Abstract
Background
Noninvasive prenatal screening (NIPS) by next-generation sequencing of cell free DNA (cfDNA) in maternal plasma is used to screen for common aneuploidies such as trisomy 21, in high risk pregnancies. NIPS can identify fetal genomic microdeletions, however sensitivity and specificity have not been systematically evaluated. Commercial companies have begun to offer expanded panels including screening for common microdeletion syndromes such as 22q11.2 deletion (DiGeorge syndrome) without reporting the genomic coordinates or whether the deletion is maternal or fetal. Here we describe a phenotypically normal mother and fetus that tested positive for atypical 22q deletion via maternal plasma cell free DNA testing.
Methods
We performed cfDNA sequencing on saved maternal plasma obtained at 11 weeks of gestation from a phenotypically normal woman with a singleton pregnancy whose earlier screening at a commercial laboratory was reported to be positive for a 22q11.2 microdeletion. FISH and chromosomal microarray diagnostic genetic tests were done postnatally.
Conclusion
NIPS detected a 22q microdeletion that upon diagnostic work up, did not include the DiGeorge critical region. Diagnostic prenatal or postnatal testing with chromosomal microarray and appropriate parental studies to determine precise genomic coordinates and inheritance should follow a positive microdeletion NIPS result.
Introduction
Noninvasive prenatal screening (NIPS) has become available over the last few years and become increasingly requested in lieu of diagnostic procedures. Fetal aneuploidies such as trisomy 21 as well as chromosomal microdeletions can be identified by analysis of fetal cell-free DNA (cfDNA) in the maternal plasma1–3. For many pregnant women the availability of NIPS offers more accurate option over the conventional prenatal methods for screening of fetal chromosomal abnormalities and removes the fear of miscarriage associated with invasive diagnostic testing. NIPS is currently offered exclusively through commercial entities. Some commercial companies have begun to offer microdeletion testing for common conditions such as DiGeorge syndrome. DiGeorge or 22q11.2 deletion syndrome (MIM 188400) affects 1 in 4,000 individuals and is associated with multiple anomalies including congenital heart disease, palatal defects, and immune deficiency. About 10% of 22q11.2 deletions are inherited from a mildly affected parent. DiGeorge syndrome in infants commonly requires an acute management of hypocalcemia, surgery for conotruncal cardiac defects, and treatment of immunodeficiency. Therefore, prenatal detection plays an important role in pregnancy management and postnatal interventions. Here we present the clinical management and diagnostic testing initiated by positive NIPS screen for 22q deletion.
Methods
Patient was enrolled into the NIPS research study in our institution as part of quality control, under an IRB approved protocol. Maternal blood was obtained at 11 weeks of gestation and banked, independent of the blood sent to commercial lab as part of clinical care. cfDNA was extracted from maternal plasma and the whole genome sequencing of cfDNA was performed as previously described4 postnatally (Figure 1). Peripheral blood sample was obtained from the newborn child of the patient for confirmatory fluorescence in situ hybridization (FISH) and microarray analyses. FISH was completed using a commercial TUPLE1 (HIRA) assay (Vysis). A 180K CGH+SNP (SurePrint G3 ISCA design, Agilent) platform was used for microarray analysis according to the manufacturer’s protocol.
Figure 1.
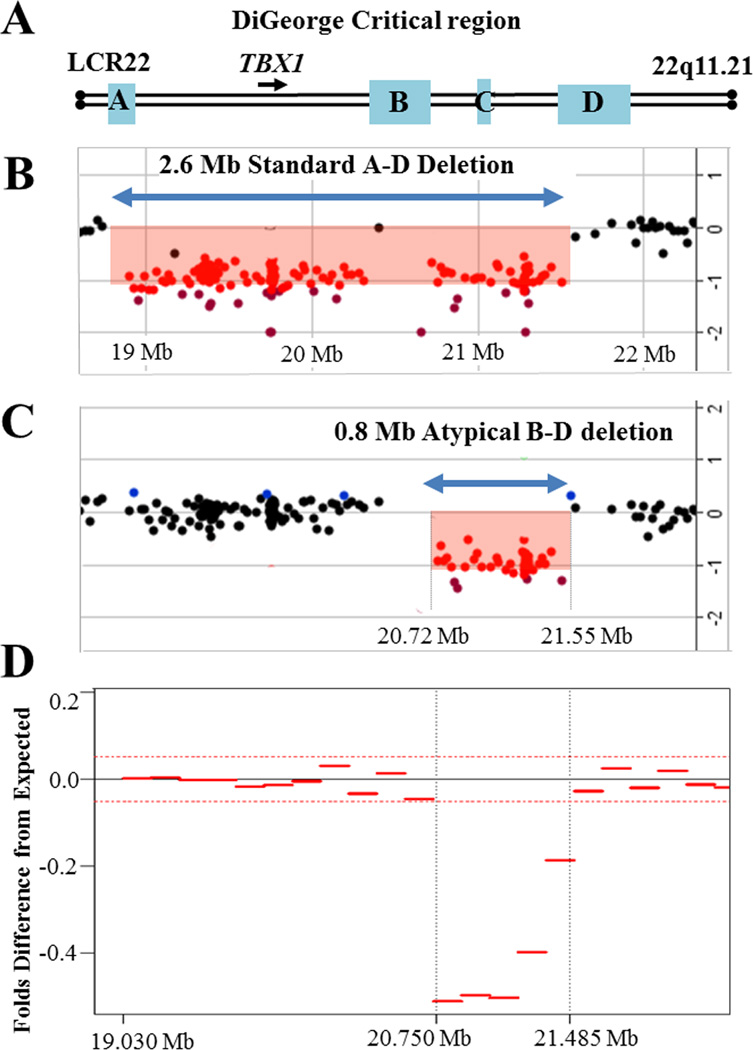
