Abstract
In addition to its actions outside the cell, cellular uptake and nuclear import of insulin-like growth factor binding protein-3 (IGFBP-3) has been recognized for almost two decades, but knowledge of its nuclear actions has been slow to emerge. IGFBP-3 has a functional nuclear localization signal and interacts with the nuclear transport protein importin-β. Within the nucleus IGFBP-3 appears to have a role in transcriptional regulation. It can bind to the nuclear receptor, retinoid X receptor-α and several of its dimerization partners, including retinoic acid receptor, vitamin D receptor (VDR), and peroxisome proliferator-activated receptor-γ (PPARγ). These interactions modulate the functions of these receptors, for example inhibiting VDR-dependent transcription in osteoblasts and PPARγ-dependent transcription in adipocytes. Nuclear IGFBP-3 can be detected by immunohistochemistry in cancer and other tissues, and its presence in the nucleus has been shown in many cell culture studies to be necessary for its pro-apoptotic effect, which may also involve interaction with the nuclear receptor Nur77, and export from the nucleus. IGFBP-3 is p53-inducible and in response to DNA damage, forms a complex with the epidermal growth factor receptor (EGFR), translocating to the nucleus to interact with DNA-dependent protein kinase. Inhibition of EGFR kinase activity or downregulation of IGFBP-3 can inhibit DNA double strand-break repair by nonhomologous endjoining. IGFBP-3 thus has the ability to influence many cell functions through its interactions with intranuclear pathways, but the importance of these interactions in vivo, and their potential to be targeted for therapeutic benefit, require further investigation.
Keywords: apoptosis, DNA damage repair, IGFBP-3, nuclear receptor, transcriptional regulation
1. Key features of IGFBP-3
Insulin-like growth factor binding protein-3 (IGFBP-3) is a multifunctional protein with a strong evolutionary link to the five other members of the IGFBP family (Daza et al., 2011). These proteins share the properties of high-affinity IGF binding and a high degree of structural conservation in their cysteine-rich amino- and carboxyterminal domains (Forbes et al., 2012). The terminal domains each comprise about one-third of the mature 28.7-kDa IGFBP-3 protein (Baxter, 2000). The central or linker domain shows little or no structural similarity among the six IGFBPs.
Like the other IGFBPs, IGFBP-3 is secreted by many cell types and is found in the circulation where, in adults, it is by far the most abundant IGFBP, with typical levels of 3–5 mg/L (Baxter, 1993; Friedrich et al., 2014). IGFBP-3 is the main circulating transport protein for IGF-I and IGF-II, which compete for a single binding site with similar affinities around 1010 L/mol (Martin and Baxter, 1986). Circulating IGF-IGFBP-3 complexes are found almost entirely bound to another protein, the acid-labile subunit (ALS), to form ternary complexes (Baxter et al., 1989). Among the other IGFBPs, only IGFBP-5 forms similar complexes with ALS (Twigg and Baxter, 1998). The site of interaction of IGFBP-3 with ALS includes a basic motif in the carboxyterminal domain (Figure 1); mutation of residues 228–232 to the corresponding residues of IGFBP-1 decreased ALS affinity by >90% while having little effect on IGF-binding affinity (Firth et al., 1998). Note: in this review, amino acid residues are numbered for mature IGFBP-3 containing 264 amino acids, excluding the 27-residue signal peptide.
Figure 1.
Aminoacid sequence of mature human IGFBP-3 (residues 1–264), omitting the 27-residue signal peptide. Conserved cysteine residues are boxed in yellow, and hydrophobic residues involved in IGF binding, in pink. Residues assumed (by comparison with IGFBP-5) to be involved in transcriptional regulation (Zhao et al., 2006), are in red font; residues shown by mutagenesis to be involved in nuclear receptor binding (Schedlich et al., 2007a), in green font. The bipartite nuclear localization signal that includes residues that bind ALS and RXRα is boxed in blue, and its basic residues are shown in blue font. The three N-glycosylation motifs are boxed in green. Putative overlapping hydrophobic nuclear export sequences are boxed in orange. The canonical 264-residue mature sequence shown above has been termed isoform 1 or isoform b (NCBI Reference Sequence: NP_000589.2, UniProt: P17936-1). The 270-residue mature product of an alternatively spliced transcript, termed isoform 2 or isoform a (NCBI Reference Sequence: NP_001013416.1, UniProt: P17936-2) has a 6-residue insertion after Pro107.
By binding the anabolic and mitogenic peptides IGF-I and IGF-II with high affinity and restricting their access to their shared receptor, the type 1 IGF receptor (IGF1R), IGFBP-3 is growth-inhibitory in many systems in vitro and in vivo (Firth and Baxter, 2002). High-affinity binding of the IGFs appears to be achieved by their interaction with hydrophobic residues in the aminoterminal domain (Figure 1), as well as carboxyterminal residues (Payet et al., 2003; Forbes et al., 2012), which may act cooperatively to maintain IGF binding even if the protein is partially proteolyzed (Yan et al., 2009), as observed in pregnancy and some other conditions (Hughes et al., 1995). Although isolated IGFBP-3 fragments have greatly reduced IGF-binding affinity, there are reports that they retain growth-inhibitory activity in a variety of cell systems (Lalou et al., 1997; Booth et al., 1999).
1.1. Post-translational modification
IGFBP-3 has three potential sites of N-linked glycosylation (Figure 1), of which either two (Asn89, Asn109) or all three are normally occupied by glycans, resulting in a doublet of approximately 40 kDa when analyzed by SDS-PAGE (Martin and Baxter, 1986; Firth and Baxter, 1999). Decreased glycosylation has little effect on IGF or ALS binding by IGFBP-3 but enhances its cell-surface binding (Firth and Baxter, 1999), as well as its interaction with glucose-regulated protein 78 (GRP78) (Grkovic et al., 2013). Since this interaction has been implicated in the induction of autophagy by IGFBP-3, it has been proposed that IGFBP-3 hypoglycosylation under conditions of nutritional deprivation might be a signal for enhanced autophagy (Grkovic et al., 2013).
The other well-characterized post-translational modification of IGFBP-3 is phosphorylation. The IGFBP-3 sequence contains multiple consensus sites for Ser/Thr phosphorylation (Coverley and Baxter, 1997), and appears to be constitutively phosphorylated at the protein kinase CK2 sites, Ser111 and Ser113 (Hoeck and Mukku, 1994). CK2 phosphorylation of IGFBP-3 has no effect on IGF binding but decreases its binding to ALS and the cell surface, and makes it relatively resistant to proteolysis (Coverley et al., 2000). In contrast to the lack of effect of CK2, and several other Ser/Thr kinases, on IGF binding, phosphorylation of IGFBP-3 by DNA-dependent protein kinase, catalytic subunit (DNA-PKcs) abolishes IGF binding (Schedlich et al., 2003). The role of this phosphorylation is further discussed below. IGFBP-3 also contains several predicted tyrosine kinase consensus sites (http://www.hprd.org/PhosphoMotif_finder), including sites for Src kinase and EGF receptor (EGFR) kinase. Phosphotyrosine residues identified by mass spectrometry include the putative EGFR kinase sites Tyr163 and Tyr183 (http://www.phosphosite.org/).
This review will focus on nuclear actions of IGFBP-3, and takes many examples from cancer cell biology, where most of the relevant discoveries have been made. For a broader view of IGFBP-3 physiology, genetics, and signaling, the reader is referred to other recent reviews (Jogie-Brahim et al., 2009; Yamada and Lee, 2009; Baxter, 2014; Johnson and Firth, 2014).
2. Nuclear import and export of IGFBP-3
2.1. Cell uptake and nuclear import
Radulescu (Radulescu, 1994) first identified a putative nuclear localization signal (NLS) in the IGFBP-3 sequence, although its activity was not tested. It was postulated that IGFBP-3 might associate in the nucleus with IGF-I, which was shown in an earlier electron microscopy study to translocate to the nucleus of chicken lens epithelial cells (Soler et al., 1990). Subsequently it was shown that cells at the growing edge of a monolayer could internalize both IGF-I and IGFBP-3 to the nucleus, where they co-localized (Li et al., 1997). In contrast, in resting cells the two proteins co-localized in endosome-like structures. IGF-I nuclear transport, demonstrated in digitonin-permeabilized Chinese hamster ovary cells, was dependent on IGFBP binding, since LR3-IGF-I, an IGF-I analog with greatly reduced IGFBP interaction, showed little nuclear translocation (Schedlich et al., 2003). The presence of nuclear IGFBP-3 is well-recognized in clinical histopathology (Hunziker et al., 2008; Seligson et al., 2013) and in cell lines (Jaques et al., 1997; Wraight et al., 1998), and extensive cell biology studies suggest important biological roles for IGFBP-3 in the nucleus.
