Abstract
Improved understanding of natural history of hepatitis C virus (HCV) RNA levels in chronic infection provides enhanced insights into immunopathogenesis of HCV and has implications for the clinical management of chronic HCV infection. This study assessed factors associated with HCV RNA levels during early chronic infection in a population with well-defined early chronic HCV infection. Data were from an international collaboration of nine prospective cohorts studying acute HCV infection (InC3 Study). Individuals with persistent HCV and detectable HCV RNA during early chronic infection (one year [±4 months] post-infection) were included. Distribution of HCV RNA levels during early chronic infection were compared by selected host and virological factors. A total of 308 individuals were included. Median HCV RNA levels was significantly higher among males (vs. females; 5.15 vs. 4.74 log IU/mL; P<0.01) and among individuals with HIV co-infection (vs. no HIV; 5.89 vs. 4.86; P=0.02). In adjusted logistic regression, male sex (vs. female, adjusted odds ratio [AOR]: 1.93; 95%CI: 1.01, 3.69), interferon lambda 4 (IFNL4) rs12979860 CC genotype (vs. TT/CT; AOR: 2.48; 95%CI: 1.42, 4.35), HIV co-infection (vs. no HIV; AOR: 3.27; 95%CI: 1.35, 7.93), and HCV genotype G2 (vs. G3; AOR: 5.40; 95%CI: 1.63, 17.84) were independently associated with high HCV RNA levels (>5.6 log IU/mL=400,000 IU/mL). In conclusion, this study demonstrated that IFNL4 rs12979860 CC genotype, male sex, HIV co-infection, and HCV genotype G2 are associated with high HCV RNA levels in early chronic infection. These factors exert their role as early as one year following infection.
Keywords: Viral load; IFNL4 genotype; IL28B genotype; sex, HCV genotype; HIV; Cohort study
Introduction
Hepatitis C virus (HCV) infection is an escalating global health problem with increasing morbidity (e.g. cirrhosis and hepatocellular carcinoma) and mortality worldwide (reviewed in (1, 2)). Among people with chronic HCV infection, a pre-treatment high HCV RNA level is predictive of lower sustained virological responses (SVR) following treatment with pegylated interferon and ribavirin, either alone (3-5), or in combination with boceprevir (6, 7), or telaprevir (8). While HCV RNA levels have limited impact on treatment responsiveness of new interferon-free direct-acting antiviral (DAA) therapies (9, 10) and potentially diminishing clinical relevance, they remain informative for HCV immunopathogenesis.
In the setting of chronic HCV infection, previous studies have demonstrated that age, gender, ethnicity, HIV co-infection, HCV genotype, and interferon lambda 4 (IFNL4) rs12979860 genotype (formerly known as interferon lambda 3 [IFNL3] and interleukin-28B [IL28B]) are associated with HCV RNA levels (11-22). However, the majority of studies assessing factors associated with HCV RNA during chronic infection have been cross-sectional. As such, the estimated duration of HCV infection is often unknown or imprecise, making it difficult to measure the longitudinal effect of various factors on HCV RNA levels in chronic infection.
The International Collaboration of Incident HIV and Hepatitis C in Injecting Cohorts (InC3) Study, is a collaboration of pooled data from nine prospective international cohorts prominently comprised of people who inject drugs (PWID) (23), including a large number of well-defined participants with acute and early chronic HCV infection and longitudinal follow-up. This current study assessed factors associated with HCV RNA levels in early chronic infection (one year post-infection).
Materials & Methods
Study population and design
The InC3 Study, a collaboration of nine prospective cohorts evaluating HIV and HCV infection outcomes, has been previously described (23). All cohorts follow participants at regular intervals using standardized methods. Participants were recruited and followed between 1985 and 2010. The InC3 Study includes both: 1) participants without HCV infection; and 2) participants with documented acute or early chronic HCV infection. All participants provided written informed consent. The study was performed according to the World Medical Association Declaration of Helsinki and the cohort protocols were approved by local ethics committees (23).
For the current study, only individuals with documented acute or early chronic HCV were included. Documented acute or early chronic HCV is defined as either: 1) HCV seroconversion with an HCV antibody (anti-HCV) or HCV RNA positive test within two years of the anti-HCV negative test; or 2) evidence of symptomatic HCV infection. Symptomatic HCV infection was defined as a) a positive anti-HCV/HCV RNA test; b) jaundice or Alanine aminotransferase [ALT] elevation >400 U/L; and c) detection of HCV RNA or history of high-risk exposure within three months of clinical manifestation of acute HCV.
HCV RNA levels in early chronic infection were assessed among InC3 participants with persistent HCV infection and detectable HCV RNA levels at one year post-infection. A window of ±4 months around the one year time-point was permitted for the inclusion of HCV RNA levels in this analysis (between 8 and 16 months post-infection). The time period of 12±4 months post-infection was chosen for defining early chronic infection given our initial analysis showed that fluctuations in HCV RNA levels which are the characteristic of acute infection mainly observed during the first eight to ten months following infection and a stable pattern of HCV RNA levels with minor fluctuations was observed after that (24, 25).
