Abstract
Ovarian cancer is known to be composed of distinct populations of cancer cells, some of which demonstrate increased capacity for cancer initiation and/or metastasis. The study of human cancer cell populations is difficult due to long requirements for tumor growth, inter-patient variability and the need for tumor growth in immune-deficient mice. We therefore characterized the cancer initiation capacity of distinct cancer cell populations in a transgenic murine model of ovarian cancer. In this model, conditional deletion of Apc, Pten, and Trp53 in the ovarian surface epithelium (OSE) results in the generation of high grade metastatic ovarian carcinomas. Cell lines derived from these murine tumors express numerous putative stem cell markers including CD24, CD44, CD90, CD117, CD133 and ALDH. We show that CD24+ and CD133+ cells have increased tumor sphere forming capacity. CD133+ cells demonstrated a trend for increased tumor initiation while CD24+ cells vs CD24− cells, had significantly greater tumor initiation and tumor growth capacity. No preferential tumor initiating or growth capacity was observed for CD44+, CD90+, CD117+, or ALDH+ versus their negative counterparts. We have found that CD24+ cells, compared to CD24− cells, have increased phosphorylation of STAT3 and increased expression of STAT3 target Nanog and c-myc. JAK2 inhibition of STAT3 phosphorylation preferentially induced cytotoxicity in CD24+ cells. In vivo JAK2 inhibitor therapy dramatically reduced tumor metastases, and prolonged overall survival. These findings indicate that CD24+ cells play a role in tumor migration and metastasis and support JAK2 as a therapeutic target in ovarian cancer.
Keywords: Cancer Initiating Cells, CD24, JAK2, STAT3, ovarian cancer, metastasis
Introduction
Ovarian cancer is the fifth leading cause of cancer-related death among women (1). Most ovarian cancer patients present with advanced stage disease such that treatment with surgery and chemotherapy results in a median progression-free survival of only 16–22 months and a 5-year survival rate of only 27% (2). Contributing to the poor outcome in ovarian cancer may be the significant cellular heterogeneity in ovarian cancer.
Our group and others have characterized distinct ovarian cancer cell populations in human ovarian cancer cell lines and patient specimens. Human cancer cell populations defined by various combinations of Hoechst dye exclusion, CD133, aldehyde dehydrogenase (ALDH), CD44, CD117, CD24 and Epcam, have all been reported to have increased ability to initiate cancer and/or promote metastasis in vivo (3–11). The study of the distinct cancer cell populations that make up human cancer is very challenging due to inter-patient variability, the need for testing tumor initiation in immunosuppressive mice, and long-term growth requirement for in vivo studies. Murine tumor models provide means to study the biology of heterogeneous cancer cell populations in tumors arising as a consequence of well-defined genetic mutations in an immunocompetent microenvironment. There are currently several genetic murine models of ovarian cancer, which utilize ovarian bursal injection of an adenovirus expressing Cre recombinase (AdCre) to induce Cre-mediated deletion of specific tumor suppressor genes in the OSE (12–14). Wu et al. developed a model of a high-grade endometrioid ovarian carcinoma, which develops after inactivation of Apc, Pten, Trp53. In this model, the addition of Trp53 mutation appears to be associated with a Type I- to Type-II ovarian cancer progression (15), with tumor bearing mice dying rapidly (within weeks) due to widely metastatic disease in a manner similar to that of patients with advanced stage ovarian cancer patients (16, 17). Genetic analysis of these tumors demonstrated gene expression patterns similar to human disease.
In this study we characterized cell lines and primary tumors from the Apc−; Pten−; Trp53− ovarian tumor model for cells with ovarian cancer initiating cell (CIC) activity. Tumors generated in this model have an endometriod histology, but in the presence of a p53 mutation, have a high grade metastatic phenotype reminiscent analogous to that seen in patients with high grade serous cancer (15). We demonstrate that cells with expression of the cell surface marker CD24 have greater sphere forming capacity, ability to passage, and ability to initiate tumors in vivo. Similar to the observation in hepatocellular carcinomas, CD24+ CIC demonstrate preferential phosphorylation of STAT3 and expression of Nanog and CD24+ cells are preferentially sensitive to inhibition of STAT3 phosphorylation with a JAK2 inhibitor. Finally, we show that JAK2 therapy in vivo using this tumor model prevents tumor metastasis. This study supports other work demonstrating CD24+ cells as a CIC population with increased metastatic potential and suggests that targeting JAK2 could reduce ovarian tumor metastasis.
Materials and Methods
Cell Culture
Murine ovarian endometrioid adenocarcinoma cell lines were derived as previously described (18). Briefly, the W2476T tumor cell line was established by mechanically dispersing ovarian tumor tissues with sterile scalpels followed by digestion at 37° C with 0.05% Trypsin-EDTA for 20 minutes. Cells were cultured for five passages in DMEM containing 10% FBS and 1% penicillin/streptomycin (p/s) in an incubator with 3% O2; 5% CO2. During the first five passages of primary culture, non-adherent cells were discarded, and only adherent cells were passaged. W2476T cells display epithelial (cobblestone) morphology in culture. Cells were maintained and grown in RPMI containing 10% of FBS and 1% of p/s (Gibco, Grand Island, NY) at 37° C and 5% CO2. To create W2476T-Luciferase expressing cells, W2476T cells were transduced with Luciferase-expressing lentiviral construct (provided by the UMCC Vector core).
Isolation of Cancer Initiating Cells from W2476T cell line and primary Apc−; Pten−; Trp53− tumors
Primary Apc−; Pten−; Trp53− tumors were mechanically dissected into single cell suspension as previously described (5). Cells from primary tumor suspensions or the W2476T cell lines were then isolated using fluorescence activated cell sorting (FACS). Briefly, primary ovarian tumor or W2476T cell line single cell suspensions were counted and incubated with primary antibodies CD24-PerCP Cy5.5, CD117-APC and CD133-PE (eBioscience, San Diego, CA), CD44-Pacific Blue (Biolegend, San Diego, CA), CD90-PE (BD Pharmingen, San Jose, CA) for 30 min at 4° C. Cells were then stained with propidium iodide (PI) or DAPI as a viability stain. For ALDH+ samples, ALDH enzymatic activity was defined using the ALDEFLUOR kit (Stem Cell Technologies, Canada) as previously described (5). FACS was performed with ~ 1 ×106 cells using FACSAria (Becton Dickinson, Franklin Lakes, NJ) under low pressure in the absence of UV light. Live cells were selected based upon both forward vs. side-scatter profiles and absence of PI or DAPI stain and ALDEFLUOR/DEAB treated cells were used to define negative gates for ALDH.
