Abstract
Iron is an essential element for the growth and development of virtually all living organisms. As iron acquisition is critical for the pathogenesis, a host defense strategy during infection is to sequester iron to restrict the growth of invading pathogens. To counteract this strategy, bacteria such as Vibrio parahaemolyticus have adapted to such an environment by developing mechanisms to obtain iron from human hosts. This review focuses on the multiple strategies employed by V. parahaemolyticus to obtain nutritional iron from host sources. In these strategies are included the use of siderophores and xenosiderophores, proteases and iron-protein receptor. The host sources used by V. parahaemolyticus are the iron-containing proteins transferrin, hemoglobin, and hemin. The implications of iron acquisition systems in the virulence of V. parahaemolyticus are also discussed.
Keywords: Vibrio parahaemolyticus, iron, virulence, host iron proteins, mechanism of acquisition
Introduction
Vibrio parahaemolyticus is a halophilic Gram-negative bacterium that naturally inhabits marine and estuarine environments (Gavilan et al., 2013). The more recent distribution of this species has been driven by climatic conditions and the eutrophication of regional waters throughout the world (Baumann et al., 1984). V. parahaemolyticus can survive in a wide variety of niches in a free-swimming state, and its motility is conferred by a single polar flagellum. Alternatively, this bacterium can be found in a sessile state, attached to inert or animate surfaces, such as suspended particulates, zooplankton, fish, and shell-fish (McCarter, 1999). Many strains of V. parahaemolyticus are strictly environmental, though small subpopulations can be opportunistic pathogens that may cause gastroenteritis, wound infection, and septicemia (Joseph et al., 1982; McCarter, 1999). As with all bacteria, V. parahaemolyticus reproduces via binary fission without a systematic exchange of genes with other individuals of the same species, leading to essentially clonal reproduction (Garcia et al., 2013); however, only some of these clones cause diarrhea in humans. Strains of V. parahaemolyticus belong to different serogroups and can produce a number of different lipopolysaccharide (O) and capsular (K) antigens, constituting the primary basis for strain classification (Nair et al., 2007; Garcia et al., 2013).
Since its discovery in 1950 by Tsunesaburo Fujino of Osaka University after an outbreak due to the ingestion of contaminated seafood, V. parahaemolyticus has been recognized as a leading cause of intestinal infection throughout coastal countries worldwide (Fujino et al., 1953). Following the consumption of raw or undercooked seafood, individuals infected with virulent strains may present clinical symptoms that include diarrhea, vomiting, nausea, abdominal cramping, and low-grade fever. Exposure to V. parahaemolyticus can also lead to wound infection and septicemia in certain medical conditions, such as immune deficiency (Su and Liu, 2007).
The infections and outbreaks caused by V. parahaemolyticus prior to 1996 were geographically isolated and associated with a diversity of serotypes (Wong et al., 2000; Chowdhury et al., 2004). However, the later increase in outbreaks was linked to the occurrence of gastroenteritis throughout Asia associated with a new and unique clone of serotype O3:K6 that emerged in Kolkata, India, in 1996 (Matsumoto et al., 2000; Ansede-Bermejo et al., 2010; Pazhani et al., 2014). This clone rapidly spread throughout the majority of Southeast Asian countries in the same year (Okuda et al., 1997b; Ansede-Bermejo et al., 2010; Gavilan et al., 2013). This clone, referred to as the pandemic clone, has since spread globally from Southeast Asia to Europe, Africa, and the Americas (Okuda et al., 1997a; Nair et al., 2007; Paranjpye et al., 2012; Yano et al., 2014). In particular, some countries on the American continent have reported cases of gastroenteritis due to the pandemic O3:K6 strain and its serovariants; the pandemic strain was first detected in Peru, subsequently spread to Chile in 1998, to the U. S. in the same year, to Brazil in 2001 and to Mexico in 2004 (Velazquez-Roman et al., 2012, 2013). Based on various molecular methods, these widespread pandemic O3:K6 strains have been found to be genetically closely related and appear to constitute a clone that differs significantly from the non-pandemic O3:K6 strains that were isolated prior to 1996 (Okuda et al., 1997b; Wong et al., 2000; Lan et al., 2009), with several serovariants apparently emerging since 1996 (Nair et al., 2007; Velazquez-Roman et al., 2012, 2013; Al-Othrubi et al., 2014; Pazhani et al., 2014; Letchumanan et al., 2015). The global occurrence of V. parahaemolyticus strains emphasizes the importance of understanding their many virulence factors as well as the mechanisms used to acquire nutrients from the environment and the effects on human hosts.
Vibrio parahaemolyticus possesses a wide variety of virulence factors that cause damage such as adhesins, toxins, and secreted effectors involved in attachment, cytotoxicity, and enterotoxicity (Broberg et al., 2011; Zhang and Orth, 2013). The two main factors are a thermostable direct hemolysin (TDH) and a thermostable direct hemolysin-related hemolysin (TRH) encoded by the tdh and trh genes, respectively (Honda et al., 1982; Nishibuchi et al., 1992; Vongxay et al., 2008). Nonetheless, the isolation of V. parahaemolyticus strains lacking functional tdh and trh genes from human infection cases and the analysis of the genome sequence of V. parahaemolyticus strain RIMD2210633 suggest that other virulence factors also play a role in the disease caused by this bacterium (Makino et al., 2003; Okada et al., 2009). The genome of V. parahaemolyticus contains two sets of type III secretion system (T3SS) gene clusters that function in the secretion and translocation of virulence factors into eukaryotic cells. These appear on each of the two chromosomes (Makino et al., 2003; Caburlotto et al., 2009). T3SSs utilize a needle-like apparatus to translocate into host cells effectors that target and hijack multiple eukaryotic signaling pathways. Indeed, T3SSs are essential virulence machines used by numerous bacterial pathogens, including Yersinia, Salmonella, Shigella, and pathogenic Escherichia coli (Makino et al., 2003).
Other systems or mechanisms that play an important role as virulence factors in Vibrio and all pathogenic microorganisms are those that confer the ability to acquire nutrients from the environment in which they live. These systems ensure that pathogens successfully reproduce and become established in a host. For example, the capacity to acquire nutrients such as iron (Fe) from a host is an ability obtained by pathogenic microorganisms during evolution. In fact, it has been speculated that the evolutionary pressure for microbes to develop pathogenic characteristics was to access the nutrient resources supplied by animals (Tanabe et al., 2012). The environment inside the colonized host has led to the evolution of new bacterial characteristics to maximize such new nutritional opportunities (Rohmer et al., 2011; Tanabe et al., 2012).
Currently, access to host nutrients is regarded as a fundamental aspect of an infectious disease. During the invasion of the human host, pathogens encounter complex nutritional microenvironments that could change, for example; the increase in inflammatory response due to the infection, local hypoxia in some tissues (Payne, 1993; Cassat and Skaar, 2013). The host can limit microbial access to nutrient supplies as a defense mechanism against the pathogens, however, the pathogens can counteract this by developing metabolic adaptations or improved mechanisms of nutrient acquisition to successfully exploit available host nutrients for their proliferation (Payne, 1993). Recent studies have pointed out an emerging paradigm that has been designated as ‘nutritional virulence’ (Abu Kwaik and Bumann, 2013). Although this term is applied to the acquisition of amino acids and carbon sources, certain nutritional ions or metals that are essential for cellular growth and other metabolic processes could be considered as part of this paradigm. As one of the most fundamental aspects of infectious diseases is the microbial acquisition of nutrients in vivo, which positively impacts in virulence as well as antibiotic resistance (Santic and Abu Kwaik, 2013), we suggest that the process of iron acquisition systems used by pathogenic microorganisms may be considered in the concept of ‘nutritional virulence.’
Iron (Fe) is an essential element for almost all cells, including most bacteria because it serves as a cofactor for metabolic processes, such as redox reactions, nucleic acid synthesis, and electron transfer (Tanabe et al., 2012). Iron is the fourth most abundant element on the Earth’s crust. In nature, there are two states of iron: (1) ferrous iron (Fe2+), which is toxic because it leads to the production of hazardous reactive oxygen species (ROS), including superoxide, in the presence of oxygen; and (2) ferric iron (Fe3+), which is insoluble under normal physiological conditions. Fe is bound to ligands, primarily proteins, in iron-dependent organisms, and trace Fe concentrations are necessary for all organisms, ranging between 0.4 and 4 μM in the majority of both eukaryotic and prokaryotic cells (Weinberg, 1974). However, there are bacteria such as some lactobacilli that are iron independent because they utilize manganese (Mn) and other cations as cofactors in their enzymes (Imbert and Blondeau, 1998).
The pathogenic microorganisms that infect mammalian hosts encounter diverse and changing environments. For example; the pH within the human body is usually neutral (7.4), but it can range from 1.0 in the stomach to 8.0 in urine. Also, if they move deeper into host tissues at mucosal surfaces, such as those from the lumen, the multilamellar mucus, and the epithelial cells of the stomach, pathogens confront drastically different or hostile environments. Some mucosal surfaces are well oxygenated, but others possess areas of low oxygen tension, for example the oral cavity, large intestine, female genital tract, abscesses and damaged tissues (Rohmer et al., 2011). The level of free Fe in mammalian bodily fluids is variable (∼10-18 M) but always far below the concentrations required for optimal bacterial growth (10-6 M), causing bacteria to rely on their own strategies or mechanisms for obtaining this metal. In an infected site there are numerous physiologically specialized environments that bacteria might encounter or colonize. For example, within the small intestine, there are variable conditions different from those found between caecum and colon (Rohmer et al., 2011). For all these reasons mentioned above, pathogens move through multiple diverse environments throughout their life cycle, and to accomplish this they require the regulation, coordination, and utilization of multiple bacterial metabolic pathways. Bacteria often use metabolic cues in order to regulate their metabolism and virulence functions to be successful as pathogens (Rohmer et al., 2011). Because they depend upon Fe as a vital cofactor that enables a wide range of key metabolic activities, bacteria must therefore ensure a balanced supply of this essential metal; accordingly, they invest considerable resources into its acquisition and employ elaborate control mechanisms to alleviate both iron-induced toxicity and Fe deficiency (Rangel et al., 2008).
