Abstract
13C Methyl TROSY NMR spectroscopy has emerged as a powerful method for studying the dynamics of large systems such as macromolecular assemblies and membrane proteins. Specific 13C labeling of aliphatic methyl groups and perdeuteration has been limited primarily to proteins expressed in E. coli, preventing studies of many eukaryotic proteins of physiological and biomedical significance. We demonstrate the feasibility of efficient 13C isoleucine δ1-methyl labeling in a deuterated background in an established eukaryotic expression host, Pichia pastoris, and show that this method can be used to label the eukaryotic protein actin, which cannot be expressed in bacteria. This approach will enable NMR studies of previously intractable targets.
Keywords: Methyl labeling, TROSY, Eukaryotic expression system, Pichia pastoris, Deuteration, Actin
Well-resolved multi-dimensional NMR spectra are essential for obtaining structural and dynamic information on backbone and sidechain moieties within proteins. However, obtaining such spectra of large macromolecules is complicated by poor peak dispersion and line broadening due to rapid transverse relaxation of nuclear magnetization and spectral crowding. To overcome this problem for aliphatic sidechains, proteins can be specifically labeled with 13C in the methyl groups of isoleucine, leucine, and valine residues using 13C α-ketoacid precursors in E. coli (Gardner and Kay 1997; Goto et al. 1999). When paired with selective protonation in an otherwise deuterated background (Rosen et al. 1996), this approach takes advantage of the favorable relaxation properties of 13C-methyl groups with the application of Transverse Relaxation Optimized Spectroscopy (TROSY) (Pervushin 1997; Ollerenshaw 2003). However these methods have remained unavailable for many eukaryotic proteins due to poor expression and folding in E. coli resulting from lack of required chaperones, lack of proper post-translational modifications, or improper membrane composition.
Several different eukaryotic hosts, including fungi (Miyazawa-Onami et al. 2013), insect cells (Nygaard et al. 2013; Kofuku et al. 2014), and mammalian cells (Werner et al. 2008), have been used to overexpress proteins for NMR. While these systems have succeeded in producing amino acid-specific and uniformly 15N or 13C labeled material (Chen et al. 2005; Fan et al. 2011; Gossert et al. 2011; Strauss et al. 2005; Hansen et al. 1992), the high expense and difficulties in perdeuteration have limited their widespread use for larger proteins. The methylotrophic yeast Pichia pastoris is a well established expression host (Cereghino and Cregg 2000) for proteins that cannot be made in E. coli - eukaryotic membrane proteins such as ATP transporters (Lee et al. 2002), ion pumps (Strugatsky et al. 2003) and G-protein coupled receptors (Shimamura et al. 2011; Hino et al. 2012) have all been successfully overexpressed in and purified from this organism. Genetic manipulation, transformation, and growth of P. pastoris are more rapid than for higher eukaryotes such as insect cells and mammalian cells.
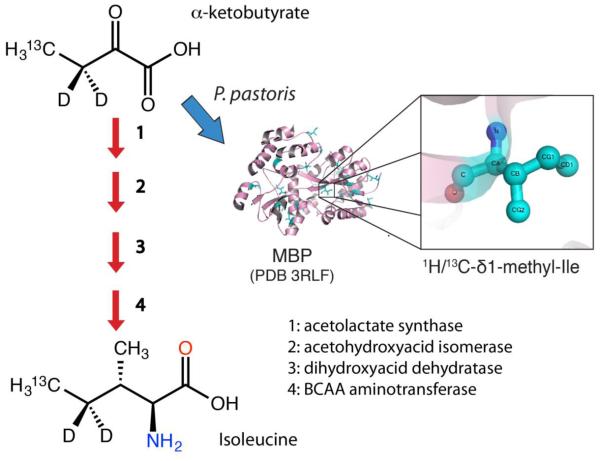
Overexpression using the tightly regulated AOX1 promoter often yields milligram quantities of recombinant protein per liter of P. pastoris suspension culture (Cereghino and Cregg 2000). P. pastoris is also favorable for NMR studies given its ability to grow on defined minimal media, uptake isotope-containing precursors, and efficiently incorporate deuterium at non-exchangeable sites (Morgan et al. 2000). Despite conservation of branched-chain amino acid biosynthesis pathways from E. coli (Figure 1), site-specific methyl labeling using α-ketoacid precursors has not been reported in. P. pastoris.