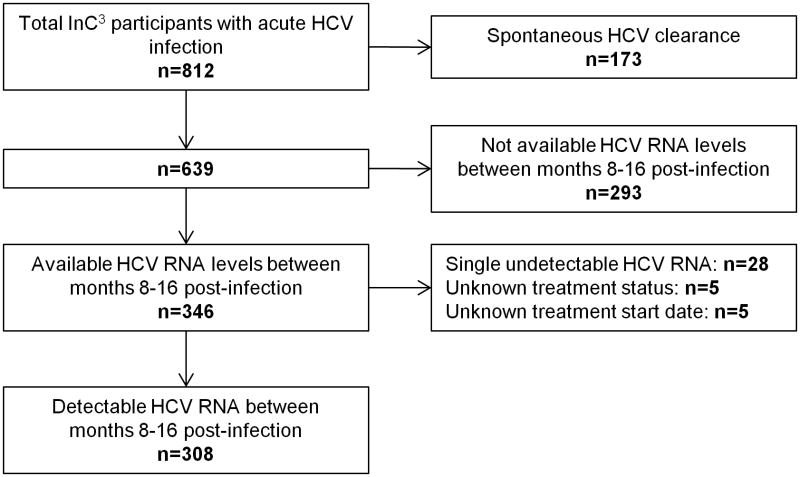
From the total population with acute or early chronic HCV infection included in the InC3 study (n=812), individuals with spontaneous HCV clearance (n=173) or unavailable HCV RNA levels between months 8-16 post-infection (n=293) were excluded. Individuals with single undetectable HCV RNA between months 8-16 post-infection (n=28) were also excluded given that enough data was not available to exclude the possibility of spontaneous clearance followed by HCV re-infection. The inclusion of these individuals could have led to misclassification bias. Among individuals receiving HCV antiviral treatment, only HCV RNA data before treatment commencement were included in the analysis. Accordingly, individuals with unknown treatment status (n=5) or unknown treatment commencement date (n=5) were also excluded. As such, factors associated with HCV RNA levels were assessed among 308 people with detectable HCV RNA between months 8-16 post-infection (Figure 1).
Figure 1. Overview of InC3 study population and participants with persistent HCV infection and a detectable HCV RNA in early chronic infection (i.e. between months 8-16 post-infection).
The estimated date of HCV infection was calculated based on a hierarchy using all serological (anti-HCV), virological (HCV RNA) and clinical (symptoms and liver function tests) data to arrive at the most precise estimate of infection date:
Among individuals with HCV RNA positive and anti-HCV negative at acute HCV detection, date of infection was four weeks prior to HCV RNA detection (26, 27).
Among individuals with symptomatic acute HCV, date of infection was six weeks prior to its onset (jaundice or ALT >400 IU/L) (28).
Among individuals with a negative anti-HCV test followed by either a positive anti-HCV or HCV RNA test, seroconversion was assumed to occur at the mid-point between the last negative and the first positive test. HCV seroconversion generally occurs about 30-60 days following infection (26, 27, 29). Date of infection in this group was six weeks prior to estimated seroconversion date if the first positive test was anti-HCV test and four weeks prior to estimated seroconversion date if the first positive test was only HCV RNA test.
Laboratory testing
Choice of qualitative and quantitative HCV RNA testing varied by cohort but was consistent at each site. Qualitative HCV RNA testing was performed using the following assays: Versant TMA [Bayer, Australia;<10 IU/ml], COBAS AmpliPrep/COBAS TaqMan (Roche, Branchburg, NJ, USA;<15 IU/ml), COBAS AMPLICOR HCV Test v2.0 (Roche Diagnostics, Mannheim, Germany; <50 IU/ml) or discriminatory HCV transcription-mediated amplification component of the Procleix HIV-1/HCV (Gen-Probe, San Diego, CA, USA; <12 copies/mL). Quantitative HCV RNA testing was performed using the Versant HCV RNA 3.0 (Bayer, Australia;<615 IU/ml), COBAS AMPLICOR HCV MONITOR 2.0 (Roche Diagnostics, Mannheim, Germany; <600 IU/ml), COBAS AmpliPrep/COBAS TaqMan (Roche, Branchburg, NJ, USA;<15 IU/ml) or an in-house PCR (<1000 IU/ml) (30, 31). HCV genotype was determined by line-probe assay (Versant LiPa1/LiPa2, Bayer, Australia) or HCV sequencing at acute HCV detection. Among those with undetectable HCV RNA (no genotype) and available samples, Murex HCV serotyping was performed to determine HCV genotype (Murex Biotech Limited, Dartford, UK). IFNL4 genotyping was determined by sequencing of the rs12979860 single nucleotide polymorphism, as previously described in (32-35). While rs12979860 has also been referred to as IFNL3 and IL28B, a recent study demonstrated that is located within intron 1 of the IFNL4 gene (36).
Study outcomes and statistical analyses
The study outcome was HCV RNA levels at one year post-infection (early chronic infection, window 8-16 months). For participants with more than one HCV RNA level during this period (n=220), the median value was used. Nonparametric statistical tests were used for analyses, given that HCV RNA levels (IU/mL) and log10 transformation of HCV RNA levels (log IU/mL) were not normally distributed. The top tertile HCV RNA levels (≥5.6 log IU/mL equals to ≥400,000 IU/mL) was defined as a high HCV RNA level. This threshold has also a clinical significance given HCV RNA levels≥400,000 is associated with lower response to interferon-based treatment for chronic HCV infection (3).
Factors hypothesized to be associated with HCV RNA levels were determined a priori based on factors shown to be associated with HCV RNA levels during chronic HCV infection and included age (11, 17, 18), sex (11, 15, 16, 22), ethnicity (11), IFNL4 rs12979860 genotype (CC vs. CT/TT) (11-13, 22), HIV co-infection (11, 17, 18), and HCV genotype (11, 14-17, 19, 21).
Median HCV RNA levels were compared between groups using the Wilcoxon-Mann-Whitney (or Kruskal Wallis) test. Logistic regression models were also used to assess factors associated with high HCV RNA levels (≥5.6 log IU/mL). The adjusted model included variables with P<0.20 in unadjusted analyses. To account for potential unmeasured confounders introduced by cohort sites, multivariate regression analysis was performed using mixed modelling, with a random intercept for cohort site. Statistically significant differences were assessed at P<0.05 (P-values are two-sided). All analyses were performed using Stata v12.0 (College Station, TX, United States).
