Abstract
Only a minority of those exposed to human papillomavirus (HPV) develop HPV-related cervical and oropharyngeal cancer. Because host immunity affects infection and progression to cancer, we tested the hypothesis that genetic variation in immune-related genes is a determinant of susceptibility to oropharyngeal cancer and other HPV-associated cancers by performing a multitier integrative computational analysis with oropharyngeal cancer data from a head and neck cancer genome-wide association study (GWAS). Independent analyses, including single-gene, gene-interconnectivity, protein–protein interaction, gene expression, and pathway analysis, identified immune genes and pathways significantly associated with oropharyngeal cancer. TGFβR1, which intersected all tiers of analysis and thus selected for validation, replicated significantly in the head and neck cancer GWAS limited to HPV-seropositive cases and an independent cervical cancer GWAS. The TGFβR1 containing p38–MAPK pathway was significantly associated with oropharyngeal cancer and cervical cancer, and TGFβR1 was overexpressed in oropharyngeal cancer, cervical cancer, and HPV+ head and neck cancer tumors. These concordant analyses implicate TGFβR1 signaling as a process dysregulated across HPV-related cancers. This study demonstrates that genetic variation in immune-related genes is associated with susceptibility to oropharyngeal cancer and implicates TGFβR1/TGFβ signaling in the development of both oropharyngeal cancer and cervical cancer. Better understanding of the immunogenetic basis of susceptibility to HPV-associated cancers may provide insight into host/virus interactions and immune processes dysregulated in the minority of HPV-exposed individuals who progress to cancer.
Introduction
Human papillomavirus (HPV) is a necessary cause of cervical cancer, and a major cause of anal, vulvar, and penile cancer as well as oropharyngeal cancer (1–3). Although tobacco-associated squamous cell carcinomas of the head and neck (HNSCC) have declined in the United States and other Western countries, the incidence of HPV-positive oropharyngeal cancer has sharply increased in the United States, rising by 225% since 1988, with more than 70% of all newly diagnosed oropharyngeal cancer believed to be HPV positive (4, 5). The National Cancer Institute predicts that HPV-positive oropharyngeal cancer will likely surpass cervical cancer as the most common HPV-associated cancer in the United States by 2020 (5).
Although HPV infection is common (6, 7), only a small fraction of infected individuals develops cancer (8). The pattern of increasing relative risk with increasing degree of relatedness found in cervical cancer suggests that susceptibility to HPV-induced cancer is modulated by genetic factors (9). At the same time, the viral etiology of HPV-induced cancers implicates host immunity as a potential susceptibility factor (10), and the higher prevalence of cervical HPV infections, HPV-positive cervical cancer (11), and HPV-positive oropharyngeal cancer (12) in HIV/AIDS individuals suggests that the host immune response is an important determinant of HPV-induced cancer risk. Thus, we hypothesize that genetic variation in immune-related genes is a determinant of susceptibility to oropharyngeal cancer and other HPV-associated cancers.
In the present study, we investigate the immunogenetics of susceptibility to HPV-associated head and neck cancer using a genome-wide association study (GWAS) of upper aerodigestive (UADT) cancers that was organized by the International Agency for Research on Cancer (IARC) and the Centre National de Genotypage (CNG) in Paris and whose replication was conducted through the International Head and Neck Cancer Epidemiology (INHANCE) consortium (13). Although the conventional unbiased GWAS approach can identify genetic variants associated with complex diseases (14), (15), many risk-modifying alleles are missed by this strategy due to the very stringent significance criteria required to minimize false discovery (16, 17). Alternate analytic approaches focused on combined effects of many loci, each contributing a small effect to the overall disease susceptibility, can successfully identify signals that individually do not meet the threshold for genome-wide significance but are in fact associated with the disease (18, 19). To test the hypothesis that variants in immune-related genes modulate susceptibility to oropharyngeal cancer, an HPV-associated head and neck cancer, we developed a multitier integrative computational approach incorporating four distinct modes of analysis, including individual single-nucleotide polymorphism (SNP)/gene, pathway, gene–gene interconnectivity (GGI), and protein–protein interaction (PPI) analyses. Resulting hits that were supported across all four dimensions of analysis were subsequently validated in replication cohorts and gene expression studies (Fig. 1). Our results are consistent with an important role for genetic variation in multiple immune-related genes as modifiers of HPV-related cancer risk, particularly those related to transforming growth factor beta (TGFβ) signaling.
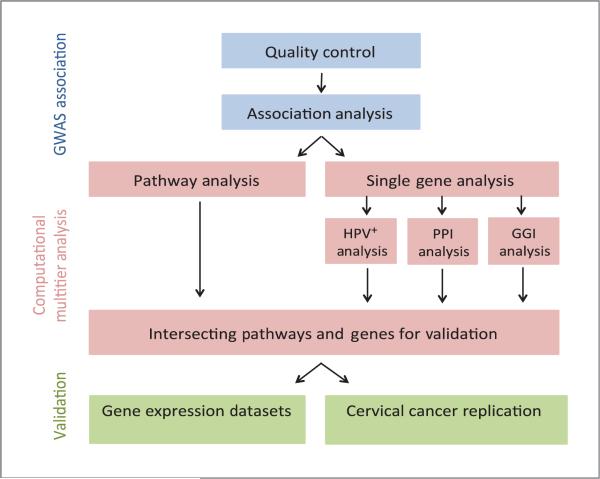
Figure 1.
Work flow for the multitier computational project. QC and association analysis was performed on the GWAS raw data, which was then subjected to several independent modes of analysis. Validation was performed on genes and pathways found to be significant in all modes of analysis.
Materials and Methods
IARC head and neck cancer GWAS and quality control of GWAS data
IARC conducted a large two-phased pooled case/control GWAS including 2,091 UADT cancer patients (primarily head and neck cancer with esophageal cancers included) and 8,334 cancer-free controls genotyped using the Illumina Sentrix HumanHap300 BeadChip. We requested the raw GWAS data from IARC, and subjected these data to additional cleaning and quality control (QC) before analysis. Details of the UADT GWAS including the study population and initial QC are reported in detail elsewhere (13). In brief, for this study, systematic QC was performed on the raw Illumina HumanHap300 genotyping data. We excluded samples with gender discrepancies, low call rates (<95%), and outliers in population structure from principle component analyses. Variants with a genotype call rate <95%, minor allele frequency (MAF) <5%, and deviation from Hardy–Weinberg equilibrium (P < 10−7) were removed. The extremely limited extent of admixture and population stratification was confirmed using multidimensional scaling analysis. The association between each genetic variant and the disease risk was estimated by the odds ratio (OR) per allele and 95% confidence interval (CI) using multivariate unconditional logistic regression assuming a log-additive genetic model with sex and country of recruitment included as covariates. All analyses were performed using PLINK (20). We conducted separate analyses on data restricted to oropharyngeal cancer and laryngeal cancer subsites only.
Immune-related gene ranking and significance threshold
SNP identifiers were mapped to associated genes using the SNP location from the human genome assembly hg18. Gene boundary extensions, extending the transcription start and stop codon by 110 kb and 40 kb, respectively, did not significantly change the results, so they were not used in the final gene ranking to minimize error. The intersection between genes annotated from the set of SNPs and the 1324 genes defined by Gene Ontology (GO) network classification (21) as immune-related genes was found. Ranking was based on the most significant P value for each SNP. Because immune-related genes are not independent of one another and operate in common pathways and networks, in our gene-based analysis we used a false discovery rate (FDR) <0.05 for a significance threshold.
In silico replication of oropharyngeal cancer GWAS results in serologically HPV-positive head and neck cancer patients
A Luminex-based multiplex immunosorbent method was used to determine each subject's antibody response to HPV infection by measuring serologic reactivity to the HPV16 E6 protein, as previously described (22). The cutoff was defined as 5 standard deviations above the mean of the final distribution. The serology data were then used to perform a validation analysis on 131 serologically HPV-positive head and neck cancer patients and 2,919 HPV-negative controls using the same methodologies applied in the oropharyngeal cancer GWAS (described above). A significance cutoff of P < 0.05 was used in the validation.