Fig 1.
Incorporation of 13C-methyl groups at the δ1 position of isoleucine residues in proteins expressed in P. pastoris.
We initially explored the use of 13C-methyl α-ketobutyrate in P. pastoris cultures to label maltose binding protein (MBP) with 13C at the δ1-methyl groups of isoleucine (Ile) residues. MBP has well-characterized 1H-13C 2D NMR spectra (Gardner et al. 1998) and is highly expressed in P. pastoris (Li et al. 2010). We collected 1H-13C heteronuclear single quantum coherence (HSQC) spectra on MBP that was labeled by addition of 13C-methyl α-ketobutyrate to the culture media (Figure 2a).
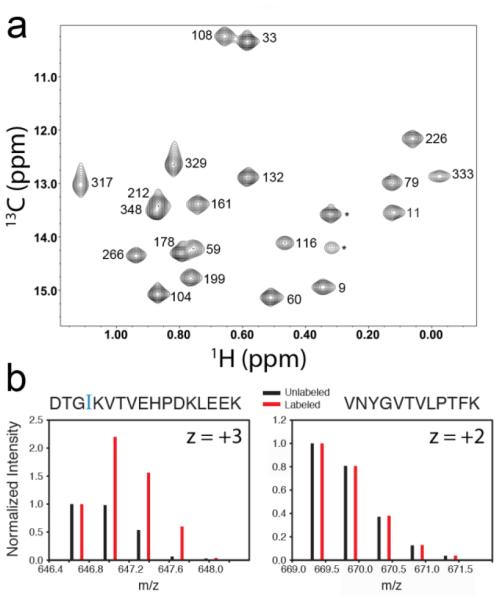
Fig. 2.
Labeling of δ1-methyl groups of MBP expressed in Pichia pastoris. (a) 1H-13C HSQC spectrum of 225 μM MBP labeled with α-ketobutyrate. Spectrum was recorded at 25°C on a Varian 800 MHz spectrometer. Peaks corresponding to Ile δ1-methyl groups are labeled in reference to assigned spectra (Gardner et al. 1998). Two unassigned peaks likely arising from differences in constructs are denoted with an asterisk. (b) Mass spectra of tryptic peptides containing Ile (left) and not containing Ile (right).
Resonances for all 22 Ile δ1-methyl groups of MBP (Gardner et al. 1998) are observed in our spectrum (Fig. 2a, Fig. S1), while little signal is present in other regions (indicating lack of “bleed-through” of the isotope into other amino acids - see Fig. 3). Based on tryptic peptide mass spectra (Figure 2b), we estimate the efficiency of incorporation for the α-ketobutyrate-derived 13C methyl probe to be 51±7% in a protonated background (see Supporting Information). The power of TROSY to obtain spectra of high-molecular weight species can only be exploited in the context of partial or full deuteration (Gardner et al. 1997; Wider and Wüthrich 1999; Ruschak and Kay 2010), which eliminates dipolar relaxation effects of surrounding protons on a given 13C-methyl spin system. To assess simultaneous 13C methyl labeling and perdeuteration in our system, we made samples of MBP in both P. pastoris and E. coli. We quantified the level of Ile δ1-methyl labeling in P. pastoris-derived deuterated MBP by comparing intensities to a concentration-matched E. coli sample (with assumed full incorporation at Ile δ1-methyl sites), yielding a labeling efficiency of 45±6% (Figure S2). The total deuteration level of P. pastoris-expressed MBP was estimated at 90% through ESI-LC-MS analysis (Figure S3; a comparison of labeling efficiency and yields of recombinant MBP from P. pastoris vs. E. coli is shown in Figure S4). Addition of α-ketoisovalerate led to very modest labeling of leucine δ- and valine γ-methyl groups (< 5%, not shown), suggesting that labeling of these sites would require significant optimization, perhaps through cytoplasmic overexpression of branched-chain-amino-acid aminotransferase as reported for a K. lactis expression system (Miyazawa-Onami et al., 2013).