Results
Participant characteristics
HCV RNA levels were assessed among 308 people with detectable HCV RNA in early chronic infection (Figure 1). Background characteristics of study population were summarized in Table 1. The median age was 26 years, 32% were female, 79% were Caucasian, 9% were HIV co-infected, and 95% had a history of injecting drug use. Among those with data on infecting HCV genotype (n=294; 96%), 54% had genotype 1. Among those with data on IFNL4 rs12979860 genotype (n=285; 93%), 41% were CC genotype. Background characteristics of InC3 participants not included in the current analysis were summarized in Supplementary Table 1.
Table 1. Background characteristics of participants with detectable HCV RNA between months 8-16 post-infection in the InC3 Study.
| Number (%) Total n=308 |
|
|---|---|
| Site | |
| ACS (the Netherlands) | 23 (7) |
| ATAHC (Australia) | 87 (28) |
| BAHSTION (United States) | 20 (6) |
| BBAASH (United States) | 54 (17) |
| HEPCO (Canada) | 2 (1) |
| HITS-c (Australia) | 6 (2) |
| HITS-p (Australia) | 53 (17) |
| N2 (Australia) | 0 (0) |
| UFO (United States) | 63 (20) |
|
| |
| Median age at the time of HCV infection, yrs (IQR) | 26 (23, 33) |
|
| |
| Sex | |
| Female | 99 (32) |
| Male | 208 (67) |
| Unknown | 1 (<1) |
|
| |
| Ethnicity | |
| Caucasian | 243 (79) |
| Black | 15 (5) |
| Indigenous | 18 (6) |
| Other | 23 (7) |
| Unknown | 9 (3) |
|
| |
| History of injecting drug use | 292 (95) |
|
| |
| Symptomatic HCV infection | |
| No | 80 (26) |
| Yes | 36 (12) |
| Unknown | 192 (62) |
|
| |
| IFNL4 genotype (rs12979860) | |
| TT | 42 (14) |
| CT | 126 (41) |
| CC | 117 (38) |
| Unknown | 23 (7) |
|
| |
| HIV infection at the time of HCV infection | |
| No | 264 (86) |
| Yes | 28 (9) |
| Unknown | 16 (5) |
|
| |
| HCV genotype | |
| Genotype 1 | 159 (52) |
| Genotype 2 | 19 (6) |
| Genotype 3 | 99 (32) |
| Genotype 4 | 6 (2) |
| Genotype 6 | 3 (1) |
| Mixed genotype | 8 (3) |
| Unknown | 14 (4) |
Abbreviations: ACS: Amsterdam Cohort Studies; ATAHC: Australian Trial in Acute Hepatitis C; BAHSTION: Boston Acute HCV Study: Transmission, Immunity and Outcomes Network; BBAASH: Baltimore Before and After Acute Study of Hepatitis; HEPCO: St. Luc Cohort, HEPCO; HITS-c: Hepatitis C Incidence and Transmission Study-Community; HITS-p: Hepatitis C Incidence and Transmission Study-Prison; N2: Networks 2; UFO: UFO STUDY; IQR: Inter-quartile range; IFNL4: interferon lambda 4
HCV RNA levels in early chronic infection
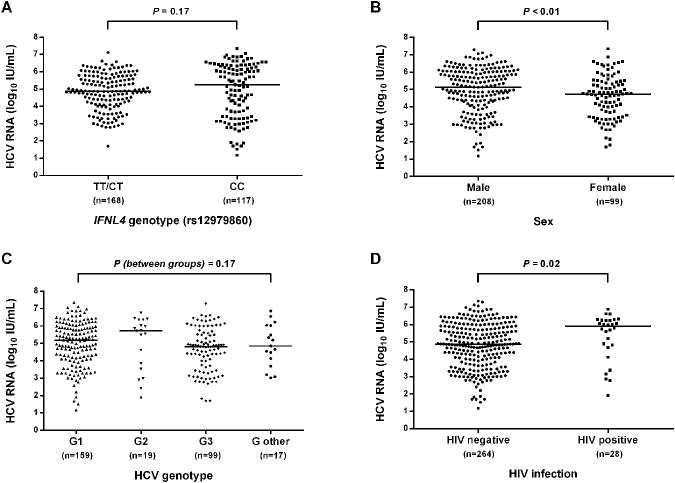
Median HCV RNA levels during early chronic infection was 4.90 log IU/mL (Inter-quartile range [IQR]: 3.99, 5.89) with the top tertile being from 5.59 to 7.35 log IU/mL. Significantly higher median HCV RNA levels were observed among males compared to females (5.15 log IU/mL vs. 4.74 log IU/mL, respectively; P<0.01) and among individuals with HIV co-infection compared to those with no HIV (5.89 log IU/mL vs. 4.86 log IU/mL, respectively; P=0.02). There was no significant difference in median HCV RNA levels by age, ethnicity, IFNL4 rs12979860 genotype and HCV genotype (Table 2 and Figure 2).
