Abstract
Myh9 and Myh10, which encode two major isoforms of non-muscle myosin II expressed in the brain, have emerged as risk factors for developmental brain disorders. Myosin II motors regulate neuronal cytoskeletal dynamics leading to optimization of synaptic plasticity and memory formation. However, the role of these motor complexes in brain development remains poorly understood. Here, we disrupted the in vivo expression of Myh9 and/or Myh10 in developing hippocampal neurons to determine how these motors contribute to circuit maturation in this brain area important for cognition. We found that Myh10 ablation in early postnatal, but not mature, CA1 pyramidal neurons reduced excitatory synaptic function in the Schaffer collateral pathway, while more distal inputs to CA1 neurons were relatively unaffected. Myh10 ablation in young neurons also selectively impaired the elongation of oblique dendrites that receive Schaffer collateral inputs, while the structure of distal dendrites was normal. We observed normal spine density and spontaneous excitatory currents in these neurons, indicating that Myh10 KO impaired proximal pathway synaptic maturation through disruptions to dendritic development rather than postsynaptic strength or spine morphogenesis. To address possible redundancy and/or compensation by other Myosin II motors expressed in neurons, we performed similar experiments in Myh9 null neurons. In contrast to findings in Myh10 mutants, evoked synaptic function in young Myh9 KO hippocampal neurons was normal. Data obtained from double Myh9/Myh10 KO neurons largely resembled the MyH10 KO synaptic phenotype. These data indicate that Myosin IIB is a key molecular factor that guides input-specific circuit maturation in the developing hippocampus.
INTRODUCTION
Actin filaments are the main structural components of dendritic spines and serve essential roles for both function and plasticity of synapses (Cingolani and Goda, 2008; Hotulainen and Hoogenraad, 2010; Luo, 2002; Matus, 2000; Racz and Weinberg, 2013). Spine actin dynamics are regulated by a multitude of actin binding proteins. Non-muscle myosin (NM) II is an actin binding protein with three isoforms in the brain, IIA, IIB and IIC, which are encoded by the Myh9, Myh10 and Myh14 genes, respectively. NM IIB is the most abundant isoform in the brain (Cheng et al., 2006; Jordan et al., 2004; Kawamoto and Adelstein, 1991; Peng et al., 2004), and studies using electron microscopy show that it is mostly localized to the necks of dendritic spines (Korobova and Svitkina, 2010). Regulation of Myosin II expression in neurons disrupts synaptic function, plasticity, and memory. However, the roles of Myosin II motors, particularly during early postnatal brain development when neural circuits are first forming, are not clearly understood.
Due to the fundamental nature of NM II in cognition (Gavin et al., 2012; Rex et al., 2010) and the recent link of these motors to neurodevelopmental disorders (O'Roak et al., 2012; Prasad et al., 2012), it is critical to understand how these motors impact the development of brain circuits. However, how these motors regulate synapse maturation and/or activity-dependent neuronal plasticity remains elusive, primarily due to several conflicting findings related to the role of these motors in the development and maintenance of ongoing synaptic function and dendritic spine morphogenesis. For instance, RNAi knockdown of Myosin IIB and/or blebbistatin perfusion disrupts spine morphology and baseline synaptic transmission associated with altered AMPAR function (Hodges et al., 2011; Rubio et al., 2011; Ryu et al., 2006). Other studies have found no effect of treatments on NM II function on baseline synaptic transmission (Peng et al., 2012; Rex et al., 2010). Instead, these studies found that NM II is involved in either pre- or post-synaptic plasticity processes (Peng et al., 2012; Rex et al., 2010), but not involved in maintaining synapse stability. It is possible that these discrepant findings could be explained, in part, by a unique developmental role of Myosin II in neurons to guide their maturation. Indeed, an investigation into Myosin II function in developing neurons from the intact hippocampus has not been performed.
To address the possibility that Myosin II has a unique role in developing hippocampal neurons, we utilized conditional knockout mice paired with recombinant adeno-associated virus (rAAV) particles that drive Cre expression in order to spatiotemporally regulate Myosin II expression in the developing hippocampus. Using a combination of functional and structural neuronal analyses, we discovered that the cell autonomous deletion of Myh10 gene causes a pathway specific impairment in the postnatal development of dendritic arborization. This structural defect was associated with a reduction in the number of functional synapses. Interestingly, mEPSCs and dendritic spine morphology were largely normal, indicating that Myosin IIB ablation did not disrupt the stability of already formed excitatory synapses. To address the possibility that other Myosin II isoforms could compensate for Myosin IIB ablation, we performed similar studies in Myh9 and Myh9/Myh10 double KO neurons. We found no effect of Myh9 deletion on neuronal function, while combined deletion of Myh9 and Myh10 largely resembled the Myh10 single KO phenotype. We conclude that Myosin IIB supports hippocampal circuit maturation by promoting the growth of certain types of dendrites.
MATERIALS AND METHODS
Mice
All animal procedures were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and protocols approved by The Scripps Institutional Animal Care and Use Committee. Myh10 conditional knock out (ko) mice (Myh10fl/fl) and Myh9 conditional ko mice (Myh9fl/fl) of both sexes, described previously (Jacobelli et al., 2010; Ma et al., 2009), were obtained from the Mutant Mouse Regional Resource Center, a NIH funded strain repository (#16981 and #32096). Both lines of mice were crossed with an Ai9 tdTomato reporter line, which expresses this fluorescent protein upon activation of Cre recombinase (stock number: 007909, Jackson Laboratory (Bar Harbour, ME)) (Madisen et al., 2010). Experiments are conducted on mice homozygous for either one or both of the motors and heterozygous for the Ai9 allele. For Sholl analysis (Fig. 3) and in order to control for non-specific effects of Cre recombinase/TdTomato expression (Fig. 2 I-K), we used mice heterozygous for the reporter allele in the wild-type background as controls (designated as Myh10+/+).
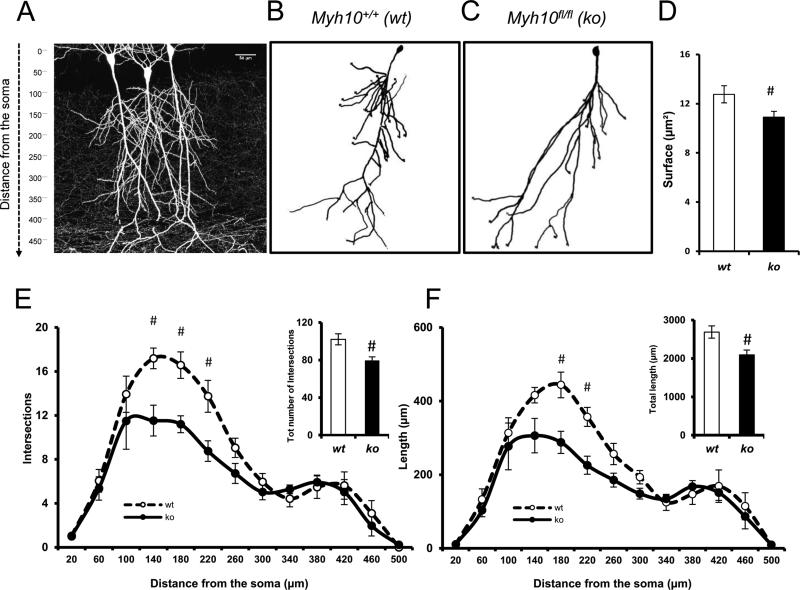
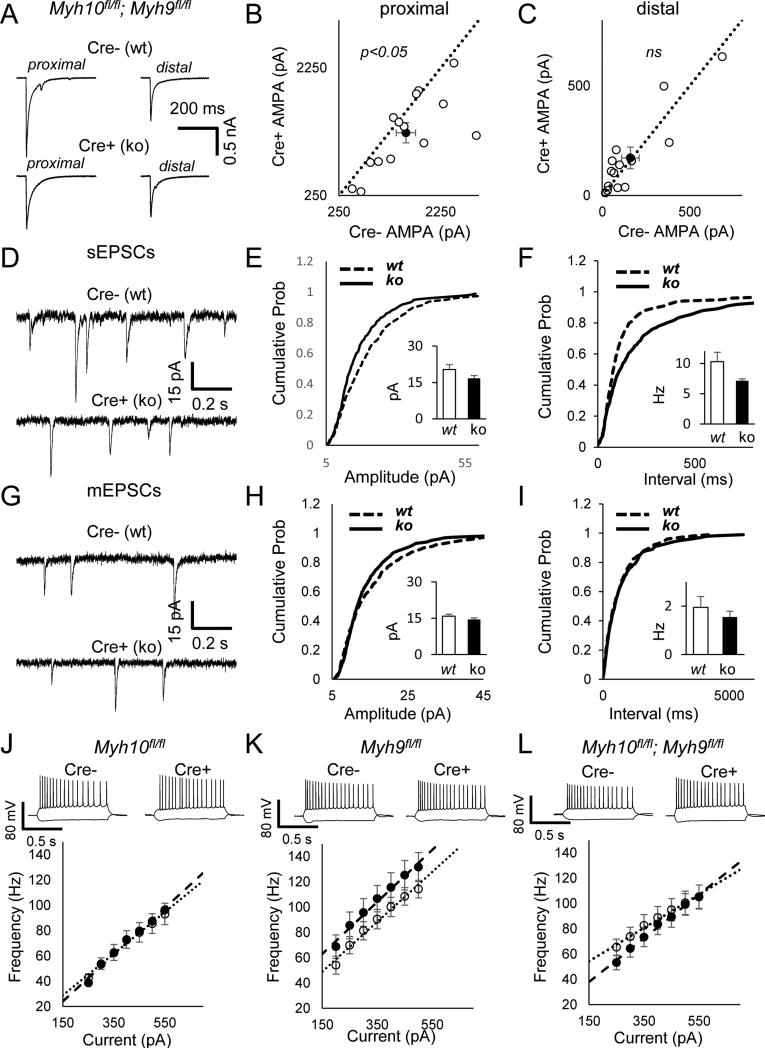
Figure 3. NM IIB ablation selectively disrupts the development of proximal dendritic branches in CA1 neurons.
(A) Photomicrograph multiphoton image of a portion of hippocampal CA1 showing labeled pyramidal neurons. (B-C) Representative 3D pyramidal hippocampal cell reconstruction using Neurolucida software. Neuronal complexity was measured applying the Sholl ring analysis method; (D) Bar graphs showing surface extension of traced apical arbors in both wt (14 neurons, n= 7 slices, n=5 mice) and ko (12 neurons, n= 6 slices, n=4 mice); p < 0.05. (E-F) Graphs exhibiting arbor complexity of pyramidal neurons in both wt and ko, measured as a number of intersections [RMANOVA, F1,12 = 2.4170; p < 0.01]) or cumulative length [RMANOVA, F(1,12) = 2.0649; p < 0.05] in relation to the distance from soma (insets depicting the total number of intersections and the total length, respectively); A LSD post-hoc test was applied where appropriate; # p<0.05 wt vs. ko comparisons. Error bars represent SEM.