The Fe concentration in coastal waters ranges from 1.3–35.9 nM up to 23.1–573.2 nM (Gledhill and Buck, 2012), a concentration that may be sufficient to support the growth of V. parahaemolyticus; however, V. parahaemolyticus also infects humans. The human body contains 3.8 g (in men) and 2.3 g (in women) of Fe. 20 mg of Fe is required daily for the production of hemoglobin (Hb) for new erythrocytes in order to preserve Fe homeostasis. Iron absorption from the diet, however, supplies only 1–2 mg daily, and the remaining Fe is derived by recycling Fe from senescent red blood cells (Ganz and Nemeth, 2006). Most bodily Fe is found in heme proteins (Hb, myoglobin, cytochromes, and multiple enzymes), and the second largest Fe pool is found in ferritin (also in hemosiderin). The remaining Fe is found in other proteins, such as iron-sulfur cluster enzymes, Fe-chelating proteins (Tf and Lf), and a pool of accessible Fe ions called the labile Fe pool (LIP), all of them constitutes the iron-containing proteins involved in metabolic pathways from hosts.
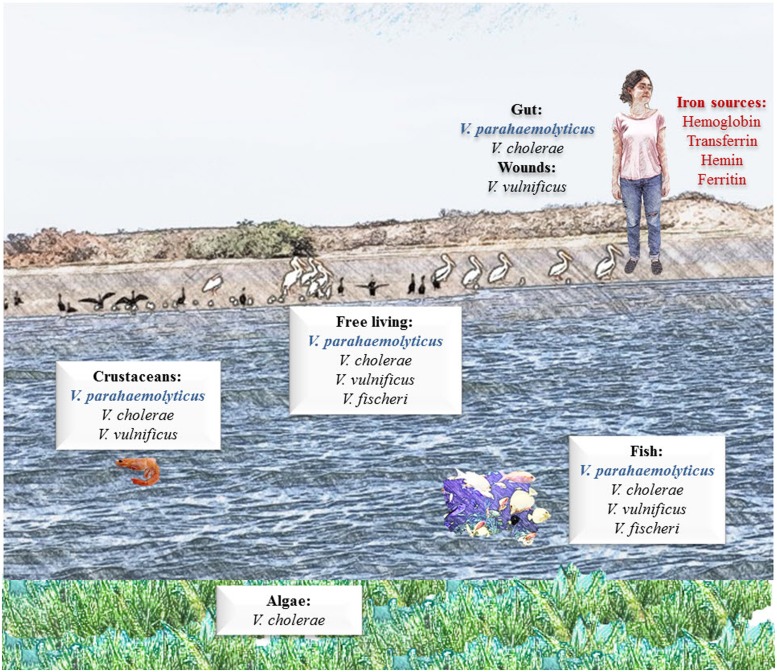
Inside the human body the solubility of iron is extremely low, because the Fe exists in insoluble mineral complexes, or under aerobic, aqueous, and neutral pH conditions, that difficult the access of bacteria to this element. Besides, Fe is bound to mammalian high-affinity iron-binding proteins such as Tf, Lf, and Ft and in consequence, many bacteria have developed high-affinity Fe transport systems to acquire Fe from sources in their niches (Rangel et al., 2008; Jin et al., 2009; Tanabe et al., 2012). The Fe sources available in the different environmental niches of V. parahaemolyticus are described and discussed in Figure 1.
FIGURE 1.
Iron sources available in the environmental niches of Vibrio parahaemolyticus. V. parahaemolyticus is an obligate halophilic organism, meaning that it requires salt to live. This organism is naturally occurring and found worldwide. It can commonly be found free swimming or attached to underwater surfaces and is found at high concentrations in areas of significant seafood consumption. Humans can acquire V. parahaemolyticus infection from infected seafood; once infected, established V. parahaemolyticus can acquire iron from different iron-containing proteins, such as hemoglobin (Hb) and transferrin (Tf).
Role of Iron in the Virulence of Vibrio parahaemolyticus
Iron regulates virulence factors of V. parahaemolyticus and almost all pathogenic bacteria. Inside a host the Fe concentration is very low, so many pathogens uses this (low-iron conditions) for inducing expression of genes involved in the virulence (Litwin and Calderwood, 1993). The presence of ferric Fe in bacterial growth media has been found to increase the adherence intensities of virulent V. parahaemolyticus strains to human fetal intestinal (HFI) cells in vitro (Hackney et al., 1980). Intraperitoneally injection with V. parahaemolyticus in the presence of ferric ammonium citrate in mice increased the bacterial proliferation, thus enhancing the lethality toward infected mice. V. parahaemolyticus cultures in low-iron conditions showed better proliferation than iron-rich cultures in response to the addition of supplementary Fe. Also, the production of thermostable direct toxin (TDH) by the hemolytic strains of V. parahaemolyticus was higher in iron-limited cultures than in iron-rich cultures, though the production of TDH by both iron-limited and iron-rich cultures was inhibited by the addition of Fe. In conclusion, the enhancement of V. parahaemolyticus virulence in the model mice likely occurred through the increase of bacterial proliferation in vivo and not the stimulation of TDH production. The V. parahaemolyticus precultured under iron-limited conditions may be more adaptable to the in vivo environment (Wong and Lee, 1994; Funahashi et al., 2003; Gode-Potratz et al., 2010).
The effect of lysed blood on the virulence of V. parahaemolyticus in mice was also investigated, and a factor released by erythrocyte lysis was found to greatly reduce the 50% lethal dose of V. parahaemolyticus in mice. Similar effects were observed with ferric ammonium citrate and Mn sulfate. Authors conclude that Fe from the lysed blood is involved in the virulence of V. parahaemolyticus (Karunasagar et al., 1984).
In a recent work, Gode-Potratz et al. (2010) demonstrated that metal ions play distinct roles in modulating gene expression and behavior in V. parahaemolyticus. In this work, high-calcium and low-iron growth conditions stimulated the induction of swarming and T3SS regulons from V. parahaemolyticus (Gode-Potratz et al., 2010). Swarming is a particular adaptation of many bacteria to grow in surfaces. In V. parahaemolyticus swarming is done by lateral flagella that enable the bacteria move over and colonize surfaces (Gode-Potratz et al., 2010). The authors concluded that swarming plays a signaling role with global consequences on the regulation of gene sets that are relevant for surface colonization and infection and that stimulation depends on the level of Fe present in the environment (Gode-Potratz et al., 2010).
Iron also regulates virulence factors in other members of the genus Vibrio. Based on the description of the virulence-enhancing effect of ferric ammonium citrate on V. cholerae by Joo (1975), other work has supported the role and importance of Fe in the virulence of Vibrio sp. (Joo, 1975). It has been demonstrated that the iron overload in humans increases the virulence of pathogens in vivo. For example in patients with liver diseases and hemochromatosis, iron overload states are common. It is known that these conditions predispose patients to recurrent infections, septicemia, and high mortality due to pathogens such as V. vulnificus (Bullen et al., 1991; Barton and Acton, 2009). This was corroborated by injecting mice with V. vulnificus and Fe, resulting in a lower 50% lethal dose and in a reduction in the time to death post-infection (Wright et al., 1981). In addition, elevated serum Fe levels were also produced by liver damage due to injections. Because the infections with V. vulnificus are acquired through the consumption of contaminated seafood, the role of iron on infections acquired by the oral route was also studied in mice. On the other hand, the role of iron on the growth of V. vulnificus in human and rabbit sera with injections of Fe or Tf were also studied. In the results, lethality and growth of V. vulnificus were more efficient in rabbit and human sera; respectively (Wright et al., 1981). Next, an induced peritonitis model was employed in mice to determine whether heme-containing molecules enhance the lethality of infections of V. vulnificus (Helms et al., 1984). In this model, the lethality toward mice inoculated intraperitonally with the bacteria and treated with methemoglobin, or hematin but not by myoglobin, was increased compared with those untreated (Helms et al., 1984). These results indicated that V. vulnificus has the capacity to produce fatal human infections because of its ability to use human proteins that bind Fe (Helms et al., 1984). Additionally, V. vulnificus strains isolated from different sources of Cuddalore coastal waters were tested for their virulence activity based on their LD50 values in mice. The LD50 was in the range of 104–107 cells in normal mice, but 101–102 cells in iron-injected mice, thus reinforcing the idea that Fe (acquired from human sources in vivo) may play a major role in the pathogenesis of V. vulnificus (Jayalakshmi and Venugopalan, 1992). Additionally, the virulence mechanisms of V. vulnificus biotype 1 and biotype 2 were studied and compared in mice. Both strains presented several properties in common, including capsule expression, the uptake of various Fe sources, and the production of exoproteins (Wright et al., 1981; Amaro et al., 1994). Taken together, data support the importance of iron in the pathogenesis of Vibrio sp. and V. parahaemoluticus in vivo and in vitro.
Iron regulatory Proteins and Mechanisms in V. parahaemolyticus
The study of Fe acquisition systems in E. coli led to the discovery of Fur, a DNA binding protein of 16.8 kDa, product of the fur (ferric uptake regulation) gene, that represses the transcription of genes involved in Fe uptake systems in iron replete conditions (Litwin and Calderwood, 1993). When the intracellular Fe concentration increases, Fur forms a dimer together with ferrous Fe (Fe2+) and binds to a consensus sequence (Fur-box), which overlaps the promoters of Fur-target genes, resulting in the inhibition of transcription. Although the role of Fur as a repressor is well-documented, emerging evidence demonstrates that Fur can function as an activator (Troxell and Hassan, 2013). Additionally to E. coli, Fur has been identified in other Gram-negative and Gram-positive bacteria. An interesting finding was that Fur also participates in functions different to the Fe metabolism for example; defense against oxygen radicals, metabolic pathways, bioluminescence, chemotaxis, swarming and production of toxins, and other virulence factors (Litwin and Calderwood, 1993). We can speculate about the importance of these Fe dependent mechanisms for bacterial virulence inside a host in vivo.