Table 2. Median HCV RNA levels in early chronic infection by selected demographic and virologic variables in participants with HCV infection in the InC3 study.
| Number Total n=308 |
Median HCV RNA levels a (IQR) | P | |
|---|---|---|---|
| Age | 0.52 | ||
| <30 years | 193 | 4.92 (3.90, 5.90) | |
| 30-39 years | 54 | 4.74 (3.56, 5.70) | |
| ≥40 years | 31 | 5.19 (4.11, 6.09) | |
|
| |||
| Sex | <0.01 | ||
| Female | 99 | 4.74 (3.52, 5.46) | |
| Male | 208 | 5.15 (4.15, 6.03) | |
|
| |||
| Ethnicity | 0.74 | ||
| Caucasian | 243 | 4.92 (3.90, 5.91) | |
| Black | 15 | 4.83 (4.26, 5.55) | |
| Indigenous | 18 | 4.34 (3.14, 6.12) | |
| Other | 23 | 4.77 (4.35, 5.91) | |
|
| |||
| IFNL4 genotype (rs12979860) | 0.17 | ||
| TT/CT | 168 | 4.88 (4.17, 5.67) | |
| CC | 117 | 5.26 (3.43, 6.28) | |
|
| |||
| HIV status | 0.02 | ||
| Negative | 264 | 4.86 (3.84, 5.82) | |
| Positive | 28 | 5.89 (4.74, 6.24) | |
|
| |||
| HCV genotype | 0.17 | ||
| Genotype 1 | 159 | 5.17 (4.11, 5.91) | |
| Genotype 2 | 19 | 5.71 (3.52, 6.36) | |
| Genotype 3 | 99 | 4.81 (3.63, 5.59) | |
| Other b | 17 | 4.85 (4.12, 6.03) | |
Abbreviations: IQR: Inter-quartile range; IFNL4: interferon lambda 4
log IU/mL
Included genotype 4 (n=6), genotype 6 (n=3), and mixed genotype (n=8).
Figure 2.
Distribution of HCV RNA levels during months 8-16 following infection, stratified by IFNL4 rs12979860 genotype, sex, HCV genotype, and HIV infection status in participants with acute HCV infection in the InC3 study. (A) Stratified by IFNL4 rs12979860 genotype; (B) Stratified by sex; (C) Stratified by HCV genotype; (D) Stratified by HIV infection status. Horizontal lines represent the medians HCV RNA levels in each subgroup.
In unadjusted logistic regression analysis (Table 3), factors associated with high HCV RNA levels (≥5.6 log IU/mL) in early chronic infection were sex, IFNL4 rs12979860 genotype, HIV co-infection, and HCV genotype. In adjusted analysis, male sex (vs. female; AOR: 1.93; 95%CI: 1.01, 3.69; P=0.04), IFNL4 rs12979860 CC genotype (vs. TT/CT; AOR: 2.48; 95%CI: 1.42, 4.35; P<0.01), HIV co-infection (vs. no HIV; AOR: 3.27; 95%CI: 1.35, 7.93; P<0.01), and HCV genotype 2 (vs. genotype 3; AOR: 5.40; 95%CI: 1.63, 17.84; P<0.01) were independently associated with high HCV RNA levels (≥5.6 log IU/mL). HCV genotype 1 (vs. genotype 3; AOR: 1.87; 95%CI: 0.99, 3.55; P=0.05) and HCV genotype 2 (vs. genotype 1; AOR: 2.88; 95%CI: 0.92, 8.97; P=0.07) trended towards also being associated with high HCV RNA levels. Interactions between covariates were not statistically significant on the multiplicative scale.
Table 3. Unadjusted and adjusted model assessing factors associated with high HCV RNA levels (≥5.6 log IU/mL) in early chronic infection in the InC3 study.
| HCV RNA levels ≥5.6 log IU/mL n (%) a | HCV RNA levels <5.6 log IU/mL n (%)a | Unadjusted model | Adjusted model b | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| OR (95% CI) | P | P overall | AOR (95% CI) | P | |||
| Age | 0.33 | ||||||
| <30 years | 63 (33) | 130 (67) | 1.00 | ||||
| 30-39 years | 16 (30) | 38 (70) | 0.87 (0.45, 1.68) | 0.67 | |||
| ≥40 years | 14 (45) | 17 (55) | 1.70 (0.79, 3.66) | 0.18 | |||
|
| |||||||
| Sex | |||||||
| Female | 20 (20) | 79 (80) | 1.00 | 1.00 | |||
| Male | 80 (38) | 128 (62) | 2.47 (1.40, 4.34) | <0.01 | 1.93 (1.01, 3.69) | 0.04 | |
|
| |||||||
| Ethnicity | 0.60 | ||||||
| Caucasian | 81 (33) | 162 (67) | 1.00 | ||||
| Black | 3 (20) | 12 (80) | 0.50 (0.14, 1.82) | 0.29 | |||
| Indigenous | 5 (28) | 13 (72) | 0.77 (0.26, 2.23) | 0.63 | |||
| Other | 9 (39) | 14 (61) | 1.28 (0.53, 3.10) | 0.57 | |||
|
| |||||||
| IFNL4 genotype (rs12979860) | |||||||
| TT/CT | 45 (27) | 123 (73) | 1.00 | 1.00 | |||
| CC | 52 (44) | 65 (56) | 2.19 (1.33, 3.60) | <0.01 | 2.48 (1.42, 4.35) | <0.01 | |
|
| |||||||
| HIV status | |||||||
| Negative | 80 (30) | 184 (70) | 1.00 | 1.00 | |||
| Positive | 17 (61) | 11 (39) | 3.55 (1.59, 7.93) | <0.01 | 3.27 (1.35, 7.93) | <0.01 | |
|
| |||||||
| HCV genotype c | 0.07 | ||||||
| Genotype 3 | 24 (24) | 75 (76) | 1.00 | 1.00 | |||
| Genotype 1 | 56 (35) | 103 (65) | 1.70 (0.98, 2.98) | 0.06 | 1.87 (0.99, 3.55) | 0.05 | |
| Genotype 2 | 10 (53) | 9 (47) | 3.47 (1.26, 9.54) | 0.02 | 5.40 (1.63, 17.84) | <0.01 | |
| Other | 5 (29) | 12 (71) | 1.30 (0.42, 4.07) | 0.65 | 1.40 (0.42, 4.67) | 0.58 | |
Abbreviations: OR: Odds Ratio; CI: Confidence Interval; AOR: Adjusted Odds Ratio; IFNL4: interferon lambda 4
The percentages represent row percentages.