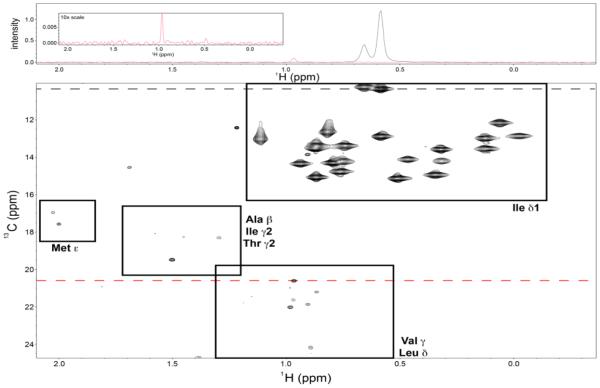
Fig. 3.
Expanded view of the 1H-13C HSQC spectrum of isoleucine δ1-methyl labeled maltose binding protein shown in Fig. 2a. Top panel shows horizontal slices of the 2D dataset (bottom panel), taken at approximately 13C = 10.3 δ1) and 20.6 ppm (red; Val/Leu) to show representative signal-to-noise in the spectrum for the labeled Ile δ1 methyl groups versus the unlabeled (natural abundance 13C) methyl groups of other amino acids. Inset of the top panel shows the 13C = 20.6 ppm trace at 10x vertical scale of the surrounding panel to provide a clearer sense of signal-to-noise. Signal-to-noise measurements for all 22 Ile δ1-methyl peaks resulted in an average S/N ratio of 280.
The impetus for using P. pastoris for 13C methyl labeling is to access proteins that are not amenable to expression and purification from E. coli – for example, the eukaryotic cytoskeletal protein actin. Actin’s capacity to change between monomeric and polymeric states arises from its conformational dynamics between distinct globular and filamentous forms (Oda et al. 2010; Pollard and Cooper 1986). NMR dynamics measurements would represent a significant new tool to study the biophysics of actin polymerization and interactions with regulatory molecules (Schmid et al. 2004; Kudryashov and Reisler 2013). While structures of actin monomers have been determined by X-ray crystallography (Otterbein et al. 2001; Rould et al. 2006; Nair et al. 2008) and actin filaments have been characterized by electron microscopy (Fujii et al. 2010; Ecken et al. 2015), expression of isotopically labeled actin for NMR has not been reported. Actin cannot be expressed at high levels in E. coli because of the lack of eukaryotic chaperone systems that are necessary for folding.
Biophysical characterization of actin is intrinsically difficult because actin polymerizes at concentrations above 100 nM. We therefore attempted to express a non-polymerizable Drosophila 5C actin (51.5 kDa, 94% identity to human actin) mutant in P. pastoris with mutations that impair the fast growing “barbed- end” of the filament (Zahm et al. 2013). However, the mutant proved toxic, presumably because it interferes with the polymerization of endogenous actin. To solve this problem, we generated a C-terminal fusion to human thymosin β4, an actin binding protein that blocks the intact, slow-growing “pointed-end” and thus ameliorates toxicity (Noguchi et al. 2007). This strategy resulted in high expression levels (10 mg/L of culture) and enabled purification to homogeneity (Supporting Information and Fig. S5).
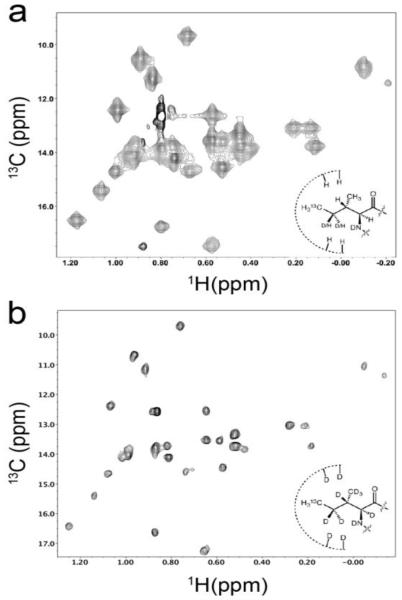
A representative HMQC (methyl TROSY) spectrum of 13C-Ile-δ1-methyl actin is shown in Figure 4a. Notably, for a protein with 30 Ile residues, we observe 33 peaks in the 1H-13C spectrum, likely reflecting slow chemical exchange processes at some sites. Taking advantage of the ability to highly deuterate proteins in P. pastoris, we repeated expression of Drosophila 5C actin in cultures where cells were adapted to D2O-containing media prior to induction, resulting in 2.5 mg/L of 13C-Ile-δ1-methyl perdeuterated actin. Lines in the 1H-13C HMQC spectra of the deuterated sample were much narrower than those in the HMQC spectrum of non-deuterated actin (Figures 4a and 4b).