Sphere Assays
Sphere culture was performed as previously described (5, 19, 20) with FACS-isolated CD24+/−, CD44+/−, CD90+/−, CD117+/−, CD133+/− and ALDH+/− cell populations plated in triplicate in either 6-well or 24-well ultra-low attachment plates in serum-free DMEM/F12, epidermal growth factor (EGF) 20ng/mL, gentamycin 20μg/mL, insulin 5μg/mL, 1% p/s (Gibco, Grand Island, NY), hydrocortisone 1ng/mL, β-Mercaptoethanol 100μM (Sigma, St. Lois, MO), fibroblast growth factor (bFGF) 10ng/mL (Millipore, Billerica, MA). Either 300 or 2000 cells (depending on the availability of cells to perform the experiment triplicates) were seeded in ultralow attachment plates (Sigma, St. Louis, MO). Primary spheres were counted on day 5 and dissociated enzymatically with warm 0.025% Trypsin-EDTA (Gibco, Grand Island, NY) for 2 minutes at 37° C and then passed 5 times through a sterile Pasteur pipette to isolate single cells. The cells obtained from dissociation were analyzed microscopically for single cellularity and then re-plated ultralow attachment plates. Secondary spheres were counted on day 5 after passaging.
Tumor Initiation Studies with Cell lines and Primary Tumor Cells
Mice were housed and maintained in the University of Michigan Unit for Laboratory Animal Medicine and all studies were performed with the approval of the University Committee on the Use and Care of Animals. For screening cell line experiments, 5×104 W2476T-Luc cells were FACS sorted cells for each cancer stem cell marker were injected subcutaneously into the contralateral axilla of CIEA NOG (NOD/Shi-scid/IL-2Rγ null mice) (Taconic, Germantown, NY). Two weeks after injection, tumor growth was monitored weekly using in vivo bioluminescence imaging and tumor measurement using calipers as previously described (19). Mice were imaged using an IVIS Image System 200 Series (Xenogen Corporation) approximately 10 min after injection of D-luciferin. The bioluminescence images were collected using sequential mode until reaching peak values and analyzed by LivingImage 3.0 sofware (Xenogen Corporation). Mice were followed until tumors met euthanasia guidleines. Tumors were then harvested, weighed and snap frozen for histologic analysis, extraction of RNA and protein. Tumor volumes were calculated using the LxW xW/2 formula.
For CD24 cell limiting dilution studies, 1500, 600, or 200 CD24+ or CD24− W2476T cells were injected into the contralateral flanks of mice euthanized as above. Confirmatory experiments were then performed with 200 CD24+ or CD24− W2476T cells injected into the flanks of separate mice. Animals were monitored, as indicated above, for tumor development for up to 6 months.
For primary murine ovarian cancer cells experiments, Apc−; Pten−; Trp53− tumor bearing mice were generated via intra-bursal AdCre injection as previously described (16, 17). Apc−; Pten−; Trp53− murine ovarian tumors were harvested and cells were dissociated and sorted using FACS for CD24+, CD24− as stated above. 5×104 or 5×103rd CD24+ and CD24− primary murine ovarian cancer cells were injected into individual NOG mice. Tumor growth was then monitored weekly using calipers for up to 5 months.
In vivo Therapeutic Response
Apc−; Pten−; Trp53− tumor bearing mice were generated via intra-bursal AdCre injection as previously described (16, 17). Mice were monitored following AdCre injection until ovarian tumors were ~500mm3 (~4weeks). Mice were paired by tumor size and then randomized to treatment with cisplatin alone (once a week 2mg/Kg × 2 weeks and vehicle daily) or cisplatin + daily TG101209 (50 mg/kg in 40ul DMSO) (n=6/group). We chose a reduced dose of TG101209 (a 4-fold reduction versus previous studies) as IP dosing would increase local tumor concentrations (21). A 21 day dosing scheme was used as our experimental data indicated dosing of 28 days increased drug related toxicity. Mice were monitored in conjunction with staff in the Unit of Laboratory and Animal Management and euthanized when the animal appeared (i) ill with signs of weight loss, (ii) had significant bowel distention, (iii) or primary ovarian tumors were felt to be ~1000mm3. For treatment of ‘early stage’ tumors, therapy was initiated 7 days after injection of AdCre. Apc−; Pten−; Trp53− mice were treated with 40μL of DMSO alone or TG101209 single agent therapy daily for 21 days, (n=11 controls, n=14 TG101209). A subset of animals (n=4/group) were sacrificed 1 hour after the final TG101209 therapy and tumors harvested for the assessment of p-Stat3.
RNA Isolation and qRT-PCR
Subpopulation of W2476T cells were isolated by FACS and RNA was extracted using PureLink RNA Mini kit, (Ambion, Grand Island, NY) per manufacturer recommendations. RNA integrity was confirmed on the Agilent 2100 BioAnalyzer. Quantitative real-time PCR was performed for 40 cycles using SYBR green (Applied Biosystems, Carlsbad, CA) as recommended by the manufacturer, with primers at 100nM concentrations. Expression was normalized to Transferrin. All transcripts were confirmed using 3% agarose gel electrophoresis.
Western Blotting
To detect differential expression of pSTAT3 in CD24+ versus CD24− cells, ~3×105 W2476T cells and CD24+/− sorted cells were incubated overnight at 37°C and 5% CO2. Protein lysates were obtained after 1 hour incubation with TG10209 or Stattic (22) (Sigma, St. Louis, MO). Protein concentrations were determined using the Bradford Protein Assay (Bio-Rad) using Spectra max (Molecular Devices, Downingtown, PA) and SofMaxPro V5 sofware. 20μg of protein lysates were separated on 4–12% NuPAGE SDS gel (Invitrogen, Grand Island, NY) and detected using total STAT3 and pSTAT3 (1:1000 dilution, Cell Signaling Technology, Danvers, MA). Bands were visualized using the ECL Kit Pierce (Thermo Scientific, Waltham, MA).
Drug treatment assays
5×105 W2476T cells were seeded and treated with increasing concentrations of TG101209 (0, 0.6, 1 and 1.3μM) were administered on day 1 only. Cell number as a percentage of untreated control was assessed with a Cell Countess after 72 hours or FACs evaluated for CD24 expression as above.
Cells in suspension
For drug treatment in spheres, W2476T cells were first sorted for CD24 positive and negative populations then subjected for sphere assay. 24 hours later, cells were treated with Stattic or TG101209 for three consecutive days and then sphere were counted and subjected to passaging studies as above on day 5 after sorting.
p-Stat3 quantification
p-Stat3 density was assessed as previously described (23). Briefly, 5 independent sections from TG101209 and DMSO treated tumors (n=4 each) were stained with anti-p-Stat3 (NEED AB 1:50) and signal detected with DAB as above. Ten high powered fields from each section were then photographed and DAB stain quantified as previously described using Olympus biologic suites software. Results were compated using a two-sided student’s t-test.