Includes 259 participants in the model. The model was adjusted for site using a random intercept model.
Genotype 2 vs. genotype 1: AOR: 2.88; 95%CI: 0.92, 8.97; P=0.07
Discussion
This study assessed factors associated with HCV RNA levels during early chronic infection in a large sample of individuals with well-defined early chronic HCV infection, the majority of whom were PWID. Among individuals with persistence and detectable HCV RNA at one year post-infection, male sex, IFNL4 rs12979860 CC genotype, HIV co-infection, and HCV genotype 2 were independently associated with high HCV RNA levels (>5.6 log IU/mL).
The findings in the current study are consistent with previous studies demonstrating that IFNL4 CC genotype (11-13, 22), male sex (11, 15, 16, 22), HIV co-infection (11, 17, 18), and HCV genotype 2 infection (11, 16, 19) are associated with higher HCV RNA levels during chronic infection. Although other studies have investigated factors associated with HCV RNA levels during chronic infection, there is considerable uncertainty and heterogeneity with the timing of HCV RNA levels post-infection, given the cross-sectional nature of studies and/or unavailable data on the estimation time of infection (11-21). However, the current study is unique given the well-defined nature of early chronic HCV infection in the InC3 study participants. Our findings suggest that during chronic infection, factors associated with HCV RNA levels exert their roles as early as one year following infection, providing a better understanding of the pathogenesis of HCV infection.
IFNL4 rs12979860 CC genotype was associated with high HCV RNA levels (>5.6 log IU/mL) during early chronic infection in this study. Previous data has demonstrated that IFNL4 CC genotype is associated with both higher initial HCV RNA levels in acute infection (37, 38) and enhanced spontaneous clearance (32, 33, 35, 39). One hypothesis is that those with IFNL4 CC genotype who fail to clear virus carry higher viraemia into chronic infection. The somewhat paradoxical association between IFNL4 CC genotype and both higher HCV RNA levels and enhanced interferon-based treatment responsiveness in chronic infection may be driven by linkages with hepatic interferon-stimulated genes (ISG), which are predictive of treatment response (40, 41). Further, rs12979860 C allele has been shown to be in high linkage disequilibrium with the IFNL4 rs368234815 TT allele, which results in a non-functional IFNL4 gene (36). IFNL4 gene encodes an active type III interferon, named IFNλ4, which has HCV antiviral activity mainly through inducing ISGs (42).
Male sex was associated with higher HCV RNA levels during early chronic infection in this study. This finding is consistent with previous data demonstrating higher HCV RNA levels among males compared to females (11, 15, 16, 22), although some studies did not find any association between sex and HCV RNA levels (17, 18). It is also well established that males are more likely to develop chronic HCV infection than females (34, 35, 43, 44). Mechanisms behind the association of sex and HCV RNA levels may be linked to sex-based differences in immunity. Compared to males, females have an increased number and magnitude of immune and inflammatory responses (45), and a higher prevalence of several autoimmune diseases (46). These differences are hypothesized to be a function of differential binding of sex steroids to specific receptors expressed in lymphoid tissue cells, macrophages, dendritic cells, and lymphocytes, thereby influencing the function of immune cells (45). However, there are little data on sex-based differences in immunological profiles in those with HCV infection although it has been shown in HIV (47). Another hypothesis to explain the difference in HCV viral control between males and females suggests a role for female hormones, such as estrogens and progesterone in viral control. This hypothesis is supported by the evidence of a lower rate of HCV clearance in postmenopausal women compared to premenopausal women (48, 49), and estrogen receptor alpha promoting HCV replication by interaction with the NS5B protein (50). Further studies should focus on mechanisms explaining the sex-based immune response to HCV infection.
HIV infection was associated with higher HCV RNA levels during early chronic infection in this study. Higher HCV RNA levels in the setting of HIV co-infection has been described previously (11, 17, 18) and may reflect impaired control of HCV replication in the setting of HIV-related immunodeficiency. HIV has been shown to interact directly with hepatocytes and hepatic stellate cells leading to increased HCV replication via transforming growth factor β1 (TGF-β1) (51, 52).
HCV genotype was associated with HCV RNA levels during early chronic infection in this study. Compared to HCV genotype 3, HCV genotype 2 was significantly associated with high levels of HCV RNA (≥5.6 IU/mL). The influence of HCV genotype on HCV RNA levels in chronic infection has been assessed by several studies, but results have been conflicting (11, 14-21). This is partly due to HCV genotype non-1 infections being grouped together, given limited genotypic diversity in some investigated populations (14, 17, 20, 21). These studies reported higher (20, 21) or lower (17) HCV RNA levels in genotype 1 compared to genotype non-1, while one study reported higher HCV RNA levels in genotype 1/4 compared to genotype 2/3 (14). Our data, consistent with most other studies assessing HCV RNA levels among HCV genotypes 1, 2 and 3 separately (11, 16, 19), revealed heterogeneous influence of genotypes non-1 on HCV RNA levels with higher HCV RNA levels being observed in genotype 2. Further research is needed to better understand the impact of viral factors, particularly HCV genotype on the pathogenesis of HCV infection.
While the current study is unique given the large sample size and well-defined nature of early chronic HCV infection, there are some limitations. Nine cohorts of individuals with acute or early chronic HCV were combined. Participating cohorts bring a range of data types and structures presenting issues surrounding both inconsistent measurement and biological data testing protocols (e.g. HCV RNA assays differed across cohorts with different sensitivity, specificity and lower limit of detection). HCV genotypes 4, 6 and mixed genotype were also grouped together due to small numbers in each category. As such, the current analysis was not able to explore the potential effect of these individual genotypes on HCV RNA levels. The date of HCV infection was estimated based on the best available data, but broad HCV seroconversion intervals among some individuals would have had an impact on some HCV RNA levels included in the analyses.