Fig. 4.
NMR spectra of Drosophila actin labeled and overexpressed in Pichia pastoris. (a) 1H-13C HMQC spectrum of 13C-Ile δ1-methyl-labeled actin (180 μM). (b) TROSY-HMQC spectrum of perdeuterated, 13C-Ile δ1-methyl-labeled actin (150 μM). Spectra were recorded at 25°C on a Varian 800 MHz spectrometer.
Future use of TROSY NMR methods to study the dynamics of high-MW mammalian protein complexes and membrane proteins will depend on the tractability of isotope incorporation. We have demonstrated efficient incorporation of 13C at the Ile δ1-methyl groups of proteins expressed in P. pastoris, a robust eukaryotic expression host. In conjunction with perdeuteration, we acquired high-quality 1H-13C methyl TROSY spectra on Drosophila actin, which were unobtainable before. This development, along with similar approaches using other yeast systems (Miyazawa-Onami et al. 2013), will allow 2D NMR spectroscopy to be applied to many previously intractable proteins.
Supplementary Material
Acknowledgements
Funding was provided by a National Science Foundation Predoctoral Fellowship (Grant No. 1000136529 to L.C.), the Welch Foundation (I-1770 to D.M.R, I-1544 to M.K.R., I-1424 to K.H.G.), the Searle Scholars Program (D.M.R), a Packard Foundation Fellowship (D.M.R), the National Institutes of Health (T32 GM008297 supporting J.Z., R01 GM106239 to K.H.G., R01-GM56322 to M.K.R) and the Howard Hughes Medical Institute (M.K.R.)
References
- Cereghino JL, Cregg JM. Heterologous expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev. 2000;24(1):45–66. doi: 10.1111/j.1574-6976.2000.tb00532.x. [DOI] [PubMed] [Google Scholar]
- Chen C-Y, Cheng C-H, Chen Y-C, Lee J-C, Chou S-H, Huang W, Chuang W-J. Preparation of amino-acid-type selective isotope labeling of protein expressed in Pichia pastoris. Proteins. 2005;62(1):279–287. doi: 10.1002/prot.20742. [DOI] [PubMed] [Google Scholar]
- Ecken J, Müller M, Lehman W, Manstein DJ, Penczek PA, Raunser S. Structure of the F-actin-tropomyosin complex. Nature. 2015;519(7541):114–117. doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Shi L, Ladizhansky V, Brown LS. Uniform isotope labeling of a eukaryotic seven-transmembrane helical protein in yeast enables high-resolution solid-state NMR studies in the lipid environment. J Biomol NMR. 2011;49(2):151–161. doi: 10.1007/s10858-011-9473-9. [DOI] [PubMed] [Google Scholar]
- Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467(7316):724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- Gardner KH, Kay LE. Production and incorporation of 15N, 13C, 2H (1H-δ1 methyl) isoleucine into proteins for multidimensional NMR studies. J Am Chem Soc. 1997;119(32):7599–7600. [Google Scholar]
- Gardner KH, Rosen MK, Kay LE. Global folds of highly deuterated, methyl-protonated proteins by multidimensional NMR. Biochemistry. 1997;36(6):1389–401. doi: 10.1021/bi9624806. [DOI] [PubMed] [Google Scholar]
- Gardner KH, Zhang X, Gehring K, Kay LE. Solution NMR Studies of a 42 KDa Escherichia C oli Maltose Binding Protein/β-Cyclodextrin Complex: Chemical Shift Assignments and Analysis. J Am Chem Soc. 1998;120(45):11738–11748. [Google Scholar]
- Gossert AD, Hinniger A, Gutmann S, Jahnke W, Strauss A, Fernández C. A simple protocol for amino acid type selective isotope labeling in insect cells with improved yields and high reproducibility. J Biomol NMR. 2011;51(4):449–456. doi: 10.1007/s10858-011-9570-9. [DOI] [PubMed] [Google Scholar]
- Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE. A robust and cost-effective method for the production of Val, Leu, Ile (δ1) methyl-protonated 15N−, 13C−, 2H-labeled proteins. J Biomol NMR. 1999;13(4):369–74. doi: 10.1023/a:1008393201236. [DOI] [PubMed] [Google Scholar]
- Hansen AP, Petros AM, Mazar AP, Pederson TM, Rueter A, Fesik SW. A practical method for uniform isotopic labeling of recombinant proteins in mammalian cells. Biochemistry. 1992;31(51):12713–8. doi: 10.1021/bi00166a001. [DOI] [PubMed] [Google Scholar]
- Hino T, Arakawa T, Iwanari H, Yurugi-Kobayashi T, Ikeda-Suno C, Nakada-Nakura Y, Kusano-Arai O, Weyand S, Shimamura T, Nomura N, et al. G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature. 2012;482(7384):237–240. doi: 10.1038/nature10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofuku Y, Ueda T, Okude J, Shiraishi Y, Kondo K, Mizumura T, Suzuki S, Shimada I. Functional Dynamics of Deuterated β2 -Adrenergic Receptor in Lipid Bilayers Revealed by NMR Spectroscopy. Angew Chem Int Ed. 2014;53(49):13376–13379. doi: 10.1002/anie.201406603. [DOI] [PubMed] [Google Scholar]
- Kudryashov DS, Reisler E. ATP and ADP actin states. Biopolymers. 2013;99(4):245–256. doi: 10.1002/bip.22155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-Y, Urbatsch IL, Senior AE, Wilkens S. Projection Structure of P-glycoprotein by Electron Microscopy: Evidence for a Closed Conformation of the Nucleotide Binding Domains. J Biol Chem. 2002;277(42):40125–40131. doi: 10.1074/jbc.M206871200. [DOI] [PubMed] [Google Scholar]
- Li Z, Leung W, Yon A, Nguyen J, Perez VC, Vu J, Giang W, Luong LT, Phan T, Salazar KA, et al. Secretion and proteolysis of heterologous proteins fused to the Escherichia coli maltose binding protein in Pichia pastoris. Protein Expression Purif. 2010;72(1):113–124. doi: 10.1016/j.pep.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa-Onami M, Takeuchi K, Takano T, Sugiki T, Shimada I, Takahashi H. Perdeuteration and methyl-selective (1)H, (13)C-labeling by using a Kluyveromyces lactis expression system. J Biomol NMR. 2013;57(3):297–304. doi: 10.1007/s10858-013-9789-8. [DOI] [PubMed] [Google Scholar]
- Morgan WD, Kragt A, Feeney J. Expression of deuterium-isotope-labelled protein in the yeast pichia pastoris for NMR studies. J Biomol NMR. 2000;17(4):337–47. doi: 10.1023/a:1008313530207. [DOI] [PubMed] [Google Scholar]
- Nair UB, Joel PB, Wan Q, Lowey S, Rould MA, Trybus KM. Crystal Structures of Monomeric Actin Bound to Cytochalasin D. J Mol Biol. 2008;384(4):848–864. doi: 10.1016/j.jmb.2008.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi TQP, Kanzaki N, Ueno H, Hirose K, Uyeda TQP. A Novel System for Expressing Toxic Actin Mutants in Dictyostelium and Purification and Characterization of a Dominant Lethal Yeast Actin Mutant. J Biol Chem. 2007;282(38):27721–27727. doi: 10.1074/jbc.M703165200. [DOI] [PubMed] [Google Scholar]