Extracellular IGFBP-3 has been shown to enter the cell through a variety of endocytic mechanisms that may involve both caveolin 1 and clathrin-coated pits (Lee et al., 2004; Micutkova et al., 2012). Its nuclear import appears to share a common pathway with IGFBP-5 (Schedlich et al., 1998), both mediated by binding to importin-β (also known as karyopherin-β) (Schedlich et al., 2000; Micutkova et al., 2012), part of the importin-α/β nuclear transport complex. This contrasts with IGFBP-6, which interacts more strongly with importin-α (Iosef et al., 2008). The different specificities for importin binding suggest the possibility of different nuclear localization and/or function (Marfori et al., 2011). All three IGFBPs interact with the nuclear transport complex through a basic 18-residue bipartite NLS domain in their carboxyterminus (Figure 1), concordant with the NLS originally proposed by Radulescu. This basic domain corresponds to the ALS-binding domain described above (Firth et al., 1998). Mutation of key basic residues within the NLS domains of all three IGFBPs greatly inhibited their nuclear localization (Schedlich et al., 2000; Iosef et al., 2008). In contrast to the basic domain bipartite NLS common to IGFBP-3, IGFBP-5, and IGFBP-6, a short (monopartite) NLS motif has been characterized in the central domain of IGFBP-2 (Azar et al., 2014).
Using digitonin-permeabilized cells it was shown that both the kinetics of IGFBP-3 nuclear import, and its retention within the nucleus, were enhanced by IGFBP-3 phosphorylation by the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) (Schedlich et al., 2003). This phosphorylation was also inhibitory to IGF binding and prevented the IGFBP-3-mediated nuclear import of IGF-I. DNA-PKcs has been shown by co-immunoprecipitation and proximity ligation assay to form a nuclear complex with IGFBP-3 in breast cancer cells exposed to etoposide, but it is not known whether IGFBP-3 phosphorylation by DNA-PKcs is necessary for this interaction (Lin et al., 2014). However, this phosphorylation is reported to be an essential step in the induction of apoptosis by IGFBP-3 transfected into prostate cancer cells (Cobb et al., 2006).
DNA-PKcs preferentially phosphorylates Ser/Thr residues followed by Gln, and preceded by Gln, Glu or Asp (Lees-Miller and Anderson, 1991). By mutating the three consensus IGFBP-3 sites for DNA-PKcs (Ser156, Ser165, and Thr170, numbered excluding the signal peptide), Ser156 was shown to be essential for IGFBP-3 mediated caspase activation, which did not occur in cells expressing the S156A-IGFBP-3 mutant. NU7026, a specific DNA-PKcs inhibitor, also blocked apoptosis (Cobb et al., 2006). In contrast, a study in retinal endothelial cells found that cells expressing S156A-IGFBP-3 had increased apoptosis, and concluded that DNA-PKcs phosphorylation of Ser156 was a critical step in the ability of IGFBP-3 to prevent apoptosis (Zhang and Steinle, 2013). A mechanistic explanation for this discrepancy is currently lacking.
2.2. Nuclear export and stability
Hydrophobic nuclear export signals (NES) that interact with the export protein CRM1 (exportin 1) typically have the sequence Φ1-X2,3-Φ2-X2,3-Φ3-X-Φ4, where Φ represents Leu, Val, Ile, Phe, or Met and X can be any amino acid (Xu et al., 2012). The IGFBP-3 sequence includes several putative NES motifs in its central and C-terminal domains. One such motif, starting at Met190 (excluding the signal peptide) has been reported to serve as a functional NES (Paharkova-Vatchkova and Lee, 2010). In fact this sequence overlaps another putative NES starting at Leu194 (Figure 1). Mutation of Leu197 and Leu200, which are included in both of the overlapping sequences, increased the nuclear retention of IGFBP-3, and abolished the apoptotic effect of IGFBP-3 in 22RV1 prostate cancer cells (Paharkova-Vatchkova and Lee, 2010). Since nuclear entry of IGFBP-3 is believed to precede, and have a role in, its induction of apoptosis (Lee and Cohen, 2002; Santer et al., 2006; Leibowitz et al., 2013), this result suggests that IGFBP-3 may first enter, and then leave, the nucleus for its pro-apoptotic activity. A possible mechanism for this nucleocytoplasmic shuttling, involving the orphan nuclear receptor Nur77, is discussed below. IGFBP-3 in the nucleus has been reported to be quite unstable, with a shorter half-life than cytoplasmic IGFBP-3. It is stabilized by proteasome inhibitors, and its degradation follows polyubiquitination at multiple sites in the carboxyterminal domain [39].
In contrast to the proposed obligatory nuclear entry of IGFBP-3 as part of its apoptotic effect, a mutant form of IGFBP-3 in which a five-residue motif in the NLS domain was altered to the corresponding IGFBP-1 residues (Schedlich et al., 2000) was still able to induce apoptosis in T47D breast cancer cells despite being excluded from the nucleus (Butt et al., 2002), suggesting that an alternative pathway of apoptosis induction by IGFBP-3, independent of nuclear entry, may also exist.
3. IGFBP-3 and nuclear receptors
3.1. Interactions with retinoid receptors
In a landmark study, retinoid X receptor-α (RXRα, NR2B1) was identified by yeast 2-hybrid screen as an IGFBP-3 binding partner, and confirmed as a functional interacting protein by a variety of biochemical methods (Liu et al., 2000). RXRα is a ligand-dependent transcription factor that is a member of the nuclear receptor (NR) family (Evans and Mangelsdorf, 2014), specifically the type II NR family. It functions by dimerizing with itself or several other NR family members including retinoic acid receptor-α (RARα, NR1B1), vitamin D receptor (VDR, NR1I1), peroxisome proliferator-activated receptor-γ (PPARγ, NR1C3), thyroid hormone receptor-α (NR1A1), and Nur77 (NR4A1). In a luciferase reporter assay, IGFBP-3 was shown to enhance transcriptional activity stimulated by the RXR ligand SR11235 (RXR homodimer signaling), and to increase the apoptotic activity of another RXRα agonist, LG1069, in prostate cancer cell lines (Liu et al., 2000). The IGFBP-3-RXRα interaction involves residues in the basic carboxyterminal domain of IGFBP-3 Figure 1), but residues Thr58 and Arg60 in the aminoterminal domain are also involved (Schedlich et al., 2007a). Important RXRα residues involved in the interaction include RXRα Gln49 and Arg52 in the second Zn-finger loop of the DNA-binding domain. IGF-I binding to IGFBP-3, but not 9-cis-RA binding to RXRα, inhibited IGFBP-3-RXRα interaction (Schedlich et al., 2007a).
In contrast to the stimulatory effect of IGFBP-3 on RXRα homodimer signaling, IGFBP-3 was inhibitory to RXR-RAR heterodimer signaling stimulated by all-trans retinoic acid (atRA) (Liu et al., 2000). IGFBP-3 inhibits atRA-stimulated transcription through direct binding to RARα as well as RXRα (Schedlich et al., 2004), and immunoneutralization of IGFBP-3 in breast cancer cells enhanced the atRA transcriptional response in a reporter assay. Similarly, immunoneutralizing IGFBP-3 was able to sensitize the relatively atRA-resistant triple-negative breast cancer cell lines, MDA-MB-231 and Hs578T, to growth inhibition by atRA, suggesting that endogenous IGFBP-3 produced by these cell lines was limiting their responsiveness to atRA (Schedlich et al., 2004). IGFBP-3 interaction with RARα has been shown to involve the same amino- and carboxyterminal IGFBP-3 residues as the IGFBP-3-RXRα interaction (Schedlich et al., 2007a).