In vitro invasion assays
5×105 W2476T cells were plated in T75 flasks and pre-treated with TG101209 0.6 μM or DMSO for 24 hours. Cells were trypsinized and plated 25 × 103 per chamber in RPMI containing 2% of FBS and TG101209 (0.6 μM) or DMSO in rehydrated BD BioCoat Matrigel Chamber (BD Biosciences, San Jose, CA). RPMI containing 10% of FBS was used as chemoattractant. Cells were incubated for 24 hours at 37° C and 5% CO2 and then cells were fixed with cold methanol and stained with Vectashield Mounting Medium for fluorescence with DAPI H-1200 (Vector Laboratories, Burlingame, CA). Five sections from each matrigel chamber were photographed and then scored for DAPI pixel density using Olympus Microsuite Biological Suite Software. DAPI labeling for TG101209 treated cells and DMSO controls were compared using a two-sided student t-test.
Migration Assay
W2476T cells were treated for 24hrs with 0.6uM of TG101209. Next day cells were sorted for CD24+ and CD24− cells as above and then resuspended in RPMI with 2% of FBS and 0.6uM of TG101209 or vehicle control. Then cells were loaded in the left end of the migration chip; chips contain 20, 1mm-long channels with a concentration of ~1 cell/per channel. The right side of the channel was loaded with RPMI-5% FBS (to create a chemoattractant gradient) containing 0.6uM of TG101209. 24hr later were measured the movement of cells in the migration channels. Three devices were used per condition in two independent experiments.
Biostatistics
All in vitro assays, including FACS analysis of CIC markers, tumor sphere assays, qRT-PCR, cytotoxicity and invasion assays are a result triplicate assays in three independent experiments. Data were compared using a two sided student’s t-test with Microsoft Excel software. For murine tumor studies, final tumor volumes/weights were also compared using a two-sided student’s t-test as above. For survival studies, Kaplan-Meier curves were generated and compared using the Mantel-Cox test and p-values obtained using Prism Graphpad (GraphPad Software La Jolla, CA).
Results
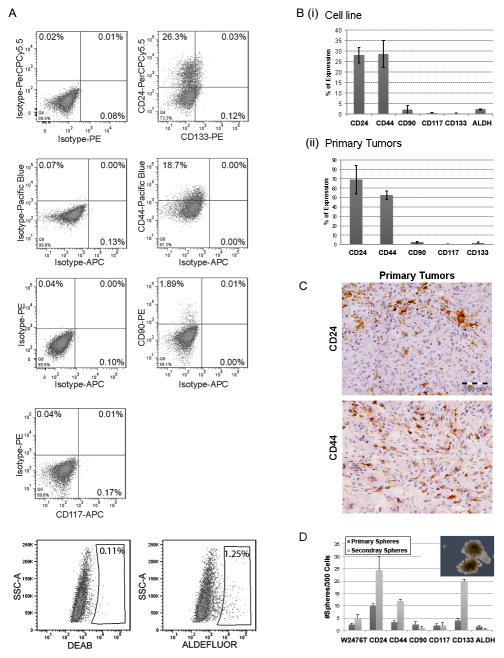
To characterize the various cell populations in the murine Apc−; Pten−; Trp53− ovarian cancer tumor model we first used flow cytometry to analyze the expression of commonly reported CIC markers in a tumor cell line (W2476T) derived from the Apc−; Pten−; Trp53− ovarian cancer tumor model (Fig. 1A and Bi). We detected substantial expression of CD24 (20–30%), CD44 (22–35%), and more limited expression of CD90, ALDH, CD117, and CD133 (0.5–2% each). We also evaluated mechanically dissected Apc−; Pten−; Trp53− primary tumors and performed FACS analysis. Primary tumors cells had low viability (20%). Surviving primary cells had higher expression of CD24 (55–85%), CD44 (48–68%) and low expression of CD90 (1–3%), CD117 (0–1%) and CD133 (0.5–1%) (Fig. 1Bii). However, immunohistochemical analysis of primary tumors demonstrated significantly lower numbers of CD24+ and CD44+ cells with each labeling 5–10% of tumor cells (Fig. 1C). The discrepancy between FACS results and IHC analysis suggest that the process of mechanically dissecting tumors into single cells may enrich for cells expressing CD24 and CD44.
Figure 1. Analysis of stem cell marker expression in cell line W2476T and primary tumors fresh isolated cells derived from the Apc−; Pten−; Trp53− ovarian mouse model.
(A) FACS plots of the indicated markers and isotype controls of W2476T cell line. (B) Summary of average percentage of CIC marker expressing cells in (i) W2476T cell line and (ii) cell freshly isolated for primary tumors (n=5). (C) Immunohistochemistry indicating CD24 and CD44 levels in Apc−; Pten−; Trp53− primary tumors. (D) Summary of average numbers of primary tumor spheres and primary tumor spheres passaged and formation of secondary spheres formed from 300 FACS isolated W2476T cells expressing the indicated CIC marker. Results are representative of at least 3 assays with 3 replicates for each group.
The ability to grow in suspension is an important attribute of ovarian cancer cells. Ovarian cancer cells commonly present in cancer associated ascites are believed to be a source of both chemotherapy resistance and metastases (24). We next tested the ability of FACS isolated cells expressing the specific cell markers characterized above to generate tumor spheres. CD24+ and CD133+ cells generated more primary tumor spheres, and demonstrated a greater ability to passage to form secondary spheres (Fig. 1D).
CD24 Identifies a Population of Cells Enriched for Tumor Initiation Capacity
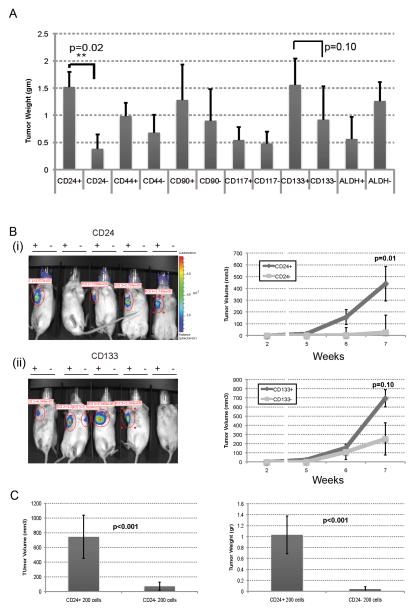
In vivo tumor initiation remains the gold standard for the identification of CIC. We therefore sorted luciferase W2476T-Luc cells for each marker and their negative counterparts and assessed tumor initiation capacity. Specifically, CD24+ vs CD24−, CD44+ vs CD44−, CD90+ vs CD90−, CD117+ vs CD117− and ALDH+ vs ALDH− cells were FACS isolated and then 5,000 cells of each population were injected subcutaneously in the axilla of NOG mice (n=5) and tumor formation monitored by bioluminescence. In each mouse, marker positive cells were injected in the right axilla and marker negative cells in the left axilla. CD24+ cells demonstrated tumor formation earlier and formed statistically larger tumors than CD24− cells (Fig 2A and B). CD133+ vs CD133− cells demonstrated a trend for increased tumor growth (Fig 2A and Bii). Neither CD44, CD90, CD117 nor ALDH marker positive vs. negative populations were noted to have significantly different tumor growth capacity between marker positive vs. marker negative populations (p>0.20, Fig. 2A and Supplemental Fig. 1).