- Nygaard R, Zou Y, Dror RO, Mildorf TJ, Arlow DH, Manglik A, Pan AC, Liu CW, Fung JJ, Bokoch MP, et al. The Dynamic Process of β2-Adrenergic Receptor Activation. Cell. 2013;152(3):532–542. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Iwasa M, Aihara T, Maéda Y, Narita A. The nature of the globular-to fibrous-actin transition. Nature. 2009;457(7228):441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- Ollerenshaw JE, Tugarinov V, Kay LE. Methyl TROSY: explanation and experimental verification. Magn Reson Chem. 2003;41(10):843–852. [Google Scholar]
- Otterbein LR, Graceffa P, Dominguez R. The crystal structure of uncomplexed actin in the ADP state. Science. 2001;293(5530):708–11. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- Pervushin K, Riek R, Wider G, Wüthrich K. Transverse relaxation-optimized spectroscopy (TROSY) for NMR studies of aromatic spin systems in 13C-labeled proteins. Proc Natl Acad Sci USA. 1997;94(23):12366–71. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu. Rev. Biochem. 1986;55:987–1035. doi: 10.1146/annurev.bi.55.070186.005011. [DOI] [PubMed] [Google Scholar]
- Rosen MK, Gardner KH, Willis RC, Parris WE, Pawson T, Kay LE. Selective methyl group protonation of perdeuterated proteins. J Mol Biol. 1996;263(5):627–636. doi: 10.1006/jmbi.1996.0603. [DOI] [PubMed] [Google Scholar]
- Rould MA, Wan Q, Joel PB, Lowey S, Trybus KM. Crystal Structures of Expressed Non-polymerizable Monomeric Actin in the ADP and ATP States. J Biol Chem. 2006;281(42):31909–31919. doi: 10.1074/jbc.M601973200. [DOI] [PubMed] [Google Scholar]
- Ruschak AM, Kay LE. Methyl groups as probes of supra-molecular structure, dynamics and function. J Biomol NMR. 2010;46(1):75–87. doi: 10.1007/s10858-009-9376-1. [DOI] [PubMed] [Google Scholar]
- Schmid MF, Sherman MB, Matsudaira P, Chiu W. Structure of the acrosomal bundle. Nature. 2004;431(7004):104–7. doi: 10.1038/nature02881. [DOI] [PubMed] [Google Scholar]
- Shimamura T, Shiroishi M, Weyand S, Tsujimoto H, Winter G, Katritch V, Abagyan R, Cherezov V, Liu W, Han GW, et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475(7354):65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss A, Bitsch F, Fendrich G, Graff P, Knecht R, Meyhack B, Jahnke W. Efficient uniform isotope labeling of Abl kinase expressed in Baculovirus-infected insect cells. J Biomol NMR. 2005;31(4):343–349. doi: 10.1007/s10858-005-2451-3. [DOI] [PubMed] [Google Scholar]
- Strugatsky D, Gottschalk K-E, Goldshleger R, Bibi E, Karlish SJD. Expression of Na+,K+-ATPase in Pichia pastoris: Analysis of Wild Type and D369N Mutant Proteins by Fe2+-Catalyzed Oxidative Cleavage and Molecular Modeling. J Biol Chem. 2003;278(46):46064–46073. doi: 10.1074/jbc.M308303200. [DOI] [PubMed] [Google Scholar]
- Werner K, Richter C, Klein-Seetharaman J, Schwalbe H. Isotope labeling of mammalian GPCRs in HEK293 cells and characterization of the C-terminus of bovine rhodopsin by high resolution liquid NMR spectroscopy. J Biomol NMR. 2008;40(1):49–53. doi: 10.1007/s10858-007-9205-3. [DOI] [PubMed] [Google Scholar]
- Wider G, Wüthrich K. NMR spectroscopy of large molecules and multimolecular assemblies in solution. Curr Opin Struct Biol. 1999;9(5):594–601. doi: 10.1016/s0959-440x(99)00011-1. [DOI] [PubMed] [Google Scholar]
- Zahm JA, Padrick SB, Chen Z, Pak CW, Yunus AA, Henry L, Tomchick DR, Chen Z, Rosen MK. The Bacterial Effector VopL Organizes Actin into Filament-like Structures. Cell. 2013;155(2):423–434. doi: 10.1016/j.cell.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.