3.2. Interaction with PPARγ
The interaction between IGFBP-3 and RXRα prompted an investigation of its possible effects on PPARγ activity. PPARγ has often been described as the master regulator of adipogenesis, impacting on the complex cascade of events that lead to the formation of mature adipocytes (Lefterova et al., 2014). RXRα-PPARγ heterodimers are known to bind to thousands of sites in the adipocyte genome (Nielsen et al., 2008). IGFBP-3 was found to bind directly to PPARγ and to co-immunoprecipitate with PPARγ from lysates of mouse 3T3-L1 adipocytes (Chan et al., 2009). It also inhibited the formation of RXRα-PPARγ dimers and inhibited PPARγ transcriptional activity stimulated by the thiazolidinedione PPARγ agonist rosiglitazone in 3T3-L1 cells (Chan et al., 2009). These results suggested that IGFBP-3 was likely to affect adipogenic differentiation, and it was found that IGFBP-3 could inhibit 3T3-L1 preadipocyte maturation as indicated by decreased induction of the adipocyte markers resistin and adiponectin (de Silva et al., 2012), reversal of the loss of the preadipocyte marker plasminogen activator inhibitor-1, and a decrease in the generation of triglycerides detected by Nile red staining. Further, treatment of mature adipocytes with IGFBP-3 caused a partial reversion to a less differentiated phenotype (Chan et al., 2009). The inhibitory effect of IGFBP-3 on adipogenesis was not simply due to sequestration of stimulatory IGF-I, since IGFBP-2, another high-affinity IGFBP, had no inhibitory effect on 3T3-L1 differentiation. Finally, a basic-domain mutant of IGFBP-3 that retains IGF-binding activity was also without inhibitory activity (Chan et al., 2009). Endogenous IGFBP-3 expression increases through the course of adipogenesis (de Silva et al., 2012), and it may be speculated that this serves as a termination signal once differentiation has been achieved.
PPARγ agonists have been extensively investigated as possible anticancer agents in either mono-or combination therapies, but evidence of clinical utility is limited (Robbins and Nie, 2012). PPARγ agonists are typically growth-inhibitory when tested in cancer cell lines in vitro, but in at least some cases their effects may not be mediated by activation of PPARγ-dependent transcription (Wei et al., 2009; Robbins and Nie, 2012). Since IGFBP-3 affects PPARγ function in preadipocytes, its role in PPARγ-dependent anti-cancer activity was investigated in breast cancer cell lines. Surprisingly in view of its PPARγ-antagonistic role in adipogenesis, IGFBP-3 was not only growth-inhibitory itself, but potentiated growth inhibition caused by the synthetic and natural PPARγ ligands, rosiglitazone and 15-deoxyΔ12,14prostaglandin J2 (15dPG), both of which were acting in a PPARγ-dependent manner as indicated by blockade of their effect by the PPARγ antagonist GW9662 (Pon et al., 2015). Growth inhibition by rosiglitazone or 15dPG was found to depend on the presence of IGFBP-3, since the effect of either agent was significantly attenuated in three breast cancer cell lines when IGFBP-3 was downregulated by siRNA. Further, two mutant forms of IGFBP-3, with reduced PPARγ binding, had no inhibitory effect when tested alone, and entirely blocked the inhibitory effect of rosiglitazone (Pon et al., 2015). These results indicate that breast cancer cell growth inhibition by PPARγ requires its interaction with endogenous IGFBP-3, and non-binding IGFBP-3 mutants can exert a dominant-negative blocking effect on the inhibition. Since endogenous IGFBP-3 is required for PPARγ ligands to exert anti-tumor effect, this study suggests that tumor IGFBP-3 expression may serve as a biomarker for the efficacy of these agents.
3.3. Interaction with Nur77
As alluded to above, nuclear export of IGFBP-3 may involve its functional interaction with the nuclear receptor Nur77, a RXRα dimerization partner. The induction of apoptosis by IGFBP-3 in prostate cancer cells was reported to be initiated by IGFBP-3-dependent translocation of Nur77 from the nucleus to the mitochondria, an effect requiring RXRα as it was not seen in RXRα-deficient cells (Lee et al., 2005). In contrast to the inhibitory effect of IGFBP-3 on the RXRα interaction with RARα and PPARγ, described above, IGFBP-3 was found to enhance RXRα-Nur77 interaction, and mitochondrial RXRα accumulation (Lee et al., 2005). Although a direct IGFBP-3-Nur77 interaction was not demonstrated, IGFBP-3 could be co-immunoprecipitated with Nur77 in the cytoplasm of prostate cancer cells, perhaps associated through their common binding partner RXRα (Lee et al., 2007). The importance of these interactions was suggested by the loss of IGFBP-3-induced apoptosis in Nur77-deficient cells. As noted earlier, mutation of a putative CRM1-binding NES motif on IGFBP-3 also prevented IGFBP-3-induced apoptosis, and also resulted in increased nuclear RXRα-Nur77 retention (Paharkova-Vatchkova and Lee, 2010). Together these date indicate a role for nuclear IGFBP-3 in the CRM1-mediated export of RXRα-Nur77 complexes during IGFBP-3-mediated apoptosis in prostate cancer cells.
3.4. Interaction with other nuclear receptors
The IGFBP3 gene is a transcriptional target of the VDR (Krishnan et al., 2003), its induction by 1,25(OH)2D3 leading to growth inhibition through cell cycle arrest in prostate cancer cells. IGFBP-3 also binds directly to VDR, and the two proteins have been shown to co-localize in the cell nucleus (Li et al., 2013). IGFBP-3 appears to compete for the binding of IGFBP-5, another VDR binding partner (Schedlich et al., 2007b). IGFBP-3 has been shown to inhibit VDR-dependent transcriptional activity (Li et al., 2013) and 1,25(OH)2D3-stimulated CD11b induction in HL-60 human promyelocytic leukemia cells (Ikezoe et al., 2004). In osteoblasts, 1,25(OH)2D3-dependent differentiation is inhibited by overexpression of IGFBP-3 (Li et al., 2013), similar to the inhibitory effect of IGFBP-5 (Schedlich et al., 2007b). The thyroid hormone receptor-α, another RXRα heterodimerization partner, is also reported to bind to IGFBP-3 in vitro and co-localize with it in the nucleus of HEK-293 cells (Qiu et al., 2011). IGFBP-3 was inhibitory to T3-mediated gene transcription. Whether other type II NRs will be shown to bind to IGFBP-3, and their transcriptional activity to be regulated by it, remains to be determined.
3.5. Interaction with histone-DNA complex
A study of the domains of IGFBP-5 with transcriptional regulatory activity reported direct interaction of IGFBP-5 with the nuclear histone-DNA complex, and highlighted amino-terminal residues that contribute to this interaction (Zhao et al., 2006). Notably, the N-terminal domain of IGFBP-3 showed transcriptional activity as strong as that of the corresponding domain of IGFBP-5. Among the IGFBP-5 residues shown by mutagenesis to be involved in transcriptional activation, residues corresponding to Glu8, Asp11, Glu30, Pro31, Glu43, and Glu52 of IGFBP-5 are all conserved in IGFBP-3 (Figure 1), strongly suggesting that the N-terminal domain of IGFBP-3 is also involved in histone-DNA interaction. As discussed earlier, it is also notable that Thr58 and Arg60, that flank Glu59 of IGFBP-3 (equivalent to Glu52 of IGFBP-5), influence the IGFBP-3 interaction with both RXRα and RARα (Schedlich et al., 2007a), suggesting overlap between residues involved in nuclear hormone receptor binding by IGFBP-3, and those involved in histone-DNA binding – or indeed that IGFBP-3 binds to DNA as part of a nuclear hormone receptor complex.
In addition to direct IGFBP-3 protein interactions with the histone-DNA complex, chromosomal interactions of the IGFBP3 gene have also been examined. Long-range chromosomal interactions of IGFBP3 have been shown to differ between normal and cancerous mammary epithelial cell lines (Zeitz et al., 2013). Among a multitude of IGFBP3-interacting genes, an interaction with EGFR was notable, suggesting a possible coordination of these genes at the genomic level (Zeitz et al., 2013), reinforcing evidence for the functional interaction of their encoded proteins in cancer (Martin et al., 2014).