GGI analysis
The highly ranked immune-related genes, corresponding to SNPs with P values suggestive of association (1 × 10−3 > P> 1 × 10−7) were analyzed in Gene Relationships Across Implicated Loci, GRAIL (23), a literature-based text-mining program that infers biologic interconnectivity between and among genes. In summary, genes were identified using the linkage disequilibrium (LD) structure of HapMap2 Release 22 CEU and text mining was performed using a text-based similarity measure that scores two genes for relatedness to each other based on text similarity in PubMed abstracts last curated in 2012. Genes in regions of interest were clustered on the basis of keyword similarity. These clusters were then scored on the basis of ranked similarity, adjusting for gene size, to generate P values evaluating the strength of the functional interconnectivity of genes in the regions of interest. A significant P value for a gene region indicates that a gene within it is more related to genes in other disease regions through PubMed abstracts than expected by chance. P values for these functional clusters were FDR adjusted to correct for multiple testing with the FDR < 0.05 considered the threshold of significance.
PPI analysis
PPI analysis was completed using Disease Association Protein-Protein Link Evaluator, DAPPLE. The details of the algorithm and methods used in DAPPLE can be found in Rossin and colleagues (24). Briefly, the top 200 ranked immune-related SNPs (maximum number allowed by the program) were input into DAPPLE as seed SNPs and converted into genes. DAPPLE then built interaction networks from proteins encoded by the seed genes that have direct connections reported in the literature. The statistical significance was assessed by a number of network connectivity parameters as well as of the connectivity of the individual proteins to other seed proteins using a within-degree node-label permutation method. The individual P values for seed proteins represent the probability that by chance the seed protein would be as connected to other seed proteins at the level observed in the network. Clusters were assigned on the basis of statistically significant connections of genes that participate in common biologic functions.
Pathway analysis
We identified pathway-based associations in the oropharyngeal cancer GWAS with the Meta-Analysis Gene-set Enrichment of variant Associations (MAGENTA) platform. The gene set enrichment (GSEA)–based methodology is described by Diaz-Sanchez and colleagues (25). In summary, all SNP from the GWAS analysis limited to oropharyngeal cancer were used in the pathway analysis. Genes in the human genome were mapped to a single index SNP with the lowest P value. This P value became the gene score and was corrected for confounding factors such as gene size, SNP density, and LD-related properties in a regression model to determine gene-wise adjusted gene score. Genes were then ranked by the adjusted gene scores. At a significance threshold of 95th and 75th percentile of all gene scores, the observed number of gene scores in a given predefined biologic pathway, with a ranked score above the specified threshold percentile, was calculated. This observed statistic was then compared with 1,000,000 randomly permuted pathways of identical size and generates an empirical GSEA P value for each pathway. Significance was determined when a pathway reached a FDR < 0.05 for either threshold. Predefined biologic pathways were taken from several publicly available databases, including GO (N = 9431), PANTHER (N = 634), KEGG (N = 185), Ingenuity (N = 92), BIOCARTA (N = 217), REACTOME (N = 427), and WikiPathways (N = 43).
In silico replication of oropharyngeal cancer results in a cervical cancer GWAS
An in silico replication was performed using a GWAS of 617 unrelated cervical cancer patients and 512 cancer-free controls generated on the Affymetrix Genome-wide Human SNP array 5.0 with approximately 440,794 SNP markers, which uses mostly different SNP markers compared with the Illumina arrays used in the UADT cancer GWAS. The details of population and QC have been described by Ivansson and colleagues (26). The data we received included for each SNP, the SNP identifier, the genomic location, and the P value from the association analysis that was estimated by ORs and 95% CI using multivariable unconditional logistic regression assuming a log-additive genetic model. Single gene analysis was completed as described above. A significance cutoff of P < 0.05 was used in the validation. A combined P value was calculated for each significant gene using the Fisher method. Using MAGENTA, a pathway analysis limited to pathways found to be significant in the oropharyngeal cancer dataset was also performed on the cervical cancer GWAS dataset.
Gene expression analysis
Oncomine was used for analysis and visualization of the tumor gene expression data (27). Oncomine is an online tool that aggregates mRNA expression data from a large number of cancer gene expression microarrays, and allows the user to investigate the relative expression of genes across various datasets. The relative expression of significant genes was investigated across oropharyngeal cancer and cervical cancer datasets. The overall P values were determined by simultaneously considering available data within Oncomine for the cancer versus normal comparisons. P values < 0.05 were considered significant. Only studies based on human samples were included in the analysis.
A parallel analysis was performed using normalized level 3 gene expression data for all HNSCC samples extracted from The Cancer Genome Atlas (TCGA) RNASeqV2 protocol (28, 29). Sixty-nine HPV-positive cases were identified, with HPV status determined by cross-referencing the corresponding TCGA clinical dataset for samples whose HPV status had been verified by in situ hybridization or PCR testing. Forty-two unmatched samples corresponding to surrounding nonneoplastic tissue were also identified. A two-tailed t test was used to assess statistically significant differential expression of TGFβR1 for all samples. P value < 0.05 was considered significant.
Results
Single-SNP and single-gene association analysis
After stringent quality control, data from 317 oropharyngeal cancer cases and 3,707 controls were tested for association with 296,728 SNPs. The QQ plot (Supplementary Fig. S1) shows minimal evidence of genomic inflation (λ = 1.01539). To enhance the power to detect associations, we dramatically reduced the number of SNPs tested by limiting analysis to the 12,258 SNPs that map to GO (21) categorized immune-related genes to test the hypothesis that germline DNA variants in these genes modulate risk of oropharyngeal cancer. Of the 1,324 GO-annotated immune-related genes, 1,304 could be assigned to an SNP. Of the top ranked immune-genes corresponding to SNPs with P values suggestive of association (1 × 10−3 > P > 1 × 10−7), the topmost 10 were significant at a gene-based level after correction for multiple testing (FDR < 0.05), whereas all have been previously associated in the literature with cancer, viral infection, or both (Table 1).
Table 1.
