S, Wu Y, Rostoks N, Ramsay L,et al. Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics 2009, 10:582 10.1186/1471-2164-10-582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Updegraff DM. Semimicro determination of cellulose in biological materials. Anal. Biochem. 1969, 32:420–424 [DOI] [PubMed] [Google Scholar]
- 28. Pettolino FA, Walsh C, Fincher GB, Bacic A. Determining the polysaccharide composition of plant cell walls. Nat Protoc 2012, 7:1590–1607 10.1038/nprot.2012.081 [DOI] [PubMed] [Google Scholar]
- 29. Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012, 28(19):2537–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000, 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 2005, 14(8): 2611–2620. [DOI] [PubMed] [Google Scholar]
- 32. Earl DA. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012, 4(2): 359–361. [Google Scholar]
- 33. Barrett JC, Fry B, Maller J, Daly MJ. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 2: 263–265. [DOI] [PubMed] [Google Scholar]
- 34. Dabney A, Storey JD. qvalue: Q-value estimation for false discovery rate control. R package version 1.38.0. 2010. [Google Scholar]
- 35. Szucs P, Blake VC, Bhat PR, Close TJ, Cuesta-Marcos A, Muehlbauer GJ,et al. An Integrated Resource for Barley Linkage Map and Malting Quality QTL Alignment. Plant Gen. 2009, 2:134–140. [Google Scholar]
- 36. Comadran J, Kilian B, Russell J, Ramsay L, Stein N, Ganal MW,et al. Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat Genet. 2012, 44:1388–1392. 10.1038/ng.2447 [DOI] [PubMed] [Google Scholar]
- 37. Mayer KF, Martis M, Hedley PE, Simkova H, Liu H, Morris JA, et al. Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell. 2011, 23: 1249–1263. 10.1105/tpc.110.082537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mascher M, Muehlbauer GJ, Rokhsar DS, Chapman J, Schmutz J, Barry K,et al. Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ). Plant J. 2013, 76(4): 718–727. 10.1111/tpj.12319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. IBSC. A physical genetic and functional sequence assembly of the barley genome. Nature 2012, 491 (7426):711–716. 10.1038/nature11543 [DOI] [PubMed] [Google Scholar]
- 40. Druka A, Franckowiak J, Lundqvist U, Bonar N, Alexander J, Houston K,et al. (2011). Genetic dissection of barley morphology and development. Plant Physiol. 2011, 155(2): 617–627. 10.1104/pp.110.166249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Richmond T, Somerville C. The cellulose synthase superfamily. Plant Physiol. 2000, 124: 495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hazen SP, Scott-Craig JS, Walton JD. Cellulose synthase-like genes of rice. Plant Physiol. 2002, 128: 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, et al. Cellulose Synthase-like CslF Genes Mediate the Synthesis of Cell Wall (1,3;1,4)-β-d-Glucans. Science 2006, 311: 1940–1942. [DOI] [PubMed] [Google Scholar]
- 44. Zhong R, Richardson EA, Ye ZH. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 2007. 19(9):2776–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang H, Smith KP, Combs E, Blake T, Horsley RD, Muehlbauer GJ. Effect of population size and unbalanced data sets on QTL detection using genome-wide association mapping in barley breeding germplasm. Theor Appl Genet 2012, 124(1):111–124. 10.1007/s00122-011-1691-8 [DOI] [PubMed] [Google Scholar]
- 46. Pauli D, Muehlbauer GJ, Smith KP, Cooper B, Hole D, Obert D E, et al. Association Mapping of Agronomic QTLs in US Spring Barley Breeding Germplasm. The Plant Genome 10.3835/plantgenome2013.11.0037 21566722 [DOI] [Google Scholar]
- 47. González-Martínez SC, Wheeler NC, Ersoz E, Nelson CD, Neale DB. Association Genetics in Pinus taeda L. I. Wood Property Traits. Genetics 2007, 175:399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thumma BR, Matheson BA, Zhang D, Meeske C, Meder R, Downes GM, Southerton SG. Identification of a Cis-Acting Regulatory Polymorphism in a Eucalypt COBRA-Like Gene Affecting Cellulose Content. Genetics 2009, 183(3):1153–64 10.1534/genetics.109.106591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thumma BR, Southerton SG, Bell JC, Owen JV, Henery ML, Moran GF. Quantitative trait locus (QTL) analysis of wood quality traits in Eucalyptus nitens. Tree Genetics & Genomes 2010, 6(2), 305–317. [Google Scholar]
- 50. Sewell MM, Davis MF, Tuskan GA, Wheeler NC, Elam CC, Bassoni DL, Neale DB. Identification of QTLs influencing wood property traits in loblolly pine (Pinus taeda L.). II. Chemical wood properties. Theor. Appl Genet. 2002, 104(2–3):214–222. [DOI] [PubMed] [Google Scholar]
- 51. Chagné D, Brown G, Lalanne C, Madur D, Pot D, Neale D, Plomion C. Comparative genome and QTL mapping between maritime and loblolly pines. Mol Breeding 2003, 12:185–195. [Google Scholar]
- 52. Markussen T, Fladung M, Achere V, Favre JM, Faivre-Rampant P, Aragones A,et al. Identification of QTLs controlling growth, chemical and physical wood property traits in Pinus pinaster (Ait.). Silvae genetica 2003, 52: 8–14. [Google Scholar]
- 53. Murray SC, Rooney WL, Mitchell SE, Sharma A, Klein PE, Mullet JE, Kresovich S. Genetic improvement of sorghum as biofuel feedstock II QTL for stem and leaf structural carbohydrates. Crop Sci 2008, 48:2181–2193. [Google Scholar]
- 54. Shiringani AL, Friedt W.QTL for fibre-related traits in grain×sweet sorghum as a tool for the enhancement of sorghum as a biomass crop. Theor Appl Genet 2011, 123:999–1011. 10.1007/s00122-011-1642-4 [DOI] [PubMed] [Google Scholar]