4. Role of IGFBP-3 in the DNA damage response
4.1. IGFBP-3 and p53
In response to genotoxic stress, the tumor suppressor p53 is activated and stabilized by the primary DNA damage sensor, ataxia-telangiectasia mutated (ATM) (Shiloh and Ziv, 2013). IGFBP-3 is a transcriptional target of p53 (Buckbinder et al., 1995), and its expression is induced by the topoisomerase II poison, doxorubicin, and other DNA-damaging chemotherapy drugs in many cell lines. The IGFBP3 gene contains putative p53-responsive sites both within introns 1 and 2 (Buckbinder et al., 1995), and in the upstream promoter region (Bourdon et al., 1997). Hypermethylation of p53 regulatory regions in the upstream promoter can suppress p53-dependent IGFBP-3 induction (Hanafusa et al., 2005) and could conceivably contribute to radio- or chemoresistance of some tumors, since p53-dependent IGFBP-3 induction appears to account for at least some of the pro-apoptotic activity of p53 (Hollowood et al., 2000; Grimberg et al., 2002).
The C-terminal basic domain of p53 has been reported to be inhibitory to IGFBP-3 induction, such that a p53 form lacking both this domain and the N-terminal activation domain was able to induce IGFBP-3 expression under conditions where full-length p53 did not (Harms and Chen, 2005). However, this effect appears to depend on histone deacetylase (HDAC) activity, since in the presence of an HDAC inhibitor the ability of full-length p53 to induce IGFBP-3 was restored (Harms and Chen, 2005). Some point mutations in the p53 DNA binding domain also impair its ability to induce IGFBP-3, even while remaining transcriptionally active as determined by p21 induction (Ludwig et al., 1996). Indeed, some p53 “hot-spot” mutations, such as Arg175, Arg273 and Arg282, that appear to be associated with gain-of-function activity (Freed-Pastor and Prives, 2012), can suppress IGFBP3 promoter activity below its basal level, an effect also reversible by HDAC inhibition (Vikhanskaya et al., 2007).
IGFBP-3 is also strongly downregulated by ΔNp63α (Barbieri et al., 2005), a variant of the p63α splicing isoform that lacks the N-terminal transactivation domain (Deyoung and Ellisen, 2007) and has been shown to oppose p53-dependent transcriptional activity (Yang et al., 1998). ΔNp63α is induced by doxorubicin (Petitjean et al., 2008), so in cells where this isoform predominates, it might contribute to IGFBP-3 downregulation rather than induction of IGFBP-3 in response to similar chemotherapy drugs.
4.2. IGFBP-3 and DNA damage-induced apoptosis or survival
When subjected to ionizing radiation, T47D breast cancer cells transfected to express IGFBP-3 show significantly worse survival over 14 days than cells expressing empty vector (Butt et al., 2000). Cell cycle distribution between the two cell lines does not differ, but the IGFBP-3-expressing cells show a marked increase in apoptosis. Other studies have similarly shown an enhanced apoptotic response to radiation in the presence of increased IGFBP-3 (Hollowood et al., 2000; Yoshino et al., 2011). IGFBP-3 is reported to be more highly expressed in radiosensitive than radioresistant cell lines (Achary et al., 2000; Zhao et al., 2012), and IGFBP-3 downregulation can reduce radiosensitivity (Yoshino et al., 2011). Similar effects of IGFBP-3 are reported in modulating tumor or cell line chemosensitivity (Granata et al., 2003; Ibanez de Caceres et al., 2010).
A dual role for IGFBP-3 has been described in human umbilical vein endothelial cells, in which IGFBP-3 potentiated doxorubicin-induced apoptosis but also enhanced survival during serum starvation (Granata et al., 2004). The proapototic response to doxorubicin was accompanied by an increase in the cell content of ceramide, a sphingolipid that has also been shown in other studies to act with IGFBP-3 in enhancing apoptosis (Gill et al., 1997). The survival effect of IGFBP-3 was associated with activation of sphingosine kinase 1 (SphK1), which generates the pro-survival lipid, sphingosine-1-phosphate (S1P), a product of ceramide metabolism (Granata et al., 2004). Activation of SphK1 by IGFBP-3, leading to S1P-dependent transactivation of EGFR, has also been described in normal mammary and breast cancer cell lines (Martin et al., 2009; Martin et al., 2014). SphK1 downregulation by shRNA has been shown to increase doxorubicin-induced DNA damage as indicated by enhanced phosphorylation of histone H2AX (Huwiler et al., 2011); conversely SphK1 activation by IGFBP-3 may be predicted to attenuate chemotherapy-induced DNA damage.
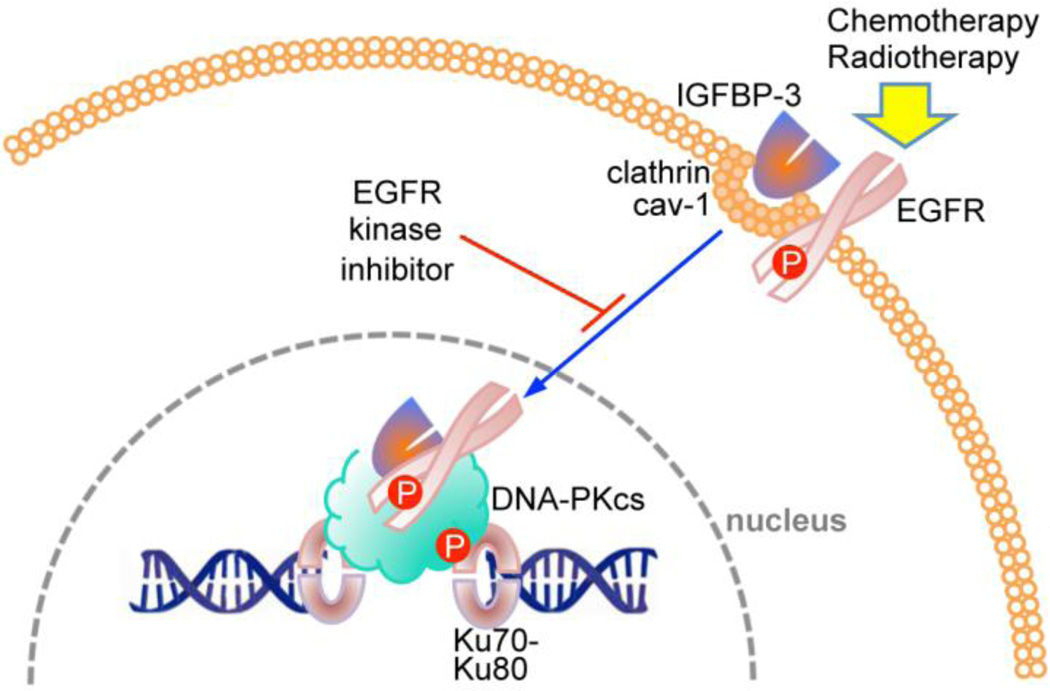
IGFBP-3 has recently been shown to have an active role in DNA repair in response to chemotherapy (Figure 2), perhaps surprising in view of its well-established pro-apoptotic role. Analyzed by co-immunoprecipitation and proximity ligation assay, IGFBP-3 was found to form nuclear complexes with EGFR and DNA-PKcs in response to chemotherapy, an effect blocked by EGFR kinase inhibition (Lin et al., 2014). When IGFBP-3 was downregulated by siRNA, DNA-PKcs autophosphorylation and nuclear EGFR-DNA-PKcs complex formation were inhibited. Non-homologous end-joining activity in nuclear extracts was also attenuated when IGFBP-3 was downregulated, indicating that the repair of chemotherapy-induced DNA double strand breaks requires the formation of nuclear complexes between IGFBP-3, EGFR and DNA-PKcs (Lin et al., 2014). It may be speculated that this effect of IGFBP-3 also involves SphK1 activation, but this was not demonstrated.
Figure 2.
Involvement of IGFBP-3 in DNA damage repair. In response to DNA damaging therapy, IGFBP-3 interacts with EGFR at the plasma membrane, and is transported to the nucleus where it binds to DNA-PKcs (complexed with Ku70/Ku80 at DNA double-strand breaks), leading to DNA-PKcs autophosphorylation (Lin et al., 2014). EGFR kinase inhibition prevents the nuclear complex from forming.
5. Summary and Conclusions
There is clearly a great deal remaining to be understood about nuclear actions of IGFBP-3. In the two decades since IGFBP-3 was first recognized as having the potential for nuclear translocation, there has been only a gradual advance in revealing how this translocation occurs and what nuclear functions IGFBP-3 is involved in. This relatively slow progress parallels a growing realization that many other regulatory proteins that were initially identified as extranuclear (e.g. tyrosine kinase receptors like EGFR and IGF1R), also have significant functions within the nucleus, and indeed that some other IGFBPs (IGFBP-2, IGFBP-5, and IGFBP-6) are also active intranuclearly.