In conclusion, the current study identified that IFNL4 rs12979860 CC genotype, male sex, HIV co-infection, and HCV genotype G2 were independently associated with high HCV RNA levels (≥5.6 IU/mL) at one year post-infection. These findings enhance our understandings of the immunopathogenesis of HCV during early chronic infection and have implications for the clinical management of chronic HCV infection with interferon-based regimens. Further research is needed to understand the mechanism of IFNL4 genotype, sex and HCV genotype on HCV replication and whether higher HCV RNA levels are associated with increased HCV-related liver disease progression.
Supplementary Material
Supplementary Table 1. Characteristics of participants with HCV in the InC3 Study who were not included in analysis
Acknowledgments
Funding: The InC3 Study is supported by the National Institute on Drug Abuse Award Number R01DA031056. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Drug Abuse or the National Institutes of Health. The Kirby Institute is funded by the Australian Government Department of Health and Ageing. The views expressed in this publication do not necessarily represent the position of the Australian Government. BH is supported by an Australian Postgraduate PhD Award. JG is supported by a National Health and Medical Research Council (NHMRC) Career Development Fellowship. GD and AL are supported by NHMRC Practitioner Research Fellowships. MH and LM are supported by NHMRC Senior Research Fellowships and MH additionally by a VicHealth Senior Research Fellowship. RSD was supported by an NHMRC postgraduate scholarship and a Centre for Research Excellence into Injecting Drug Use postgraduate top-up scholarship. Other research support includes NIH U19 AI088791 (AC), NIH U19 AI066345 (AK, BM), R01 DA033541 (AK), MOP-103138, MOP-210232 (JB), the Netherlands National Institute for Public Health and the Environment (MP), and NHMRC Project Grant #630483 (LM).
List of abbreviations (in alphabetical order)
- ACS
Amsterdam Cohort Studies
- ALT
Alanine aminotransferase
- anti-HCV
HCV-specific antibodies
- AOR
Adjusted odds ratio
- ATAHC
Australian Trial in Acute Hepatitis C
- BAHSTION
Boston Acute HCV Study, Transmission, Immunity and Outcomes Network
- BBAASH
Baltimore Before and After Acute Study of Hepatitis
- DAA
Direct-acting antiviral
- HCV
Hepatitis C virus
- HEPCO
St. Luc Cohort, HEPCO
- HITS-c
Hepatitis C Incidence and Transmission Study-community
- HITS-p
Hepatitis C Incidence and Transmission Study-prison
- HIV
Human immunodeficiency virus
- IFNL3
Interferon lambda 3
- IFNL4
Interferon lambda 4
- IL28B
Interleukin-28B
- InC3
The International Collaboration of Incident HIV and Hepatitis C in Injecting Cohorts
- IQR
Interquartile range
- ISGs
Interferon stimulated genes
- N2
Networks 2
- OR
Odds ratio
- PWID
People who inject drugs
- SVR
Sustained virological response
- UFO
UFO STUDY
Footnotes
Conflict of Interest: The authors do not have any commercial or other association that might pose a conflict of interest
References
- 1.Hajarizadeh B, Grebely J, Dore GJ. Epidemiology and natural history of HCV infection. Nature Review Gastroenterology Hepatology. 2013;10(9):553–62. doi: 10.1038/nrgastro.2013.107. [DOI] [PubMed] [Google Scholar]
- 2.Grebely J, Dore GJ. What Is Killing People with Hepatitis C Virus Infection? Semin Liver Dis. 2011;31(04):331–9. doi: 10.1055/s-0031-1297922. [DOI] [PubMed] [Google Scholar]
- 3.Zeuzem S, Rodríguez-Torres M, Rajender Reddy K, et al. Optimized threshold for serum HCV RNA to predict treatment outcomes in hepatitis C patients receiving peginterferon alfa-2a/ribavirin. Journal of Viral Hepatitis. 2012;19(11):766–74. doi: 10.1111/j.1365-2893.2012.01624.x. [DOI] [PubMed] [Google Scholar]
- 4.Manns MP, McHutchison JG, Gordon SC, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. The Lancet. 2001;358(9286):958–65. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 5.McHutchison JG, Lawitz EJ, Shiffman ML, et al. Peginterferon Alfa-2b or Alfa-2a with Ribavirin for Treatment of Hepatitis C Infection. New England Journal of Medicine. 2009;361(6):580–93. doi: 10.1056/NEJMoa0808010. [DOI] [PubMed] [Google Scholar]
- 6.Poordad F, McCone J, Bacon BR, et al. Boceprevir for Untreated Chronic HCV Genotype 1 Infection. New England Journal of Medicine. 2011;364(13):1195–206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poordad F, Bronowicki JP, Gordon SC, et al. Factors That Predict Response of Patients With Hepatitis C Virus Infection to Boceprevir. Gastroenterology. 2012;143(3):608–18.e5. doi: 10.1053/j.gastro.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 8.McHutchison JG, Manns MP, Muir AJ, et al. Telaprevir for Previously Treated Chronic HCV Infection. New England Journal of Medicine. 2010;362(14):1292–303. doi: 10.1056/NEJMoa0908014. [DOI] [PubMed] [Google Scholar]
- 9.Lawitz E, Poordad FF, Pang PS, et al. Sofosbuvir and ledipasvir fixed-dose combination with and without ribavirin in treatment-naive and previously treated patients with genotype 1 hepatitis C virus infection (LONESTAR): an open-label, randomised, phase 2 trial. The Lancet. 2014;383(9916):515–23. doi: 10.1016/S0140-6736(13)62121-2. [DOI] [PubMed] [Google Scholar]