Figure 2. W2476T CD24+ cells demonstrate preferential tumor growth capacity in vivo.
(A) Tumor weight of tumors generated from 5000 FACS isolated W2476T cells with or without the indicated cell surface marker expression. (B) Bioluminescent images and tumor growth curves of tumors formed from 200 FACS isolated W2476T cells which express or do not express (i) CD24 or (ii) CD133 (Right axilla marker positive cells and left axilla marker negative cells). (C) Final tumor weights and tumor volume of CD24+ vs CD24− cell initiated tumors grown in contralateral flanks of NOG mice (n=5 per group in two independent experiments).
Given the increased tumor growth rate of CD24+ cells, we next performed limiting dilution tumor initiation studies with 1500, 600 and 200 CD24+ versus CD24− cells injected into contralateral flanks of mice and monitored for up to 3 months. 200 CD24+ cells had a greater rate of tumor initiation (5/5) compared to CD24− cells (2/5) (Fig. 2C) and Table 1. To confirm these studies we repeated assays with 200 CD24+ versus CD24− cells injected into separate mice and followed for 5 months. Once again, CD24+ cells demonstrated greater tumor initiation (9/10) capacity vs. CD24− cells (5/10). From the cell line tumor initiation studies using the ‘extreme limiting dilution analysis’ (25) software we estimate tumor initiating cell frequency of 1 in 133 (Range 72–238) for CD24+ cells and 1 in 668 (range 357–1252) for CD24− cells (p=0.0000517). Interestingly FACS analysis of tumors derived from both CD24+ and CD24− cell initiated tumors had both CD24− and CD24+ cells (Supplemental Figure 2). Not surprisingly, given the presence of both CD24− and CD24+ cells in all tumors, tumors from both CD24+ and CD24− cells were capable of serial passage through at least three generations.
Table 1.
Number of tumors initiated from CD24+ and CD24− cell populations from cell line and primary tumors for each cell number. Cells were in contralateral flanks of NOG mice and monitored up to 3 months.
| Cell Source | ||||
|---|---|---|---|---|
| Tumor Cell line | Primary Tumors | |||
| Number of cells injected | Tumors formed from CD24+ | Tumors formed from CD24(−) | Tumors formed from CD24+ | Tumors formed from CD24(−) |
| 50,000* | ND | ND | 11/15 | 6/15 |
| 5000* | 5/5 | 5/5 | 2/10 | 1/10 |
| 1500 | 5/5 | 3/5 | ND | ND |
| 600 | 4/5 | 3/5 | ND | ND |
| 200 | 5/5 | 2/5 | ND | ND |
| 200** | 9/10 | 5/10 | ND | ND |
Cells injected into separate mice and monitored up to 5 months.
Next, we similarly assessed the tumor initiation rates of CD24+ cells freshly isolated from Apc−; Pten−; Trp53− tumors. Likely due to the poor viability of primary cells after mechanical cell isolation, significantly higher cell numbers (50,000) were necessary to initiate tumors. However, once again CD24+ cells showed greater tumor initiation rates compared to CD24− cells counterparts (Table 1).
CD24 Cells Preferentially Express Stem Cell Genes and Phosphorylate STAT3
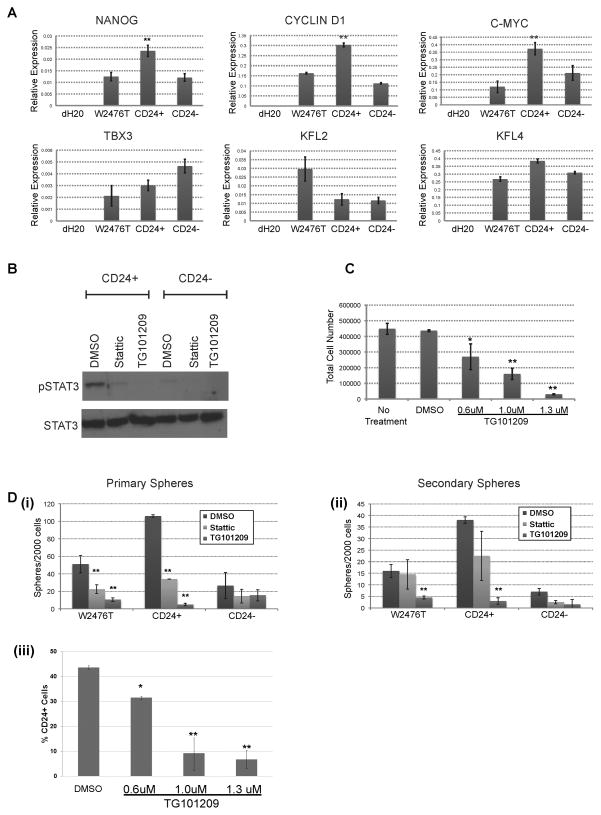
CD24 has been reported as a CIC marker in hepatic cancer and CD24+ cells in hepatic cancer had increased expression of Nanog, and Nanog promoted cellular self-renewal via phosphorylation of STAT3 (26). We therefore evaluated pSTAT3 in CD24+ and CD24− cells. CD24+ cells showed increased basal levels of STAT3 phosporylation compared to the CD24− cells (Fig 3B). Treatment with either the direct STAT3 inhibitor Stattic, or the JAK2 inhibitor TG101209 could reduce p-STAT3 levels, though TG101209 inhibited pSTAT3 more efficiently in isolated CD24+ cells than Stattic (Fig 3B). Treatment of unsorted WT2476T cells with increasing doses of TG101209 was associated with significant decreases in cell number with TD50 ~880 nM, (Fig 3C). In order to determine the functional importance of STAT3 phosphorylation, we treated CD24+ and CD24− cells in a tumor sphere formation assay either Stattic or TG101209. Both Stattic and TG101209 preferentially reduced primary tumor sphere formation of unsorted cells and preferentially decreased CD24+ primary sphere growth (Fig. 3Di). TG101209 treatment was associated with a decrease in the ability of secondary tumor sphere formation from both unsorted and CD24+ cells, and essentially eliminated passaging potential (Fig. 3Dii). Consistent with TG101209 preferentially targeting CD24+ cells, FACS evaluation of W2476T cell line with increasing doses of TG101209 demonstrated declining percentages of CD24+ cells (Fig. 3Diii).
Figure 3. W2476T CD24+ cells preferentially expresses stem cell genes and have increased levels of pSTAT3.