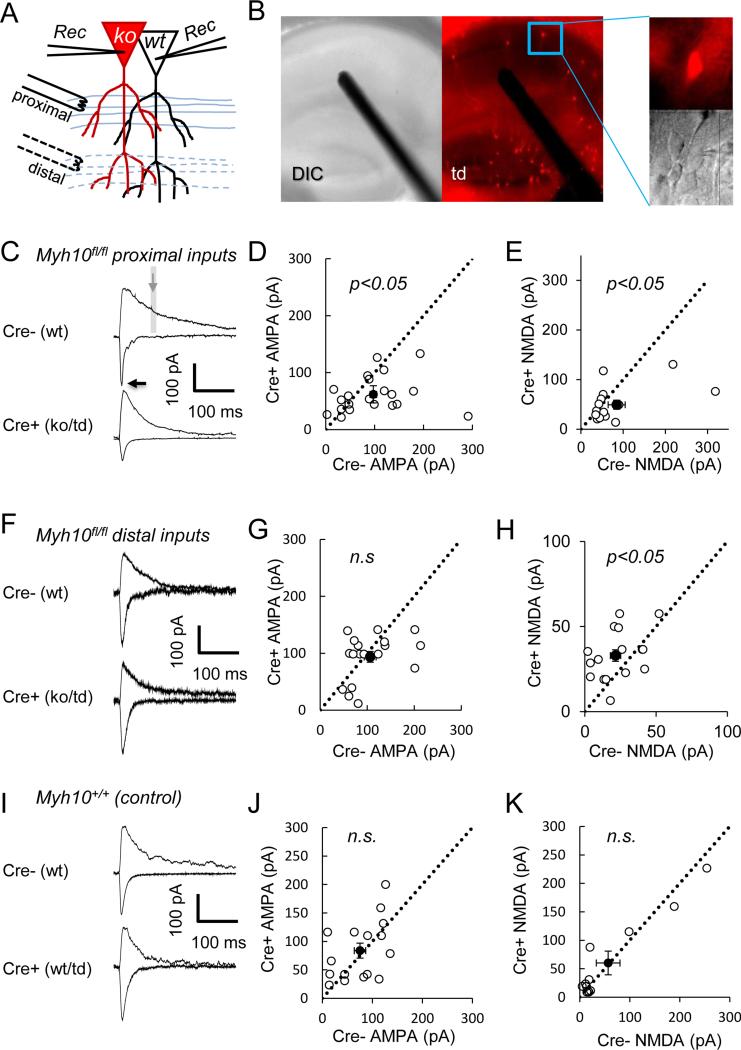
Figure 2. NM IIB ablation cell-autonomously disrupts the pathway-specific development of synaptic excitation in CA1 neurons.
(A) The illustration shows the design of these experiments with electrode replacement either in proximal (C, I) or in distal (F) stimulation locations in separate sets of experiments. (B) Representative DIC and fluorescent images of Myh10fl/fl hippocampus show sparse Cre expression. Representative AMPA (inward, recorded at −70 mV) and NMDA currents (outward, recorded at +40 mV) from (C, I) Myh10fl/fl and (F) Myh10+/+ animals injected with Cre. Arrows point to where AMPA and NMDA measurements were made. Recordings from neighboring Cre negative and Cre positive neurons (Myh10 KO) show (D) reduced AMPA (paired t(20)=2.33, p=0.03, n=21 pairs from n=9 mice ) and (E) reduced NMDA currents (paired t(14)=2.16, p=0.048, n=15 pairs from n=7 mice) at proximal stimulation locations, but (G) normal AMPA (paired t(17)=0.99, p=0.33, n=18 pairs from n=3 mice) and (H) increased NMDA currents (paired t(15)=2.57, p=0.021, n=16 pairs from n=3 mice) at distal stimulation locations. (I-K) Control experiments at proximal locations show that Cre positive neurons (wt/td) have (G) normal AMPA (paired t(15)=0.50, p=0.63, n=16 pairs from n=5 mice) and (H) normal NMDA currents (paired t(11)=0.11, p=0.92, n=12 pairs from n=5 mice) from Myh10+/+ animals. Dashed black line is identity line. Filled circle is the sample mean. Error bars represent SEM.
rAAV-Cre infusions
For postnatal day 1 (PND1) injections, mice were anesthetized for 3 min in ice and infused with 1 μl of a SynCreVenus construct (Clement et al., 2012) packaged in rAAV8 or rAAV9 (~2 ×1013 virions/ml; referred to as AAV-Cre) undiluted (for field recordings and immunoblots) or diluted 1:20 (for sparse labeling experiments) with sterile Hyclone DPBS/Modified solution (Thermo Scientific SH30264.02) using a neonatal stereotaxic adaptor (Stoelting Model 51625) in conjunction with a Kopf small animal stereotaxic with the following coordinates relative to lambda for the purpose of Cre recombinase expression in hippocampal cells: A/P: +1.5; M/L: −1.0; D/V: −2.0 (Pilpel et al., 2009). A 33 gauge needle was used for the infusions at a rate of 0.33 μl/min after which the needle was left in place for 3 min and pups transferred to a 30-32°C incubator until active and returned to their respective dam.
In addition, for some sparse labeling experiments, we delivered an rAAV9-packaged Cre-expressing virus via the superficial temporal vein (STV) of PND1 mouse pups as described by others (Foust et al., 2009; Kienstra et al., 2007). Both methods produced comparable results, i.e. a sparse labeling of less than 5% of the neuronal population, though the STV method is less invasive. Briefly, pups were sedated by covering them with ice for 3 minutes. Superficial temporal vein is visualized using a handheld transilluminator (WeeSight, Respironics, Murrysville, PA), and a pair of standard reading glasses. Virus solution is prepared by 1:50 dilution of stock solution (final ~1×1012 GC/ml) in Dulbecco's Phosphate-Buffered Saline, also supplemented with 0.001% pluronic-F68. Virus solution (50 μl) was injected using a 100 μl Nanofil syringe attached with a 34 gauge Nanofil beveled needle (World Precision Instruments). A correct injection was verified by noting blanching of the vein. After the injection, pups were returned to the incubator until active and then returned to their mother. Brain slices were cut 21-28 days after infusions for electrophysiological or structural measurements.
For PND42 injections (Fig 1G-J), six to eight week old mice were anesthetized with isoflurane and infused with AAV-Cre into the right hippocampus (CA1) using the following coordinates relative to bregma: A/P: −2.3, M/L: −1.5, D/V: −1.5. Infusions (1 μl) were delivered at a rate of 0.1 ul/min with a 33 gauge needle that was left in place 10 min after infusion. Brain slices were cut 21-28 days after infusions for electrophysiological recordings.
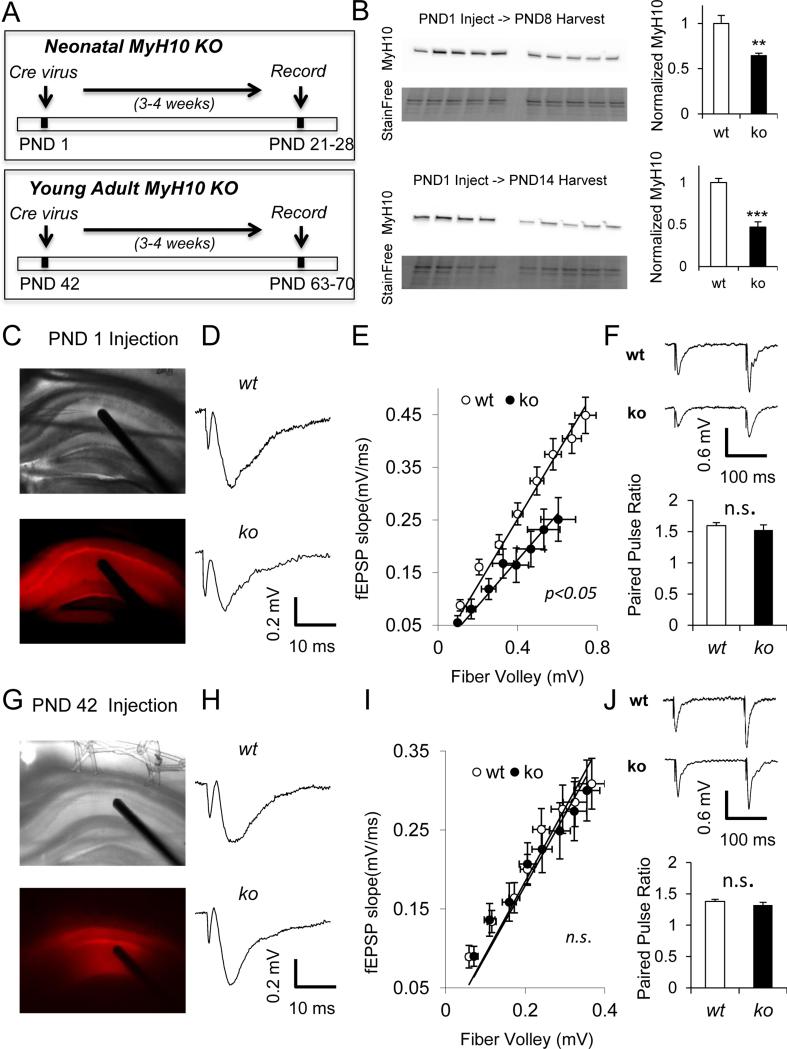
Figure 1. NM IIB ablation disrupts baseline excitatory synaptic function in developing, but not in adult CA1.
(A) Experimental design of the study. (B) Representative western blots from hippocampi of PND8 and PND14 Myh10fl/fl animals injected with 1μl AAV-Cre at PND1 show reduction in relative Myh10 expression compared to control animals ( PND8 t(8)=3.85, p=0.005; PND14 t(7)=6.32, p=0.0004). Representative DIC and fluorescent images of (C) PND1 and (G) PND42 injections show td tomato expression in the hippocampus which reports Cre expression. (D) Representative field recording traces and (E) I/O curves show reduction in synaptic transmission in PND1-injected Myh10fl/fl hippocampus compared to wild-type (t(36)=2.29, p=0.028; n=22 wt, n=16 ko slices from n=5 wt, n=4 ko mice) . (F) Normal paired pulse ratio (interpulse interval 100 ms) in PND1 injected animals (t(28)=0.71, p=0.48, n=16 wt, n=14 ko slices from n=5 wt, n=4 ko mice). (H) Representative field recording traces and (I) I/O curves show normal synaptic transmission in PND42-injected Myh10fl/fl hippocampus compared to wild-type (t(21)=0.41, p=0.69, n=11 wt, n=12 ko slices from n=3 wt, n=3 ko mice). (J) Normal paired pulse ratio (interpulse interval 100 ms) in PND42-injected animals (t(21)=0.50, p=0.62, n=11 wt, n=12 ko slices from n=3 wt, n=3 ko mice ). Traces reflect responses at 30% of maximal stimulation intensity. Error bars represent SEM.