Vibrio parahaemolyticus contains a Fur protein that is 81% identical with the Fur protein from E. coli and over 90% identical with those of the Vibrio sp. (Yamamoto et al., 1997). Funahashi et al. (2002) reported that V. parahaemolyticus psuA and pvuA genes (which encode the TonB-dependent outer membrane receptors for a putative ferric siderophore and ferric-vibrioferrin), are regulated by Fur (Funahashi et al., 2002). Additionally, a homolog of the iutA gene in V. parahaemolyticus (which encodes for the receptor of ferric aerobactin) is apparently regulated by Fur (Funahashi et al., 2003). Furthermore, Fur regulates V. parahaemolyticus peuA gene (which encodes for an alternative ferric-enterobactin receptor; Tanabe et al., 2014).
Fur has been involved in the regulation of other virulence factors from Vibrio sp. Lee et al. (2013) demonstrated that in V. vulnificus Fur regulates hemolysin production at the transcriptional (vvhBA operon) and post-translational (by regulating the expression of two VvhA-degrading exoproteases, VvpE, and VvpM) levels (Lee et al., 2013). In contrast, other transcriptional regulators such as AraC-type family members and LysR-type family members, have been shown to activate transcription initiation of genes involved in the synthesis and utilization of siderophores in bacteria (Balado et al., 2009; Tanabe et al., 2012). In V. cholerae Fur regulates the expression of a number of genes in response to changes in the level of available iron. Fur usually acts as a repressor, but it has been shown that Fur positively regulates the expression of ompT, which encodes a major outer membrane porin, involved in the virulence of V. cholerae (Craig et al., 2011). It has been reported that Fur also represses the synthesis of RyhB, which negatively regulates genes for iron-containing proteins involved in the tricarboxylic acid cycle and respiration as well as genes for motility and chemotaxis (Wyckoff et al., 2007). Mey et al. (2005) reported the effects of iron and Fur on gene expression in V. cholerae. According with this work, nearly all of the known iron acquisition genes were repressed by Fur under iron-replete conditions, and also those genes involved in the transport of iron inside of pathogens (Mey et al., 2005). The iron transport systems regulated negatively by Fur in iron-replete conditions were feo and fbp genes (involved in the transport of ferrous and ferric iron inside cells; respectively). Both were found to be negatively regulated by iron and Fur (Mey et al., 2005). This is consistent with others genes involved in iron acquisition; in high concentrations of this nutrient the genes for iron acquisition systems are repressed.
Iron acquisition Systems Used by Pathogenic Microorganisms
An obligate question is how does V. parahaemolyticus acquire iron? This theme is complex; however, it has been well established in other pathogens. To acquire Fe from host sources, microorganisms generally use the iron-acquisition systems described below.
Receptors for Host Iron-Containing Proteins
Transferrins (Tfs) are a family of iron-binding glycoproteins that chelate free ferric Fe in biological fluids (Crichton and Charloteaux-Wauters, 1987). Bacteria such as Neisseria gonorrhoeae and Haemophilus influenzae are able to use the Fe in these proteins by binding Tf and Lf iron directly through the use of membrane receptors or binding proteins for host iron-glycoproteins (Tbp and Lbp, respectively). These receptors have been reported to be induced in some bacteria in the absence of Fe (Gray-Owen and Schryvers, 1996). Some receptors are specific for one iron-containing protein. On the contrary, other receptors that recognizes Tf, also can recognize Lf or other iron proteins, these receptors have been found in Neisseria (Morgenthau et al., 2013). Tf binding protein (TbpA) and lactoferrin binding protein (LbpA) are receptors that share 25% amino acid identity and a high degree of similarity. These Tf and Lf receptors can be promiscuous; for example, TbpA can recognizes Lf in some bacteria. Moreover, Tbps also exhibit low homology to other transport proteins and siderophore receptors. The homology among the members of this family of transporters suggests that the ancestral meningococcal Tf and Lf receptors may have been a single-unit transporter, similar to siderophore receptors (Perkins-Balding et al., 2004). The best characterized Tfbps and Lfbps to date belong to Neisseria species (Schryvers et al., 1998; Schryvers and Stojiljkovic, 1999).
Production and Secretion of Siderophores/Hemophores
Siderophores and hemophores are relatively small (<1 kDa) compounds produced and secreted by some species of bacteria and fungi to acquire Fe. Once produced and secreted from microorganisms, their function is to chelate ferric Fe with very high affinity (formation constant up to 1050 M; Wandersman and Delepelaire, 2004; Saha et al., 2013). In fact, these compounds can remove ferric Fe from the host proteins Tf, Lf, and ferritin. Siderophores are produced as common products of microbial metabolism under Fe stress conditions and facilitate the solubilization of ferric Fe and transport Fe via a specific receptor expressed in the cell plasma membrane. In addition, Bacillus subtilis and Mycobacterium smegmatis that are unable to synthesize siderophores resolve this problem by using the Fe from siderophores derived from other microorganisms (xenosiderophores; Schumann and Mollmann, 2001; Miethke et al., 2013). The capacity of these systems to acquire Fe from various environmental or biological sources is an evident advantage for organisms that may exist in several niches. Hemophores, which are less well studied and only found thus far in Gram-negative bacteria, are employed via the following strategy: they are secreted into the extracellular medium where they scavenge heme from various hemoproteins due to their higher affinity for this compound and then return the heme to hemophore-specific outer membrane receptors (Wandersman and Delepelaire, 2012). These proteins are secreted under conditions of iron depletion (Faraldo-Gomez and Sansom, 2003; Wandersman and Delepelaire, 2012).
Ferrireductases/Proteases
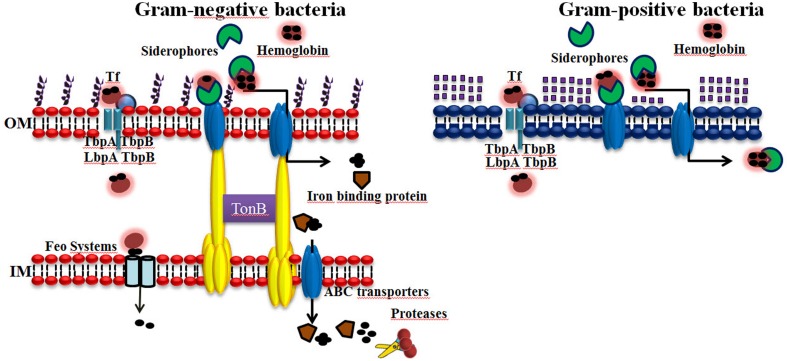
Some microorganisms secrete ferrireductases or produce membrane-associated proteins that reduce the ferric iron in holoTf, holoLf, or ferritin to the more accessible ferrous form. The reduction of ferric iron destabilizes the host iron-containing protein, and the ferrous iron is thus released. It has been reported that some pathogens are able to produce and secrete proteases that cleave host iron-containing proteins, and iron is easily acquired by pathogens in this form, for example, Entamoeba histoytica has hemoglobinases (Payne, 1993; Serrano-Luna et al., 1998; Ortiz-Estrada et al., 2012). A scheme for Fe acquisition systems in bacteria is represented in Figure 2.
FIGURE 2.
Iron acquisition systems used by bacteria that infect humans. Iron (Fe) is an essential element for virtually all forms of cellular life, including most bacteria, because it serves as a cofactor for several key enzymes required for many metabolic processes. Gram-negative and Gram-positive bacteria can acquire iron using elaborated mechanisms. (1) Receptors for host iron-containing proteins. (2) The production and secretion of siderophores/hemophores. (3) Ferrireductases/Proteases. The abilities of bacterial pathogens to adapt to the environment within a host are essential to their virulence.
Human Iron Sources and Iron Acquisition Systems in V. parahaemolyticus
Vibrio parahaemolyticus and other bacteria from the genus Vibrio including V. cholerae, V. vulnificus, require Fe for their growth and have developed systems during evolution to acquire Fe for sustaining metabolism and replication. As mentioned above, the Fe concentration in coastal waters ranges from 1.3–35.9 to 23.1–573.2 nM, which is sufficient for the growth of V. parahaemolyticus. In the human host, V. parahemolyticus can utilize host sources, such as the iron-proteins Hb, heme, hemin, and Tf (Yamamoto et al., 1994), and possibly others, such as Lf and Ft (Figure 1).