(A) qRT-PCR Comparison of stem cell gene expression in W2476T whole cell line and W2476T sorted CD24+ and CD24− cells. (B) Western blot showing differential phosphorylation of STAT3 in CD24+ and CD24− W2476T cells and inhibition by Stattic and TG101209. (C) Percent viable W2476T cells treated with increasing doses of TG101209. (Di) Tumor sphere formation of W2476T cells treated with Stattic and TG101209 for three consecutive days and counted on day 5. (Dii) Passage and formation of secondary spheres from spheres grown in Di. Results show preferential inhibition of spheres formed from CD24+ W2476T cells. (Diii) Percentage of CD24+ W2476T cells after treatment with increasing doses of TG101209. * indicates p≤0.05, ** indicates p≤0.01.
We also characterized the expression of several known stem cell gene/targets of STAT3 phosphorylation in CD24+ and CD24− populations (Fig. 3A). Unsorted W2476T cells, and sorted CD44+ and CD44− cells were used as controls (Supplemental Fig. 3). While no differential expression of any tested marker was noted between CD44+ and CD44− cells, qRT-PCR demonstrated that CD24+ cells, compared to CD24− cells, had increased expression of Nanog, c-myc and Cyclin D1.
Therapy with TG101209 Prevents Metastases and Improves Survival
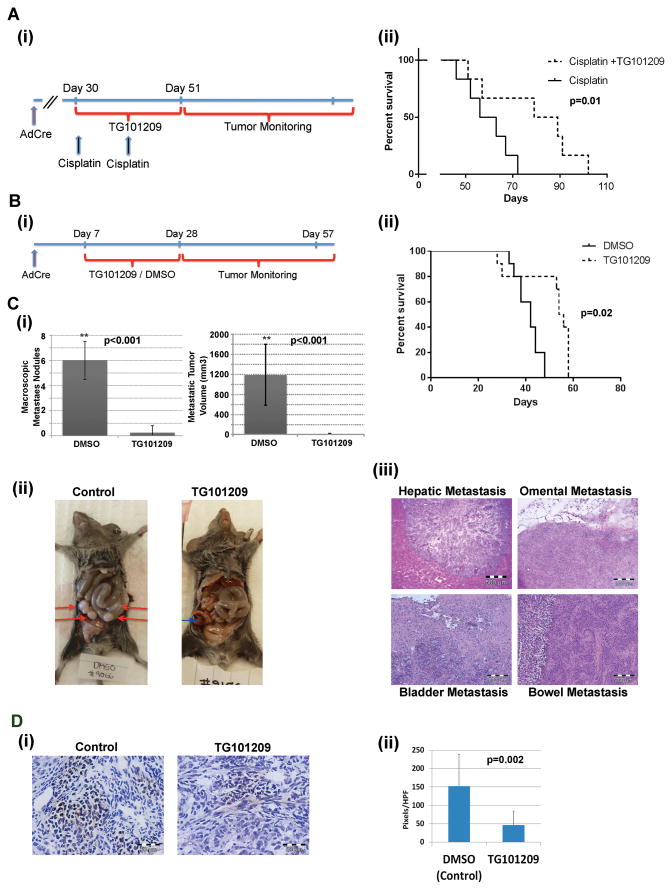
In order to confirm anti-tumor activity of inhibiting STAT3 phosphorylation in vivo we assessed the impact of TG101209 in growth of established Apc−; Pten−; Trp53− ovarian tumors. Tumors were initiated and monitored until they reached ~500 mm3. Tumors were paired based on size and then randomized to cisplatin vs. cisplatin plus TG101209 for 21 days. Combinatory treatment of cisplatin plus TG101209 improved survival of mice compared with mice treated with cisplatin only (Fig. 4A).
Figure 4. TG101209 treatment in Apc−; Pten−; Trp53− mice improves survival and reduces metastasis.
(Ai) Timeline of cisplatin and TG101209 administration and (ii) Survival curves for mice treated with cisplatin (2mg/Kg once per week × 2 weeks) vs cisplatin + TG101209 (50mg/kg daily for 21 days) p=0.04 (n=12 total combined results from two (n=6) independent experiments). (Bi) Timeline of TG101209 treatment and (ii) survival curves of mice treated with control (DMSO, n=9) and TG101209 (n=14) combined results from three independent experiments. (C) Evaluation of metastases in mice demonstrates significant reduction in the number of (i) macroscopic metastatic tumor nodules and metastatic tumor volume. C (ii and iii) images of metastatic nodules and histologic confirmation of metastases; red arrows indicate metastasis while blue arrow indicates primary ovarian tumor. (D) Representative IHC and quantification of p-STAT3 stain in control TG101209 treated tumors. ** indicates p<0.01.
We next treated mice with ‘early stage’ ovarian cancers with TG101209 alone. AdCre was injected into the ovarian bursa to initiate tumors and then treatment with TG101209 or vehicle was initiated one week after injection and maintained for 21 days (Fig. 4Bi). A subset of animals was sacrificed during treatment and IHC confirmed significant reductions in STAT3 phosphorylation in TG101209 treated animals (Fig. 4D). The remaining mice n= 12 were monitored until they became ill or primary tumor volumes reached humane endpoints for euthanasia per institutional guidelines (~1000 mm3). TG101209-treated mice demonstrated significantly increased survival p=0.02 (Fig. 4Bii). In addition, at the time of sacrifice, control mice were ill appearing with evidence of ascites and bowel obstructions. In contrast, TG101209 treated mice were well at the time of euthanasia but were sacrificed because primary tumor volumes met euthanasia guidelines. At necropsy, vehicle treated mice had widespread metastatic disease with metastatic nodules in the intestine, liver, peritoneum, adipose and bladder (Fig. 4Ci–iii). In contrast, only 1 of 14 mice treated with TG101209 had demonstrable metastases. IHC evaluation of tumors suggested that TG101209 treated tumors a slight increase in microvascular density however, no difference of CD45, CD31, or CD14 infiltrates, or SMA staining was noted.
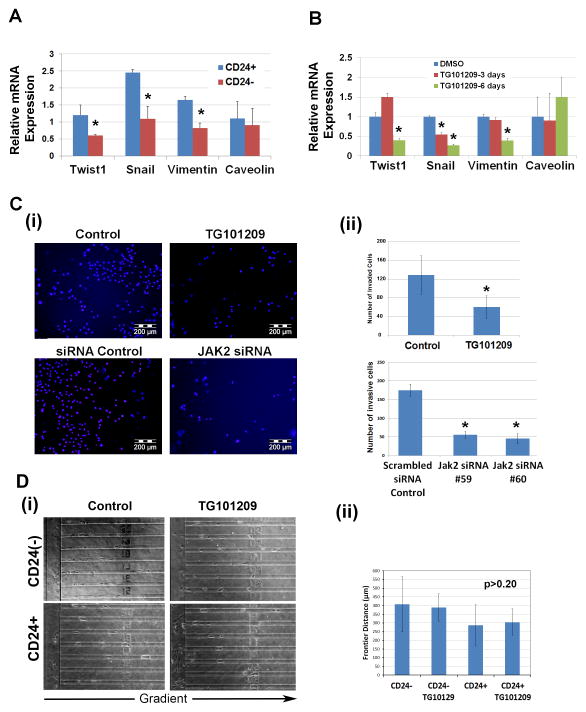
Given the decrease in metastasis observed with TG101209 treatment, and the preferential toxicity of TG101209 on CD24+ CIC, we evaluated epithelial mesenchymal transition (EMT) gene status of CD24+ and CD24− cells. CD24+ cells demonstrated statistically significant increases in expression of the EMT associated genes Twist1, Snail, and Vimentin (Fig. 5A). No significant difference was observed for epithelial genes (Fig. 5), epithelial genes such as E-cadherin and cytokeratin-19 were expressed at relatively low levels. Consistent with CD24+ cell specific depleting effects of TG101209, treatment of W2476T cells with TG101209 resulted in significant reduction in expression of Twist1, Snail, and Vimentin (Fig. 5B).