Immune genes with P values suggestive of association
| SNP | Gene | OPC P value | OR | CI | Rank | HPV+ P value | Larynx P value | Comments |
|---|---|---|---|---|---|---|---|---|
| rs9922120 | CBFA2T3 | 8.10E–07 | 1.646 | 1.35–2.01 | 6 | 4.52E–03 | 1.16E–02 | Candidate breast cancer tumor suppressor (44) |
| Associated with AML (45) | ||||||||
| rs4240847 | MAPKAPK2 | 3.39E–05 | 1.518 | 1.25–1.85 | 24 | 6.75E–04 | 3.21E–01 | Required for Flu A infection/propagation (46) |
| Activated by inflammatory cytokines (46) | ||||||||
| Major Stabilizer of p38 signaling (47) | ||||||||
| rs895437 | ZAP70 | 6.64E–05 | 1.653 | 1.29–2.12 | 40 | 6.90E–03 | 7.25E–01 | Mutations cause selective T-cell defect (48) |
| Used as a prognostic marker for CLL in B cells (49) | ||||||||
| Mutations results in immune deficiency (50) | ||||||||
| rs3107638 | CRTAM | 8.75E–05 | 1.551 | 1.25–1.93 | 51 | 5.15E–04 | 2.87E–01 | KO mice have reduced viral immunity (51) |
| NK activation and tumor surveillance (52) | ||||||||
| Associated with cervical cancer (37) | ||||||||
| rs1780365 | PBX1 | 2.09E–04 | 0.711 | 0.59–0.85 | 113 | 8.28E–04 | 6.81E–01 | Associated with esophageal SCCA (53) |
| Induces apoptosis (54) | ||||||||
| Retards ovarian tumor growth (55) | ||||||||
| rs2869433 | MAPK10 | 2.80E–04 | 0.708 | 0.59–0.85 | 134 | 4.66E–02 | 9.66E–01 | Activated by pro-inflammatory cytokines (56) |
| Associated with cancer and inflammation (57) | ||||||||
| rs6778945 | CD80 | 3.75E–04 | 0.590 | 0.44–0.79 | 171 | 5.85E–03 | 8.01E–01 | Necessary for T-cell activation and survival (58) |
| Associated with cancer including cervical cancer (38) | ||||||||
| rs3024498 | IL10 | 3.95E–04 | 1.421 | 1.17–1.73 | 181 | 6.75E–04 | 3.03E–01 | Involved in immunoregulation and inflammation (59) |
| Released during immune response to viral infection (60) | ||||||||
| Associated with cervical cancer [PMID: 18341210] | ||||||||
| rs10167561 | SOCS5 | 4.23E–04 | 0.710 | 0.59–0.86 | 192 | 2.00E–02 | 3.32E–01 | Involved in regulating T helper cell differentiation (61) |
| Associated with HIV replication (62) | ||||||||
| Suppresses EGFR signaling (63) | ||||||||
| rs2026811 | TGFβR1 | 4.29E–04 | 1.414 | 1.17–1.72 | 196 | 1.85E–03 | 5.23E–01 | Prooncogenic role (64) |
| Advanced carcinomas and tumor invasiveness (65) | ||||||||
| Polymorphism associated with several cancers (65) | ||||||||
| rs11135045 | EBF1 | 6.71E–04 | 1.437 | 1.16–1.77 | 289 | 1.68E–02 | 2.68E–01 | Associated with cervical cancer (40) |
| Associated with HNSCCA (66) | ||||||||
| Associated with breast cancer (67) | ||||||||
| rs11633294 | IGF1R | 7.03E–04 | 0.725 | 0.60–0.87 | 310 | 4.09E–03 | 5.27E–01 | Antiapoptotic (68) |
| Associated with cancer (68) | ||||||||
| rs2667975 | LYN | 7.56E–04 | 0.706 | 0.58–0.86 | 331 | 1.57E–03 | 1.78E–01 | Associated with myeloid progenitor and monocyte tumors (69) |
| Associated with breast cancer (70) | ||||||||
| rs3816375 | ITGAV | 7.75E–04 | 1.335 | 1.13–1.58 | 338 | 4.90E–02 | 3.11E–01 | Associated with HCC (71);In HIV, associated with Kaposi sarcoma lesions (72) |
NOTE: The table lists the top 14 ranked immune genes and their corresponding SNP and includes the SNP identifier (rs#), the official gene symbol, the overall rank in the oropharyngeal cancer GWAS, the genes most significant SNP's P value from the oropharyngeal cancer, HPV+, and laryngeal cancer association analysis and connection of each gene to cancer, progressive viral infection, or both described in existing scientific literature. The top 10 ranked genes are significant at a gene-based threshold (FDR < 0.05).
Abbreviations: CI, confidence interval; LC, laryngeal cancer; OPC, oropharyngeal cancer; OR, odds ratio.
To confirm that the oropharyngeal subsite is a valid surrogate for HPV-related head and neck cancer, we performed a validation analysis on serologically HPV positive head and neck cancer patients. As expected, HPV E6 seropositivity was predominantly found in patients with oropharyngeal cancer (Supplementary Table S1, Supplementary Fig. S2). Even with the statistical significance limited by the low numbers of HPV-positive cases and limited sensitivity of serologic testing (30, 31), the overall results were strikingly supportive of the association analysis restricted to oropharyngeal cancer. As shown in Table 1 and Supplementary Table S2, all 14 immune-related genes identified in the oropharyngeal cancer GWAS with SNP P values suggestive of association replicated significantly in the HPV+ cohort (P value < 0.05).
To determine whether immune-related genes are uniquely associated with oropharyngeal cancer rather than all types of HNSCC, we performed a parallel analysis of laryngeal cancer, a head and neck cancer strongly associated with tobacco and alcohol use rather than HPV infection (32). As shown in Table 1, out of the 14 highly ranked genes in the oropharyngeal cancer analysis that replicated in the HPV+ cohort, only one replicated in laryngeal cancer. The Two-sample Kolmogorov–Smirnov test demonstrated that the distribution of P values of the candidate immune-related genes was much more significant (P value = 6.64E—04) in the oropharyngeal cancer analysis compared with the laryngeal cancer analysis, demonstrating a unique association of candidate immune genes with oropharyngeal cancer that is not seen with tobacco/alcohol-associated head and neck cancer. Association of candidate genes was also much higher for oropharyngeal cancer than for all HNSCC aggregated (data not shown).
In summary, the suggestive association of oropharyngeal cancer with multiple candidate genes plausibly linked to viral immunity, the robustness of these associations when applied to HPV seropositive head and neck cancer patients, and the lack of similar strong association of immune related genes with non–HPV-related head and neck cancer supports our hypothesis that there is an immunogenetic basis for oropharyngeal cancer risk due to its unique association with HPV. We then further tested this hypothesis by applying further tiers of validation to the genes most strongly associated with oropharyngeal cancer risk.
GGI and PPI analysis
Analysis of functionally related gene combinations can unmask cumulative/cooperative risk associations that would be missed by single-gene analysis. To explore functional and biologic processes driving susceptibility to oropharyngeal cancer, we performed GGI and PPI analyses on immune-related genes from the oropharyngeal cancer single-gene association analysis. GGI analysis demonstrated a high degree of significant interconnectedness among these genes, suggesting their involvement in common biologic processes (Fig. 2A and Supplementary Table S3). PPI analysis performed on the top 200 ranked SNPs in immune-related genes from the oropharyngeal cancer analysis revealed multiple statistically significant first-degree interactions (Fig. 2B and Supplementary Table S4). The PPI network contained 81 direct connections between seed proteins from different loci with an expected value of only 24.58 (P value = 9.99E—04) and the average seed protein direct binding degree of 2.7 with an expected value of only 1.42 (P value = 9.99E—04). In randomly selected subsets of 200 genes from the set of 1304 GO-annotated immune-related genes, the connectivity in the PPI network was significantly reduced and both the direct connections between seed proteins and the average seed protein direct binding degree were not significant (Supplementary Fig. S3A).
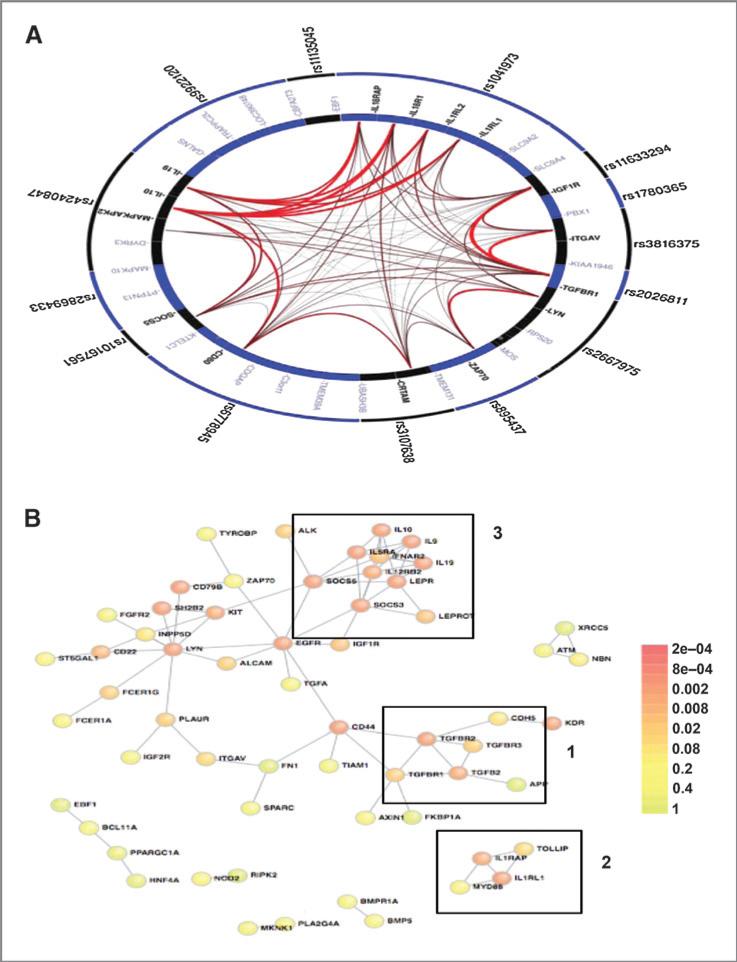
Figure 2.