Siderophores as a Mechanism of Iron Acquisition Used by V. parahaemolyticus
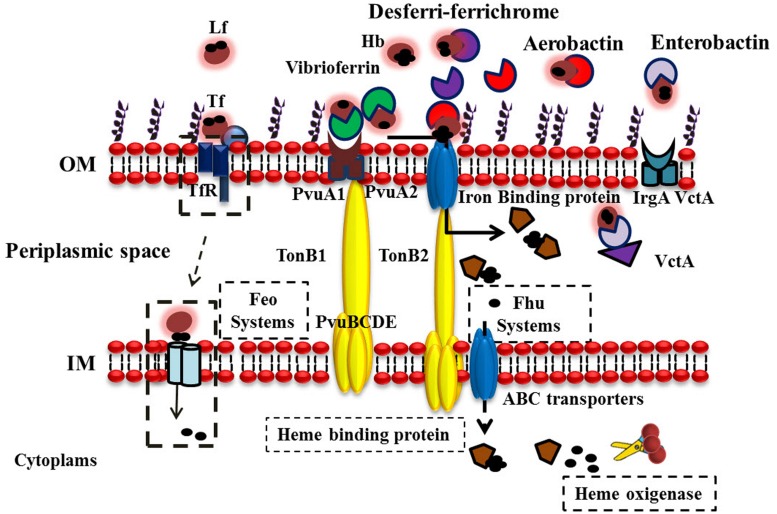
One of the strategies of V. parahaemolyticus to obtain the iron from the host iron-proteins is the use of siderophores. Under iron-limited conditions V. parahaemolyticus secretes Vibrioferrin to facilitate Fe acquisition. Fe-charged Vibrioferrin is recognized by an outer-membrane receptor composed of the proteins PvuA1 and PvuA2, which also recognizes heme and Hb. This receptor is coupled with the ABC transport system PvuBCDE which is located in the inner membrane and whose function is import the ferric-charged vibrioferrin to the inner membrane (Tanabe et al., 2003, 2011, 2012). V. parahaemolyticus also contains the TonB system which consists of three proteins designated TonB1, TonB2, TonB3 (Kuehl and Crosa, 2010). The energy for the transport of ferric-vibrioferrin is provided by the TonB2 system for PvuA1 and both the TonB1 and TonB2 systems for PvuA2 (Tanabe et al., 2011, 2012).
The V. parahaemolyticus also is able to utilize siderophores produced by other bacteria, for example, exogenous aerobactin, desferri-ferrichrome, and enterobactin (Funahashi et al., 2003, 2009; Tanabe et al., 2012). We have compared and analyzed in silico the mechanism used by V. parahaemolyticus with V. cholerae; the most studied member of the genus Vibrio. V. cholerae has multiple strategies for iron acquisition, including the endogenous siderophore vibriobactin and several siderophores that are produced by other microorganisms (Wyckoff et al., 2007). In general the Vibrio species, including V. cholerae and V. anguillarum can use catecholate-type siderophores as their cognate siderophores (Griffiths et al., 1984; Actis et al., 1986; Balado et al., 2009; Tanabe et al., 2012). Furthermore, some Vibrio sp. use the xenosiderophore enterobactin (Ent) that is produced mainly by members of the Enterobacteriaceae family (Griffiths et al., 1984; Naka and Crosa, 2012; Tanabe et al., 2012). On the other hand, Tan et al. (2014) described the use of the siderophore vulnibactin, essential in Fe uptake from host proteins. The importance of the vulnibactin in V. vulnificus pathogenicity was clinically demonstrated (Ceccarelli et al., 2013; Tan et al., 2014).
Once the iron has been acquired, V. cholerae has two systems for the transport of free iron: the Feo system, which transports ferrous iron, and the Fbp system, which transports ferric iron (Occhino et al., 1998; Wyckoff et al., 2007). It has been speculated that V. cholerae contains one additional high affinity iron transport system. Apparently, iron transport genes are regulated by Fur (Occhino et al., 1998; Wyckoff et al., 2007).
Transferrin Receptors as an Iron Acquisition System Used by V. parahaemolyticus
Using a basic local alignment search tool (BLAST) analysis of the sequences in V. parahaemolyticus, we identified putative genes that could encode for Tf receptors, but not for Lf receptors. The putative V. parahaemolyticus Tf receptor gene (tbpA) had 88% of identity with for those reported for N. meningitidis (not shown; unpublished data). However, apparently V. parahaemolyticus also can use LF as in Fe source (Wong et al., 1996). This probably could be due to the siderophore utilization by this bacterium. Because of Lf is one of the main iron transporters at intestinal level, the V. parahaemolyticus capacity of to use Lf as an iron source and the mechanism from iron-Lf acquisition must to be determined. Other members of the genus Vibrio have receptors for the use of Tf as iron source. For example, recently; Pajuelo et al. (2015) demonstrated that pVvbt2 from V. vulnificus, which causes vibriosis in fish (mainly eels), encodes a host-specific Fe acquisition system that depends on an outer membrane receptor called Vep20. This protein recognizes eel Tf and belongs to a new family of plasmid-encoded fish-specific Tf receptors (Pajuelo et al., 2015). Furthermore, it was found that Vep20 is encoded by an iron-regulated gene that is overexpressed in eel blood during artificially induced vibriosis with V. vulnificus both in vitro and in vivo (Pajuelo et al., 2015). The Vep20 gene homologs have been identified on the transferable plasmids of two species of fish pathogens with broad host ranges: V. harveyi (pVh1) and Photobacterium damselae subsp. damselae (pPHDD1; Pajuelo et al., 2015). It has been hypothesized that V. cholerae contains three proteins that could be Tf receptors. These proteins were shown to be involved in the binding of LF, Hemin, Ft, and Hb (Ascencio et al., 1992).
Binding and Transport of Iron or Iron-Charged compounds in V. parahaemolyticus
The expression of two proteolytic proteins of 43 and 90 kDa from V. parahaemolyticus were identified. Apparently the protease of 43 kDa is capable of degrading Hb and it has been speculated that this could be one of the strategies of V. parahaemolyticus to acquire iron from the human host (Wong and Shyu, 1994). By using the BLAST, we also identified genes that could play a role in Fe acquisition (Table 1). Although they have not reported and their biological functions have not been described, these genes likely encode proteins involved in Fe acquisition for V. parahaemolyticus in different niches. The probable functions of putative Fe acquisition genes and homologies are described in Table 1. Additionally; a schematic view of Fe acquisition systems with putative proteins used by V. parahaemolyticus is shown in Figure 3.
Table 1.
Genes related to Iron acquisition systems in Vibrio cholerae and putative genes in Vibrio parahaemolyticus.
|
V. cholerae |
Homology V. parahaemolyticus (%) |
||||||
|---|---|---|---|---|---|---|---|
| Protein/Gene | Function | Reference for source | Reference for source | Accession number | Vibrio sp. | Protein identity (%) | Gene identity (%) |
| VctA∗ | Outer membrane receptor for enterobactin (VctA) Vct(PGD) participate in the transport of vibriobactin and enterobactin | Wyckoff et al. (2009) | Tanabe et al. (2012) | (CP006005.1) | V. parahaemolyticus O1:Kuk | 65 | 67 |
| VctP | (BA000032.2) | V. parahaemolyticus RIMD 2210633 | 56 | 66 | |||
| VctG | (CP006005.1) | V. parahaemolyticus O1:Kuk | 71 | 66 | |||
| VctD | (BA000038.2) | V. parahaemolyticus RIMD 2210633 | 75 | 71 | |||
| IrgA∗ | Major membrane receptor for ferric enterobactin | Goldberg et al. (1990) | (CP001805.1) | Vibrio sp | 69 | 69 | |
| TonB1∗ | Energy transduction system, provides energy for transport of Enterobactin across the outer membrane | Occhino et al. (1998) | O’Malley et al. (1999) | (CP007005.1) | V. parahaemolyticus UCM-V493 | 51 | 70 |
| TonB2∗ | Energy transduction system, provides energy for the transport of vibrioferrin across the outer membrane | Wyckoff et al. (2004) | (CP006007.1) | V. parahaemolyticus 13-028/A3 | 54 | 72 | |
| FhuA∗ | Transport of ferrichrome across the outer membrane | Rogers et al. (2000) | Funahashi et al. (2009) | (CP003973.1) | V. parahaemolyticus BB22OP | 64 | 67 |
| FhuB∗ | (BA000032.2) | V. parahaemolyticus RIMD 2210633 | 77 | 70 | |||
| FhuC∗ | (AB300920.1) | V. parahaemolyticus W-9175 | 77 | 65 | |||
| FhuD∗ | (AB119276.1) | V. parahaemolyticus | 62 | 69 | |||
| HutA∗ | Outer membrane receptors for heme and transporters | Mey and Payne (2001) | O’Malley et al. (1999) | (CP003973.1) | V. parahaemolyticus BB22OP | 68 | 69 |
| HutR | (CP006007.1) | V. parahaemolyticus 13-028/A3 | 54 | 67 | |||
| HutB | (CP003973.1) | V. parahaemolyticus BB22OP | 68 | 64 | |||
| HutC∗ | O’Malley et al. (1999) | (CP006008.1) | V. parahaemolyticus O1:K33 | 67 | 70 | ||
| HutD | (CP006008.1) | V. parahaemolyticus O1:K33 | 68 | 68 | |||
| FeoA | Ferrous iron transporter | Cartron et al. (2006) | (CP007004.1) | V. parahaemolyticus UCM-V493 | 73 | 68 | |
| FeoB | (CP006004.1) | V. parahaemolyticus O1:Kuk | 79 | 73 | |||
| FeoC | - | V. parahaemolyticus | 51 | - | |||
| FbpA | Ferric iron transporter | Kirby et al. (1998) | (CP006008.1) | V. parahaemolyticus O1:K33 | 81 | 75 | |
| FbpB | Periplasmic ferric iron binding protein | (BA000031.1) | V. parahaemolyticus | 78 | 70 | ||
*indicate proteins that were studied and reported in V. parahaemoyticus.
Column 6 contain the V. parahaemolyticus strain with the highest identity.
FeoC was not found in the V. parahaemolyticus strains.
FIGURE 3.
Iron acquisition systems Used by V. parahaemolyticus to acquire iron. V. parahaemolyticus can be found free swimming or attached to underwater surfaces, seafood products, and humans. The bacterium produces and chelates iron from different sources (depending on the habitat) and transports ferric-charged vibrioferrin into cells via the outer-membrane receptors PvuA1 and PvuA2 and the inner-membrane ABC transport system PvuBCDE. Three sets of TonB systems (TonB1, TonB2, and TonB3) are present in this bacterium, and the energy required for PvuA1 and PvuA2 to transport ferric vibrioferrin is provided by the TonB2 system for PvuA1 and both the TonB1 and TonB2 systems for PvuA2. V. parahaemolyticus can also utilize hydroxamate-type xenosiderophores, such as desferri-ferrichrome and aerobactin. Discontinuous lines indicate possible iron sources and iron acquisition mechanisms in V. parahaemolyticus.