Figure 5. CD24+ W2476T cells have an EMT phenotype.
(A) qRTPCR demonstrating preferential expression of the EMT associated genes Twist1, Snail, and Vimentin in CD24+ vs. CD24− W2476T cells. (B) qRTPCR demonstrating treatment of W2476T cells with TG101209 reduces expression of Twist1, Snail, and Vimentin. (C) DAPI immunofluorescence and quantification of matrigel invasive W2476T cells in control and TG101209 W2476T treated cells, as well as scrambled siRNA control, and Jak2 siRNA transfected W2476T cells. * p<0.05. (D) (i) Representative photomicrographs and (ii) summation of migration distance for CD24+ vs CD24− W2476T cells with and without TG101209 treatment in a microfluidic cell migration device.
We next evaluated the impact of TG101209 on cancer cell line invasion capacity using boyden chamber Matrigel invasion assays. Treatment of W2476T cells with TG101209 was associated with a 2.2 fold decrease in cellular invasion (Fig. 5C). To confirm that reduction in invasion was secondary to JAK2 inhibition and not likely due to off target effects, we performed JAK2 siRNA knockdown (Fig. 5C). qRT-PCR with JAK2 siRNA treated cells demonstrated a 3.5 fold decrease in JAK2 mRNA (Supplemental Fig. 4). JAK2 siRNA knockdown resulted in a 3.5 fold decrease in cellular invasion (Fig. 5C). JAK2 siRNA had no impact on cellular viability relative to controls, suggesting JAK2 knockdown was impacting invasion and not just killing migratory cells (Supplemental Fig. 4). Finally, we assessed the impact of TG101209 on cellular migration in vitro using a microfluidic cell migration device (Fig. 5D). In this device using a serum gradient, cells migrated along a narrow channel and the migrated distance was evaluated via microscopy. Interestingly, TG101209 had no impact on migration of either whole cell line or isolated CD24+ cells. Taken together, these data support a critical role for JAK2 in ovarian cancer invasion and ultimately metastasis.
Discussion
Genetic models of cancer provide a homogeneous and reproducible system to study cancer cell biology in an immunocompetent host. The cancer initiating capacity of distinct populations of cancer cells have been studied in some murine tumor cell lines (27, 28), and in a K-rasG12D mutation/Pten deletion driven tumor model (29). These studies found cells enriched for cancer initiating capacity could be identified by Hoechst dye exclusion (side population cells) or expression of CD24, CD44, and Epcam. We characterized cancer cells in a Apc−; Pten−; Trp53− murine ovarian cancer model. We found CD24+ cells represent a subset of cells with increased tumor sphere forming and tumor-initiating capacity. CD24+ cells preferentially have increased pSTAT3 and express the stem cell genes Nanog, cyclin-D1, and c-Myc all of which have been previously reported to be regulated by STAT3 (30, 31) (32, 33). Cyclin D1 and c-Myc are also upregulated in primary human ovarian tumor spheroids (34).
Interestingly, while CD24+ cells have increased tumor initiation capacity compared to CD24− cells, CD24− cells were capable of initiating tumors. In vitro and in vivo lineage tracking experiments demonstrated that CD24− cells could generate CD24+ cells. There are several possibilities to explain these findings. It is possible that there is a subset of CD24− cells that acts as a cancer stem-like cell that has a slower proliferation rate than the CD24+ cells and thus initiates tumors more slowly than CD24+ cells. Another possible explanation could stochastic events leading to CD24- cell production of CD24+ cells. Finally, the ability CD24− cells to generate CD24+ cells could be the result of FACS contamination. We have found, and others have similarly reported (35), that FACS sorting has 0.2–2% impurity (Sup Fig 5). Double sort purification improves purity, but unfortunately decreases cellular viability. Ultimately, single cell studies with confirmed purity will be necessary to determine if this tumor model follows a hierarchical/stem cell model.
In this study we found that in vitro blockade of pSTAT3 was associated with reduced cell numbers and reductions in invasive capacity. This is consistent with prior studies in ovarian cancer indicating a critical STAT3 phosphorylation in ovarian cancer cell survival (36). Blockade of STAT3 phosphorylation with a JAK2 inhibitor preferentially induced cytotoxicity in CD24+ CICs. This is analogous to studies in liver, breast and colon cancer where cancer initiating cells demonstrate preferential phosphorylation of STAT3 (30, 37, 38). Interestingly, it has been reported that the JAK-STAT pathway is upregulated in metastatic ovarian cancer cells versus primary tissue (39) and we observed that in vivo inhibition of STAT3 phosphorylation with a JAK2 inhibitor was associated with near complete loss of metastasis. This strongly implicates this pathway, in ovarian cancer metastasis.
Given the preferential phosphorylation of STAT3 in CD24+ cells, it indirectly implicates CD24+ cells in metastasis. Consistent with this observation we found CD24+ cells demonstrated a more ‘mesenchymal’ phenotype, with higher expression of Twist1, Snail, and Vimentin. Our work is consistent with numerous reports linking CD24 as a marker of ovarian CIC with an EMT phenotype; Gao and colleagues reported that CD24+ primary human ovarian cancer cells in addition to having the stem cell characteristics of quiescence, chemoresistance, and tumor initiation (9) also exhibited an EMT phenotype, with high invasive capacity (40). Suggesting a direct role for CD24 in EMT, shRNA depletion of CD24 suppressed cell invasion and reversed the EMT phenotype (41). In addition, CD24 expression is reported to be a negative prognostic marker in ovarian cancer (42, 43). While there is increasing evidence of the role of CD24 in metastasis, the exact mechanism remains uncertain.
CD24 also appears to play an important role in signal-transduction to promote metastasis. CD24 is reported to physically interact with c-src in both breast and ovarian cancer to regulate STAT3 (44–46). In hepatocellular carcinoma, CD24 has been shown to promote self-renewal through Nanog mediated by STAT3 phosphorylation (30). Our studies suggest that the activation of STAT3 plays a critical role in CD24+ cell metastasis. STAT3 phosphorylation can be mediated by either Src or JAK2 (30, 47). We did not see significant phosphorylation of Src in these tumor cells. Thus our studies implicate a critical role for JAK2.