Systemic injection of rAAV9 particles (such as by STV injection method) can transduce many tissues depending on the promoter type (Inagaki et al., 2006; Pacak et al., 2008). In order to knockdown protein levels in liver, STV injections of rAAV9 virus expressing CMV driven Cre were performed without modifying any of the methods outlined above.
Electrophysiology
All signals were amplified using Multiclamp 700B (Molecular Devices, Sunnyville, CA), filtered at 2 KHz, digitized (10 KHz), and stored on a personal computer for off-line analysis. Analog to digital conversion was performed using the Digidata 1440A system (Molecular devices). Data acquisitions and analyses were performed using pClamp 10.2 software (Molecular devices). Acute brain slices were prepared from male or female mice 3-4 weeks after virus injections. Following decapitation, brains were rapidly removed and placed in ice-cold cutting solution, which consisted of (mM): 110 choline-Cl, 25 NaHCO3, 1.25 NaH2PO4, 2.5 KCl, 0.5 CaCl2, 5 MgCl2, 25 Glucose, 10 Ascorbic acid, 5 Pyruvic acid, pH 7.4. The tissue was hemisected, blocked on the ventral surface, then mounted on a vibrating microtome (Leica VT1200S, Germany), and 350 μm thick sections were cut. The slices were then warmed to 35°C for 40 minutes in standard artificial cerebrospinal fluid (aCSF), composed of (mM): 125 NaCl, 2.5 KCl, 24 NaHCO3, 2 CaCl2, 1.25 NaH2PO4, 2 MgSO4, and 10 D-Glucose, and equilibrated with 95% O2 and 5% CO2. Following this, slices were maintained in gassed aCSF at room temperature until transferred to a submerged-type recording chamber (Warner Instruments, Hamden, CT). All experiments were performed at 32°C±2.
Extracellular field recordings
Field EPSPs (fEPSPs) were elicited by a concentric bipolar stimulating electrode (inner diameter (ID): 25 μm; outer diameter (OD): 125 μm FHC Inc., USA) connected to a constant current isolated stimulator unit (A-M sytems, Model 2100). fEPSPs were recorded from stratum radiatum of CA1 area of the hippocampus. These fEPSPs were recorded using low resistance (3-5 MΩ) glass pipette (ID:1.16 mm, OD:1.50 mm, Warner Instruments) filled with aCSF. Stimulation frequency was set to 0.1 Hz. Input-output (I/O) curves were generated by adjusting the stimulus intensity by 20 μA per sweep with increments from 0-250 μA. A regression analysis has been performed on the I/O curves and slope has been determined (Zaman et al., 2000). Means of slopes have been compared using student's t-test. Paired pulse ratio (PPR) was assessed using a succession of two pulses separated by 100 ms. The degree of facilitation was determined by taking the ratio of the initial slope of the second fEPSP to the slope of the first fEPSP.
Whole-cell patch clamp
Whole-cell patch clamp experiments were conducted from visually identified neighboring Cre positive (tdTomato positive) and Cre negative neurons. For recording evoked excitatory postsynaptic currents (EPSCs) and miniature excitatory postsynaptic currents (mEPSCs), neurons were voltage clamped with the following internal solution (mM): 110 cesium gluconate, 20 CsCl, 10 HEPES, 0.5 EGTA, 10 Phosphocreatine disodium, 0.5 Na-GTP and 2 Mg-ATP (pH 7.3, 285-290 mOsm). For proximal spine analysis, internal solution also contained 200 μM Alexa 488 hydrazide sodium (Invitrogen). 5 μM bicuculline was used to isolate excitatory synaptic currents. AMPA receptor-mediated currents were recorded at −70 mV. To isolate NMDA currents, neurons were voltage-clamped at +40 mV, and mean response size was measured between 80-120 ms after the peak current (Kamenetz et al., 2003). Student's paired t-test was used to compare AMPA and NMDA currents between neighboring neurons in the same slice. In our initial experiments (Fig 2 and Fig 5), we aimed at obtaining an EPSC peak amplitude of approximately 150 pA in the first recorded neuron before patching the second neuron. In later experiments (Fig 6), we measured the maximum EPSC from each neuron, as this approach was less variable. mEPSCs were recorded at −75 mV. In order to isolate mEPSCs, we used 1μM tetrodotoxin, 50μM picrotoxin and 50μM AP5. Pharmacologically isolated NMDA currents were recorded at +40 mV in the presence of 50μM picrotoxin and 10μM NBQX. For current clamp recordings, the following internal solution was used (mM): 130 potassium gluconate, 5 KCl, 10 HEPES, 0.25 EGTA, 10 Phosphocreatine disodium, 0.5 Na-GTP and 4 Mg-ATP (pH 7.3, 285-290 mOsm). Intrinsic properties of neurons were recorded at the resting membrane potential and measured as described (Ozkan et al., 2014). Spontaneous excitatory postsynaptic currents (sEPSCs) were also recorded using potassium gluconate-based solution, but switching to voltage clamp mode at −65 mV, thereby electrically isolating excitatory potentials (reversal potential for Cl− ~-86 mV).
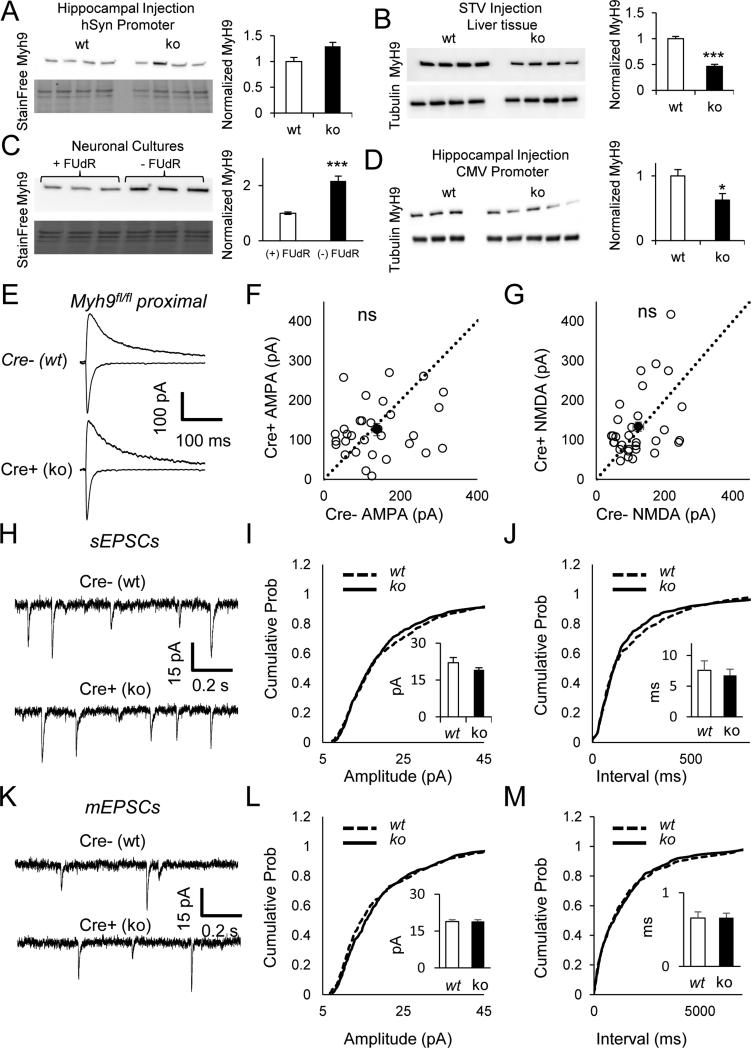
Figure 5. NM IIA ablation has no impact on excitatory synaptic function in CA1.
(A-D) Representative western blots show no reduction in Myh9 levels in hippocampus (A) (t(6)=0.80, p=0.45)(A), but robust reduction in liver (B) (t(6)=8.95, p=0.0001). Glial enriched cultures show higher levels of Myh9 (C) (t(4)=6.42, p=0.003). Driving Cre expression in hippocampus using CMV promoter results in Myh9 knockdown (D) (t(4)=4.21, =0.014). (E) Representative AMPA (inward, recorded at −70 mV) and NMDA currents (outward, recorded at +40 mV) from Myh9fl/fl animals injected with Cre. Recordings from neighboring Cre negative and Cre positive neurons (Myh9 KO) show (F) normal AMPA (paired t(30)=0.68, p=0.50, n=31 pairs from n=8 mice) and (G) normal NMDA currents (paired t(30)=0.70,p=0.49, n=31 pairs from n=8 mice) at proximal stimulation locations. Dashed black line is identity line. Filled circle is the sample mean. (H-J) Example traces (H), cumulative probability distribution of amplitude (I) and frequency (J) of spontaneous EPSC recordings indicate normal synaptic function in the absence of NMIIA (K-S test on amplitude Z=0.97,p=0.30; Bar graph inset for amplitude, Student's t-test t(23)=1.42, p=0.17; K-S test on frequency Z=1.34,p=0.054; Bar graph inset for frequency, Student's t-test t(23)=0.47, p=0.64; n=11 cre−, n=14 cre+ from n=4 mice). (K-M) Example traces (K), cumulative probability distribution of amplitude (L) and frequency (M) of miniature EPSC recordings (+TTX) indicate normal synaptic function in the absence of NMIIA (K-S test on amplitude Z=1.28,p=0.08; Bar graph inset for frequency, Student's t-test t(30)=0.1, p=0.94; K-S test on frequency Z=0.60, p=0.87; Bar graph inset for frequency, Student's t-test t(30)=0.1, p=0.95; n=15 cre−, n=17 cre+ from n=5 mice). Error bars represent SEM.
Figure 6. Ablation of both NM IIB and NM IIA in developing CA1 neurons results in a phenotype similar to that of NMIIB null neurons.