A. Schematic presentation of the DiGeorge critical region on the proximal long arm of chromosome 22. The location of the LCR22-A, LCR22-B, LCR22-C and LCR22-D repeats are shown as blue boxes. Arrowhead represents the location and orientation of the TBX1 gene transcription. B. The common ~2.6 – 3 Mb deletion (~85% of patients with DiGeorge/ Velocardiofacial syndrome) occurs between the LCR22-A and LCR22-D repeats (A–D deletion) and includes TBX1. An array CGH plot from an individual with a standard A–D deletion is shown. Each dot represents an oligonucleotide probe arranged according to their physical map locations from the proximal (left) to the distal (right) long arm of the chromosome 22 (X-axis), placed on a log2 scale (Y-axis). At the bottom, genomic coordinates (in megabases, Mb) are given according to the Human Genome Browser map (GRCh37/hg19). Black dots indicate DNA probes with normal copy number (log2 is between +0.3 and −0.5). Red dots designate chromosomal segment with a negative log2 ratio (below −0.5), depicting the deletion region. C. Array-CGH plot from the newborn baby showing an atypical deletion of the DiGeorge region with the breakpoints between LCR22-B and LCR22-D repeats (blue double arrowhead). The B–D deletion does not involve TBX1, and therefore cannot be detected by the commercially available FISH probes. D. Maternal plasma at 11 weeks of gestation was used to isolate and whole genome sequence cell free DNA1. Relative difference between the observed tag count in part of chromosome 22 in the patient’s sample and the expected tag count for a normal sample was performed as previously described4. The whole genome sequencing of cell free DNA shows a deletion between 20.75 and 21.485 Mb, which corresponds to the array CGH findings.
Results
A 40-year-old gravida 6, para 2 underwent noninvasive cfDNA testing in her private physician’s office for advanced maternal age. In addition to the negative aneuploidy result, the test was reported as positive for a fetal DiGeorge syndrome microdeletion. However, the report did not indicate genomic coordinates. The patient was counseled that the baby may be affected with DiGeorge syndrome and referred to tertiary care center at 18 weeks of gestation for fetal echocardiography and ultrasound screen. Prenatal ultrasound demonstrated no apparent anomalies. The location and volume of fetal thymus and fetal echocardiogram were normal. Fetal growth restriction was detected in the third trimester and likely related to chronic hypertension. The patient declined diagnostic genetic testing during pregnancy. Due to uncertainty of whether NIPS result was a true or false positive, and screening nature of the prenatal ultrasound, recommendation was made to deliver at a tertiary care center for detailed postnatal examination. She developed severe preeclampsia and delivered by repeat cesarean section at 32 weeks, 5 days of a 1053 grams male with Apgars of 8 and 9. The baby was discharged to home at 5 weeks of life. Physical examination did not reveal craniofacial abnormalities and postnatal echocardiogram did not identify heart defects. Because of the abnormal NIPS result, the DiGeorge critical region was evaluated by FISH for the commonly deleted region (chr22:19,318,224–19,419,219). No deletion was detected in this region. A chromosomal microarray on the newborn showed a 790 kb microdeletion on chr22:20,719,112–21,505,417(hg19) (Figure 1A–C). This deletion involves the distal part of the classical DiGeorge region (B–D atypical), which does not include the TBX1 gene, thought to be responsible for the DiGeorge syndrome characteristic craniofacial features and heart malformations5. Microarray analysis on parental samples showed that the deletion was inherited from the mother, whose physical and echocardiographic examination was unremarkable. The repeated whole-genome sequencing in our academic laboratory of maternal plasma DNA, obtained at 11 weeks gestation, discovered that coordinates of the deleted region match closely those found by microarray (Figure 1D).