It is important to note that TG101209, in addition to inhibiting JAK2, also inhibits FLT3 and RET kinases (48). While further studies will be necessary to determine if inhibition of either FLT3 or RET is also contributing to the CD24+ cell targeting/metastasis-inhibiting role of TG101209, several lines of evidence suggest a direct role for JAK2/STAT3; similar results were obtained in vitro with both Stattic, a direct pSTAT3 inhibitor and TG101209. Furthermore, siRNA studies knocking down the expression of JAK2, mimicked TG101209 therapy.
Finally, while we did not see a significant change in the numbers of tumor infiltrating white blood cells in our in vivo studies, we cannot rule out an important contribution of TG101209 activity on the tumor microenvironment contributing to inhibition of metastasis.
In conclusion, we show that in a well-established murine model of ovarian cancer the CD24+ cell population is enriched for cancer initiation capacity. These findings should be very useful for future therapeutic studies targeting ovarian CIC. Using this model we find that CD24+ cells have increased susceptibility to JAK2 inhibition and JAK2 inhibition in vivo significantly restricts metastasis. These data indicate JAK2 as an important therapeutic target in epithelial ovarian carcinoma.
Supplementary Material
Acknowledgments
Financial Support: RJ Buckanovich received support for this work from a grant from the National Functional Genomics Center (W81XWH-10-2-0013) and the Ovarian Cancer Research Fund (PPD/UM/01/2011-KRC). D. Burgos-Ojeda was supported by NIHT32-GM007315
We would like to thank the Members of the Flow Cytometry Core and members of the University Laboratory Animal Management for technical assistance with experiments.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to report
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Kipps E, Tan DS, Kaye SB. Meeting the challenge of ascites in ovarian cancer: new avenues for therapy and research. Nat Rev Cancer. 2013;13:273–82. doi: 10.1038/nrc3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrandina G, Martinelli E, Petrillo M, Prisco MG, Zannoni G, Sioletic S, et al. CD133 antigen expression in ovarian cancer. BMC Cancer. 2009;9:221. doi: 10.1186/1471-2407-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba T, Convery PA, Matsumura N, Whitaker RS, Kondoh E, Perry T, et al. Epigenetic regulation of CD133 and tumorigenicity of CD133+ ovarian cancer cells. Oncogene. 2009;28:209–18. doi: 10.1038/onc.2008.374. [DOI] [PubMed] [Google Scholar]
- 5.Silva IA, Bai S, McLean K, Yang K, Griffith K, Thomas D, et al. Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer Res. 2011;71:3991–4001. doi: 10.1158/0008-5472.CAN-10-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kryczek I, Liu S, Roh M, Vatan L, Szeliga W, Wei S, et al. Expression of aldehyde dehydrogenase and CD133 defines ovarian cancer stem cells. Int J Cancer. 2012;130:29–39. doi: 10.1002/ijc.25967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM, et al. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008;68:4311–20. doi: 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alvero AB, Chen R, Fu HH, Montagna M, Schwartz PE, Rutherford T, et al. Molecular phenotyping of human ovarian cancer stem cells unravels the mechanisms for repair and chemoresistance. Cell Cycle. 2009;8:158–66. doi: 10.4161/cc.8.1.7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao MQ, Choi YP, Kang S, Youn JH, Cho NH. CD24+ cells from hierarchically organized ovarian cancer are enriched in cancer stem cells. Oncogene. 2010;29:2672–80. doi: 10.1038/onc.2010.35. [DOI] [PubMed] [Google Scholar]
- 10.Burgos-Ojeda D, Rueda BR, Buckanovich RJ. Ovarian cancer stem cell markers: prognostic and therapeutic implications. Cancer Lett. 2012;322:1–7. doi: 10.1016/j.canlet.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei X, Dombkowski D, Meirelles K, Pieretti-Vanmarcke R, Szotek PP, Chang HL, et al. Mullerian inhibiting substance preferentially inhibits stem/progenitors in human ovarian cancer cell lines compared with chemotherapeutics. Proc Natl Acad Sci U S A. 2010;107:18874–9. doi: 10.1073/pnas.1012667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kostourou V, Lechertier T, Reynolds LE, Lees DM, Baker M, Jones DT, et al. FAK-heterozygous mice display enhanced tumour angiogenesis. Nat Commun. 2013;4:2020. doi: 10.1038/ncomms3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connolly DC, Bao R, Nikitin AY, Stephens KC, Poole TW, Hua X, et al. Female mice chimeric for expression of the simian virus 40 TAg under control of the MISIIR promoter develop epithelial ovarian cancer. Cancer Res. 2003;63:1389–97. [PubMed] [Google Scholar]
- 14.Hu X, Han W, Li L. Targeting the weak point of cancer by induction of necroptosis. Autophagy. 2007;3:490–2. doi: 10.4161/auto.4592. [DOI] [PubMed] [Google Scholar]
- 15.Wu R, Baker SJ, Hu TC, Norman KM, Fearon ER, Cho KR. Type I to type II ovarian carcinoma progression: mutant Trp53 or Pik3ca confers a more aggressive tumor phenotype in a mouse model of ovarian cancer. Am J Pathol. 2013;182:1391–9. doi: 10.1016/j.ajpath.2012.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu R, Hendrix-Lucas N, Kuick R, Zhai Y, Schwartz DR, Akyol A, et al. Mouse model of human ovarian endometrioid adenocarcinoma based on somatic defects in the Wnt/beta-catenin and PI3K/Pten signaling pathways. Cancer Cell. 2007;11:321–33. doi: 10.1016/j.ccr.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–9. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Galban S, Wu R, Bowman BM, Witte A, Vetter K, et al. Molecular imaging reveals a role for AKT in resistance to cisplatin for ovarian endometrioid adenocarcinoma. Clin Cancer Res. 2013;19:158–69. doi: 10.1158/1078-0432.CCR-12-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLean K, Gong Y, Choi Y, Deng N, Yang K, Bai S, et al. Human ovarian carcinoma-associated mesenchymal stem cells regulate cancer stem cells and tumorigenesis via altered BMP production. J Clin Invest. 2011;121:3206–19. doi: 10.1172/JCI45273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–70. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pardanani A, Hood J, Lasho T, Levine RL, Martin MB, Noronha G, et al. TG101209, a small molecule JAK2-selective kinase inhibitor potently inhibits myeloproliferative disorder-associated JAK2V617F and MPLW515L/K mutations. Leukemia. 2007;21:1658–68. doi: 10.1038/sj.leu.2404750. [DOI] [PubMed] [Google Scholar]