(A) Representative EPSCs from proximal and distal stimulation locations in Myh10fl/fl; Myh9fl/fl animals injected with Cre. Recordings from neighboring Cre negative and Cre positive neurons (Myh10/Myh9 double KO) show (B) reduced proximal (paired t(13)=2.49,p=0.026) and (C) normal distal EPSCs (paired t(13)=0.49, p=0.62) (n=14 pairs from n=6 mice). Dashed black line is identity line. Filled circle is the sample mean. (D-F) Example traces (D), cumulative probability distribution of amplitude (E) and frequency (F) of spontaneous EPSC recordings indicate reduced synaptic function in the absence of NMIIA and NMIIB (K-S test on amplitude Z=2.41, p<0.001; Bar graph inset for amplitude, Student's t-test t(20)=1.61, p=0.12; K-S test on frequency Z=3.23, p<0.001; Bar graph inset for frequency, Student's t-test t(20)=1.51, p=0.14; n=11 cre−, n=11 cre+ from n=5 mice). (G-I) Example traces (G), cumulative probability distribution of amplitude (H) and frequency (I) of miniature EPSC recordings (+TTX) indicate reduced quantal content in the absence of NMIIA and NMIIB(K-S test on amplitude Z=1.57, p=0.014; ; Bar graph inset for amplitude, Student's t-test t(24)=1.30, p=0.21, K-S test on frequency Z=0.78, p=0.58; Bar graph inset for frequency, Student's t-test t(24)=0.79, p=0.44; n=13 cre−, n=13 cre+ from n=7 mice). (J-L) Current injection spike frequency response curves indicate normal spiking responses in Myh10 ko neurons (J), slightly non-significant depolarized responses in Myh9 neurons (K) and altered slope of spiking responses in Myh10/Myh9 double ko neurons (L)(Myh10 RM ANOVA genotype F(1,37)=0,p=0.98, genotype*current F(6,222)=0.83,p=0.55, n=21 cre−, n=18 cre+ from n=7 mice; Myh9 RM ANOVA genotype F(1,25)=1.63,p=0.21, genotype*current F(6,150)=0.24,p=0.96 n=12 cre−, n=15 cre+ from n=4 mice, Myh10/Myh9 double ko RMANOVA genotype F(1,28)=0.24, p=0.63 genotype*current F(6,168)=3.67, p=0.002, n=15 cre−, n=15 cre+ from n=7 mice. Error bars represent SEM.
Analysis of miniature or spontaneous events was performed using Clampfit 10.4 software (Molecular devices). Template search method was used using a template match threshold of 2 and manually selecting responses. Experimenter was always blind to genotypes during this analysis. 50-100 responses were obtained from each recording. Events in each genotype were pooled in order to construct cumulative percentage plots for amplitude and frequency. Kolmogorov–Smirnov test was used for statistical analyses. NMDA current responses were fit with a single exponential using Clampfit in order to calculate tau. Due to the smaller and noisier nature of NMDA responses in distal dendrites, a meaningful fit (a positive tau) was obtained in a very few cases, and therefore tau for distal NMDA responses was not analyzed.
Sholl Analysis
For dendritic tracing experiments, we fixed acute slices (4% PFA in PBS overnight; mounted in Prolong Gold) derived from Myh10fl/fl (Fig. 2C-E) or Myh10+/+ (FIG. 2I-K) after the completion of physiology experiments. We exploited the extremely bright tdTomato signal emanating from sparsely expressing Cre+ CA1 neurons in both genotypes. Complete dendritic trees in both ko (Myh10fl/fl, Cre injected) and wt (Myh10 +/+,Cre injected) hippocampal pyramidal neurons (wt, no. slices = 7; ko, no. slices = 6) were selected and analyzed. x-y-z image stacks (0.497 μm/Pixel, 1.5 μm/z; x1 zoom; 1 AU) were acquired using a multiphoton laser-scanning microscope tuned to 940nm (Olympus FV1000MPE-TWIN), equipped with a water immersion objective lens (ULTRA 25x, numerical aperture 1.05, Olympus) and analyzed using FluoView software. Two neurons per slice were analyzed for a total of 14 wt and 12 ko neurons. Measurements of dendritic complexity contingent on dendritic distances from their somas were obtained using concentric-circle Sholl analysis performed with Neurolucida and NeuroExplorer (MicroBrightField, Inc.), a 3D visualization and morphometric analysis program designed for data analyses. Surface area (μm2), total length of dendritic branches (μm), and total intersections were used to represent the degree of complexity for both ko and wt pyramidal cells. Student's t-test was used to compare surface area, total number of intersections, and total length in wt vs ko neurons. Repeated measures analysis of variance (RMANOVA) was used to analyze intersections and lengths from the soma. Data collection and analysis were performed blind by different experimenters.
Dendritic Spine Density
Acute slices were prepared from Myh10fl/fl mice. Cre+ (td; ko) or Cre- (non-fluorescent; wt) neurons were patch-clamped and rapidly filled with 200μM Alexa-Fluor 488 hyrdazide (Invitrogen). Slices were then fixed (4% PFA in PBS overnight; mounted in Prolong Gold) and cleared by ScaleA2 for two weeks (Hama et al., 2011) to increase depth penetration. Because it was not possible to visualize distal spines using this methodology, we conducted an additional set of experiments where mice (again sparsely labeled for TdTomato expression) were transcardially perfused with 4% PFA, cryoprotected with 30% sucrose and then processed for ScaleA2 for 6 months for increased clearance. Neurons were imaged by standard confocal methods (Olympus FV1000MPE-TWIN), equipped with a water immersion objective lens (ULTRA 25x, numerical aperture 1.05, Olympus) and analyzed using FluoView software (x-y-z image stacks; 0.124 μm/Pixel, 1 μm/z; x2 zoom; 2 AU). Spine density and morphology were measured for fluorescent CA1 pyramidal neurons, tracing proximal and distal segments. At least 200 μm per branch category were collected and considered for analyses. During tracing, each individual spine was marked and measured for spine head diameter determination. Detection and quantification of dendritic spine density and morphology was performed using Neurolucida 3D reconstruction software (MicroBrightField, Inc.). Statistical analyses were performed with Student's t-test for spine density and Kolmogorof-Smirnoff for spine width and spine length distribution. All analyses were performed by an experimenter blind to the experimental conditions.
Primary cortical culture preparation
Cultured cortical cell experiments were performed based on prior studies with some modifications (Clement et al., 2012). Briefly, cortices were dissected from P0 pups and dissociated in papain-treated dissection media and further processed with plating media for addition to poly L lysine-treated 6-well plates. Cells were treated with FUdR DIV4 and then twice a week with feeding media + B27 until collected. Cells were collected in RIPA buffer (Cell Signaling Technology, Danvers, MA) containing Phosphatase Inhibitor Cocktails 2 and 3 (Sigma-Aldrich, St. Louis, MO) and mini-Complete Protease Inhibitor Cocktail (Roche Diagnostics) on DIV10, transferred to tubes in dry ice, and stored at −80 °C.
Western Blots
Hippocampi from mutant mice injected with a hSyn promoter-driven Cre construct in rAAV9 on PND1 were dissected at indicated time points (PND7 to PND35) in ice-cold PBS and immediately homogenized in RIPA buffer (Cell Signaling Technology, Danvers, MA) containing Phosphatase Inhibitor Cocktails 2 and 3 (Sigma-Aldrich, St. Louis, MO) and mini-Complete Protease Inhibitor Cocktail (Roche Diagnostics), transferred to tubes in dry ice, and stored at −80 °C. In a minority of experiments, CMV promoter-driven Cre rAAV9 was used, and these experiments are described in the results section.
Sample protein levels were measured (Pierce BCA Protein Assay Kit, Thermo Scientific, Rockford, IL), and volumes were adjusted to normalize microgram per microliter protein content. 10 ug of protein per sample were loaded and separated by SDS-PAGE on 4-15% Tris-glycine gels (Mini Protean TGX or Mini-Protean TGX Stain Free, BioRad, Hercules, CA) and transferred to PVDF membranes (88518, Thermo Scientific). Membranes were incubated with blocking buffer for 1hr (5% Blotting Grade Blocker in TBS, cat # 170-6404, Biorad, Hercules, CA) followed by overnight incubation with antibodies directed against rabbit polyclonal nonmuscle myosin heavy chain II-A (1:1,000 PRB-440P, Covance, Princeton, NJ), rabbit polyclonal nonmuscle myosin heavy chain II-B (1:25,000 PRB-445P, Covance, Princeton, NJ), rabbit polyclonal nonmuscle myosin heavy chain II-C (1:1,000 #3405, Cell Signaling, Danvers, MA), glial fibrillary acidic protein (1:500 ab10062, Abcam, Cambridge,MA), anti-β-Tubulin (1:10,000 clone AA2; 05-661; Millipore, Billerica, MA) or Histone 3 (1:1000 #3638, Cell Signaling, Danvers, MA). Membranes were then incubated for 1hr using appropriate species-specific secondary antibodies (W4011-rabbit; W4021-mouse; Promega, Madison, WI) followed by signal amplification and chemiluminescence detection (SuperSignal West Pico Chemiluminescent Substrate; Thermo Scientic, Rockford, IL). Blots were imaged using a Biorad Universal Hood III imager. Levels of immunoreactivity were assessed by densitometric analysis of the images using ImageJ (NIH, Bethesda, MD) as described previously (Rumbaugh and Miller, 2011). Statistical significance was determined by unpaired t test.
Total protein detection with Stain Free Gels
Stain-free gels (Mini-Protean TGX Stain Free, BioRad, Hercules, CA) were imaged with 2.5 minutes of UV exposure following electrophoresis using a Biorad Universal Hood III. After protein transfer, blots were subsequently imaged, and levels of total protein were assessed by densitometric analysis using ImageJ (NIH, Bethesda, MD) taking integrated density measurements of each lane. The densitometric values were used to normalize the protein of interest for each relevant blot. In a few experiments, normalization was performed with respect to Histone 3 or tubulin. In repeated experiments, the results obtained using these two normalization techniques were in general agreement.
Statistics
For extracellular recordings, the means of slopes in I/O curves were compared using student's unpaired t-test. Student's paired t-test was used to compare AMPA and NMDA response amplitude in pairs of neurons in the same slice. Student's unpaired t-test was used to compare surface area, total number of intersections, and total length in dendritic tracings. Repeated measures analysis of variance (RMANOVA) was used to analyze intersections and lengths of dendrites from the soma. Student's unpaired t-test was used to compare the means of spine density, spine width, spine length, mEPSC amplitude, sEPSC amplitude, mEPSC frequency, sEPSC frequency and normalized protein intensity. In comparing spine width, spine length, mEPSC amplitude, sEPSC amplitude, mEPSC frequency, sEPSC frequency we also constructed cumulative probability (or frequency) plots to show the whole data distribution and performed Kolmogorov–Smirnov (K-S) test on these distributions. However because of the highly powered nature of this test (Huang et al., 2007), we took a more conservative approach and inferred the differences to be biologically meaningful only if both student's t-test and K-S indicates statistical significance.