Literature-based connectivity analyses. A, GGI analysis demonstrates the high degree of significant connectivity amongst the highly ranked immune SNPs in the oropharyngeal cancer GWAS. The figure was generated using the GRAIL platform, which determined the corresponding corrected P values for all gene–gene connections. The outer ring contains the input SNPs and the inner ring shows all possible genes corresponding to each SNP locus. Genes with significant intergene connections, determined by the GRAIL P value (Supplementary Table S2), are in bold and the thickness of the connection corresponds to strength of their interconnections. B, PPI network for the top ranked 200 immune genes in the oropharyngeal cancer GWAS generated using the DAPPLE platform. Nodes, proteins; links, direct interactions reported in the literature; colors, significance of the interaction as defined by the color key. Visual analysis of the oropharyngeal cancer GWAS immune network reveals three distinct immune clusters: 1, TGFβ signaling; 2, innate immunity; 3, Th1/Th2 balance.
Both the GGI and PPI analyses indicate that the top-ranked immune-related genes display a high degree of interconnectivity and are likely to be involved in related biologic processes. Visual inspection of the PPI plot identified three distinct clusters related to TGFβ signaling, Th1/Th2 balance, and innate immunity. Even when we widened the gene margins to be less stringent by extending the transcription start and stop codon by 110 kb and 40 kb, respectively (Supplementary Fig. S3B), these clusters are apparent, demonstrating their robustness.
Pathway analysis
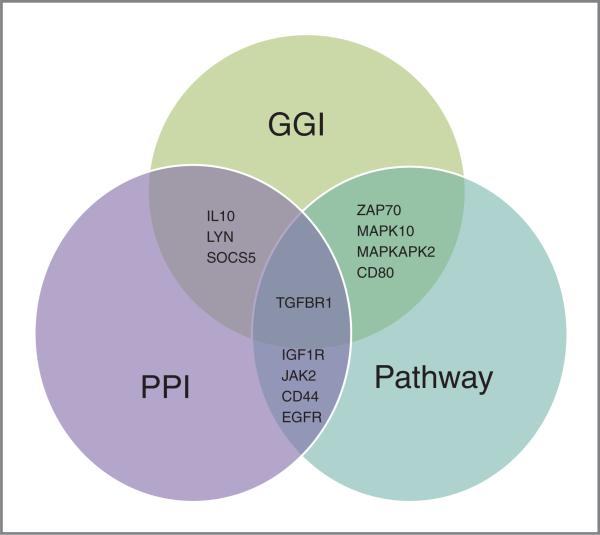
To identify specific immune pathways that may play a role in the etiology of HPV-mediated cancer, we performed a GSEA of 11,029 pathways associated with a broad array of biologic processes. We identified 21 immune-related pathways significantly associated with oropharyngeal cancer, organized into six categories, including five toll-like receptor (TLR) and innate immunity pathways, four NFKB-related pathways, two T-cell activation pathways, four cell death pathways, four TGFβ/TGFBR-related pathways, and two miscellaneous immune pathways (Table 2). Many of these pathways contained one or more of the top ranked genes from the oropharyngeal cancer single-gene association analysis, most notably the p38 MAPK pathway, which contains MAPKAPK2 and for which TGFβR1 is the most significant gene. Most strikingly, many of the highly ranked immune-related genes found in the single-gene analysis participated in significant PPI and/or significant pathways, including TGFβR1, IL10, SOCS5, ZAP70, MAPKAPK2, CD80, LYN and MAPK10. Several immune-related genes that displayed significant PPI were also the most significant genes in many of the significant pathways from GSEA analysis, including TGFβR1, EGFR, CD44, IGF1R, and JAK2 (Fig. 3). The density of participation of highly ranked immune-related genes in functional pathways and protein–protein and gene–gene interaction networks is consistent with our hypothesis that multiple functionally related genes participating in key immune processes contribute incrementally to risk of oropharyngeal cancer.
Table 2.
Significant pathways
| Immune category | Database | Pathway | FDR | Pathway GS | Effective GS |
|---|---|---|---|---|---|
| Cell death | GOTERM | Negative regulation of caspase activity | 2.98E–02 | 16 | 16 |
| Cell death | PANTHER | Other apoptosis | 1.54E–02 | 14 | 10 |
| Cell death | REACTOME | P75ntr signals via NFKB | 2.01E–02 | 13 | 12 |
| Cell death | REACTOME | NRIF signals cell death from the nucleus | 3.23E–02 | 13 | 13 |
| Misc immune | PANTHER | B cell and antibody-mediated immunity | 3.66E–02 | 97 | 65 |
| Misc immune | GOTERM | Initiation of viral infection | 3.74E–02 | 11 | 11 |
| NFKB | BIOCARTA | EPONFKB pathway | 5.20E–03 | 11 | 11 |
| NFKB | Ingenuity | NFKB signaling | 3.05E–02 | 43 | 39 |
| NFKB | REACTOME | Human TAK1 activates NFKB by phosphorylation and activation of IKKS complex | 4.00E–02 | 15 | 13 |
| NFKB | BIOCARTA | NFKB pathway | 3.90E–03 | 23 | 20 |
| T-cell activation | BIOCARTA | CD40 pathway | 2.21E–02 | 15 | 13 |
| T-cell activation | Panther | T-cell activation | 2.23E–02 | 31 | 22 |
| TGFβ signaling | wiki pathway | P38 MAPK signaling pathway | 4.56E–02 | 54 | 52 |
| TGFβ signaling | KEGG | MAPK signaling pathway | 2.19E–02 | 267 | 241 |
| TGFβ signaling | wiki pathway | MAPK signaling pathway | 4.80E–02 | 34 | 33 |
| TGFβ signaling | wiki pathway | TGFβ signaling pathway | 4.62E–02 | 162 | 154 |
| TLR and innate immunity | REACTOME | Toll-like receptor 3 cascade | 1.61E–02 | 59 | 55 |
| TLR and innate immunity | Ingenuity | LPSIL-1 mediated inhibition of RXR function | 1.80E–02 | 59 | 55 |
| TLR and innate immunity | KEGG | NOD-like receptor signaling pathway | 4.91E–02 | 62 | 52 |
| TLR and innate immunity | Panther | Toll receptor signaling pathway | 4.26E–02 | 30 | 26 |
| TLR and innate immunity | REACTOME | Toll receptor cascades | 2.03E–02 | 86 | 78 |
NOTE: The 21 significant immune pathways assigned to one of six immune categories. For each pathway, the table lists the immune category, the database the pathway was found in, the official name of the pathway, FDR, and the gene size (GS, the number of genes included in the pathway) and the effective gene size (EGS, the number of genes found in the GWAS dataset).
Figure 3.
The intersections of the integrative analysis. The results of each mode of analysis are listed in the Venn diagram. Multiple genes were found to overlap the results of two analyses but only TGFβR1 intersected the results of all three independent analyses. (Single-gene analysis was not included in this diagram as results informed the GGI analysis and thus would be redundant).
Replication of oropharyngeal cancer results in a cervical cancer GWAS
Our hypothesis that risk of oropharyngeal cancer, as an HPV-related cancer, is uniquely associated with variation in immune genes, would be strongly supported by validation in cervical cancer, the cancer most strongly associated with HPV. Because TGFβR1 was significant across all four tiers of independent analyses, we attempted to replicate it in the cervical cancer GWAS. First, we sought to confirm that variants in TGFβR1 are associated with cervical cancer, and found that the TGFβR1-associated SNP rs334356 was ranked the 11th most significant immune gene in the cervical cancer GWAS with a P value of 2.00E—03. Because a SNP-based replication was unable to be performed because the oropharyngeal cancer and cervical cancer GWAS each used different genotyping platforms, we attempted a gene-based replication. The combined P value of TGFβR1 across the oropharyngeal cancer and cervical cancer GWAS met the significance threshold for gene-based analysis (using the nearest gene and most significant SNP in each gene loci) even after standard Bonferroni correction (P < 3.8E—05) with a P combined = 1.25E—05.