The most significant advance in understanding the nuclear role of IGFBP-3 has resulted from the recognition that it interacts with RXRα and several of its nuclear receptor heterodimerization partners. This has revealed the potential for IGFBP-3 to be directly involved in transcriptional regulatory complexes, in one case (RXRα-dependent transcription) exerting a stimulatory effect, but in others (e.g. RARα- and VDR-dependent transcription) being inhibitory, apparently by disrupting heterodimeric RXRα complexes. In its apoptosis-inducing complex with the orphan receptor Nur77, its major role may be extranuclear, and in its complex with PPARγ, current evidence suggests that PPARγ activity is opposed by IGFBP-3 during adipogenesis, but requires IGFBP-3 in tumor suppression.
Involvement of IGFBP-3 in the DNA damage response may be another key intranuclear role, but once again, some of the observations are contradictory and the relevant mechanisms have not been fully elicidated. There is evidence to support roles for IGFBP-3 both in mediating, or at least supporting, cell cycle arrest and induction of apoptosis in response to DNA damage, while at the same time participating in DNA repair through interaction with EGFR and DNA-PKcs. These opposing actions have been termed, respectively, the gatekeeper and caretaker responses (van Heemst et al., 2007), and the involvement of IGFBP-3 in both suggests that it may have a fundamental role in supporting genomic integrity.
To date, all of the above observations have been restricted to a variety of cell culture systems, and it is unclear how important they will turn out to be in vivo, notwithstanding clear evidence that IGFBP-3 can be seen intranuclearly on histological examination of cancer and other tissues. Continuing investigation of these phenomena in vitro, and the development of suitable in vivo models, should be an important research goal, since a better understanding of IGFBP-3 actions in the nucleus may open new opportunities for therapeutic intervention in cancer and other diseases.
Table 1.
Nuclear receptors known to interact with IGFBP-3
| Nuclear Receptor | Abbreviation | Systematic Name* |
Ligand | References |
|---|---|---|---|---|
| Retinoid X receptor-α | RXRα | NR2B1 | 9-cis-retinoic acid | (Liu et al., 2000; Schedlich et al., 2007a) |
| Retinoic acid receptor-α | RARα | NR1B1 | All-trans-retinoic acid | (Liu et al., 2000; Schedlich et al., 2004) |
| Peroxisome proliferator-activated receptor-γ | PPARγ | NR1C3 | Fatty acids, prostaglandin J2, thiazolidinediones | (Chan et al., 2009; Pon et al., 2015) |
| Vitamin D receptor | VDR | NR1I1 | 1,25-dihydroxy-vitamin D3 | (Schedlich et al., 2007b; Li et al., 2013) |
| Nuclear receptor-77/Nerve growth factor-inducible receptor B | Nur77/NGFI-B | NR4A1 | Orphan | (Lee et al., 2007) |
| Thyroid receptor-α | TRα | NR1A1 | Thyroid hormones | (Qiu et al., 2011) |
Highlights.
IGFBP-3 is a multifunctional protein with roles both outside and inside the cell.
IGFBP-3 binds to nuclear hormone receptors, regulating transcriptional activity.
Nuclear transport of IGFBP-3 may be required for its pro-apoptotic activity.
Nuclear IGFBP-3 also influences cell survival by promoting DNA damage repair.
Acknowledgements
This review and the corresponding Gene Wiki article are written as part of the Gene Wiki Review series – a series resulting from a collaboration between the journal GENE and the Gene Wiki Initiative. The Gene Wiki Initiative is supported by National Institutes of Health (GM083924). Additional support for Gene Wiki Reviews is provided by Elsevier, the publisher of GENE. This work was supported by Grant No. DP140100137 to RCB from the Australian Research Council.
Abbreviations
- IGFBP-3
insulin-like growth factor binding protein-3
- ALS
acid-labile subunit
- IGF1R
type 1 IGF receptor
- GRP78
glucose-regulated protein 78
- DNA-PKcs
DNA-dependent protein kinase catalytic subunit
- EGFR
epidermal growth factor receptor
- NLS
nuclear localization signal
- NES
nuclear export signal
- RXRα
retinoid X receptor-α
- NR
nuclear receptor
- RARα
retinoic acid receptor-α
- VDR
vitamin D receptor
- PPARγ
peroxisome proliferator-activated receptor-γ
- atRA
all-trans retinoic acid
- 15dPG
15-deoxyΔ12,14prostaglandin J2
- ATM
ataxia-telangiectasia mutated
- HDAC
histone deacetylase
- SphK1
sphingosine kinase 1
- S1P
sphingosine-1-phosphate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achary MP, Jaggernauth W, Gross E, Alfieri A, Klinger HP, Vikram B. Cell lines from the same cervical carcinoma but with different radiosensitivities exhibit different cDNA microarray patterns of gene expression. Cytogenet Cell Genet. 2000;91:39–43. doi: 10.1159/000056815. [DOI] [PubMed] [Google Scholar]
- Azar WJ, Zivkovic S, Werther GA, Russo VC. IGFBP-2 nuclear translocation is mediated by a functional NLS sequence and is essential for its pro-tumorigenic actions in cancer cells. Oncogene. 2014;33:578–588. doi: 10.1038/onc.2012.630. [DOI] [PubMed] [Google Scholar]
- Barbieri CE, Perez CA, Johnson KN, Ely KA, Billheimer D, Pietenpol JA. IGFBP-3 is a direct target of transcriptional regulation by DeltaNp63alpha in squamous epithelium. Cancer Res. 2005;65:2314–2320. doi: 10.1158/0008-5472.CAN-04-3449. [DOI] [PubMed] [Google Scholar]
- Baxter RC. Circulating binding proteins for the insulinlike growth factors. Trends Endocrinol Metab. 1993;4:91–96. doi: 10.1016/1043-2760(93)90085-s. [DOI] [PubMed] [Google Scholar]
- Baxter RC. Insulin-like growth factor (IGF)-binding proteins: interactions with IGFs and intrinsic bioactivities. Am J Physiol Endocrinol Metab. 2000;278:E967–E976. doi: 10.1152/ajpendo.2000.278.6.E967. [DOI] [PubMed] [Google Scholar]
- Baxter RC. IGF binding proteins in cancer: mechanistic and clinical insights. Nat Rev Cancer. 2014;14:329–341. doi: 10.1038/nrc3720. [DOI] [PubMed] [Google Scholar]
- Baxter RC, Martin JL, Beniac VA. High molecular weight insulin-like growth factor binding protein complex. Purification and properties of the acid-labile subunit from human serum. J Biol Chem. 1989;264:11843–11848. [PubMed] [Google Scholar]
- Booth BA, Boes M, Dake BL, Bar RS. Isolation and characterization of plasmin-generated bioactive fragments of IGFBP-3. Am J Physiol. 1999;276:E450–E454. doi: 10.1152/ajpendo.1999.276.3.E450. [DOI] [PubMed] [Google Scholar]
- Bourdon JC, Deguin-Chambon V, Lelong JC, Dessen P, May P, Debuire B, May E. Further characterisation of the p53 responsive element--identification of new candidate genes for trans-activation by p53. Oncogene. 1997;14:85–94. doi: 10.1038/sj.onc.1200804. [DOI] [PubMed] [Google Scholar]
- Buckbinder L, Talbott R, Velasco-Miguel S, Takenaka I, Faha B, Seizinger BR, Kley N. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377:646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- Butt AJ, Firth SM, King MA, Baxter RC. Insulin-like growth factor-binding protein-3 modulates expression of Bax and Bcl-2 and potentiates p53-independent radiation-induced apoptosis in human breast cancer cells. J Biol Chem. 2000;275:39174–39181. doi: 10.1074/jbc.M908888199. [DOI] [PubMed] [Google Scholar]