RESULTS
We were interested in understanding the role of non-muscle myosins during early postnatal development. All three isoforms, NMIIA, NMIIB and NMIIC, are expressed in hippocampus during this time period (Fig S1). NM IIB, which is encoded principally by Myh10, is the highest-expressed among the three isoforms, and it is found in dendritic spines (Cheng et al., 2006; Peng et al., 2004). Therefore, we initially focused our studies on the role of this gene in the development of hippocampal circuit function. To probe NM II function during development, we paired Myh10 conditional knockout mice (Myh10fl/fl) (Ma et al., 2009) with rAAV particles that drive Cre expression (Clement et al., 2012) (Fig 1A). We first determined the time course of NM IIB knockdown in hippocampal tissue (Fig. 1B). Maximal reduction in NM IIB protein levels could be obtained just 7 days after AAV-Cre injections, allowing us to examine the role Myh10 during early postnatal development. In addition, there was no gross compensation by other (off-target) non-muscle myosins at the protein level in WTs, although we cannot exclude more subtle forms of compensation, such as changes in sub-cellular localization or motor phosphorylation (Fig S2). Viral particles were injected into hippocampus at either PND1 or PND42 and field recordings from Str. radiatum were performed 3-4 weeks later. In all experiments, mice were also heterozygous for the Ai9 tdTomato reporter allele (Madisen et al., 2010), which reports Cre activity at the cellular level. We chose the PND1 injection as our early postnatal time point, because the period between PND1 and PND21 is that of extensive dendrite growth and spinogenesis (De Simoni et al., 2003). When Myh10 was disrupted at PND1, there was a prominent reduction in I/O curves of field EPSPs (Fig. 1C-E). This was most likely due to a postsynaptic effect, as there was no change in paired pulse ratio (Fig. 1F). In contrast, when the same procedure was carried out at PND42, no difference was observed in either field EPSP, I/O or the paired pulse ratio (Fig. 1G-J). Furthermore, the bath application of (-)-blebbistatin (50μM) in adult mouse hippocampal slices, which inhibits motor activity of all three non-muscle myosins, did not affect the magnitude of field EPSP or paired pulse ratio (Fig S3). These experiments suggested that Myh10 is necessary for the proper development of hippocampal synaptic function but is not required once functional circuits are formed and stabilized.
To better understand how NM IIB contributes to the maturation of hippocampal function, Myh10fl/fl mice were injected at PND1 with low titer AAV-Cre to induce sparse expression of Cre in the hippocampus. This method enabled whole-cell recordings of neighboring Cre-positive (ko/td+) and Cre-negative neurons (wt/td-)(Fig. 2A-B) (Clement et al., 2012; Lu et al., 2009; Marie et al., 2005). First, we evoked synaptic responses from the Schaefer collateral pathway, which are proximal inputs that relay information from CA3 to CA1. Myh10 ko neurons displayed a reduction in both AMPA and NMDA mediated components of excitatory synaptic transmission compared to neighboring wt neurons (Fig. 2C-E). We next probed distal synaptic inputs that receive information from areas outside CA3 (Fig. 2A). For these experiments, the stimulating electrode was placed in the distal molecular layer, which allowed stimulation of the temporoammonic (TA) pathway. Surprisingly, we did not observe any difference in AMPAR-mediated synaptic transmission between neighboring wt and ko neurons in this distal pathway (Fig. 2F-G). We did, however, observe an increase in NMDA-mediated synaptic transmission (Fig. 2H). In addition, there was a non-significant trend for faster decay in NMDA responses in proximal dendrites (wt tau=117.9±5.0 ms, ko tau=145.9±16.1 ms, paired t-test, t(12)=1.82, p=0.09), which could confound our method for determining the peak of NMDAR responses. Therefore, we next measured pharmacologically isolated NMDA responses in both proximal and distal dendrites. We did not observe significant differences in these measures (Fig S4). There was also no difference in decay time constant of isolated NMDA responses in proximal dendrites (wt tau=112.0 ms, ko tau=97.5 ms, paired t-test, t(12)=1.09, p=0.29). In order to formally test the differential effects of Myh10 deletion on proximal and distal pathways, we also ran a repeated measures ANOVA with pathway as a between-subjects variable, as well as genotype and types of responses (AMPA or NMDA) as a within subjects variable (from pairs of neurons in the same slice). This analyses revealed a significant genotype by types of responses interaction (F(1,29)=5.78, p=0.023) and a significant pathway by genotype by types of responses interaction (F(1,29)=5.03, p=0.033) confirming the differential effects of Myh10 deletion on proximal and distal pathways as well as on AMPA and NMDA responses. Importantly, there was no change in either AMPA- or NMDA-mediated excitatory synaptic transmission when comparing Cre+ and Cre- neurons in the Myh10+/+ background (Fig. 2I-K), indicating that both viral transduction and Cre transgene expression do not interfere with these measures of neuronal function. The combined synaptic physiology results in Myh10 mutants clearly demonstrate a pathway-specific disruption in postsynaptic responses in developing CA1 neurons, with proximal synaptic excitation principally disrupted (e.g. reduced) in NM IIB null neurons.
A decrease in excitatory synaptic currents can be explained by several possible cellular mechanisms. Because our methodology resulted in sporadic Myh10 disruption in the brain, the inputs onto recorded CA1 neurons were largely unimpaired. Thus, our findings most likely arise from a postsynaptic mechanism, such as a defect in dendritic structure, which would indirectly disrupt synaptogenesis, or from disruptions originating within synapses themselves, which would directly impair post-synaptic strength independent of dendritic structure. In order to distinguish between these possibilities, we first analyzed the structural complexity of apical dendrites using Sholl analysis (Fig. 3A-C). We focused on apical dendrites because these structures are the postsynaptic targets of the inputs evoked in Figure 2. Analysis of td+ CA1 neurons in Myh10fl/fl mice demonstrated a decrease in the total dendritic surface area compared to td+ neurons in Myh10wt/wt mice (Fig. 3D). We observed reductions in number of dendritic branch intersections (Fig. 3E) and total dendritic length (Fig. 3F). A closer analysis of intersections and dendritic length showed that reduced complexity of arborization was limited to proximal oblique segments (Fig. 3E, F), which receive Schaffer collateral inputs. Furthermore, these dendrites were shorter compared to similar dendrites in wt animals of the same developmental age (Fig. 3F). Interestingly, distal dendrites were normal both in terms of complexity and overall length (Fig. 3E, F), suggesting that the reduction in synaptic efficacy in the proximal pathway was caused, at least in part, by a reduction in synaptogenesis resulting from incomplete elongation of proximal oblique dendrites.
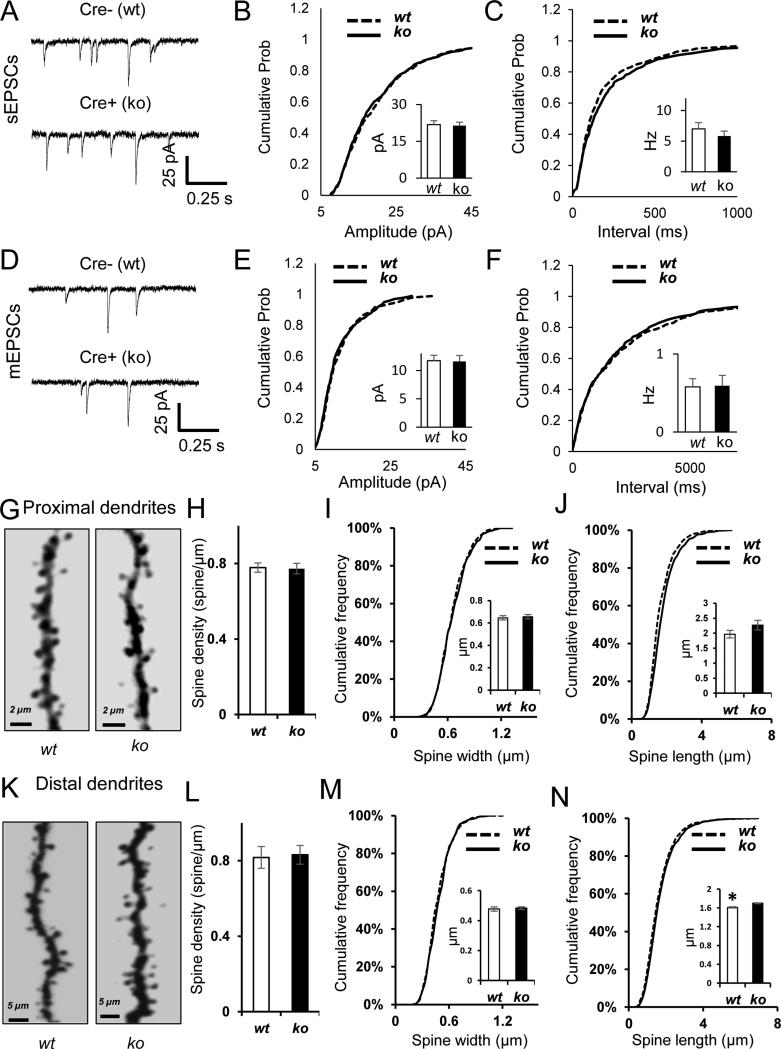
If the disruption to proximal postsynaptic excitatory function largely occurred from a reduction in synapses formed due to shorter dendrites, then excitatory synapses that did form on these neurons should be relatively unimpaired. To test this idea, we first recorded spontaneous excitatory synaptic events in both wt and Myh10 ko neurons. We did not observe an effect of genotype on sEPSC amplitude or sEPSC frequency (Fig. 4A-C). The small change in cumulative probability distributions of sEPSC frequency were not biologically meaningful as there was no statistically significant change in comparing sample means (see methods). To look more directly at individual events at postsynapses, we next recorded mEPSCs in wt and Myh10 ko neurons. In these studies, we did not observe an effect of genotype in either amplitude or frequency of mEPSCs (Fig. 4D-F). Dendritic spines mark the presence of excitatory synapses in forebrain pyramidal neurons. As a result, spine density is a proxy measure for the density of excitatory synapses on glutamatergic neurons. In general, dendritic spine morphology was relatively unaffected by Myh10 ablation. We did not observe a change in dendritic spine density in either proximal or distal Myh10 ko neurons (Fig. 4G-H, K-L), a finding consistent with the sEPSC and mEPSC studies. We did, however, observe a small change in spine length (Fig. 4J, N) in ko neurons compared to wts, though this defect would not be expected to significantly impair baseline synaptic function, a viewpoint supported by our mEPSC and sEPSC data. Together, these data indicate that Myh10 ablation in vivo has a minimal direct impact on postsynaptic function and spine morphogenesis, but instead reduces pathway-specific synaptic transmission by reducing dendritic complexity.