- 10.Jacobson IM, Gordon SC, Kowdley KV, et al. Sofosbuvir for Hepatitis C Genotype 2 or 3 in Patients without Treatment Options. New England Journal of Medicine. 2013;368(20):1867–77. doi: 10.1056/NEJMoa1214854. [DOI] [PubMed] [Google Scholar]
- 11.Uccellini L, Tseng FC, Monaco A, et al. HCV RNA levels in a multiethnic cohort of injection drug users: human genetic, viral and demographic associations. Hepatology. 2012;56(1):86–94. doi: 10.1002/hep.25652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarthy JJ, Li JH, Thompson A, et al. Replicated Association Between an IL28B Gene Variant and a Sustained Response to Pegylated Interferon and Ribavirin. Gastroenterology. 2010;138(7):2307–14. doi: 10.1053/j.gastro.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 14.Labarga P, Soriano V, Caruz A, et al. Association between IL28B gene polymorphisms and plasma HCV-RNA levels in HIV/HCV-co-infected patients. AIDS. 2011;25(6):761–6. doi: 10.1097/QAD.0b013e32834488e7. [DOI] [PubMed] [Google Scholar]
- 15.Rong X, Lu L, Wang J, et al. Correlation of Viral Loads with HCV Genotypes: Higher Levels of Virus Were Revealed among Blood Donors Infected with 6a Strains. PLoS ONE. 2012;7(12):e52467. doi: 10.1371/journal.pone.0052467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McMahon BJ, Hennessy TW, Christensen C, et al. Epidemiology and risk factors for hepatitis C in Alaska Natives. Hepatology. 2004;39(2):325–32. doi: 10.1002/hep.20046. [DOI] [PubMed] [Google Scholar]
- 17.Fishbein DA, Lo Y, Netski D, Thomas DL, Klein RS. Predictors of Hepatitis C Virus RNA Levels in a Prospective Cohort Study of Drug Users. JAIDS Journal of Acquired Immune Deficiency Syndromes. 2006;41(4):471–6. doi: 10.1097/01.qai.0000218360.28712.f3. [DOI] [PubMed] [Google Scholar]
- 18.Thomas DL, Astemborski J, Vlahov D, et al. Determinants of the Quantity of Hepatitis C Virus RNA. The Journal of infectious diseases. 2000;181(3):844–51. doi: 10.1086/315314. [DOI] [PubMed] [Google Scholar]
- 19.Rauch A, Gaudieri S, Evison J, et al. Low current and nadir CD4+ T-cell counts are associated with higher hepatitis C virus RNA levels in the Swiss HIV cohort study. Antiviral Therapy. 2008;13(3):455–60. [PubMed] [Google Scholar]
- 20.Grint D, Peters L, Reekie J, et al. Stability of hepatitis C virus (HCV) RNA levels among interferon-naïve HIV/HCV-coinfected individuals treated with combination antiretroviral therapy. HIV medicine. 2013;14(6):370–8. doi: 10.1111/hiv.12033. [DOI] [PubMed] [Google Scholar]
- 21.Yoo TW, Donfield S, Lail A, et al. Effect of Hepatitis C Virus (HCV) Genotype on HCV and HIV-1 Disease. Journal of Infectious Diseases. 2005;191(1):4–10. doi: 10.1086/426513. [DOI] [PubMed] [Google Scholar]
- 22.Grady BPX, Prins M, Rebers S, Molenkamp R, Geskus RB, Schinkel J. BMI, male sex and IL28B genotype associated with persistently high hepatitis C virus RNA levels among chronically infected drug users up to 23 years following seroconversion. Journal of Viral Hepatitis. 2014 doi: 10.1111/jvh.12303. in press. [DOI] [PubMed] [Google Scholar]
- 23.Grebely J, Morris MD, Rice TM, et al. Cohort Profile: The International Collaboration of Incident HIV and Hepatitis C in Injecting Cohorts (InC3) Study. International Journal of Epidemiology. 2012;42(6):1649–59. doi: 10.1093/ije/dys167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hajarizadeh B, Grebely J, Applegate T, et al. Dynamics of HCV RNA levels during acute hepatitis C virus infection. Journal of Medical Virology. 2014;86(10):1722–9. doi: 10.1002/jmv.24010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hajarizadeh B, Page K, Kim AY, et al. Early HCV RNA dynamics and factors associated with high early HCV RNA level during acute HCV infection. Journal of Hepatology. 2013;58(Supplement 1):S470–S1. Abstract. [Google Scholar]
- 26.Page-Shafer K, Pappalardo BL, Tobler LH, et al. Testing strategy to identify cases of acute hepatitis C virus (HCV) infection and to project HCV incidence rates. Journal of Clinical Microbiology. 2008;46(2):499–506. doi: 10.1128/JCM.01229-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox AL, Netski DM, Mosbruger T, et al. Prospective Evaluation of Community-Acquired Acute-Phase Hepatitis C Virus Infection. Clinical Infectious Diseases. 2005;40(7):951–8. doi: 10.1086/428578. [DOI] [PubMed] [Google Scholar]
- 28.Hofer H, Watkins-Riedel T, Janata O, et al. Spontaneous viral clearance in patients with acute hepatitis C can be predicted by repeated measurements of serum viral load. Hepatology. 2003;37(1):60–4. doi: 10.1053/jhep.2003.50019. [DOI] [PubMed] [Google Scholar]
- 29.Busch MP, Page Shafer KA. Acute-phase hepatitis C virus infection: Implications for research, diagnosis, and treatment. Clinical Infectious Diseases. 2005;40(7):959–61. doi: 10.1086/428583. [DOI] [PubMed] [Google Scholar]
- 30.Badr G, Bédard N, Abdel-Hakeem MS, et al. Early Interferon Therapy for Hepatitis C Virus Infection Rescues Polyfunctional, Long-Lived CD8+ Memory T Cells. Journal of Virology. 2008;82(20):10017–31. doi: 10.1128/JVI.01083-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van de Laar TJW, Molenkamp R, van den Berg C, et al. Frequent HCV reinfection and superinfection in a cohort of injecting drug users in Amsterdam. Journal of Hepatology. 2009;51(4):667–74. doi: 10.1016/j.jhep.2009.05.027. [DOI] [PubMed] [Google Scholar]