Apparently, the V. parahaemolyticus contain the proteins VctA, VctP, VctG, and VctD (Tanabe et al., 2012). These proteins are involved in the use of the siderophore ferric-enterobactin (Wyckoff et al., 2007, 2009). VctA and IrgA are receptors for enterobactin in V. cholerae and V. parahaemolyticus (Goldberg et al., 1990; Tanabe et al., 2012). Also, in V. parahaemolyticus were identified the TonB1 and TonB2 system similar to V. cholerae (Occhino et al., 1998; Seliger et al., 2001; Wyckoff et al., 2004). The proteins that belong to the TonB systems are involved in the transport of Fe3+ in Gram-negative bacteria. The system includes outer membrane receptors, all of which are connected with a complex of proteins located at the inner membrane such as TonB, ExbB, and ExbD. This system also includes a periplasmic binding protein associated with an ABC transporter (Larsen et al., 1996; Koster, 2005). The first step is the binding of the ferric-siderophore to the receptor, it has been speculated that this binding induces a conformational change and then the interaction of the receptor with TonB is enhanced. The proteins ExbB and ExbD provide the energy for the transport of the ferric-siderophore to the inner membrane, in this second step a periplasmic binding protein associated with an ABC transporter delivers the ferric-siderophore into the cytoplasm, in the third step the Fe dissociates from the siderophore (Larsen et al., 1996; Occhino et al., 1998; O’Malley et al., 1999). Little is known about the recycling, storage, and modification of the siderophore. The secretion of E. coli enterobactin is mediated by the membrane exporter protein EntS (Furrer et al., 2002; Danese et al., 2004).
In V. parahaemolyticus the genes encoding both for the TonB1 and TonB2 systems are located on the small chromosome, and the TonB3 system on the large chromosome (O’Malley et al., 1999; Kuehl and Crosa, 2010; Tanabe et al., 2012). In V. parahaemolyticus TonB2 is most active than TonB1 in providing the energy necessary for the transport of ferric-enterobactin via the receptors IrgA and VctA (Tanabe et al., 2012). However, this is different in V. cholerae, during the transport of ferric-enterobactin the energy required for IrgA and VctA receptors is provided by the TonB2 system (Seliger et al., 2001; Tanabe et al., 2012). The TonB3 system is not implicated in the transport of iron either V. parahaemolyticus or V. cholerae (Kuehl and Crosa, 2010; Tanabe et al., 2012), has been reported that the TonB3 system from V. vulnificus is induced when the bacterium grows in human serum (Alice and Crosa, 2012). The V. cholerae has two TonB systems, which are present on small chromosome (this is different from V. parahaemolyticus), and those encoding the TonB2 system are located on the large chromosome. They have unique as well as common functions (Seliger et al., 2001). Both mediate the transport of hemin, vibriobactin, and ferrichrome. However, only TonB1 participates in the use of the siderophore schizokinen, but TonB2 is required for the transport of enterobactin (Seliger et al., 2001).
With respect to FhuA, this protein is the receptor for the siderophores desferri-ferrichrome and aerobactin in V. parahaemolyticus (Funahashi et al., 2009). In addition, FhuB, FhuC, and FhuD apparently are involved in the transport of the siderophores, and also are present in V. cholerae (Rogers et al., 2000; Funahashi et al., 2009). Regarding the hut genes, it has been reported that HutA is the receptor for the uptake of heme. In addition, HutR has significant homology to HutA as well as to other outer membrane heme receptors (Occhino et al., 1998; Mey and Payne, 2001). In the V. cholerae the presence of hutBCD stimulated growth when hemin was the iron source, but these genes were not essential for hemin utilization (Occhino et al., 1998; Mey and Payne, 2001; Wyckoff et al., 2004). Other genes found in the V. parahaemolyticus genome were the feo system. The feo system consists of genes that encoded proteins involved in the transport of ferrous iron (Fe2+), which is expected to be a major iron source in the intestine (Cartron et al., 2006). This Fe2+ iron transport system feo is widely distributed among bacterial species such as V. cholerae. In this bacterium, the feo operon consists of three genes, feoABC. FeoB is an 83-kDa protein involving in the pore formation for iron transport (Weaver et al., 2013). FeoA and FeoC are all required for iron acquisition; however, their functions have not been described in detail. Apparently, in the genome of V. parahaemolyticus there are genes that encode for this Fe2+ transport. Moreover, the V. parahaemolyticus contains other iron transporters such as FbpA and FbpB. In Pasteurella haemolytica the presence of FbpABC family of iron uptake systems has been documented (Kirby et al., 1998). This family of proteins is involved in the utilization and transport of the ferric-xenosiderophore of the bacterium N. gonorrhoeae, and is independent of the TonB system (Strange et al., 2011). We speculate that V. parahaemolyticus bacteria could have this family of proteins in order to acquire ferric iron from xenosiderophores, in a TonB-independient manner. All of these Fe acquisition systems could be likely involved in the survival of V. parahaemolyticus and other Vibrio sp. in the different environments that they can colonize, i.e., water, humans, and several other vertebrate hosts (Fouz et al., 1997).
Conclusion
The element Fe is essential for the growth of pathogenic microorganisms, is fundamental and necessary for establishment and replication inside a host, and is required to cause infection. To this end, microbes that live in hostile environments and extracellular spaces of their host must employ different strategies for Fe acquisition to be successful in these niches. It has been postulated that such strategies were acquired during evolution and are involved in the pathogenesis and virulence of bacteria such as V. parahaemolyticus. Based on reported findings, this bacterium can utilize the Fe from the proteins Tf, Hb, and hemin by means of siderophores (vibrioferrin, aerobactin, and desferri-ferrichrome) and likely also receptors to acquire Fe from humans during infection of the gut. In low iron, V. parahaemolyticus express two proteins of 78 and 83 kDa (now called PvuA2 and PvuA1; respectively), which are the receptors for the siderophore vibrioferrin, and a protease of 43 kDa, which has been hypothesized is involved in one of the strategies of V. parahaemolyticus in order to acquire iron from the host. V. parahaemolyticus also encodes for LutA, which is the receptor for the siderophore aerobactin. Additionally, according to Table 1 and other works, this pathogen possesses genes that encode accessory proteins involved in Fe acquisition, transport and synthesis of molecules implicated in Fe acquisition systems.
The iron per se has been involved in increase the virulence of V. parahaemolyticus and other bacteria. For example, in recent works it have been demonstrated that iron uptake and Quorum sensing (QS) can act together as global regulators of bacterial virulence factors (Wen et al., 2012). QS is a regulatory mechanism used by several bacteria to regulate or modulate the production of extracellular compounds at high cell densities with the aim of establish bacterial biofilms (nowadays, the main medical problem for the control of infectious diseases). Bacterial QS serves as simple indicator of population density, by means of secreting signaling molecules called autoinducers. The link among iron and QS was reported firstly in Pseudomonas aeuruginosa (Bollinger et al., 2001). This bacterium in iron-depleted conditions, retarded biofilm formation and increased the twitching motility and expression of QS-related genes, suggesting a link between iron and QS system during biofilm formation (the most important virulence factor of P. aeruginosa; Cai et al., 2010), in contrast; Staphylococcus aureus in Fe limitation appeared to stimulate biofilm formation (Johnson et al., 2005). These controversial observations can be explained because biofilm formation QS-dependent is nutritionally conditional (Shrout et al., 2006). In other words, in the absence of an acquisition system needed for obtain nutritional iron, or other nutrients such as carbon sources, a bacterium such as P. aeuruginosa could establish thin and weak biofilms instead of mature biofilms (Banin et al., 2005; Shrout et al., 2006). At intracellular level the main regulator is Fur. Therefore, Fur regulates genes that are crucial for the iron acquisition needed for the bacteria replication and consequently; in biofilms development (Banin et al., 2005).
Until now, the association between iron and QS on biofilms formed by V. parahaemolyticus has not been studied. The results of a BLAST search indicate that some strains of V. parahaemolyticus have genes involved in QS and biofilm formation (luxP, luxQ, cqsA, luxO, hapR, aphA), with high identity for those reported in the V. cholerae (Jobling and Holmes, 1997; Skorupski and Taylor, 1999; Miller et al., 2002; Zhu and Mekalanos, 2003; Raychaudhuri et al., 2006; Table 2). We speculate that iron and QS could be involved in the virulence of V. parahaemolyticus. In the V. vulnificus the biosynthesis of the siderophore vulnibactin is regulated by Fur and QS (Wen et al., 2012; Kim et al., 2013). Once vulnibactin sequesters the iron needed for the replication of the bacterium, V. vulnificus catalizes the enzyme LuxS, which synthetizes the autoinducer (AI-2), involved in the activation of signals for QS and Biofilm formation. At high cell density, V. vulnificus enhances the expression of the gene vvpE, which encodes for the virulence factor elastase (Kim et al., 2013). It has been observed that mutations in LuxS reduce not only biofilm formation, also reduce virulence factors such as motility, production of proteases and the secretion of the V. vulnificus hemolysin, etc. (Wen et al., 2012).
Table 2.