Figure 4. NM IIB ablation has no effect on spine density or postsynaptic strength.
(A-C) Example traces (A), cumulative probability distribution of amplitude (B) and frequency (C) of spontaneous EPSC recordings indicate slightly reduced frequency of sEPSCs in the absence of NMIIB. (K-S test on amplitude Z=0.95, p=0.33; Bar graph inset for amplitude, Student's t-test, t(33)=0.22, p=0.83; K-S test on frequency Z=1.58, p=0.013; n=18 cre−, n=17 cre+ from n=6 mice; Bar graph inset for frequency, Student's t-test t(33)=0.90, p=0.38) (D-F) Example traces (D), cumulative probability distribution of amplitude (E) and frequency (F) of miniature EPSC recordings indicate normal mEPSCs in the absence of NMIIB (K-S test on amplitude Z=0.940, p=0.34; Bar graph inset for amplitude, Student's t-test, t(26)=0.1, p=0.92 ; K-S test on frequency Z=0.664, p=0.77; n15 cre−, n=13 cre+ from n=7 mice; Bar graph inset for frequency. Student's t-test, t(26)=0.1, p=0.93). (G, K) Representative examples of proximal (G) and distal (K) dendrites from wt and ko CA1 neurons. (H, L) Bar graphs show normal spine density in proximal (H) and distal (L) dendrites of Myh10 ko CA1 neurons (Proximal branches, wt = 9 cells, 4 mice, ko = 10 cells, 4 mice. [Student t-Test, t(17)=0.1397 p=0.6087]; Distal branches wt = 10 cells, 2 mice, ko = 11 cells, 3 mice [Student t-Test, t(19)=0.1909, p=0.8506]) (I,M) Cumulative frequency curves show normal spine width in proximal (I) and in distal (M) dendrites of Myh10 ko neurons. (Proximal branches, wt = 1512 spines, 4 mice; ko = 1512 spines, 4 mice. [K-S test, p > 0.05]; Bar graph inset for spine width, Student t-test [t(6)=0.370, p=0.7237]; Distal branches wt = 2507 spines, 2 mice; ko = 2507 spines, 3 mice [K-S test, p < 0.005]; Bar graph inset for spine width, Student t-test [t(3)=0.389, p=0.7225]). (J, N) Cumulative frequency curves show normal spine length in proximal (J) and increased spine length in distal (N) dendrites of Myh10 ko neurons. (Proximal branches, wt = 1512 spines, 4 mice; ko = 1512 spines, 4 mice. [K-S test, p < 0.001]; Bar graphs inset for spine length, Student t-test [t(6)=1.469, p=0.192]) (Distal branches wt = 2507 spines, 2 mice; ko = 2507 spines, 3 mice [K-S test, p < 0.001]; Bar graph inset for spine length, Student t-test [t(3)=3.681, p=0.034]). Error bars represent SEM.
The relatively nuanced impact of in vivo Myh10 ablation in CA1 neurons was somewhat surprising considering past findings in cultured neurons demonstrating the necessity of this motor to maintain both spine morphology as well as synaptic strength (Rubio et al., 2011; Ryu et al., 2006). One possible explanation for the unique phenotype observed in Myh10 ko neurons in this study could stem from functional redundancy of distinct NM II motors expressed in the developing hippocampus. Indeed, NM IIA complexes are also expressed in developing neurons and these motors are localized within dendritic spines (Cheng et al., 2006; Peng et al., 2004). Furthermore, past studies have shown that certain functions of NM IIA and NM IIB are redundant (Bao et al., 2005; Rochlin et al., 1995). To address basic aspects of redundancy and compensation by NM II motors, we performed synaptic physiology experiments in Myh9 null neurons. Interestingly, we could not observe any knockdown in Myh9fl/fl brain slices infected with our standard Cre virus (hSyn promoter) (Fig 5A). This was not due to inefficiency of homologous recombination because robust knockdown could be obtained in liver tissue (Fig 5B). Relatively lower expression of Myh9 compared to Myh10 was previously observed in neurons (Rochlin et al., 1995). Interestingly, we also detected more Myh9 protein in glial enriched hippocampal cultures (Fig 5C). This finding prompted us to drive Cre expression in the developing hippocampus by a less cell-specific CMV promoter, which would be expected to lead to an increase in Cre expression in non-neuronal cells. Under these conditions, we could now detect robust Myh9 knockdown (Fig 5D), supporting the view that neurons express low levels of Myosin IIA in development. Importantly, this perturbation did not cause gross changes in expression of Myh10 or Myh14 (Fig S5A, B). As expected from relatively lower expression of Myh9 in neurons, we did not detect any significant disruptions to excitatory synaptic function or measures of intrinsic excitability in Myh9 ko neurons. Indeed, proximally-evoked excitatory synaptic function in Myh9 ko neurons was normal (Fig. 5E-G), indicating that the proximal pathway-specific damage observed in Myh10 ko neurons is a specific function of NM IIB in developing neurons. In addition, we did not observe any disruptions to spontaneous or miniature excitatory events (Fig. 5H-M) or firing properties of these neurons (Table 1).
Table 1.
Intrinsic neuronal properties of Myh10, Myh9 and Myh10/Myh9 double knockout neurons
| Myh10 control | Myh10 KO | P value | Myh9 control | Myh9 KO | P value | Double control | Double KO | P value | |
|---|---|---|---|---|---|---|---|---|---|
| n | 21 | 18 | 12 | 15 | 15 | 15 | |||
| Rinput(MΟ) | 71.1±2.7 | 70.0±3.3 | 0.81 | 84.2±6.2 | 94.2±4.8 | 0.2 | 94.1±3.4 | 91.1±4.1 | 0.57 |
| Cm(pF) | 370±12 | 366±11 | 0.81 | 355±37 | 299±23 | 0.19 | 385±22 | 340±19 | 0.13 |
| Vrest(mV) | −61.3±1.5 | −61.8±2.2 | 0.87 | −64.9±1.4 | −62.4±1.1 | 0.16 | −61.7±1.5 | −62.9±1.4 | 0.58 |
| Tau(ms) | 26.3±1.4 | 25.6±1.3 | 0.69 | 27.7±1.0 | 27.2±1.4 | 0.45 | 35.5±1.7 | 30.7±1.9 | 0.07 |
| AP height(mV) | 103.9±1.1 | 104.3±1.6 | 0.82 | 103.4±1.8 | 102.4±1.2 | 0.63 | 99.1±1.1 | 103.8±1.7 | 0.03 |
| AP width(ms) | 0.99±0.02 | 1.03±0.02 | 0.20 | 0.99±0.03 | 0.99±0.02 | 0.90 | 1.0±0.02 | 1.1±0.02 | 0.31 |
| AP threshold(mV) | 51.3±0.7 | 52.3±0.7 | 0.16 | −50.3±1.0 | −50.0±0.9 | 0.81 | −48.8±1.0 | −50.7±1.0 | 0.17 |
| Rhobase(pA) | 155±13 | 133±8 | 0.19 | 116±14 | 93±10 | 0.17 | 90±15 | 107±14 | 0.43 |
| Ih (mV) | 7.1±0.5 | 6.5±0.5 | 0.35 | 9.9±0.9 | 10.7±0.7 | 0.50 | 8.6±1.0 | 9.0±0.9 | 0.8 |
| F/I Slope | 0.16±0.01 | 0.19±0.01 | 0.26 | 0.20±0.01 | 0.20±0.01 | 0.58 | 0.13±0.01 | 0.17±0.02 | 0.04 |
To further address redundancy and compensation, we generated Myh9/Myh10 double flox mice and then performed electrophysiological studies in Cre-positive and neighboring Cre-negative neurons. As in single knockouts, we did not observe any compensation by Myh14 in double KO tissue (Fig S5C). For these experiments, we evoked proximal and distal synaptic responses from the same neurons to gain a better understanding of the pathway-specific effect of double KO on synaptic transmission. First, we reproduced the pathway-specific defects first observed in Myh10 null neurons (Fig. 6A-B). Indeed, there was a selective reduction in AMPAR-mediated currents in proximal inputs onto Myh9/Myh10 double ko neurons (Fig. 6B), though distal evoked currents were unimpaired in these same neurons (Fig. 6C). There was also a significant difference in the ratio of proximal EPSCs to distal EPSCs in pairs of wt and ko neurons, highlighting the pathway-specific effect of double knockout on synaptic transmission, an effect likely due to Myh10 deletion alone (proximal to distal ratio 18.4±3.8 wt, 12.4±2.6 ko; paired t(13)=2.34, p=0.034)(Fig S6). sEPSC amplitude and frequency , mEPSC amplitude and frequency were also normal in double ko neurons (Fig 6D-I). We also measured the intrinsic excitability in each of the single KO neurons and double KO neurons (Fig. 6J-L; Table 1), and these measures were grossly normal. These data demonstrate that disrupting both NM IIA and NM IIB in developing hippocampal neurons causes a phenotype largely similar to that of the Myh10 single ko.
DISCUSSION
There were three major conclusions from our work. First, Myh10, but not Myh9, regulates the development of hippocampal circuits. Second, Myh10 influences dendrite development selectively in the Schaeffer collateral pathway. Finally, Myh10 regulation of baseline synaptic responses is restricted to the early postnatal period.
NM IIB is the prominent isoform regulating the development of hippocampal circuits
Our studies uncovered unique functions of NM II motors that differentiate the roles of Myh9 from Myh10 in developing hippocampal pyramidal neurons. In general, Myh10 and Myh9 genes have non-overlapping patterns of expression in different tissues at different times during development, and they serve separate functions in growth cones (Wylie and Chantler, 2003), in cytokinesis (Bao et al., 2005) and during oocyte maturation (Simerly et al., 1998). Here, we found that Myh10, but not Myh9, ablation disrupted evoked-excitatory responses specifically in the Schaffer collateral pathway. Our studies indicated that Myosin IIB was more enriched in developing neurons relative to Myosin IIA, which could explain the distinctions between the Myh10 and Myh9 cellular phenotypes, respectively. The cellular mechanism driving this pathway-specific disruption in Myh10 null neurons was most likely through the stunted maturation of proximal oblique dendrites that receive excitatory synaptic input from CA3. The role of NM II on growth cone dynamics and neuronal polarity has been extensively studied (Bridgman et al., 2001; Medeiros et al., 2006; Wylie and Chantler, 2003, 2008). Indeed, both NM IIA and NM IIB can regulate neurite extension in very immature cells (Matsumoto et al., 2014). However, there are no studies that have investigated the impact of NM II on post-mitotic neuronal differentiation and maturation in vivo. Our results indicated that Myh10 is involved in the mechanisms that selectively promote the elongation of particular types of dendritic branches in developing hippocampal neurons. Several potential mechanisms could account for this regulation. NM IIB can function as a motor protein, but also as a structural component in cell adhesion complexes. Interestingly, NM IIA can replace certain structural, but not motor, functions of NMIIB (Bao et al., 2007). We observed pathway-specific synaptic disruptions only when NM IIB was ablated, suggesting that unique motor functions of NM IIB are responsible for guiding dendrite morphogenesis during early brain development.