- Butt AJ, Fraley KA, Firth SM, Baxter RC. IGF-binding protein-3-induced growth inhibition and apoptosis do not require cell surface binding and nuclear translocation in human breast cancer cells. Endocrinology. 2002;143:2693–2699. doi: 10.1210/endo.143.7.8876. [DOI] [PubMed] [Google Scholar]
- Chan SS, Schedlich LJ, Twigg SM, Baxter RC. Inhibition of adipocyte differentiation by insulin-like growth factor-binding protein-3. Am J Physiol Endocrinol Metab. 2009;296:E654–E663. doi: 10.1152/ajpendo.90846.2008. [DOI] [PubMed] [Google Scholar]
- Cobb LJ, Liu B, Lee KW, Cohen P. Phosphorylation by DNA-dependent protein kinase is critical for apoptosis induction by insulin-like growth factor binding protein-3. Cancer Res. 2006;66:10878–10884. doi: 10.1158/0008-5472.CAN-06-0585. [DOI] [PubMed] [Google Scholar]
- Coverley JA, Baxter RC. Phosphorylation of insulin-like growth factor binding proteins. Mol Cell Endocrinol. 1997;128:1–5. doi: 10.1016/s0303-7207(97)04032-x. [DOI] [PubMed] [Google Scholar]
- Coverley JA, Martin JL, Baxter RC. The effect of phosphorylation by casein kinase 2 on the activity of insulin-like growth factor-binding protein-3. Endocrinology. 2000;141:564–570. doi: 10.1210/endo.141.2.7306. [DOI] [PubMed] [Google Scholar]
- Daza DO, Sundstrom G, Bergqvist CA, Duan C, Larhammar D. Evolution of the insulin-like growth factor binding protein (IGFBP) family. Endocrinology. 2011;152:2278–2289. doi: 10.1210/en.2011-0047. [DOI] [PubMed] [Google Scholar]
- de Silva HC, Firth SM, Twigg SM, Baxter RC. Interaction between IGF binding protein-3 and TGFbeta in the regulation of adipocyte differentiation. Endocrinology. 2012;153:4799–4807. doi: 10.1210/en.2011-1444. [DOI] [PubMed] [Google Scholar]
- Deyoung MP, Ellisen LW. p63 and p73 in human cancer: defining the network. Oncogene. 2007;26:5169–5183. doi: 10.1038/sj.onc.1210337. [DOI] [PubMed] [Google Scholar]
- Evans RM, Mangelsdorf DJ. Nuclear Receptors, RXR, the Big Bang. Cell. 2014;157:255–266. doi: 10.1016/j.cell.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth SM, Baxter RC. Characterisation of recombinant glycosylation variants of insulin-like growth factor binding protein-3. J Endocrinol. 1999;160:379–387. doi: 10.1677/joe.0.1600379. [DOI] [PubMed] [Google Scholar]
- Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev. 2002;23:824–854. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- Firth SM, Ganeshprasad U, Baxter RC. Structural determinants of ligand and cell surface binding of insulin-like growth factor-binding protein-3. J Biol Chem. 1998;273:2631–2638. doi: 10.1074/jbc.273.5.2631. [DOI] [PubMed] [Google Scholar]
- Forbes BE, McCarthy P, Norton RS. Insulin-like growth factor binding proteins: a structural perspective. Front Endocrinol (Lausanne) 2012;3:38. doi: 10.3389/fendo.2012.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich N, Wolthers OD, Arafat AM, Emeny RT, Spranger J, Roswall J, Kratzsch J, Grabe HJ, Hubener C, Pfeiffer AF, Doring A, Bielohuby M, Dahlgren J, Frystyk J, Wallaschofski H, Bidlingmaier M. Age- and sex-specific reference intervals across life span for insulin-like growth factor binding protein 3 (IGFBP-3) and the IGF-I to IGFBP-3 ratio measured by new automated chemiluminescence assays. J Clin Endocrinol Metab. 2014;99:1675–1686. doi: 10.1210/jc.2013-3060. [DOI] [PubMed] [Google Scholar]
- Germain P, Staels B, Dacquet C, Spedding M, Laudet V. Overview of nomenclature of nuclear receptors. Pharmacol Rev. 2006;58:685–704. doi: 10.1124/pr.58.4.2. [DOI] [PubMed] [Google Scholar]
- Gill ZP, Perks CM, Newcomb PV, Holly JM. Insulin-like growth factor-binding protein (IGFBP-3) predisposes breast cancer cells to programmed cell death in a non-IGF-dependent manner. J Biol Chem. 1997;272:25602–25607. doi: 10.1074/jbc.272.41.25602. [DOI] [PubMed] [Google Scholar]
- Granata R, De Petrini M, Trovato L, Ponti R, Pons N, Ghe C, Graziani A, Ferry RJ, Jr, Muccioli G, Ghigo E. Insulin-like growth factor binding protein-3 mediates serum starvation- and doxorubicin-induced apoptosis in H9c2 cardiac cells. J Endocrinol Invest. 2003;26:1231–1241. doi: 10.1007/BF03349163. [DOI] [PubMed] [Google Scholar]
- Granata R, Trovato L, Garbarino G, Taliano M, Ponti R, Sala G, Ghidoni R, Ghigo E. Dual effects of IGFBP-3 on endothelial cell apoptosis and survival: involvement of the sphingolipid signaling pathways. FASEB J. 2004;18:1456–1458. doi: 10.1096/fj.04-1618fje. [DOI] [PubMed] [Google Scholar]
- Grimberg A, Liu B, Bannerman P, El-Deiry WS, Cohen P. IGFBP-3 mediates p53-induced apoptosis during serum starvation. Int J Oncol. 2002;21:327–335. [PMC free article] [PubMed] [Google Scholar]
- Grkovic S, O'Reilly VC, Han S, Hong M, Baxter RC, Firth SM. IGFBP-3 binds GRP78, stimulates autophagy and promotes the survival of breast cancer cells exposed to adverse microenvironments. Oncogene. 2013;32:2412–2420. doi: 10.1038/onc.2012.264. [DOI] [PubMed] [Google Scholar]
- Hanafusa T, Shinji T, Shiraha H, Nouso K, Iwasaki Y, Yumoto E, Ono T, Koide N. Functional promoter upstream p53 regulatory sequence of IGFBP3 that is silenced by tumor specific methylation. BMC Cancer. 2005;5:9. doi: 10.1186/1471-2407-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms KL, Chen X. The C terminus of p53 family proteins is a cell fate determinant. Mol Cell Biol. 2005;25:2014–2030. doi: 10.1128/MCB.25.5.2014-2030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeck WG, Mukku VR. Identification of the major sites of phosphorylation in IGF binding protein-3. J Cell Biochem. 1994;56:262–273. doi: 10.1002/jcb.240560220. [DOI] [PubMed] [Google Scholar]
- Hollowood AD, Lai T, Perks CM, Newcomb PV, Alderson D, Holly JM. IGFBP-3 prolongs the p53 response and enhances apoptosis following UV irradiation. Int J Cancer. 2000;88:336–341. [PubMed] [Google Scholar]
- Hughes SC, Xu S, Fernihough J, Hampton A, Mason HD, Franks S, van der Stappen J, Donnelly MJ, Holly JM. Tissue IGFBP-3 proteolysis: contrasting pathophysiology to that in the circulation. Prog Growth Factor Res. 1995;6:293–299. doi: 10.1016/0955-2235(96)00041-5. [DOI] [PubMed] [Google Scholar]
- Hunziker EB, Kapfinger E, Martin J, Buckwalter J, Morales TI. Insulin-like growth factor (IGF)-binding protein-3 (IGFBP-3) is closely associated with the chondrocyte nucleus in human articular cartilage. Osteoarthritis Cartilage. 2008;16:185–194. doi: 10.1016/j.joca.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huwiler A, Kotelevets N, Xin C, Pastukhov O, Pfeilschifter J, Zangemeister-Wittke U. Loss of sphingosine kinase-1 in carcinoma cells increases formation of reactive oxygen species and sensitivity to doxorubicin-induced DNA damage. Br J Pharmacol. 2011;162:532–543. doi: 10.1111/j.1476-5381.2010.01053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez de Caceres I, Cortes-Sempere M, Moratilla C, Machado-Pinilla R, Rodriguez-Fanjul V, Manguan-Garcia C, Cejas P, Lopez-Rios F, Paz-Ares L, de CastroCarpeno J, Nistal M, Belda-Iniesta C, Perona R. IGFBP-3 hypermethylation-derived deficiency mediates cisplatin resistance in non-small-cell lung cancer. Oncogene. 2010;29:1681–1690. doi: 10.1038/onc.2009.454. [DOI] [PubMed] [Google Scholar]