Genes related to quorum sensing in Vibrio cholerae and putative genes in Vibrio parahaemolyticus.
|
V. cholerae |
V. parahaemolyticus |
|||||
|---|---|---|---|---|---|---|
| Protein/Gen | Function | Reference | Accession number | Vibrio sp. | Protein identity | Gene identity |
| luxP | Detects the AI-2 as quorum sensing (QS) signal. | Miller et al. (2002) | CP006007.1 | V. parahaemolyticus O1:K33 | 65% | 67% |
| luxQ | Can be autophosphorylated, resulting in the transfer of a phosphate group to LuxO. | Raychaudhuri et al. (2006) | CP006005.1 | V. parahaemolyticus O1:Kuk | - | 64% |
| cqsA | Acts as an autoinducer to form biofilms. | Zhu and Mekalanos (2003) | BA000032.2 | V. parahaemolyticus RIMD 2210633 | 59% | 74% |
| luxO | Activates expression of four sRNAs that destabilize hapR mRNA repressing expression of HapR. | Jobling and Holmes (1997) | CP007004.1 | V. parahaemolyticus UCM-V493 | - | 75% |
| hapR | Master regulator of QS. | Jobling and Holmes (1997) | CP006008.1 | V. parahaemolyticus O1:K33 | 72% | 75% |
| aphA | Is a winged-helix transcription factor that controls virulence factor production in the closely related pathogen and QS. | Jobling and Holmes (1997), Skorupski and Taylor (1999) | CP007004.1 | V. parahaemolyticus UCM-V493 | 68% | |
Although the link between Fur and QS is complex, the siderophores production and the coordinated regulation by the two systems (Fur and QS) probably ensures to bacteria maintain an appropriate iron concentration to optimize its survival and propagation within the human host (Wen et al., 2012). It has been shown that blocking the nutritional support and the communication pathways of one’s adversaries serves as an effective tactic to disrupt cooperative actions among individuals or groups. Removal of iron as a therapeutic approach has been investigated in vitro for several infections, with promising results (Gorska et al., 2014). The generation of analogs that block or alternative signals involved in QS have been developed, in order to disrupt biofilm formation and other virulence factors (LaSarre and Federle, 2013). These strategies could be success in bacteria, because Fe limitation and Fe excess affect QS-dependent biofilm formation, therefore understand how these sophisticated and complex regulatory systems are regulated, is vital to predict bacterial behaviors and possibly then, develop drugs that can interfere with the iron acquisitions systems, or with the response of signal molecules involved in iron acquisition systems or QS (Bollinger et al., 2001; Wen et al., 2012; LaSarre and Federle, 2013).
When the host tries to limit infection by lowering iron, pathogens such as V. parahaemolyticus triggered increased expression of virulence factors (that are relevant for surface colonization and infection) in order to cause damage to the host. Based in these results, we conclude that unnecessary or excessive iron administration may be harmful, due the possible multiplication of bacterial growth and increase in their virulence. While it is clear that iron levels are important in infection, it is not an easy task to control their levels in the host. The complete detailed mechanism for Fe acquisition and its role in V. parahaemolyticus virulence remains to be determined.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a grant from CONACyT México (CB-2014- 236546).
References
- Abu Kwaik Y., Bumann D. (2013). Microbial quest for food in vivo: ‘nutritional virulence’ as an emerging paradigm. Cell Microbiol. 15 882–890. 10.1111/cmi.12138 [DOI] [PubMed] [Google Scholar]
- Actis L. A., Fish W., Crosa J. H., Kellerman K., Ellenberger S. R., Hauser F. M., et al. (1986). Characterization of anguibactin, a novel siderophore from Vibrio anguillarum 775(pJM1). J. Bacteriol. 167 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alice A. F., Crosa J. H. (2012). The TonB3 system in the human pathogen Vibrio vulnificus is under the control of the global regulators Lrp and cyclic AMP receptor protein. J. Bacteriol. 194 1897–1911. 10.1128/JB.06614-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Othrubi S. M., Kqueen C. Y., Mirhosseini H., Hadi Y. A., Radu S. (2014). Antibioticresistanceof Vibrio parahaemolyticus isolatedfromcockles and shrimpseafoodmarketedin Selangor, Malaysia. Clin. Microbiol. 3 148–154. 10.4172/2327-5073.1000148 [DOI] [Google Scholar]
- Amaro C., Biosca E. G., Fouz B., Toranzo A. E., Garay E. (1994). Role of iron, capsule, and toxins in the pathogenicity of Vibrio vulnificus biotype 2 for mice. Infect. Immun. 62 759–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansede-Bermejo J., Gavilan R. G., Trinanes J., Espejo R. T., Martinez-Urtaza J. (2010). Origins and colonization history of pandemic Vibrio parahaemolyticus in South America. Mol. Ecol. 19 3924–3937. 10.1111/j.1365-294X.2010.04782.x [DOI] [PubMed] [Google Scholar]
- Ascencio F., Ljungh A., Wadstrom T. (1992). Lactoferrin binding properties of Vibrio cholerae. Microbios 70 103–117. [PubMed] [Google Scholar]
- Balado M., Osorio C. R., Lemos M. L. (2009). FvtA is the receptor for the siderophore vanchrobactin in Vibrio anguillarum: utility as a route of entry for vanchrobactin analogues. Appl. Environ. Microbiol. 75 2775–2783. 10.1128/AEM.02897-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin E., Vasil M. L., Greenberg E. P. (2005). Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 102 11076–11081. 10.1073/pnas.0504266102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton J. C., Acton R. T. (2009). Hemochromatosis and Vibrio vulnificus wound infections. J. Clin. Gastroenterol. 43 890–893. 10.1097/MCG.0b013e31819069c1 [DOI] [PubMed] [Google Scholar]
- Baumann P., Furniss A. L., Lee J. V. (1984). Genus Vibrio, in Bergey’s. Baltimore, MD: Williams and Wilkins. [Google Scholar]
- Bollinger N., Hassett D. J., Iglewski B. H., Costerton J. W., Mcdermott T. R. (2001). Gene expression in Pseudomonas aeruginosa: evidence of iron override effects on quorum sensing and biofilm-specific gene regulation. J. Bacteriol. 183 1990–1996. 10.1128/JB.183.6.1990-1996.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broberg C. A., Calder T. J., Orth K. (2011). Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 13 992–1001. 10.1016/j.micinf.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen J. J., Spalding P. B., Ward C. G., Gutteridge J. M. (1991). Hemochromatosis, iron and septicemia caused by Vibrio vulnificus. Arch. Intern. Med. 151 1606–1609. 10.1001/archinte.1991.00400080096018 [DOI] [PubMed] [Google Scholar]
- Caburlotto G., Gennari M., Ghidini V., Tafi M., Lleo M. M. (2009). Presence of T3SS2 and other virulence-related genes in tdh-negative Vibrio parahaemolyticus environmental strains isolated from marine samples in the area of the Venetian Lagoon, Italy. FEMS Microbiol. Ecol. 70 506–514. 10.1111/j.1574-6941.2009.00764.x [DOI] [PubMed] [Google Scholar]
- Cai Y., Wang R., An M. M., Liang B. B. (2010). Iron-Depletion prevents biofilm formation in Pseudomonas Aeruginosa through twitching mobility and quorum sensing. Braz. J. Microbiol. 41 37–41. 10.1590/S1517-83822010000100008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartron M. L., Maddocks S., Gillingham P., Craven C. J., Andrews S. C. (2006). Feo–transport of ferrous iron into bacteria. Biometals 19 143–157. 10.1007/s10534-006-0003-2 [DOI] [PubMed] [Google Scholar]
- Cassat J. E., Skaar E. P. (2013). Iron in infection and immunity. Cell Host Microbe 13 509–519. 10.1016/j.chom.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli D., Hasan N. A., Huq A., Colwell R. R. (2013). Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front. Cell Infect. Microbiol. 3:97 10.3389/fcimb.2013.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A., Ishibashi M., Thiem V. D., Tuyet D. T., Tung T. V., Chien B. T., et al. (2004). Emergence and serovar transition of Vibrio parahaemolyticus pandemic strains isolated during a diarrhea outbreak in Vietnam between 1997 and 1999. Microbiol. Immunol. 48 319–327. 10.1111/j.1348-0421.2004.tb03513.x [DOI] [PubMed] [Google Scholar]
- Craig S. A., Carpenter C. D., Mey A. R., Wyckoff E. E., Payne S. M. (2011). Positive regulation of the Vibrio cholerae porin OmpT by iron and fur. J. Bacteriol. 193 6505–6511. 10.1128/JB.05681-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crichton R. R., Charloteaux-Wauters M. (1987). Iron transport and storage. Eur. J. Biochem. 164 485–506. 10.1111/j.1432-1033.1987.tb11155.x [DOI] [PubMed] [Google Scholar]
- Danese I., Haine V., Delrue R. M., Tibor A., Lestrate P., Stevaux O., et al. (2004). The ton system, an ABC transporter, and a universally conserved GTPase are involved in iron utilization by Brucella melitensis 16M. Infect. Immun. 72 5783–5790. 10.1128/IAI.72.10.5783-5790.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraldo-Gomez J. D., Sansom M. S. (2003). Acquisition of siderophores in gram-negative bacteria. Nat. Rev. Mol. Cell Biol. 4 105–116. 10.1038/nrm1015 [DOI] [PubMed] [Google Scholar]
- Fouz B., Biosca E. G., Amaro C. (1997). High affinity iron-uptake systems in Vibrio damsela: role in the acquisition of iron from transferrin. J. Appl. Microbiol. 82 157–167. 10.1111/j.1365-2672.1997.tb02846.x [DOI] [PubMed] [Google Scholar]