NM IIB regulates elongation of a particular class of CA1 dendrites
Although the role of Myh10 in regulating growth cone dynamics and neuronal polarity has been characterized (Bridgman et al., 2001; Medeiros et al., 2006; Wylie and Chantler, 2003, 2008), we have shown for the first time that Myh10 guides elongation of dendrites in the developing brain. It is important to note that Myh10 knockout affected only proximal oblique branches of CA1 neurons, and did not alter the length or complexity of distal tufts. Consistent with these data, Myh10 cell-autonomous ablation weakened synaptic transmission only at Schaffer collateral inputs. The overall negative findings with respect to the spontaneous synaptic and spine morphology analysis (e.g. figures 4-6) also helped us to conclude that Myosin IIB regulates pathway-specific hippocampal maturation by promoting the elongation of dendrites in developing CA1 neurons. Various mechanisms could account for a pathway-specific effect in response to Myh10 ablation. For example, Myh10 or its upstream effectors, may be preferentially localized to proximal dendrites. There are other genetic manipulations known to preferentially affect proximal oblique dendrites in CA1 region of the hippocampus. Both Euchromatin histone methyl transferase 1 (Ehmt1) heterozygous mice (Balemans et al., 2013) and Serotonin receptor 1A (Htr1a) knockout mice (Ferreira et al., 2010) have impaired proximal oblique dendrites with normal distal branches. In addition, neurotrophin signaling exerts opposite effects on proximal versus distal dendrites (Horch et al., 1999; Yacoubian and Lo, 2000). It will be interesting to determine if there are interactions among these molecules and NM IIB to enable selective dendritic elongation.
Interestingly, reduced dendritic branching has been observed in individuals diagnosed with ASD (Raymond et al., 1996). FMR1 regulates early development of dendrites (Cruz-Martin et al., 2012). Dentate gyrus granule neurons have less dendritic surface volume in Syngap1 Heterozygous KO mice (Clement et al., 2012), which model a human syndrome characterized by severe non-syndromic intellectual disability with high comorbidities of ASD and epilepsy. Hepatocyte growth factor (HGF) and its receptor tyrosine kinase MET regulate dendritic morphology of cortical pyramidal neurons (Gutierrez et al., 2004) and mutations in MET gene have been associated with ASD. Mutations in CACNA1C cause Timothy syndrome, an ASD, and neurons heterozygous for mutant Cacna1c show reduced dendritic complexity through abnormal Rho activation (Krey et al., 2013). Given that mutations in MYH10 gene have also been linked to ASD, and we show a specific effect of myh10 disruptions on dendritic branching without spine abnormalities, abnormal dendritic morphology of neurons may be an underappreciated pathway contributing to complex brain disorders (Kulkarni and Firestein, 2012).
NM IIB has distinct roles on synaptic function in young versus mature hippocampal neurons
Another important conclusion from our work was the developmental regulation of baseline responses by Myosin IIB. The lack of effect in postsynaptic baseline synaptic transmission in adult neurons agrees with other studies in acute slice preparations using RNAi or blebbistatin (Peng et al., 2012; Rex et al., 2010), but differs from those in cultured hippocampal neurons (Rubio et al., 2011; Ryu et al., 2006). It has been suggested that one possible reason for these differences is the concentration of blebbistatin used (Kneussel and Wagner, 2013). However, the results of our current study using 50 μM blebbistatin and those of others using 100 μM blebbistatin (Peng et al., 2012) argue against this possibility. In this study, we also utilized a conditional knockout approach to induce sparse ablation of the canonical neuron NM isoform, IIB. We propose that the most likely reason for discrepant results on synaptic function is a different state of the spine F-actin cytoskeleton in young (i.e. cultures) vs. mature neurons (i.e. acute hippocampal slices). It has been shown that actin filaments are present in three different states in spines [i.e. monomer, treadmilling, and stable actin (Star et al., 2002)], and the ratio of filamentous actin to these other forms is higher in larger spines (Frost et al., 2010; Honkura et al., 2008). Recycling pools of other proteins such as PSD-95 also decrease as the animal ages (Gray et al., 2006). Therefore, it is possible that spine F-actin dynamics in cultured neurons are in a state that requires constant molding by NM IIB motor activity (Haviv et al., 2008). In addition, there have been few studies that have probed spine actin dynamics in intact preparations. Svoboda and colleagues assessed F-actin turnover rates in vivo from superficial dendritic spines in cortex, though these measurements were performed in animals younger than PND21 (Grey et al., 2006). It is possible that the overall state of the spine F-actin cytoskeleton changes as dendritic spines, and neuronal networks that they subserve, mature. Further studies, carried out in vivo, are needed in order to understand how development alters the functional interactions between NM IIB and spine F-actin dynamics, and how these alterations impact spine structure and function.
Supplementary Material
Acknowledgements
This work was supported by grants to GR from The National Institute for Neurological Disorders and Stroke (R01NS064079) and The National Institute for Mental Health (R01MH096847). CAM was supported by grants from the National Institute for Drug Abuse (R01 DA034116; R03 DA033499). The authors have no conflicts of interest to declare.
The list of abbreviations
- AMPA
alpha-amino-3-hydroxy-5-methylisoxazole-4-propionate
- CMV
Cytomegalovirus
- fEPSP
field excitatory postsynaptic potential
- hSyn
human synapsin promoter
- mEPSC
miniature excitatory postsynaptic currents
- NMDA
N-Methyl-D-aspartate
- NMIIA
non-muscle myosin IIA
- NMIIB
non-muscle myosin IIB
- PPR
paired pulse ratio
- PND
postnatal day
- rAAV
recombinant adeno-associated virus
- RMANOVA
repeated measures ANOVA
- sEPSC
spontaneous excitatory postsynaptic currents
- STV
the superficial temporal vein
- td
tdTomato protein
REFERENCES
- Balemans MC, Kasri NN, Kopanitsa MV, Afinowi NO, Ramakers G, Peters TA, Beynon AJ, Janssen SM, van Summeren RC, Eeftens JM, et al. Hippocampal dysfunction in the Euchromatin histone methyltransferase 1 heterozygous knockout mouse model for Kleefstra syndrome. Human molecular genetics. 2013;22:852–866. doi: 10.1093/hmg/dds490. [DOI] [PubMed] [Google Scholar]
- Bao J, Jana SS, Adelstein RS. Vertebrate nonmuscle myosin II isoforms rescue small interfering RNA-induced defects in COS-7 cell cytokinesis. The Journal of biological chemistry. 2005;280:19594–19599. doi: 10.1074/jbc.M501573200. [DOI] [PubMed] [Google Scholar]
- Bao J, Ma X, Liu C, Adelstein RS. Replacement of nonmuscle myosin II-B with II-A rescues brain but not cardiac defects in mice. The Journal of biological chemistry. 2007;282:22102–22111. doi: 10.1074/jbc.M702731200. [DOI] [PubMed] [Google Scholar]
- Bridgman PC, Dave S, Asnes CF, Tullio AN, Adelstein RS. Myosin IIB is required for growth cone motility. J Neurosci. 2001;21:6159–6169. doi: 10.1523/JNEUROSCI.21-16-06159.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D, Hoogenraad CC, Rush J, Ramm E, Schlager MA, Duong DM, Xu P, Wijayawardana SR, Hanfelt J, Nakagawa T, et al. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Molecular & cellular proteomics : MCP. 2006;5:1158–1170. doi: 10.1074/mcp.D500009-MCP200. [DOI] [PubMed] [Google Scholar]
- Cingolani LA, Goda Y. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nature reviews Neuroscience. 2008;9:344–356. doi: 10.1038/nrn2373. [DOI] [PubMed] [Google Scholar]
- Clement JP, Aceti M, Creson TK, Ozkan ED, Shi Y, Reish NJ, Almonte AG, Miller BH, Wiltgen BJ, Miller CA, et al. Pathogenic SYNGAP1 mutations impair cognitive development by disrupting maturation of dendritic spine synapses. Cell. 2012;151:709–723. doi: 10.1016/j.cell.2012.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Martin A, Crespo M, Portera-Cailliau C. Glutamate induces the elongation of early dendritic protrusions via mGluRs in wild type mice, but not in fragile X mice. PloS one. 2012;7:e32446. doi: 10.1371/journal.pone.0032446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simoni A, Griesinger CB, Edwards FA. Development of rat CA1 neurones in acute versus organotypic slices: role of experience in synaptic morphology and activity. J Physiol. 2003;550:135–147. doi: 10.1113/jphysiol.2003.039099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira TA, Iacono LL, Gross CT. Serotonin receptor 1A modulates actin dynamics and restricts dendritic growth in hippocampal neurons. The European journal of neuroscience. 2010;32:18–26. doi: 10.1111/j.1460-9568.2010.07283.x. [DOI] [PubMed] [Google Scholar]
- Foust KD, Nurre E, Montgomery CL, Hernandez A, Chan CM, Kaspar BK. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nature biotechnology. 2009;27:59–65. doi: 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost NA, Shroff H, Kong H, Betzig E, Blanpied TA. Single-molecule discrimination of discrete perisynaptic and distributed sites of actin filament assembly within dendritic spines. Neuron. 2010;67:86–99. doi: 10.1016/j.neuron.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin CF, Rubio MD, Young E, Miller C, Rumbaugh G. Myosin II motor activity in the lateral amygdala is required for fear memory consolidation. Learning & memory. 2012;19:9–14. doi: 10.1101/lm.024042.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray NW, Weimer RM, Bureau I, Svoboda K. Rapid redistribution of synaptic PSD-95 in the neocortex in vivo. PLoS biology. 2006;4:e370. doi: 10.1371/journal.pbio.0040370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez H, Dolcet X, Tolcos M, Davies A. HGF regulates the development of cortical pyramidal dendrites. Development. 2004;131:3717–3726. doi: 10.1242/dev.01209. [DOI] [PubMed] [Google Scholar]
- Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- Haviv L, Gillo D, Backouche F, Bernheim-Groswasser A. A cytoskeletal demolition worker: myosin II acts as an actin depolymerization agent. Journal of molecular biology. 2008;375:325–330. doi: 10.1016/j.jmb.2007.09.066. [DOI] [PubMed] [Google Scholar]
- Hodges JL, Newell-Litwa K, Asmussen H, Vicente-Manzanares M, Horwitz AR. Myosin IIb activity and phosphorylation status determines dendritic spine and post-synaptic density morphology. PloS one. 2011;6:e24149. doi: 10.1371/journal.pone.0024149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkura N, Matsuzaki M, Noguchi J, Ellis-Davies GC, Kasai H. The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron. 2008;57:719–729. doi: 10.1016/j.neuron.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Horch HW, Kruttgen A, Portbury SD, Katz LC. Destabilization of cortical dendrites and spines by BDNF. Neuron. 1999;23:353–364. doi: 10.1016/s0896-6273(00)80785-0. [DOI] [PubMed] [Google Scholar]
- Hotulainen P, Hoogenraad CC. Actin in dendritic spines: connecting dynamics to function. The Journal of cell biology. 2010;189:619–629. doi: 10.1083/jcb.201003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Yeo AA, Li SD. Modification of Kolmogorov-Smirnov test for DNA content data analysis through distribution alignment. Assay and drug development technologies. 2007;5:663–671. doi: 10.1089/adt.2007.071. [DOI] [PubMed] [Google Scholar]
- Inagaki K, Fuess S, Storm TA, Gibson GA, McTiernan CF, Kay MA, Nakai H. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Molecular therapy : the journal of the American Society of Gene Therapy. 2006;14:45–53. doi: 10.1016/j.ymthe.2006.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobelli J, Friedman RS, Conti MA, Lennon-Dumenil AM, Piel M, Sorensen CM, Adelstein RS, Krummel MF. Confinement-optimized three-dimensional T cell amoeboid motility is modulated via myosin IIA-regulated adhesions. Nature immunology. 2010;11:953–961. doi: 10.1038/ni.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan BA, Fernholz BD, Boussac M, Xu C, Grigorean G, Ziff EB, Neubert TA. Identification and verification of novel rodent postsynaptic density proteins. Molecular & cellular proteomics : MCP. 2004;3:857–871. doi: 10.1074/mcp.M400045-MCP200. [DOI] [PubMed] [Google Scholar]
- Kamenetz F, Tomita T, Hsieh H, Seabrook G, Borchelt D, Iwatsubo T, Sisodia S, Malinow R. APP processing and synaptic function. Neuron. 2003;37:925–937. doi: 10.1016/s0896-6273(03)00124-7. [DOI] [PubMed] [Google Scholar]
- Kawamoto S, Adelstein RS. Chicken nonmuscle myosin heavy chains: differential expression of two mRNAs and evidence for two different polypeptides. The Journal of cell biology. 1991;112:915–924. doi: 10.1083/jcb.112.5.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienstra KA, Freysdottir D, Gonzales NM, Hirschi KK. Murine neonatal intravascular injections: modeling newborn disease. Journal of the American Association for Laboratory Animal Science : JAALAS. 2007;46:50–54. [PubMed] [Google Scholar]
- Kneussel M, Wagner W. Myosin motors at neuronal synapses: drivers of membrane transport and actin dynamics. Nature reviews Neuroscience. 2013;14:233–247. doi: 10.1038/nrn3445. [DOI] [PubMed] [Google Scholar]
- Korobova F, Svitkina T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Molecular biology of the cell. 2010;21:165–176. doi: 10.1091/mbc.E09-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krey JF, Pasca SP, Shcheglovitov A, Yazawa M, Schwemberger R, Rasmusson R, Dolmetsch RE. Timothy syndrome is associated with activity-dependent dendritic retraction in rodent and human neurons. Nat Neurosci. 2013;16:201–209. doi: 10.1038/nn.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni VA, Firestein BL. The dendritic tree and brain disorders. Molecular and cellular neurosciences. 2012;50:10–20. doi: 10.1016/j.mcn.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Lu W, Shi Y, Jackson AC, Bjorgan K, During MJ, Sprengel R, Seeburg PH, Nicoll RA. Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron. 2009;62:254–268. doi: 10.1016/j.neuron.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L. Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annual review of cell and developmental biology. 2002;18:601–635. doi: 10.1146/annurev.cellbio.18.031802.150501. [DOI] [PubMed] [Google Scholar]
- Ma X, Takeda K, Singh A, Yu ZX, Zerfas P, Blount A, Liu C, Towbin JA, Schneider MD, Adelstein RS, et al. Conditional ablation of nonmuscle myosin II-B delineates heart defects in adult mice. Circ Res. 2009;105:1102–1109. doi: 10.1161/CIRCRESAHA.109.200303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie H, Morishita W, Yu X, Calakos N, Malenka RC. Generation of silent synapses by acute in vivo expression of CaMKIV and CREB. Neuron. 2005;45:741–752. doi: 10.1016/j.neuron.2005.01.039. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Inden M, Tamura A, Hatano R, Tsukita S, Asano S. Ezrin Mediates Neuritogenesis via Down-Regulation of RhoA Activity in Cultured Cortical Neurons. PloS one. 2014;9:e105435. doi: 10.1371/journal.pone.0105435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus A. Actin-based plasticity in dendritic spines. Science. 2000;290:754–758. doi: 10.1126/science.290.5492.754. [DOI] [PubMed] [Google Scholar]
- Medeiros NA, Burnette DT, Forscher P. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nature cell biology. 2006;8:215–226. doi: 10.1038/ncb1367. [DOI] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan ED, Creson TK, Kramar EA, Rojas C, Seese RR, Babyan AH, Shi Y, Lucero R, Xu X, Noebels JL, et al. Reduced Cognition in Syngap1 Mutants Is Caused by Isolated Damage within Developing Forebrain Excitatory Neurons. Neuron. 2014;82:1317–1333. doi: 10.1016/j.neuron.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacak CA, Sakai Y, Thattaliyath BD, Mah CS, Byrne BJ. Tissue specific promoters improve specificity of AAV9 mediated transgene expression following intra-vascular gene delivery in neonatal mice. Genetic vaccines and therapy. 2008;6:13. doi: 10.1186/1479-0556-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng A, Rotman Z, Deng PY, Klyachko VA. Differential motion dynamics of synaptic vesicles undergoing spontaneous and activity-evoked endocytosis. Neuron. 2012;73:1108–1115. doi: 10.1016/j.neuron.2012.01.023. [DOI] [PubMed] [Google Scholar]
- Peng J, Kim MJ, Cheng D, Duong DM, Gygi SP, Sheng M. Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. The Journal of biological chemistry. 2004;279:21003–21011. doi: 10.1074/jbc.M400103200. [DOI] [PubMed] [Google Scholar]
- Pilpel N, Landeck N, Klugmann M, Seeburg PH, Schwarz MK. Rapid, reproducible transduction of select forebrain regions by targeted recombinant virus injection into the neonatal mouse brain. J Neurosci Methods. 2009;182:55–63. doi: 10.1016/j.jneumeth.2009.05.020. [DOI] [PubMed] [Google Scholar]
- Prasad A, Merico D, Thiruvahindrapuram B, Wei J, Lionel AC, Sato D, Rickaby J, Lu C, Szatmari P, Roberts W, et al. A discovery resource of rare copy number variations in individuals with autism spectrum disorder. G3. 2012;2:1665–1685. doi: 10.1534/g3.112.004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racz B, Weinberg RJ. Microdomains in forebrain spines: an ultrastructural perspective. Molecular neurobiology. 2013;47:77–89. doi: 10.1007/s12035-012-8345-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond GV, Bauman ML, Kemper TL. Hippocampus in autism: a Golgi analysis. Acta neuropathologica. 1996;91:117–119. doi: 10.1007/s004010050401. [DOI] [PubMed] [Google Scholar]
- Rex CS, Gavin CF, Rubio MD, Kramar EA, Chen LY, Jia Y, Huganir RL, Muzyczka N, Gall CM, Miller CA, et al. Myosin IIb regulates actin dynamics during synaptic plasticity and memory formation. Neuron. 2010;67:603–617. doi: 10.1016/j.neuron.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochlin MW, Itoh K, Adelstein RS, Bridgman PC. Localization of myosin II A and B isoforms in cultured neurons. Journal of cell science. 1995;108(Pt 12):3661–3670. doi: 10.1242/jcs.108.12.3661. [DOI] [PubMed] [Google Scholar]
- Rubio MD, Johnson R, Miller CA, Huganir RL, Rumbaugh G. Regulation of synapse structure and function by distinct myosin II motors. J Neurosci. 2011;31:1448–1460. doi: 10.1523/JNEUROSCI.3294-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh G, Miller CA. Epigenetic changes in the brain: measuring global histone modifications. Methods in molecular biology. 2011;670:263–274. doi: 10.1007/978-1-60761-744-0_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J, Liu L, Wong TP, Wu DC, Burette A, Weinberg R, Wang YT, Sheng M. A critical role for myosin IIb in dendritic spine morphology and synaptic function. Neuron. 2006;49:175–182. doi: 10.1016/j.neuron.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Simerly C, Nowak G, de Lanerolle P, Schatten G. Differential expression and functions of cortical myosin IIA and IIB isotypes during meiotic maturation, fertilization, and mitosis in mouse oocytes and embryos. Molecular biology of the cell. 1998;9:2509–2525. doi: 10.1091/mbc.9.9.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star EN, Kwiatkowski DJ, Murthy VN. Rapid turnover of actin in dendritic spines and its regulation by activity. Nat Neurosci. 2002;5:239–246. doi: 10.1038/nn811. [DOI] [PubMed] [Google Scholar]
- Wylie SR, Chantler PD. Myosin IIA drives neurite retraction. Molecular biology of the cell. 2003;14:4654–4666. doi: 10.1091/mbc.E03-03-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie SR, Chantler PD. Myosin IIC: a third molecular motor driving neuronal dynamics. Molecular biology of the cell. 2008;19:3956–3968. doi: 10.1091/mbc.E07-08-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yacoubian TA, Lo DC. Truncated and full-length TrkB receptors regulate distinct modes of dendritic growth. Nat Neurosci. 2000;3:342–349. doi: 10.1038/73911. [DOI] [PubMed] [Google Scholar]
- Zaman SH, Parent A, Laskey A, Lee MK, Borchelt DR, Sisodia SS, Malinow R. Enhanced synaptic potentiation in transgenic mice expressing presenilin 1 familial Alzheimer's disease mutation is normalized with a benzodiazepine. Neurobiology of disease. 2000;7:54–63. doi: 10.1006/nbdi.1999.0271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.