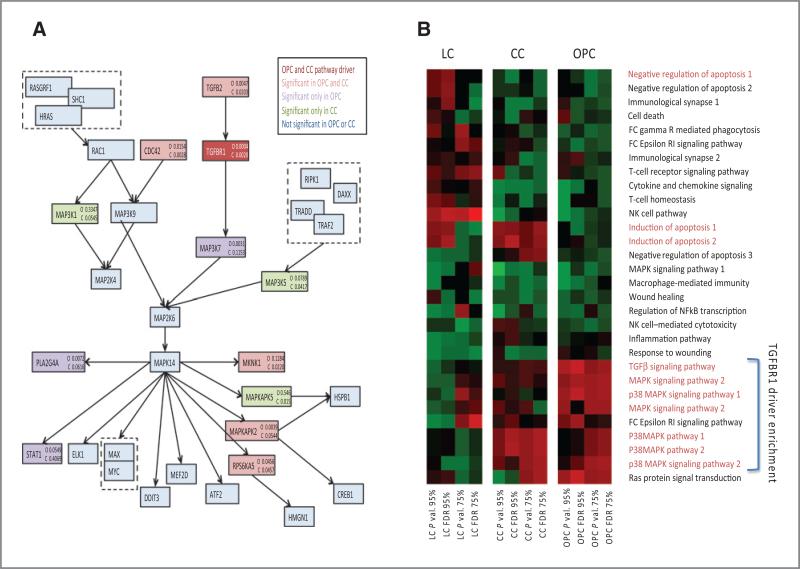
We also sought to determine whether any of the significant pathways from the oropharyngeal cancer analysis replicated in the cervical cancer dataset. The P38 MAPK pathway was the only pathway significant in both the oropharyngeal cancer and cervical cancer GWAS datasets (FDR adjusted P value < 0.05), and in both datasets TGFβR1 was the most significant gene. Of the 54 genes comprising the p38 MAPK pathway, the same seven genes had the lowest P values and were drivers of significance in both the oropharyngeal cancer and cervical cancer GWAS datasets (Fig. 4A), suggesting conservation of functional gene associations across both HPV-related cancers. To determine whether TGFβR1-related pathways are uniquely associated with HPV-related cancers, we compared the nominal P values and corresponding FDR of all immune-related pathways containing closely TGFβR1-related genes (TGFβ1, TGFβ2, TGFβ3, TGFβR1, TGFβR2, and TGFβR3) in oropharyngeal cancer and cervical cancer (HPV related) and laryngeal cancer (tobacco/alcohol related; Fig. 4B). The significance trends are more similar for the two HPV-mediated cancers, oropharyngeal cancer and cervical cancer, than either is to laryngeal cancer, which shows almost inverse significance trends. These findings support a unique and potentially functionally conserved role for TGFβ signaling across HPV-related cancers.
Figure 4.
Pathway replication. A, the P38 MAPK pathway containing TGFβR1 that is significant in both the oropharyngeal cancer and cervical cancer GWAS. Significant P values from the oropharyngeal cancer and cervical cancer datasets are listed for each gene. The colors correspond to the dataset in which the genes were significant, as described in the color key. TGFβR1 is the most significant gene in both datasets. B, the comparative significance of all TGFβ-related pathways in the two HPV-associated cancers, oropharyngeal cancer and cervical cancer, and in classic non-HPV–mediated HNSCC, laryngeal cancer. Heatmap color intensity is proportional to the P value or FDR as indicated. Pathway names highlighted in red represent those that include TGFβR1.
Expression of TGFβR1 is altered in HPV-associated tumors
Significant genetic associations with oropharyngeal cancer and other HPV-associated cancers are likely to have functional consequences, such as altered expression of target genes in cancer. We analyzed TGFβR1 gene expression in oropharyngeal cancer and cervical cancer tumor specimens in the Oncomine human cancer genomic database (27) and in HPV+ head and neck cancer samples in the TCGA database (28, 29). TGFβR1 was significantly overexpressed in oropharyngeal cancer, cervical cancer, and HPV+ head and neck cancer with respect to benign tissue (Table 3). This suggests that the genes found to be associated with HPV-related head and neck cancer are not simply markers of susceptibility, but likely to play a functional role in interactions between the host and the virus or virally transformed cancer cells.
Table 3.
TGFBR1 gene expression analysis
| Tissues | P value | Fold change | Databases |
|---|---|---|---|
| Oropharyngeal cancer | 2.00E–03 | 2.21 | Oncomine |
| Cervical cancer | 3.85E–07 | 2.88 | Oncomine |
| HPV+ head and neck cancer | 1.04E–08 | 1.69 | TCGA |
NOTE: Gene expression levels of TGFβR1 in oropharyngeal cancer, cervical cancer, and HPV+ HNSCC. The table lists the tissue type, P value, fold change, and database for all entries.
Discussion
Little is known about determinants of host susceptibility to viral carcinogenesis, and whether common biologic themes may account for the heterogeneity of progression to cancer following viral exposure. The present study is the first to exploit GWAS as a high-throughput strategy to examine immunogenetic susceptibility to oropharyngeal cancer, unique among HNSCC subsites for its strong association with HPV. We found that variation in immune-related genes is an important determinant of susceptibility to HPV-related but not HPV-unrelated HNSCC, and that this relationship is robust across multiple levels of analysis. Our findings also specifically highlight the pivotal contribution of variation in TGFβ/TGFβR signaling-associated molecules to HPV-related cancer susceptibility, which was consistent across oropharyngeal cancer, HPV seropositive HNSCC, and cervical cancer analyses. Significance of immune-related genes was much greater in the oropharyngeal cancer GWAS than that in a parallel analysis of laryngeal cancer, consistent with a unique contribution of immune-related genes to HPV-related cancer. Although this does not mean that host immunity plays no role in susceptibility to laryngeal cancer or other tobacco-associated HNSCC, it highlights the impressive and unique magnitude of association between variation in specific immune-related genes and oropharyngeal cancer, presumably due to high proportion of virally induced cancers at this subsite.
The strongest support for GWAS findings is their replication in independent datasets. For this, we focused on TGFβR1 and the TGFβ signaling pathways, which were significant across all modes of analysis. Because the association between immune genes and risk of oropharyngeal cancer is presumed to reflect enrichment at this subsite for HPV-driven cancer, we replicated our findings in cervical cancer, another HPV-related cancer. TGFβR1 was successfully validated in the cervical cancer GWAS, with a significance level in the top 99.999% of the whole GWAS. Pathway level analysis comparing significance among oropharyngeal cancer, cervical cancer, and laryngeal cancer of all pathways that include a core TGFβ-related gene, also strongly supported a role for TGFβR1 and TGFβ signaling unique to virally associated cancer. The likely functional significance of altered TGFβ signaling in HPV-related cancers is further supported by our finding that TGFβR1 is significantly overexpressed in both oropharyngeal cancer and cervical cancer.
These findings are consistent with the existing literature, in which TGFβ signaling has already been linked to HPV-associated cancer by classic methods of genetic analysis. Dysregulated TGFβ signaling is associated with malignant progression of HPV-positive cervical dysplasia (33, 34), and HPV has been shown to promote cervical cancer by attenuating TGFβR1 signaling required for epithelial homeostasis at early stages of viral infection (35). Most importantly, genetic variants in TGFβ1 have also been shown to be associated with HPV-positive oropharyngeal cancer, with people carrying genotypes with TGFβ1 variants more than twice as likely to have an HPV-positive tumor as patients with the wild-type genotype (36). Our results further support the concept that dysregulated TGFβ signaling is a key process common to multiple HPV-related cancers.