Discussion
Non-invasive prenatal screening is currently offered as an alternative to serum screening programs based on protein and hormone markers for high-risk pregnancies due to its superiority in detecting trisomy 212,6–8. Moreover, societies such as American College of Obstetrics and Gynecology support its use in high risk patients9. Despite some advantages, NIPS is a screening test. The recent addition of microdeletion detection involving 4p, 5p, 15q12, 22q11 and other regions, marks an attempt by commercial companies to show the feasibility of identifying pathogenic microdeletions, as well as differentiate from each other. Although NIPS can be used to uncover fetal microdeletions, positive results have been described in few cases1–3. The accurate detection of microdeletions requires higher number of sequencing reads, and its sensitivity and specificity are still unknown. An argument can be made that routine NIPS microdeletion screening at this time is premature and should only be offered under a research protocol, until we understand its true sensitivity, specificity, technical and biological variables that impact the detection accuracy, and relevance to phenotypic penetrance. Nonetheless, we have seen that general clinicians have begun to offer deletion screening in addition to aneuploidy testing. Fetal fraction constitutes 10% or less of the maternal plasma3, which makes it difficult to differentiate whether the deletion is fetal or maternal, or both. Positive deletion testing on NIPS should be followed by diagnostic testing to confirm the fetal origin of the deletion as well as parental studies to establish inheritance and to provide accurate counseling for conditions with variable expressivity and incomplete penetrance. Moreover, in the absence of precise genomic coordinates, it is difficult to counsel phenotypic consequences of a particular genotype, as deleted gene content is not reported. In our case, the family lived in rural area, approximately 3 hours away from the tertiary care center. Due to NIPS findings, our patient underwent multiple fetal ultrasound visits before her care was transferred to the tertiary care center for closer monitoring, but at increased cost to the patient due to transportation, time spent and delivery away from home. In our case, both the patient and her child were found to carry atypical small 22q11.2 deletion that did not include the DiGeorge critical region. Deletions of this region have been associated with a variable phenotype and incomplete penetrance. The exact genomic coordinates of the abnormality would be extremely valuable for optimized prenatal management, accurate genetic counseling of our patient, and appropriate diagnostic postnatal testing. With an increased use of NIPS in prenatal diagnosis, and current shift to also reporting microdeletions, it is of great importance to have precise genomic coordinates in order to determine affected gene content, and to appropriately counsel the patient. When NIPS report detects a fetal deletion, it is essential to counsel the mother that she is at risk of having the deletion and to perform diagnostic evaluation on the trio of fetus, mother and father for the origin of the genomic alteration. NIPS, whether for aneuploidy or copy number variant identification is a screening test and always warrants further counseling with an offer of additional diagnostic studies.
References
- 1.Peters D, Chu T, Yatsenko SA, et al. Noninvasive prenatal diagnosis of a fetal microdeletion syndrome. N Engl J Med. 2011;365(19):1847–1848. doi: 10.1056/NEJMc1106975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srinivasan A, Bianchi DW, Huang H, Sehnert AJ, Rava RP. Noninvasive detection of fetal subchromosome abnormalities via deep sequencing of maternal plasma. Am J Hum Genet. 2013;92(2):167–176. doi: 10.1016/j.ajhg.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lau TK, Jiang FM, Stevenson RJ, et al. Secondary findings from non-invasive prenatal testing for common fetal aneuploidies by whole genome sequencing as a clinical service. Prenat Diagn. 2013;33(6):602–608. doi: 10.1002/pd.4076. [DOI] [PubMed] [Google Scholar]
- 4.Chu T, Yeniterzi S, Rajkovic A, et al. High resolution non-invasive detection of a fetal microdeletion using the GCREM algorithm. Prenat Diagn. 2014;34(5):469–477. doi: 10.1002/pd.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Minaur S, Fantes J, Murray RS, et al. A novel atypical 22q11.2 distal deletion in father and son. J Med Genet. 2002;39(10):E62. doi: 10.1136/jmg.39.10.e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCullough RM, Almasri EA, Guan X, et al. Non-invasive prenatal chromosomal aneuploidy testing - clinical experience: 100,000 clinical samples. PloS one. 2014;9(10):e109173. doi: 10.1371/journal.pone.0109173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palomaki GE, Kloza EM, Lambert-Messerlian GM, et al. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genet Med. 2011;13(11):913–920. doi: 10.1097/GIM.0b013e3182368a0e. [DOI] [PubMed] [Google Scholar]
- 8.Norton ME, Brar H, Weiss J, et al. Non-Invasive Chromosomal Evaluation (NICE) Study: results of a multicenter prospective cohort study for detection of fetal trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;207(2):137, e131–e138. doi: 10.1016/j.ajog.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 9.American College of O, Gynecologists Committee on G. Committee Opinion No. 545: Noninvasive prenatal testing for fetal aneuploidy. Obstet Gynecol. 2012;120(6):1532–1534. doi: 10.1097/01.AOG.0000423819.85283.f4. [DOI] [PubMed] [Google Scholar]