- 22.Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chemistry & biology. 2006;13:1235–42. doi: 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 23.Winer I, Wang S, Lee YE, Fan W, Gong Y, Burgos-Ojeda D, et al. F3-targeted cisplatin-hydrogel nanoparticles as an effective therapeutic that targets both murine and human ovarian tumor endothelial cells in vivo. Cancer Res. 2010;70:8674–83. doi: 10.1158/0008-5472.CAN-10-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmed N, Stenvers KL. Getting to know ovarian cancer ascites: opportunities for targeted therapy-based translational research. Front Oncol. 2013;3:256. doi: 10.3389/fonc.2013.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Y, Smyth GK. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. Journal of Immunological Methods. 2009;347:70–8. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 26.Lee TK, Castilho A, Cheung VC, Tang KH, Ma S, Ng IO. CD24(+) liver tumor-initiating cells drive self-renewal and tumor initiation through STAT3-mediated NANOG regulation. Cell Stem Cell. 2011;9:50–63. doi: 10.1016/j.stem.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Szotek PP, Pieretti-Vanmarcke R, Masiakos PT, Dinulescu DM, Connolly D, Foster R, et al. Ovarian cancer side population defines cells with stem cell-like characteristics and Mullerian Inhibiting Substance responsiveness. Proc Natl Acad Sci U S A. 2006;103:11154–9. doi: 10.1073/pnas.0603672103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meirelles K, Benedict LA, Dombkowski D, Pepin D, Preffer FI, Teixeira J, et al. Human ovarian cancer stem/progenitor cells are stimulated by doxorubicin but inhibited by Mullerian inhibiting substance. Proc Natl Acad Sci U S A. 2012;109:2358–63. doi: 10.1073/pnas.1120733109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McAuliffe SM, Morgan SL, Wyant GA, Tran LT, Muto KW, Chen YS, et al. Targeting Notch, a key pathway for ovarian cancer stem cells, sensitizes tumors to platinum therapy. Proc Natl Acad Sci U S A. 2012;109:E2939–48. doi: 10.1073/pnas.1206400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kloos RT, Reynolds JD, Walsh PS, Wilde JI, Tom EY, Pagan M, et al. Does addition of BRAF V600E mutation testing modify sensitivity or specificity of the Afirma Gene Expression Classifier in cytologically indeterminate thyroid nodules? J Clin Endocrinol Metab. 2013;98:E761–8. doi: 10.1210/jc.2012-3762. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki A, Raya A, Kawakami Y, Morita M, Matsui T, Nakashima K, et al. Nanog binds to Smad1 and blocks bone morphogenetic protein-induced differentiation of embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:10294–9. doi: 10.1073/pnas.0506945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dauer DJ, Ferraro B, Song L, Yu B, Mora L, Buettner R, et al. Stat3 regulates genes common to both wound healing and cancer. Oncogene. 2005;24:3397–408. doi: 10.1038/sj.onc.1208469. [DOI] [PubMed] [Google Scholar]
- 33.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan X, Molotkov A, Manabe S, Donmoyer CM, Deltour L, Foglio MH, et al. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol Cell Biol. 2003;23:4637–48. doi: 10.1128/MCB.23.13.4637-4648.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–44. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 36.Burke WM, Jin X, Lin HJ, Huang M, Liu R, Reynolds RK, et al. Inhibition of constitutively active Stat3 suppresses growth of human ovarian and breast cancer cells. Oncogene. 2001;20:7925–34. doi: 10.1038/sj.onc.1204990. [DOI] [PubMed] [Google Scholar]
- 37.Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44(+)CD24(-) stem cell-like breast cancer cells in human tumors. J Clin Invest. 2011;121:2723–35. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin L, Liu A, Peng Z, Lin HJ, Li PK, Li C, et al. STAT3 is necessary for proliferation and survival in colon cancer-initiating cells. Cancer Res. 2011;71:7226–37. doi: 10.1158/0008-5472.CAN-10-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malek JA, Martinez A, Mery E, Ferron G, Huang R, Raynaud C, et al. Gene expression analysis of matched ovarian primary tumors and peritoneal metastasis. J Transl Med. 2012;10:121. doi: 10.1186/1479-5876-10-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bryce NS, Reynolds AB, Koleske AJ, Weaver AM. WAVE2 regulates epithelial morphology and cadherin isoform switching through regulation of Twist and Abl. PLoS One. 2013;8:e64533. doi: 10.1371/journal.pone.0064533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang KS, Choi YP, Gao MQ, Kang S, Kim BG, Lee JH, et al. CD24(+) ovary cancer cells exhibit an invasive mesenchymal phenotype. Biochem Biophys Res Commun. 2013;432:333–8. doi: 10.1016/j.bbrc.2013.01.102. [DOI] [PubMed] [Google Scholar]
- 42.Zhu J, Zhang G, Lu H. CD24, COX-2, and p53 in epithelial ovarian cancer and its clinical significance. Frontiers in bioscience (Elite edition) 2012;4:2745–51. doi: 10.2741/e580. [DOI] [PubMed] [Google Scholar]
- 43.Kristiansen G, Denkert C, Schluns K, Dahl E, Pilarsky C, Hauptmann S. CD24 is expressed in ovarian cancer and is a new independent prognostic marker of patient survival. Am J Pathol. 2002;161:1215–21. doi: 10.1016/S0002-9440(10)64398-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baumann P, Cremers N, Kroese F, Orend G, Chiquet-Ehrismann R, Uede T, et al. CD24 expression causes the acquisition of multiple cellular properties associated with tumor growth and metastasis. Cancer Res. 2005;65:10783–93. doi: 10.1158/0008-5472.CAN-05-0619. [DOI] [PubMed] [Google Scholar]
- 45.Bretz N, Noske A, Keller S, Erbe-Hofmann N, Schlange T, Salnikov AV, et al. CD24 promotes tumor cell invasion by suppressing tissue factor pathway inhibitor-2 (TFPI-2) in a c-Src-dependent fashion. Clin Exp Metastasis. 2012;29:27–38. doi: 10.1007/s10585-011-9426-4. [DOI] [PubMed] [Google Scholar]
- 46.Bretz NP, Salnikov AV, Perne C, Keller S, Wang X, Mierke CT, et al. CD24 controls Src/STAT3 activity in human tumors. Cell Mol Life Sci. 2012;69:3863–79. doi: 10.1007/s00018-012-1055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu B, Shu Y, Liu P. INF-gamma sensitizes head and neck squamous cell carcinoma cells to chemotherapy-induced apoptosis and necroptosis through up-regulation of Egr-1. Histol Histopathol. 2014;29:1437–43. doi: 10.14670/HH-29.1437. [DOI] [PubMed] [Google Scholar]
- 48.Mendoza-Naranjo A, El-Naggar A, Wai DH, Mistry P, Lazic N, Ayala FR, et al. ERBB4 confers metastatic capacity in Ewing sarcoma. EMBO Mol Med. 2013;5:1019–34. doi: 10.1002/emmm.201202343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.