In addition to TGFβR1, all the genes linked to the top-ranked SNPs are highly plausible candidates with established connection to cancer and/or viral infection (Fig. 3 and Table 1), and several have been previously reported to be associated with cervical cancer, including CRTAM, CD80, IL10, and EBF1 (37–40). More than half of the top-ranked immune genes were significantly associated with oropharyngeal cancer across multiple tiers of analysis, supporting the hypothesis that the class of immune-related genes is uniquely enriched for oropharyngeal cancer susceptibility genes. Although these associations at the single-SNP level do not meet the criteria for genome-wide significance (P< 1 × 10−8), the immune system is characterized by interconnectivity, cooperativity, and redundancy of functions. Thus, even in the absence of individual SNPs meeting genome-wide significance, immune-related genes may contribute additively and incrementally to cancer risk by participating in common pathways and networks underlying disease susceptibility. Hence, we decided not to focus on single SNPs, but to consider their significance in the context of a broad, biology-driven, pathway/network-discovery approach. The broad concordance of our results across disparate tiers of analysis, including GGI, PPI, GSEA/pathway, and gene expression analyses supports the premise that functionally related genesets rather than individual genes modulate susceptibility to oropharyngeal cancer.
Additional evidence from the literature supports the association of multiple immune-related genes with susceptibility to HPV-related cancer. In a recent GWAS of cervical cancer in a Swedish population (41), three independent loci were identified in the major histocompatibility complex (MHC) region that influence susceptibility, the first in the MHC class I polypeptide-related sequence A gene (MICA), the second between HLA-DRB1 and HLA-DQA1, and the third at HLA-DPB2. Another cervical cancer GWAS performed in China also found associations with the HLA-DPB1 and HLA-DPB2 genes (42). Further support for a role for HLA molecules as modulators of susceptibility to cervical cancer comes from a pathway analysis performed on the cervical cancer GWAS, in which a subset was used as replication cohort in this study and identified several pathways, including HLA genes, that influence risk of cervical cancer (26). However, in the oropharyngeal cancer GWAS, the pathways and SNPs associated with HLA molecules were not highly ranked, with the most significant SNP P value equal to 1 × 10−2. These differences may potentially reflect differences in the underlying biology of oropharyngeal cancer and cervical cancer, which share HPV as an etiologic agent, but are in many respects clinically and epidemiologically different. On the other hand, a recent study (43) found NFKB-related pathways to be significantly associated with cervical cancer and vulvar cancer, which is concordant with our identification of four NFkB-related pathways in oropharyngeal cancer.
One significant limitation of this study is the lack of gold-standard tumor HPV status, requiring us to use HPV serology to infer which patients have HPV-associated tumors. Although HPV serology of blood samples is an established method for identifying patients with a high likelihood of HPV-induced cancer, the methods used for serologic analysis only identify roughly 60% of HPV+ oropharyngeal cancer (30, 31). An additional limitation of the approach used is that we only considered seropositivity for HPV16, whereas other oncogenic subtypes are associated with oropharyngeal cancer and presumably interact with the immune system in a similar fashion. Thus, we based our primary analysis on oropharyngeal cancer, the cancer subsite most highly associated with HPV, and used serology results to support the unique association of oropharyngeal cancer with HPV-related cancer. Although limited by the low number of cases with positive serology, the oropharyngeal cancer and HPV-seropositive datasets had strongly overlapping highly ranked significant SNPs and immune-related genes. Also, our results showing a much stronger association of immune genes with oropharyngeal cancer as compared with laryngeal cancer and all HNSCC, and the close similarity to cervical cancer, further support our hypothesis that the association of variation in immune-related genes with oropharyngeal cancer is uniquely strong because of its association with HPV-induced cancer. Replicating our oropharyngeal cancer findings in the cervical cancer dataset further validates our findings as genetic signatures associated with HPV-driven carcinogenesis.
In summary, this article supported the hypothesis that variants in immune-related genes, pathways, and networks increase susceptibility to HPV-associated HNSCC. Using a multitier analytic approach, TGFβ signaling was found to be associated with both HPV-mediated HNSCC and cervical cancer. Although this study focused on replicating the finding with the most analytical evidence, TGFβ signaling, we plan to investigate other immune-related genes and pathways that showed striking overlap between tiers of analysis. Identifying these causal variants will provide clues to pathogenetic mechanisms against which preventative interventions might be targeted and novel immunotherapeutic strategies developed for HPV-mediated oropharyngeal cancer.
Supplementary Material
Acknowledgments
The authors thank Dr. Inga Peter (Department of Genetics and Genomic Sciences, ISMMS) and Dr. Ke Hao (Department of Genetics and Genomic Sciences, ISMMS) for their helpful input.
Grant Support
This work was partially supported by grants from the NIH National Cancer Institute (1F30CA165615-01; C. Levovitz); National Institute of Dental and Craniofacial Research (5R03DE021741-02; A.G. Sikora); the NIH National Cancer Institute (1K08CA154963-01A1; A.G. Sikora).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors' Contributions
Conception and design: C. Levovitz, E.E. Schadt, M.R. Posner, E.M. Genden, P. Boffetta, A.G. Sikora
Development of methodology: C. Levovitz, M.R. Posner, P. Boffetta, A.G. Sikora
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): C. Levovitz, U. Gyllensten, J. Finnigan, E.E. Schadt, M.R. Posner, P. Boffetta, A.G. Sikora
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): C. Levovitz, D. Chen, E. Ivansson, J. Finnigan, S. Alshawish, W. Zhang, E.E. Schadt, M.R. Posner, P. Boffetta, A.G. Sikora
Writing, review, and/or revision of the manuscript: C. Levovitz, D. Chen, E. Ivansson, U. Gyllensten, S. Alshawish, W. Zhang, M.R. Posner, P. Boffetta, A.G. Sikora
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): S. Alshawish, E.E. Schadt, M.R. Posner, P. Boffetta, A.G. Sikora
Study supervision: M.R. Posner, P. Boffetta, A.G. Sikora
References
- 1.De Vuyst H, Clifford GM, Nascimento MC, Madeleine MM, Franceschi S. Prevalence and type distribution of human papillomavirus in carcinoma and intraepithelial neoplasia of the vulva, vagina and anus: a meta-analysis. Int J Cancer. 2009;124:1626–36. doi: 10.1002/ijc.24116. [DOI] [PubMed] [Google Scholar]
- 2.Ihloff AS, Petersen C, Hoffmann M, Knecht R, Tribius S. Human papilloma virus in locally advanced stage III/IV squamous cell cancer of the oropharynx and impact on choice of therapy. Oral Oncol. 2010;46:705–11. doi: 10.1016/j.oraloncology.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Mehanna H, Beech T, Nicholson T, El-Hariry I, McConkey C, Paleri V, et al. Prevalence of human papillomavirus in oropharyngeal and nonoropharyngeal head and neck cancer–systematic review and meta-analysis of trends by time and region. Head Neck. 2013;35:747–55. doi: 10.1002/hed.22015. [DOI] [PubMed] [Google Scholar]