- Ikezoe T, Tanosaki S, Krug U, Liu B, Cohen P, Taguchi H, Koeffler HP. Insulin-like growth factor binding protein-3 antagonizes the effects of retinoids in myeloid leukemia cells. Blood. 2004;104:237–242. doi: 10.1182/blood-2003-07-2203. [DOI] [PubMed] [Google Scholar]
- Iosef C, Gkourasas T, Jia CY, Li SS, Han VK. A functional nuclear localization signal in insulin-like growth factor binding protein-6 mediates its nuclear import. Endocrinology. 2008;149:1214–1226. doi: 10.1210/en.2007-0959. [DOI] [PubMed] [Google Scholar]
- Jaques G, Noll K, Wegmann B, Witten S, Kogan E, Radulescu RT, Havemann K. Nuclear localization of insulin-like growth factor binding protein 3 in a lung cancer cell line. Endocrinology. 1997;138:1767–1770. doi: 10.1210/endo.138.4.5177. [DOI] [PubMed] [Google Scholar]
- Jogie-Brahim S, Feldman D, Oh Y. Unraveling insulin-like growth factor binding protein-3 actions in human disease. Endocr Rev. 2009;30:417–437. doi: 10.1210/er.2008-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Firth SM. IGFBP-3: a cell fate pivot in cancer and disease. Growth Horm IGF Res. 2014;24:164–173. doi: 10.1016/j.ghir.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Krishnan AV, Peehl DM, Feldman D. Inhibition of prostate cancer growth by vitamin D: Regulation of target gene expression. J Cell Biochem. 2003;88:363–371. doi: 10.1002/jcb.10334. [DOI] [PubMed] [Google Scholar]
- Lalou C, Sawamura S, Segovia B, Ogawa Y, Binoux M. Proteolytic fragments of insulin-like growth factor binding protein-3: N-terminal sequences and relationships between structure and biological activity. C R Acad Sci III. 1997;320:621–628. doi: 10.1016/s0764-4469(97)85695-8. [DOI] [PubMed] [Google Scholar]
- Lee KW, Cobb LJ, Paharkova-Vatchkova V, Liu B, Milbrandt J, Cohen P. Contribution of the orphan nuclear receptor Nur77 to the apoptotic action of IGFBP-3. Carcinogenesis. 2007;28:1653–1658. doi: 10.1093/carcin/bgm088. [DOI] [PubMed] [Google Scholar]
- Lee KW, Cohen P. Nuclear effects: unexpected intracellular actions of insulin-like growth factor binding protein-3. J Endocrinol. 2002;175:33–40. doi: 10.1677/joe.0.1750033. [DOI] [PubMed] [Google Scholar]
- Lee KW, Liu B, Ma L, Li H, Bang P, Koeffler HP, Cohen P. Cellular internalization of insulin-like growth factor binding protein-3: distinct endocytic pathways facilitate re-uptake and nuclear localization. J Biol Chem. 2004;279:469–476. doi: 10.1074/jbc.M307316200. [DOI] [PubMed] [Google Scholar]
- Lee KW, Ma L, Yan X, Liu B, Zhang XK, Cohen P. Rapid apoptosis induction by IGFBP-3 involves an insulin-like growth factor-independent nucleomitochondrial translocation of RXRalpha/Nur77. J Biol Chem. 2005;280:16942–16948. doi: 10.1074/jbc.M412757200. [DOI] [PubMed] [Google Scholar]
- Lees-Miller SP, Anderson CW. The DNA-activated protein kinase, DNA-PK: a potential coordinator of nuclear events. Cancer Cells. 1991;3:341–346. [PubMed] [Google Scholar]
- Lefterova MI, Haakonsson AK, Lazar MA, Mandrup S. PPARgamma and the global map of adipogenesis and beyond. Trends Endocrinol Metab. 2014;25:293–302. doi: 10.1016/j.tem.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz BJ, Agostini-Dreyer A, Jetzt AE, Krumm CS, Cohick WS. IGF binding protein-3 mediates stress-induced apoptosis in non-transformed mammary epithelial cells. J Cell Physiol. 2013;228:734–742. doi: 10.1002/jcp.24220. [DOI] [PubMed] [Google Scholar]
- Li J, Jin D, Fu S, Mei G, Zhou J, Lei L, Yu B, Wang G. Insulin-like growth factor binding protein-3 modulates osteoblast differentiation via interaction with vitamin D receptor. Biochem Biophys Res Commun. 2013;436:632–637. doi: 10.1016/j.bbrc.2013.04.111. [DOI] [PubMed] [Google Scholar]
- Li W, Fawcett J, Widmer HR, Fielder PJ, Rabkin R, Keller GA. Nuclear transport of insulin-like growth factor-I and insulin-like growth factor binding protein-3 in opossum kidney cells. Endocrinology. 1997;138:1763–1766. doi: 10.1210/endo.138.4.5176. [DOI] [PubMed] [Google Scholar]
- Lin MZ, Marzec KA, Martin JL, Baxter RC. The role of insulin-like growth factor binding protein-3 in the breast cancer cell response to DNA-damaging agents. Oncogene. 2014;33:85–96. doi: 10.1038/onc.2012.538. [DOI] [PubMed] [Google Scholar]
- Liu B, Lee HY, Weinzimer SA, Powell DR, Clifford JL, Kurie JM, Cohen P. Direct functional interactions between insulin-like growth factor-binding protein-3 and retinoid X receptor-alpha regulate transcriptional signaling and apoptosis. J Biol Chem. 2000;275:33607–33613. doi: 10.1074/jbc.M002547200. [DOI] [PubMed] [Google Scholar]
- Ludwig RL, Bates S, Vousden KH. Differential activation of target cellular promoters by p53 mutants with impaired apoptotic function. Mol Cell Biol. 1996;16:4952–4960. doi: 10.1128/mcb.16.9.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfori M, Mynott A, Ellis JJ, Mehdi AM, Saunders NF, Curmi PM, Forwood JK, Boden M, Kobe B. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim Biophys Acta. 2011;1813:1562–1577. doi: 10.1016/j.bbamcr.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Martin JL, Baxter RC. Insulin-like growth factor-binding protein from human plasma. Purification and characterization. J Biol Chem. 1986;261:8754–8760. [PubMed] [Google Scholar]
- Martin JL, de Silva HC, Lin MZ, Scott CD, Baxter RC. Inhibition of insulin-like growth factor-binding protein-3 signaling through sphingosine kinase-1 sensitizes triple-negative breast cancer cells to EGF receptor blockade. Mol Cancer Ther. 2014;13:316–328. doi: 10.1158/1535-7163.MCT-13-0367. [DOI] [PubMed] [Google Scholar]
- Martin JL, Lin MZ, McGowan EM, Baxter RC. Potentiation of growth factor signaling by insulin-like growth factor-binding protein-3 in breast epithelial cells requires sphingosine kinase activity. J Biol Chem. 2009;284:25542–25552. doi: 10.1074/jbc.M109.007120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micutkova L, Hermann M, Offterdinger M, Hess MW, Matscheski A, Pircher H, Muck C, Ebner HL, Laich A, Ferrando-May E, Zwerschke W, Huber LA, Jansen-Durr P. Analysis of the cellular uptake and nuclear delivery of insulin-like growth factor binding protein-3 in human osteosarcoma cells. Int J Cancer. 2012;130:1544–1557. doi: 10.1002/ijc.26149. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, Stunnenberg HG. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paharkova-Vatchkova V, Lee KW. Nuclear export and mitochondrial and endoplasmic reticulum localization of IGF-binding protein 3 regulate its apoptotic properties. Endocr Relat Cancer. 2010;17:293–302. doi: 10.1677/ERC-09-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payet LD, Wang XH, Baxter RC, Firth SM. Amino- and carboxyl-terminal fragments of insulin-like growth factor (IGF) binding protein-3 cooperate to bind IGFs with high affinity and inhibit IGF receptor interactions. Endocrinology. 2003;144:2797–2806. doi: 10.1210/en.2003-0102. [DOI] [PubMed] [Google Scholar]
- Petitjean A, Ruptier C, Tribollet V, Hautefeuille A, Chardon F, Cavard C, Puisieux A, Hainaut P, Caron de Fromentel C. Properties of the six isoforms of p63: p53-like regulation in response to genotoxic stress and cross talk with DeltaNp73. Carcinogenesis. 2008;29:273–281. doi: 10.1093/carcin/bgm258. [DOI] [PubMed] [Google Scholar]