- Fujino T., Okuno Y., Nakada D., Aoyama A., Fukai K., Mukai T. (1953). On the bacteriological examination of shirasu-food poisoning. Med. J. Osaka. Univ. 4 299–304. [Google Scholar]
- Funahashi T., Moriya K., Uemura S., Miyoshi S., Shinoda S., Narimatsu S., et al. (2002). Identification and characterization of pvuA, a gene encoding the ferric vibrioferrin receptor protein in Vibrio parahaemolyticus. J. Bacteriol. 184 936–946. 10.1128/jb.184.4.936-946.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funahashi T., Tanabe T., Aso H., Nakao H., Fujii Y., Okamoto K., et al. (2003). An iron-regulated gene required for utilization of aerobactin as an exogenous siderophore in Vibrio parahaemolyticus. Microbiology 149 1217–1225. 10.1099/mic.0.26066-0 [DOI] [PubMed] [Google Scholar]
- Funahashi T., Tanabe T., Shiuchi K., Nakao H., Yamamoto S. (2009). Identification and characterization of genes required for utilization of desferri-ferrichrome and aerobactin in Vibrio parahaemolyticus. Biol. Pharm. Bull. 32 359–365. 10.1248/bpb.32.359 [DOI] [PubMed] [Google Scholar]
- Furrer J. L., Sanders D. N., Hook-Barnard I. G., Mcintosh M. A. (2002). Export of the siderophore enterobactin in Escherichia coli: involvement of a 43 kDa membrane exporter. Mol. Microbiol. 44 1225–1234. 10.1046/j.1365-2958.2002.02885.x [DOI] [PubMed] [Google Scholar]
- Ganz T., Nemeth E. (2006). Regulation of iron acquisition and iron distribution in mammals. Biochim. Biophys. Acta 1763 690–699. 10.1016/j.bbamcr.2006.03.014 [DOI] [PubMed] [Google Scholar]
- Garcia K., Bastias R., Higuera G., Torres R., Mellado A., Uribe P., et al. (2013). Rise and fall of pandemic Vibrio parahaemolyticus serotype O3:K6 in southern Chile. Environ. Microbiol. 15 527–534. 10.1111/j.1462-2920.2012.02883.x [DOI] [PubMed] [Google Scholar]
- Gavilan R. G., Zamudio M. L., Martinez-Urtaza J. (2013). Molecular epidemiology and genetic variation of pathogenic Vibrio parahaemolyticus in Peru. PLoS Negl. Trop. Dis. 7:e2210 10.1371/journal.pntd.0002210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gledhill M., Buck K. N. (2012). The organic complexation of iron in the marine environment: a review. Front. Microbiol. 3:69 10.3389/fmicb.2012.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gode-Potratz C. J., Chodur D. M., Mccarter L. L. (2010). Calcium and iron regulate swarming and type III secretion in Vibrio parahaemolyticus. J. Bacteriol. 192 6025–6038. 10.1128/JB.00654-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M. B., Boyko S. A., Calderwood S. B. (1990). Transcriptional regulation by iron of a Vibrio cholerae virulence gene and homology of the gene to the Escherichia coli fur system. J. Bacteriol. 172 6863–6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorska A., Sloderbach A., Marszall M. P. (2014). Siderophore-drug complexes: potential medicinal applications of the ‘Trojan horse’ strategy. Trends. Pharmacol. Sci. 35 442–449. 10.1016/j.tips.2014.06.007 [DOI] [PubMed] [Google Scholar]
- Gray-Owen S. D., Schryvers A. B. (1996). Bacterial transferrin and lactoferrin receptors. Trends. Microbiol. 4 185–191. 10.1016/0966-842X(96)10025-1 [DOI] [PubMed] [Google Scholar]
- Griffiths G. L., Sigel S. P., Payne S. M., Neilands J. B. (1984). Vibriobactin, a siderophore from Vibrio cholerae. J. Biol. Chem. 259 383–385. [PubMed] [Google Scholar]
- Hackney C. R., Kleeman E. G., Ray B., Speck M. L. (1980). Adherence as a method of differentiating virulent and avirulent strains of Vibrio parahaemolyticus. Appl. Environ. Microbiol. 40 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms S. D., Oliver J. D., Travis J. C. (1984). Role of heme compounds and haptoglobin in Vibrio vulnificus pathogenicity. Infect. Immun. 45 345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda T., Sornchai C., Takeda Y., Miwatani T. (1982). Immunological detection of the Kanagawa phenomenon of Vibrio parahaemolyticus on modified selective media. J. Clin. Microbiol. 16 734–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert M., Blondeau R. (1998). On the iron requirement of lactobacilli grown in chemically defined medium. Curr. Microbiol. 37 64–66. 10.1007/s002849900339 [DOI] [PubMed] [Google Scholar]
- Jayalakshmi S., Venugopalan V. K. (1992). Role of iron in the virulence of Vibrio vulnificus isolated from Cuddalore coastal waters (India). Indian J. Med. Res. 95 294–296. [PubMed] [Google Scholar]
- Jin S. Q., Yin B. C., Ye B. C. (2009). Multiplexed bead-based mesofluidic system for detection of food-borne pathogenic bacteria. Appl. Environ. Microbiol. 75 6647–6654. 10.1128/AEM.00854-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling M. G., Holmes R. K. (1997). Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 26 1023–1034. 10.1046/j.1365-2958.1997.6402011.x [DOI] [PubMed] [Google Scholar]
- Johnson M., Cockayne A., Williams P. H., Morrissey J. A. (2005). Iron-responsive regulation of biofilm formation in staphylococcus aureus involves fur-dependent and fur-independent mechanisms. J. Bacteriol. 187 8211–8215. 10.1128/JB.187.23.8211-8215.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo I. (1975). Virulence-enhancing effect of ferric ammonium citrate on Vibrio cholerae. Prog. Drug Res. 19 546–553. 10.1007/978-3-0348-7090-0_62 [DOI] [PubMed] [Google Scholar]
- Joseph S. W., Colwell R. R., Kaper J. B. (1982). Vibrio parahaemolyticus and related halophilic Vibrios. Crit. Rev. Microbiol. 10 77–124. 10.3109/10408418209113506 [DOI] [PubMed] [Google Scholar]
- Karunasagar I., Joseph S. W., Twedt R. M., Hada H., Colwell R. R. (1984). Enhancement of Vibrio parahaemolyticus virulence by lysed erythrocyte factor and iron. Infect. Immun. 46 141–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I. H., Wen Y., Son J. S., Lee K. H., Kim K. S. (2013). The fur-iron complex modulates expression of the quorum-sensing master regulator, SmcR, to control expression of virulence factors in Vibrio vulnificus. Infect. Immun. 81 2888–2898. 10.1128/IAI.00375-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby S. D., Lainson F. A., Donachie W., Okabe A., Tokuda M., Hatase O., et al. (1998). The Pasteurella haemolytica 35 kDa iron-regulated protein is an FbpA homologue. Microbiology 144(Pt 12), 3425–3436. 10.1099/00221287-144-12-3425 [DOI] [PubMed] [Google Scholar]
- Koster W. (2005). Cytoplasmic membrane iron permease systems in the bacterial cell envelope. Front. Biosci. 10 462–477. 10.2741/1542 [DOI] [PubMed] [Google Scholar]
- Kuehl C. J., Crosa J. H. (2010). The TonB energy transduction systems in Vibrio species. Fut. Microbiol. 5 1403–1412. 10.2217/fmb.10.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan S. F., Huang C. H., Chang C. H., Liao W. C., Lin I. H., Jian W. N., et al. (2009). Characterization of a new plasmid-like prophage in a pandemic Vibrio parahaemolyticus O3:K6 strain. Appl. Environ. Microbiol. 75 2659–2667. 10.1128/AEM.02483-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen R. A., Myers P. S., Skare J. T., Seachord C. L., Darveau R. P., Postle K. (1996). Identification of TonB homologs in the family Enterobacteriaceae and evidence for conservation of TonB-dependent energy transduction complexes. J. Bacteriol. 178 1363–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSarre B., Federle M. J. (2013). Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 77 73–111. 10.1128/MMBR.00046-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Kim J. A., Lee M. A., Park S. J., Lee K. H. (2013). Regulation of haemolysin (VvhA) production by ferric uptake regulator (Fur) in Vibrio vulnificus: repression of vvhA transcription by Fur and proteolysis of VvhA by Fur-repressive exoproteases. Mol. Microbiol. 88 813–826. 10.1111/mmi.12224 [DOI] [PubMed] [Google Scholar]
- Letchumanan V., Yin W. F., Lee L. H., Chan K. G. (2015). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 6:33 10.3389/fmicb.2015.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwin C. M., Calderwood S. B. (1993). Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6 137–149. 10.1128/CMR.6.2.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino K., Oshima K., Kurokawa K., Yokoyama K., Uda T., Tagomori K., et al. (2003). Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361 743–749. 10.1016/S0140-6736(03)12659-1 [DOI] [PubMed] [Google Scholar]
- Matsumoto C., Okuda J., Ishibashi M., Iwanaga M., Garg P., Rammamurthy T., et al. (2000). Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38 578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter L. (1999). The multiple identities of Vibrio parahaemolyticus. J. Mol. Microbiol. Biotechnol. 1 51–57. [PubMed] [Google Scholar]
- Mey A. R., Payne S. M. (2001). Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 42 835–849. 10.1046/j.1365-2958.2001.02683.x [DOI] [PubMed] [Google Scholar]
- Mey A. R., Wyckoff E. E., Kanukurthy V., Fisher C. R., Payne S. M. (2005). Iron and fur regulation in Vibrio cholerae and the role of fur in virulence. Infect. Immun. 73 8167–8178. 10.1128/IAI.73.12.8167-8178.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miethke M., Kraushaar T., Marahiel M. A. (2013). Uptake of xenosiderophores in Bacillus subtilis occurs with high affinity and enhances the folding stabilities of substrate binding proteins. FEBS Lett. 587 206–213. 10.1016/j.febslet.2012.11.027 [DOI] [PubMed] [Google Scholar]