- 4.Hocking JS, Stein A, Conway EL, Regan D, Grulich A, Law M, et al. Head and neck cancer in Australia between 1982 and 2005 show increasing incidence of potentially HPV-associated oropharyngeal cancers. Br J Cancer. 2011;104:886–91. doi: 10.1038/sj.bjc.6606091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindau ST, Drum ML, Gaumer E, Surawska H, Jordan JA. Prevalence of high-risk human papillomavirus among older women. Obstet Gynecol. 2008;112:979–89. doi: 10.1097/AOG.0b013e31818b0df2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hariri S, Unger ER, Sternberg M, Dunne EF, Swan D, Patel S, et al. Prevalence of genital human papillomavirus among females in the United States, the National Health And Nutrition Examination Survey, 2003–2006. J Infect Dis. 2011;204:566–73. doi: 10.1093/infdis/jir341. [DOI] [PubMed] [Google Scholar]
- 8.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 9.Magnusson PK, Lichtenstein P, Gyllensten UB. Heritability of cervical tumours. Int J Cancer. 2000;88:698–701. doi: 10.1002/1097-0215(20001201)88:5<698::aid-ijc3>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 10.Fuentes-Gonzalez AM, Contreras-Paredes A, Manzo-Merino J, Lizano M. The modulation of apoptosis by oncogenic viruses. Virol J. 2013;10:182. doi: 10.1186/1743-422X-10-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kreitchmann R, Bajotto H, daSilva DA, Fuchs SC. Squamous intraepithelial lesions in HIV-infected women: prevalence, incidence, progression and regression. Arch Gynecol Obstet. 2013;288:1107–13. doi: 10.1007/s00404-013-2871-3. [DOI] [PubMed] [Google Scholar]
- 12.Tami-Maury IM, Willig JH, Jolly PE, Vermund S, Aban I, Hill JD, et al. Prevalence, incidence, and recurrence of oral lesions among HIV-infected patients on HAART in Alabama: a two-year longitudinal study. South Med J. 2011;104:561–6. doi: 10.1097/SMJ.0b013e318224a15f. [DOI] [PubMed] [Google Scholar]
- 13.McKay JD, Truong T, Gaborieau V, Chabrier A, Chuang SC, Byrnes G, et al. A genome-wide association study of upper aerodigestive tract cancers conducted within the INHANCE consortium. PLoS Genet. 2011;7:e1001333. doi: 10.1371/journal.pgen.1001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ioannidis JP, Thomas G, Daly MJ. Validating, augmenting and refining genome-wide association signals. Nat Rev Genet. 2009;10:318–29. doi: 10.1038/nrg2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korte A, Farlow A. The advantages and limitations of trait analysis with GWAS: a review. Plant Methods. 2013;9:29. doi: 10.1186/1746-4811-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun YV. Integration of biological networks and pathways with genetic association studies. Hum Genet. 2012;131:1677–86. doi: 10.1007/s00439-012-1198-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farber CR. Systems-level analysis of genome-wide association data. G3. 2013;3:119–29. doi: 10.1534/g3.112.004788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen D, McKay JD, Clifford G, Gaborieau V, Chabrier A, Waterboer T, et al. Genome-wide association study of HPV seropositivity. Hum Mol Genet. 2011;20:4714–23. doi: 10.1093/hmg/ddr383. [DOI] [PubMed] [Google Scholar]
- 23.Raychaudhuri S, Plenge RM, Rossin EJ, Ng AC, Purcell SM, Sklar P, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diaz-Sanchez V, Larrea F, Ulloa-Aguirre A, Garza-Flores J, Richards E, Veayra F. Absorption of dihydrotestosterone (DHT) after its intramuscular administration. Fertil Steril. 1989;51:493–7. doi: 10.1016/s0015-0282(16)60560-5. [DOI] [PubMed] [Google Scholar]
- 26.Ivansson EL, Juko-Pecirep I, Erlich HA, Gyllensten UB. Pathway-based analysis of genetic susceptibility to cervical cancer in situ: HLA-DPB1 affects risk in Swedish women. Genes Immun. 2011;12:605–14. doi: 10.1038/gene.2011.40. [DOI] [PubMed] [Google Scholar]
- 27.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Available from: http://cancergenome.nih.gov.
- 30.Smith EM, Pawlita M, Rubenstein LM, Haugen TH, Hamsikova E, Turek LP. Risk factors and survival by HPV-16 E6 and E7 antibody status in human papillomavirus positive head and neck cancer. Int J Cancer. 2010;127:111–7. doi: 10.1002/ijc.25015. [DOI] [PubMed] [Google Scholar]
- 31.Smith EM, Ritchie JM, Pawlita M, Rubenstein LM, Haugen TH, Turek LP, et al. Human papillomavirus seropositivity and risks of head and neck cancer. Int J Cancer. 2007;120:825–32. doi: 10.1002/ijc.22330. [DOI] [PubMed] [Google Scholar]
- 32.Halec G, Holzinger D, Schmitt M, Flechtenmacher C, Dyckhoff G, Lloveras B, et al. Biological evidence for a causal role of HPV16 in a small fraction of laryngeal squamous cell carcinoma. Br J Cancer. 2013;109:172–83. doi: 10.1038/bjc.2013.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iancu IV, Botezatu A, Goia-Rusanu CD, Stanescu A, Huica I, Nistor E, et al. TGF-beta signalling pathway factors in HPV-induced cervical lesions. Roum Arch Microbiol Immunol. 2010;69:113–8. [PubMed] [Google Scholar]
- 34.Deng W, Tsao SW, Kwok YK, Wong E, Huang XR, Liu S, et al. Transforming growth factor beta1 promotes chromosomal instability in human papillomavirus 16 E6E7-infected cervical epithelial cells. Cancer Res. 2008;68:7200–9. doi: 10.1158/0008-5472.CAN-07-6569. [DOI] [PubMed] [Google Scholar]
- 35.French D, Belleudi F, Mauro MV, Mazzetta F, Raffa S, Fabiano V, et al. Expression of HPV16 E5 down-modulates the TGFbeta signaling pathway. Mol Cancer. 2013;12:38. doi: 10.1186/1476-4598-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guan X, Sturgis EM, Lei D, Liu Z, Dahlstrom KR, Wei Q, et al. Association of TGF-beta1 genetic variants with HPV16-positive oropharyngeal cancer. Clin Cancer Res. 2010;16:1416–22. doi: 10.1158/1078-0432.CCR-09-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garay E, Patino-Lopez G, Islas S, Alarcon L, Canche-Pool E, Valle-Rios R, et al. CRTAM: A molecule involved in epithelial cell adhesion. J Cell Biochem. 2010;111:111–22. doi: 10.1002/jcb.22673. [DOI] [PubMed] [Google Scholar]
- 38.Manickam A, Sivanandham M. Mycobacterium bovis BCG and purified protein derivative-induced reduction in the CD80 expression and the antigen up-take function of dendritic cells from patients with cervical cancer. Eur J Obstet Gynecol Reprod Biol. 2011;159:413–7. doi: 10.1016/j.ejogrb.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 39.Bhairavabhotla RK, Verm V, Tongaonkar H, Shastri S, Dinshaw K, Chiplunkar S. Role of IL-10 in immune suppression in cervical cancer. Indian J Biochem Biophys. 2007;44:350–6. [PubMed] [Google Scholar]
- 40.Geng LY, Shi ZZ, Dong Q, Cai XH, Zhang YM, Cao W, et al. Expression of SNC73, a transcript of the immunoglobulin alpha-1 gene, in human epithelial carcinomas. World J Gastroenterol. 2007;13:2305–11. doi: 10.3748/wjg.v13.i16.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen D, Juko-Pecirep I, Hammer J, Ivansson E, Enroth S, Gustavsson I, et al. Genome-wide association study of susceptibility loci for cervical cancer. J Natl Cancer Inst. 2013;105:624–33. doi: 10.1093/jnci/djt051. [DOI] [PubMed] [Google Scholar]
- 42.Shi Y, Li L, Hu Z, Li S, Wang S, Liu J, et al. A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Nat Genet. 2013;45:918–22. doi: 10.1038/ng.2687. [DOI] [PubMed] [Google Scholar]
- 43.Bodelon C, Madeleine MM, Johnson LG, Du Q, Galloway DA, Malkki M, et al. Genetic variation in the TLR and NF-kappaB pathways and cervical and vulvar cancer risk: A population-based case-control study. Int J Cancer. 2014;134:437–44. doi: 10.1002/ijc.28364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar R, Selth LA, Schulz RB, Tay BS, Neilsen PM, Callen DF. Genome-wide mapping of ZNF652 promoter binding sites in breast cancer cells. J Cell Biochem. 2011;112:2742–7. doi: 10.1002/jcb.23214. [DOI] [PubMed] [Google Scholar]