- 32.Grebely J, Petoumenos K, Hellard M, et al. Potential role for interleukin-28B genotype in treatment decision-making in recent hepatitis C virus infection. Hepatology (Baltimore, Md) 2010;52(4):1216–24. doi: 10.1002/hep.23850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461(7265):798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Berg CHBS, Grady BPX, Schinkel J, et al. Female sex and IL28b, a synergism for spontaneous viral clearance in hepatitis c virus (HCV) seroconverters from a community-based cohort. PLoS One. 2011;6(11) doi: 10.1371/journal.pone.0027555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grebely J, Page K, Sacks-Davis R, et al. The effects of female sex, viral genotype, and IL28B genotype on spontaneous clearance of acute hepatitis C virus infection. Hepatology. 2014;59(1):109–20. doi: 10.1002/hep.26639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prokunina-Olsson L, Muchmore B, Tang W, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013;45(2):164–71. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu L, Fisher BE, Thomas DL, Cox AL, Ray SC. Spontaneous clearance of primary acute hepatitis C virus infection correlated with high initial viral RNA level and rapid HVR1 evolution. Hepatology. 2012;55(6):1684–91. doi: 10.1002/hep.25575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hajarizadeh B, Grady B, Page K, et al. Interferon lambda 3 genotype predicts hepatitis C virus RNA levels in early acute infection among people who inject drugs: The InC3 Study. Journal of Clinical Virology. 2014;61(3):430–4. doi: 10.1016/j.jcv.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tillmann HL, Thompson AJ, Patel K, et al. A Polymorphism Near IL28B Is Associated With Spontaneous Clearance of Acute Hepatitis C Virus and Jaundice. Gastroenterology. 2010;139(5):1586–92. doi: 10.1053/j.gastro.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 40.Honda M, Sakai A, Yamashita T, et al. Hepatic ISG Expression Is Associated With Genetic Variation in Interleukin 28B and the Outcome of IFN Therapy for Chronic Hepatitis C. Gastroenterology. 2010;139(2):499–509. doi: 10.1053/j.gastro.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 41.Urban TJ, Thompson AJ, Bradrick SS, et al. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 2010;52(6):1888–96. doi: 10.1002/hep.23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamming OJ, Terczyńska-Dyla E, Vieyres G, et al. Interferon lambda 4 signals via the IFNλ receptor to regulate antiviral activity against HCV and coronaviruses. The EMBO Journal. 2013;32(23):3055–65. doi: 10.1038/emboj.2013.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Page K, Hahn JA, Evans J, et al. Acute Hepatitis C Virus Infection in Young Adult Injection Drug Users: A Prospective Study of Incident Infection, Resolution, and Reinfection. Journal of Infectious Diseases. 2009;200(8):1216–26. doi: 10.1086/605947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Micallef JM, Kaldor JM, Dore GJ. Spontaneous viral clearance following acute hepatitis C infection: A systematic review of longitudinal studies. Journal of Viral Hepatitis. 2006;13(1):34–41. doi: 10.1111/j.1365-2893.2005.00651.x. [DOI] [PubMed] [Google Scholar]
- 45.Klein SL, Jedlicka A, Pekosz A. The Xs and Y of immune responses to viral vaccines. The Lancet Infectious Diseases. 2010;10(5):338–49. doi: 10.1016/S1473-3099(10)70049-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouman A, Heineman MJ, Faas MM. Sex hormones and the immune response in humans. Human Reproduction Update. 2005;11(4):411–23. doi: 10.1093/humupd/dmi008. [DOI] [PubMed] [Google Scholar]
- 47.Meier A, Chang JJ, Chan ES, et al. Sex differences in the Toll-like receptor-mediated response of plasmacytoid dendritic cells to HIV-1. Nat Med. 2009;15(8):955–9. doi: 10.1038/nm.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Floreani A, Cazzagon N, Boemo DG, et al. Female patients in fertile age with chronic hepatitis C, easy genotype, and persistently normal transaminases have a 100% chance to reach a sustained virological response. European Journal of Gastroenterology and Hepatology. 2011;23(11):997–1003. doi: 10.1097/MEG.0b013e32834ae863. [DOI] [PubMed] [Google Scholar]
- 49.Villa E, Karampatou A, Camm C, et al. Early menopause is associated with lack of response to antiviral therapy in women with chronic hepatitis C. Gastroenterology. 2011;140(3):818–29. doi: 10.1053/j.gastro.2010.12.027. [DOI] [PubMed] [Google Scholar]
- 50.Hillung J, Ruiz-López E, Bellón-Echeverría I, Clemente-Casares P, Mas A. Characterization of the interaction between hepatitis C virus NS5B and the human oestrogen receptor alpha. Journal of General Virology. 2012;93(Pt 4):780–5. doi: 10.1099/vir.0.039396-0. [DOI] [PubMed] [Google Scholar]
- 51.Lin W, Weinberg EM, Tai AW, et al. HIV increases HCV replication in a TGF-beta1-dependent manner. Gastroenterology. 2008;134(3):803–11. doi: 10.1053/j.gastro.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 52.Zhang X, Daucher M, Baeza J, Kim CW, Russell R, Kottilil S. Human immunodeficiency virus enhances hepatitis C virus replication by differential regulation of IFN and TGF family genes. J Med Virol. 2012;84(9):1344–52. doi: 10.1002/jmv.23315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Characteristics of participants with HCV in the InC3 Study who were not included in analysis