- Pon CK, Firth SM, Baxter RC. Involvement of insulin-like growth factor binding protein-3 in peroxisome proliferator activated receptor gamma-mediated inhibition of breast cancer cell growth. Mol Cell Endocrinol. 2015;399:354–361. doi: 10.1016/j.mce.2014.10.023. [DOI] [PubMed] [Google Scholar]
- Qiu J, Ma X-L, Wang X, Chen H, Huang B-R. Insulin-like growth factor binding protein-3 interacts with the thyroid hormone receptor alpha1 and modulates transcription of thyroid hormone responsive gene. Acta Acad. Med. Sin. 2011;33:156–161. doi: 10.3881/j.issn.1000-503X.2011.02.012. [DOI] [PubMed] [Google Scholar]
- Radulescu RT. Nuclear localization signal in insulin-like growth factor-binding protein type 3. Trends Biochem Sci. 1994;19:278. doi: 10.1016/0968-0004(94)90004-3. [DOI] [PubMed] [Google Scholar]
- Robbins GT, Nie D. PPAR gamma, bioactive lipids, and cancer progression. Front Biosci (Landmark Ed) 2012;17:1816–1834. doi: 10.2741/4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santer FR, Bacher N, Moser B, Morandell D, Ressler S, Firth SM, Spoden GA, Sergi C, Baxter RC, Jansen-Durr P, Zwerschke W. Nuclear insulin-like growth factor binding protein-3 induces apoptosis and is targeted to ubiquitin/proteasome-dependent proteolysis. Cancer Res. 2006;66:3024–3033. doi: 10.1158/0008-5472.CAN-05-2013. [DOI] [PubMed] [Google Scholar]
- Schedlich LJ, Graham LD, O'Han MK, Muthukaruppan A, Yan X, Firth SM, Baxter RC. Molecular basis of the interaction between IGFBP-3 and retinoid X receptor: role in modulation of RAR-signaling. Arch Biochem Biophys. 2007a;465:359–369. doi: 10.1016/j.abb.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Schedlich LJ, Le Page SL, Firth SM, Briggs LJ, Jans DA, Baxter RC. Nuclear import of insulin-like growth factor-binding protein-3 and-5 is mediated by the importin beta subunit. J Biol Chem. 2000;275:23462–23470. doi: 10.1074/jbc.M002208200. [DOI] [PubMed] [Google Scholar]
- Schedlich LJ, Muthukaruppan A, O'Han MK, Baxter RC. Insulin-like growth factor binding protein-5 interacts with the vitamin D receptor and modulates the vitamin D response in osteoblasts. Mol Endocrinol. 2007b;21:2378–2390. doi: 10.1210/me.2006-0558. [DOI] [PubMed] [Google Scholar]
- Schedlich LJ, Nilsen T, John AP, Jans DA, Baxter RC. Phosphorylation of insulin-like growth factor binding protein-3 by deoxyribonucleic acid-dependent protein kinase reduces ligand binding and enhances nuclear accumulation. Endocrinology. 2003;144:1984–1993. doi: 10.1210/en.2002-220798. [DOI] [PubMed] [Google Scholar]
- Schedlich LJ, O'Han MK, Leong GM, Baxter RC. Insulin-like growth factor binding protein-3 prevents retinoid receptor heterodimerization: implications for retinoic acid-sensitivity in human breast cancer cells. Biochem Biophys Res Commun. 2004;314:83–88. doi: 10.1016/j.bbrc.2003.12.049. [DOI] [PubMed] [Google Scholar]
- Schedlich LJ, Young TF, Firth SM, Baxter RC. Insulin-like growth factor-binding protein (IGFBP)-3 and IGFBP-5 share a common nuclear transport pathway in T47D human breast carcinoma cells. J Biol Chem. 1998;273:18347–18352. doi: 10.1074/jbc.273.29.18347. [DOI] [PubMed] [Google Scholar]
- Seligson DB, Yu H, Tze S, Said J, Pantuck AJ, Cohen P, Lee KW. IGFBP-3 nuclear localization predicts human prostate cancer recurrence. Horm Cancer. 2013;4:12–23. doi: 10.1007/s12672-012-0124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- Soler AP, Alemany J, Smith RM, de Pablo F, Jarett L. The state of differentiation of embryonic chicken lens cells determines insulin-like growth factor I internalization. Endocrinology. 1990;127:595–603. doi: 10.1210/endo-127-2-595. [DOI] [PubMed] [Google Scholar]
- Twigg SM, Baxter RC. Insulin-like growth factor (IGF)-binding protein 5 forms an alternative ternary complex with IGFs and the acid-labile subunit. J Biol Chem. 1998;273:6074–6079. doi: 10.1074/jbc.273.11.6074. [DOI] [PubMed] [Google Scholar]
- van Heemst D, den Reijer PM, Westendorp RG. Ageing or cancer: a review on the role of caretakers and gatekeepers. Eur J Cancer. 2007;43:2144–2152. doi: 10.1016/j.ejca.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Vikhanskaya F, Lee MK, Mazzoletti M, Broggini M, Sabapathy K. Cancer-derived p53 mutants suppress p53-target gene expression--potential mechanism for gain of function of mutant p53. Nucleic Acids Res. 2007;35:2093–2104. doi: 10.1093/nar/gkm099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Yang J, Lee SL, Kulp SK, Chen CS. PPARgamma-independent antitumor effects of thiazolidinediones. Cancer Lett. 2009;276:119–124. doi: 10.1016/j.canlet.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wraight CJ, Liepe IJ, White PJ, Hibbs AR, Werther GA. Intranuclear localization of insulin-like growth factor binding protein-3 (IGFBP-3) during cell division in human keratinocytes. J Invest Dermatol. 1998;111:239–242. doi: 10.1046/j.1523-1747.1998.00258.x. [DOI] [PubMed] [Google Scholar]
- Xu D, Farmer A, Collett G, Grishin NV, Chook YM. Sequence and structural analyses of nuclear export signals in the NESdb database. Mol Biol Cell. 2012;23:3677–3693. doi: 10.1091/mbc.E12-01-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada PM, Lee KW. Perspectives in mammalian IGFBP-3 biology: local vs. systemic action. Am J Physiol Cell Physiol. 2009;296:C954–C976. doi: 10.1152/ajpcell.00598.2008. [DOI] [PubMed] [Google Scholar]
- Yan X, Payet LD, Baxter RC, Firth SM. Activity of human pregnancy insulin-like growth factor binding protein-3: determination by reconstituting recombinant complexes. Endocrinology. 2009;150:4968–4976. doi: 10.1210/en.2009-0090. [DOI] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Yoshino K, Motoyama S, Koyota S, Shibuya K, Usami S, Maruyama K, Saito H, Minamiya Y, Sugiyama T, Ogawa J. IGFBP3 and BAG1 enhance radiation-induced apoptosis in squamous esophageal cancer cells. Biochem Biophys Res Commun. 2011;404:1070–1075. doi: 10.1016/j.bbrc.2010.12.115. [DOI] [PubMed] [Google Scholar]
- Zeitz MJ, Ay F, Heidmann JD, Lerner PL, Noble WS, Steelman BN, Hoffman AR. Genomic interaction profiles in breast cancer reveal altered chromatin architecture. PLoS One. 2013;8:e73974. doi: 10.1371/journal.pone.0073974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Steinle JJ. DNA-PK phosphorylation of IGFBP-3 is required to prevent apoptosis in retinal endothelial cells cultured in high glucose. Invest Ophthalmol Vis Sci. 2013;54:3052–3057. doi: 10.1167/iovs.12-11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, He LR, Xi M, Cai MY, Shen JX, Li QQ, Liao YJ, Qian D, Feng ZZ, Zeng YX, Xie D, Liu MZ. Nimotuzumab promotes radiosensitivity of EGFR-overexpression esophageal squamous cell carcinoma cells by upregulating IGFBP-3. J Transl Med. 2012;10:249. doi: 10.1186/1479-5876-10-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yin P, Bach LA, Duan C. Several acidic amino acids in the N-domain of insulin-like growth factor-binding protein-5 are important for its transactivation activity. J Biol Chem. 2006;281:14184–14191. doi: 10.1074/jbc.M506941200. [DOI] [PubMed] [Google Scholar]