- 45.Barrett CW, Fingleton B, Williams A, Ning W, Fischer MA, Washington MK, et al. MTGR1 is required for tumorigenesis in the murine AOM/DSS colitis-associated carcinoma model. Cancer Res. 2011;71:1302–12. doi: 10.1158/0008-5472.CAN-10-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ehlting C, Ronkina N, Bohmer O, Albrecht U, Bode KA, Lang KS, et al. Distinct functions of the mitogen-activated protein kinase-activated protein (MAPKAP) kinases MK2 and MK3: MK2 mediates lipopolysaccharide-induced signal transducers and activators of transcription 3 (STAT3) activation by preventing negative regulatory effects of MK3. J Biol Chem. 2011;286:24113–24. doi: 10.1074/jbc.M111.235275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holzer TR, Fulford AD, Arkins AM, Grondin JM, Mundy CW, Nasir A, et al. Ischemic time impacts biological integrity of phospho-proteins in PI3K/Akt, Erk/MAPK, and p38 MAPK signaling networks. Anticancer Res. 2011;31:2073–81. [PubMed] [Google Scholar]
- 48.Chiang J, Hodes RJ. Cbl enforces Vav1 dependence and a restricted pathway of T cell development. PLoS ONE. 2011;6:e18542. doi: 10.1371/journal.pone.0018542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morabito F, De Filippi R, Laurenti L, Zirlik K, Recchia AG, Gentile M, et al. The cumulative amount of serum-free light chain is a strong prognosticator in chronic lymphocytic leukemia. Blood. 2011;118:6353–61. doi: 10.1182/blood-2011-04-345587. [DOI] [PubMed] [Google Scholar]
- 50.Fallah-Arani F, Schweighoffer E, Vanes L, Tybulewicz VL. Redundant role for Zap70 in B cell development and activation. Eur J Immunol. 2008;38:1721–33. doi: 10.1002/eji.200738026. [DOI] [PubMed] [Google Scholar]
- 51.Takeuchi A, Itoh Y, Takumi A, Ishihara C, Arase N, Yokosuka T, et al. CRTAM confers late-stage activation of CD8+ T cells to regulate retention within lymph node. J Immunol. 2009;183:4220–8. doi: 10.4049/jimmunol.0901248. [DOI] [PubMed] [Google Scholar]
- 52.Steigerwald J, Raum T, Pflanz S, Cierpka R, Mangold S, Rau D, et al. Human IgG1 antibodies antagonizing activating receptor NKG2D on natural killer cells. MAbs. 2009;1:115–27. doi: 10.4161/mabs.1.2.7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu DB, Gu ZD, Cao XZ, Liu H, Li JY. Immunocytochemical detection of HoxD9 and Pbx1 homeodomain protein expression in Chinese esophageal squamous cell carcinomas. World J Gastroenterol. 2005;11:1562–6. doi: 10.3748/wjg.v11.i10.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shiozawa Y, Pedersen EA, Taichman RS. GAS6/Mer axis regulates the homing and survival of the E2A/PBX1-positive B-cell precursor acute lymphoblastic leukemia in the bone marrow niche. Exp Hematol. 2010;38:132–40. doi: 10.1016/j.exphem.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morgan R, Plowright L, Harrington KJ, Michael A, Pandha HS. Targeting HOX and PBX transcription factors in ovarian cancer. BMC Cancer. 2010;10:89. doi: 10.1186/1471-2407-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Biton S, Ashkenazi A. NEMO and RIP1 control cell fate in response to extensive DNA damage via TNF-alpha feedforward signaling. Cell. 2011;145:92–103. doi: 10.1016/j.cell.2011.02.023. [DOI] [PubMed] [Google Scholar]
- 57.Ying J, Li H, Cui Y, Wong AH, Langford C, Tao Q. Epigenetic disruption of two proapoptotic genes MAPK10/JNK3 and PTPN13/FAP-1 in multiple lymphomas and carcinomas through hypermethylation of a common bidirectional promoter. Leukemia. 2006;20:1173–5. doi: 10.1038/sj.leu.2404193. [DOI] [PubMed] [Google Scholar]
- 58.Nakayama M, Takeda K, Kawano M, Takai T, Ishii N, Ogasawara K. Natural killer (NK)-dendritic cell interactions generate MHC class II-dressed NK cells that regulate CD4+ T cells. Proc Natl Acad Sci U S A. 2011;108:18360–5. doi: 10.1073/pnas.1110584108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takata K, Kinoshita M, Okuno T, Moriya M, Kohda T, Honorat JA, et al. The lactic acid bacterium Pediococcus acidilactici suppresses auto-immune encephalomyelitis by inducing IL-10-producing regulatory T cells. PLoS ONE. 2011;6:e27644. doi: 10.1371/journal.pone.0027644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cui W, Liu Y, Weinstein JS, Craft J, Kaech SM. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity. 2011;35:792–805. doi: 10.1016/j.immuni.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gridley DS, Pecaut MJ, Rizvi A, Coutrakon GB, Luo-Owen X, Makinde AY, et al. Low-dose, low-dose-rate proton radiation modulates CD4(+) T cell gene expression. Int J Radiat Biol. 2009;85:250–61. doi: 10.1080/09553000902748609. [DOI] [PubMed] [Google Scholar]
- 62.Smith AJ, Li Q, Wietgrefe SW, Schacker TW, Reilly CS, Haase AT. Host genes associated with HIV-1 replication in lymphatic tissue. J Immunol. 2010;185:5417–24. doi: 10.4049/jimmunol.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Segatto O, Anastasi S, Alema S. Regulation of epidermal growth factor receptor signalling by inducible feedback inhibitors. J Cell Sci. 2011;124:1785–93. doi: 10.1242/jcs.083303. [DOI] [PubMed] [Google Scholar]
- 64.Bian Y, Hall B, Sun ZJ, Molinolo A, Chen W, Gutkind JS, et al. Loss of TGF-beta signaling and PTEN promotes head and neck squamous cell carcinoma through cellular senescence evasion and cancer-related inflammation. Oncogene. 2012;31:3322–32. doi: 10.1038/onc.2011.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goudie DR, D'Alessandro M, Merriman B, Lee H, Szeverenyi I, Avery S, et al. Multiple self-healing squamous epithelioma is caused by a disease-specific spectrum of mutations in TGFBR1. Nat Genet. 2011;43:365–9. doi: 10.1038/ng.780. [DOI] [PubMed] [Google Scholar]
- 66.Liao D. Emerging roles of the EBF family of transcription factors in tumor suppression. Mol Cancer Res. 2009;7:1893–901. doi: 10.1158/1541-7786.MCR-09-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perez-Vera P, Reyes-Leon A, Fuentes-Panana EM. Signaling proteins and transcription factors in normal and malignant early B cell development. Bone Marrow Res. 2011;2011:502751. doi: 10.1155/2011/502751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jameson MJ, Beckler AD, Taniguchi LE, Allak A, Vanwagner LB, Lee NG, et al. Activation of the insulin-like growth factor-1 receptor induces resistance to epidermal growth factor receptor antagonism in head and neck squamous carcinoma cells. Mol Cancer Ther. 2011;10:2124–34. doi: 10.1158/1535-7163.MCT-11-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gray P, Dagvadorj J, Michelsen KS, Brikos C, Rentsendorj A, Town T, et al. Myeloid differentiation factor-2 interacts with Lyn kinase and is tyrosine phosphorylated following lipopolysaccharide-induced activation of the TLR4 signaling pathway. J Immunol. 2011;187:4331–7. doi: 10.4049/jimmunol.1100890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Faccio R. Immune regulation of the tumor/bone vicious cycle. Ann N Y Acad Sci. 2011;1237:71–8. doi: 10.1111/j.1749-6632.2011.06244.x. [DOI] [PubMed] [Google Scholar]
- 71.Ali MY, Grimm CF, Ritter M, Mohr L, Allgaier HP, Weth R, et al. Activation of dendritic cells by local ablation of hepatocellular carcinoma. J Hepatol. 2005;43:817–22. doi: 10.1016/j.jhep.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 72.Ushijima H, Kunisada T, Ami Y, Tsuchie H, Takahashi I, Klocking HP, et al. Characterization of human immunodeficiency virus-1-infected cells of myeloid-monocytic lineage (ML-1, HL-60, THP-1, U-937). J Acquir Immune Defic Syndr. 1992;5:1001–4. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.