- 55. Mohammadi M, Endelman JB, Nair S, Shiaoman C, Jones SS, Muehlbauer GJ,et al. Association mapping of grain hardness, polyphenol oxidase, total phenolics, amylose content, and β-glucan in US barley breeding germplasm. Mol Breeding 10.1007/s11032-014-0112-5 [DOI] [Google Scholar]
- 56. Brady SM, Song S, Dhugga KS, Rafalski JA, Benfey PN. Combining expression and comparative evolutionary analysis. The COBRA gene family. Plant Physiol 2007, 143(1): 172–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Held MA, Penning B, Brandt AS, Kessans SA, Yong W, Scofield SR, Carpita NC. Small-interfering RNAs from natural antisense transcripts derived from a cellulose synthase gene modulate cell wall biosynthesis in barley. Proc Natl Acad Sci U S A 2008, 105(51):20534–9. 10.1073/pnas.0809408105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sánchez-Rodríguez C, Bauer S, Hématy K, Saxe F, Ibáñez AB, Vodermaier V, et al. Chitinase-like1/pom-pom1 and its homolog CTL2 are glucan-interacting proteins important for cellulose biosynthesis in Arabidopsis. Plant Cell 2012, 24(2): 589–607. 10.1105/tpc.111.094672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Van Loon LC, Rep M, Pieterse CMJ. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol 2006, 44; 135–162 [DOI] [PubMed] [Google Scholar]
- 60. Schindelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, et al. COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev 2001, 15(9):1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Roudier F, Fernandez AG, Fujita M, Himmelspach R, Borner GH, Schindelman G,et al. COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 2005, 17(6): 1749–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ching A, Dhugga KS, Appenzeller L, Meeley R, Bourett TM, Howard RJ, Rafalski A. Brittle stalk 2 encodes a putative glycosylphosphatidylinositol-anchored protein that affects mechanical strength of maize tissues by altering the composition and structure of secondary cell walls. Planta 2006, 224(5), 1174–1184. [DOI] [PubMed] [Google Scholar]
- 63. Liu L, Shang-Guan K, Zhang B, Liu X, Yan M, Zhang L,et al. Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils. PLoS Genet 2013, 9(8): e1003704 10.1371/journal.pgen.1003704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J 1991, 280: 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Park YW, Tominaga R, Sugiyama J, Furuta Y, Tanimoto E, Samejima M, et al. Enhancement of growth by expression of poplar cellulase in Arabidopsis thaliana. Plant J. 2003, 33:1099–1106. [DOI] [PubMed] [Google Scholar]
- 66. Wu B, Zhang B, Dai Y, Zhang L, Shang-Guan K, Peng Y,et al. Brittle Culm15 encodes a membrane-associated chitinase-like protein required for cellulose biosynthesis in rice. Plant Physiol 2012, 159: 1440–1452. 10.1104/pp.112.195529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hirano K, Aya K, Morinaka Y, Nagamatsu S, Sato Y, Antonio BA, et al. Survey of genes involved in rice secondary cell wall formation through a co-expression network. Plant Cell Physiol. 2013, 54:1803–1821. 10.1093/pcp/pct121 [DOI] [PubMed] [Google Scholar]
- 68. Waugh R, Flavell AJ, Russell J, Thomas W, Ramsay L, Comadran J. Exploiting Barley Genetic Resources for Genome Wide Association Scans (GWAS) Tuberosa R. et al. (eds.), Genomics of Plant Genetic Resources, 237 Springer Science+Business Media Dordrecht; 2014. [Google Scholar]
- 69. Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23:2633–2635. [DOI] [PubMed] [Google Scholar]
- 70. Komatsuda T, Pourkheirandish M, He C, Azhaguvel P, Kanamori H, Perovic D,et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci U S A 2007,104: 1424–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ramsay L, Comadran J, Druka A, Marshall DF, Thomas WTB, Macaulay M,et al. 2011. INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat Genet 43: 169–172 10.1038/ng.745 [DOI] [PubMed] [Google Scholar]
- 72. Koppolu R, Anwar N, Sakuma S, Tagiri A, Lundqvist U, Pourkheirandish M,et al. Six-rowed spike4 (Vrs4) controls spikelet determinacy and row-type in barley. Proc Natl Acad Sci U S A. 2013, 110: 13198–203. 10.1073/pnas.1221950110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mohammadi M, Blake TK, Budde AD, Chao S, Hayes PM, Horsley RD,et al. A genome-wide association study of malting quality across eight US barley breeding programs. Theor Appl Genet 2015: 1–17. 10.1007/s00122-014-2399-3 [DOI] [PubMed] [Google Scholar]
- 74. Morgan J.L., Strumillo J., and Zimmer J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 2013, 493: 181–186. 10.1038/nature11744 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Breeding program 1 = Washington (WA), 2 = Montana (MT), 3 = 2- row North Dakota (N2), 4 = Utah (UT), 5 = 6-row North Dakota (N6), and 6 = Minnesota (MN). Q value represents proportion of ancestry to a given subpopulation.
(EPS)
(EPS)
* = included in this study.
(DOCX)
The probability that an individual belongs to a population, Q = 0.6, and the number of subpopulations within the dataset, ΔK = 6. Numbers in brackets representing the number of lines from each breeding program assigned into subpopulation.
(DOCX)
(DOCX)
Marker positions are as described in Comadran et al [36].
(DOCX)
Regression analysis of the rice orthologs of these genes is included for comparison.
(DOCX)
Marker positions are as described in Comadran et al [36].
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.