- Miller M. B., Skorupski K., Lenz D. H., Taylor R. K., Bassler B. L. (2002). Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110 303–314. 10.1016/S0092-8674(02)00829-2 [DOI] [PubMed] [Google Scholar]
- Morgenthau A., Pogoutse A., Adamiak P., Moraes T. F., Schryvers A. B. (2013). Bacterial receptors for host transferrin and lactoferrin: molecular mechanisms and role in host-microbe interactions. Fut. Microbiol. 8 1575–1585. 10.2217/fmb.13.125 [DOI] [PubMed] [Google Scholar]
- Nair G. B., Ramamurthy T., Bhattacharya S. K., Dutta B., Takeda Y., Sack D. A. (2007). Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20 39–48. 10.1128/CMR.00025-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka H., Crosa J. H. (2012). Identification and characterization of a novel outer membrane protein receptor FetA for ferric enterobactin transport in Vibrio anguillarum 775 (pJM1). Biometals 25 125–133. 10.1007/s10534-011-9488-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibuchi M., Fasano A., Russell R. G., Kaper J. B. (1992). Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infect. Immun. 60 3539–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Occhino D. A., Wyckoff E. E., Henderson D. P., Wrona T. J., Payne S. M. (1998). Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol. Microbiol. 29 1493–1507. 10.1046/j.1365-2958.1998.01034.x [DOI] [PubMed] [Google Scholar]
- Okada N., Iida T., Park K. S., Goto N., Yasunaga T., Hiyoshi H., et al. (2009). Identification and characterization of a novel type III secretion system in trh-positive Vibrio parahaemolyticus strain TH3996 reveal genetic lineage and diversity of pathogenic machinery beyond the species level. Infect. Immun. 77 904–913. 10.1128/IAI.01184-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda J., Ishibashi M., Abbott S. L., Janda J. M., Nishibuchi M. (1997a). Analysis of the thermostable direct hemolysin (tdh) gene and the tdh-related hemolysin (trh) genes in urease-positive strains of Vibrio parahaemolyticus isolated on the West Coast of the United States. J. Clin. Microbiol. 35 1965–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda J., Ishibashi M., Hayakawa E., Nishino T., Takeda Y., Mukhopadhyay A. K., et al. (1997b). Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 35 3150–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley S. M., Mouton S. L., Occhino D. A., Deanda M. T., Rashidi J. R., Fuson K. L., et al. (1999). Comparison of the heme iron utilization systems of pathogenic Vibrios. J. Bacteriol. 181 3594–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Estrada G., Luna-Castro S., Pina-Vazquez C., Samaniego-Barron L., Leon-Sicairos N., Serrano-Luna J., et al. (2012). Iron-saturated lactoferrin and pathogenic protozoa: could this protein be an iron source for their parasitic style of life? Fut. Microbiol. 7 149–164. 10.2217/fmb.11.140 [DOI] [PubMed] [Google Scholar]
- Pajuelo D., Lee C. T., Roig F., Hor L. I., Amaro C. (2015). Novel host-specific iron acquisition system in the zoonotic pathogen Vibrio vulnificus. Environ. Microbiol. 6 2076–2089. 10.1111/1462-2920.12782 [DOI] [PubMed] [Google Scholar]
- Paranjpye R., Hamel O. S., Stojanovski A., Liermann M. (2012). Genetic diversity of clinical and environmental Vibrio parahaemolyticus strains from the Pacific Northwest. Appl. Environ. Microbiol. 78 8631–8638. 10.1128/AEM.01531-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne S. M. (1993). Iron acquisition in microbial pathogenesis. Trends. Microbiol. 1 66–69. 10.1016/0966-842X(93)90036-Q [DOI] [PubMed] [Google Scholar]
- Pazhani G. P., Bhowmik S. K., Ghosh S., Guin S., Dutta S., Rajendran K., et al. (2014). Trends in the epidemiology of pandemic and non-pandemic strains of Vibrio parahaemolyticus isolated from diarrheal patients in Kolkata, India. PLoS Negl. Trop. Dis. 8:e2815 10.1371/journal.pntd.0002815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins-Balding D., Ratliff-Griffin M., Stojiljkovic I. (2004). Iron transport systems in Neisseria meningitidis. Microbiol. Mol. Biol. Rev. 68 154–171. 10.1128/MMBR.68.1.154-171.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel D. E., Alston D. G., Roberts D. W. (2008). Effects of physical and nutritional stress conditions during mycelial growth on conidial germination speed, adhesion to host cuticle, and virulence of Metarhizium anisopliae, an entomopathogenic fungus. Mycol. Res. 112 1355–1361. 10.1016/j.mycres.2008.04.011 [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S., Jain V., Dongre M. (2006). Identification of a constitutively active variant of LuxO that affects production of HA/protease and biofilm development in a non-O1, non-O139 Vibrio cholerae O110. Gene 369 126–133. 10.1016/j.gene.2005.10.031 [DOI] [PubMed] [Google Scholar]
- Rogers M. B., Sexton J. A., Decastro G. J., Calderwood S. B. (2000). Identification of an operon required for ferrichrome iron utilization in Vibrio cholerae. J. Bacteriol. 182 2350–2353. 10.1128/JB.182.8.2350-2353.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer L., Hocquet D., Miller S. I. (2011). Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 19 341–348. 10.1016/j.tim.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R., Saha N., Donofrio R. S., Bestervelt L. L. (2013). Microbial siderophores: a mini review. J. Basic Microbiol. 53 303–317. 10.1002/jobm.201100552 [DOI] [PubMed] [Google Scholar]
- Santic M., Abu Kwaik Y. (2013). Nutritional virulence of Francisella tularensis. Front. Cell Infect. Microbiol. 3:112 10.3389/fcimb.2013.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schryvers A. B., Bonnah R., Yu R. H., Wong H., Retzer M. (1998). Bacterial lactoferrin receptors. Adv. Exp. Med. Biol. 443 123–133. 10.1007/978-1-4757-9068-9_15 [DOI] [PubMed] [Google Scholar]
- Schryvers A. B., Stojiljkovic I. (1999). Iron acquisition systems in the pathogenic Neisseria. Mol. Microbiol. 32 1117–1123. 10.1046/j.1365-2958.1999.01411.x [DOI] [PubMed] [Google Scholar]
- Schumann G., Mollmann U. (2001). Screening system for xenosiderophores as potential drug delivery agents in mycobacteria. Antimicrob. Agents Chemother. 45 1317–1322. 10.1128/AAC.45.5.1317-1322.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seliger S. S., Mey A. R., Valle A. M., Payne S. M. (2001). The two TonB systems of Vibrio cholerae: redundant and specific functions. Mol. Microbiol. 39 801–812. 10.1046/j.1365-2958.2001.02273.x [DOI] [PubMed] [Google Scholar]
- Serrano-Luna J. J., Negrete E., Reyes M., De La Garza M. (1998). Entamoeba histolytica HM1:IMSS: hemoglobin-degrading neutral cysteine proteases. Exp. Parasitol. 89 71–77. 10.1006/expr.1998.4258 [DOI] [PubMed] [Google Scholar]
- Shrout J. D., Chopp D. L., Just C. L., Hentzer M., Givskov M., Parsek M. R. (2006). The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62 1264–1277. 10.1111/j.1365-2958.2006.05421.x [DOI] [PubMed] [Google Scholar]
- Skorupski K., Taylor R. K. (1999). A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol. Microbiol. 31 763–771. 10.1046/j.1365-2958.1999.01215.x [DOI] [PubMed] [Google Scholar]
- Strange H. R., Zola T. A., Cornelissen C. N. (2011). The fbpABC operon is required for Ton-independent utilization of xenosiderophores by Neisseria gonorrhoeae strain FA19. Infect. Immun. 79 267–278. 10.1128/IAI.00807-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y. C., Liu C. (2007). Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24 549–558. 10.1016/j.fm.2007.01.005 [DOI] [PubMed] [Google Scholar]
- Tan W., Verma V., Jeong K., Kim S. Y., Jung C. H., Lee S. E., et al. (2014). Molecular characterization of vulnibactin biosynthesis in Vibrio vulnificus indicates the existence of an alternative siderophore. Front. Microbiol. 5:1 10.3389/fmicb.2014.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe T., Funahashi T., Nakao H., Miyoshi S., Shinoda S., Yamamoto S. (2003). Identification and characterization of genes required for biosynthesis and transport of the siderophore vibrioferrin in Vibrio parahaemolyticus. J. Bacteriol. 185 6938–6949. 10.1128/JB.185.23.6938-6949.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe T., Funahashi T., Okajima N., Nakao H., Takeuchi Y., Miyamoto K., et al. (2011). The Vibrio parahaemolyticus pvuA1 gene (formerly termed psuA) encodes a second ferric vibrioferrin receptor that requires tonB2. FEMS Microbiol. Lett. 324 73–79. 10.1111/j.1574-6968.2011.02389.x [DOI] [PubMed] [Google Scholar]
- Tanabe T., Funahashi T., Shiuchi K., Okajima N., Nakao H., Miyamoto K., et al. (2012). Characterization of Vibrio parahaemolyticus genes encoding the systems for utilization of enterobactin as a xenosiderophore. Microbiology 158 2039–2049. 10.1099/mic.0.059568-0 [DOI] [PubMed] [Google Scholar]
- Tanabe T., Kato A., Shiuchi K., Miyamoto K., Tsujibo H., Maki J., et al. (2014). Regulation of the expression of the Vibrio parahaemolyticus peuA gene encoding an alternative ferric enterobactin receptor. PLoS ONE 9:e105749 10.1371/journal.pone.0105749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell B., Hassan H. M. (2013). Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front. Cell Infect. Microbiol. 3:59 10.3389/fcimb.2013.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Roman J., Leon-Sicairos N., De Jesus Hernandez-Diaz L., Canizalez-Roman A. (2013). Pandemic Vibrio parahaemolyticus O3:K6 on the American continent. Front. Cell Infect. Microbiol. 3:110 10.3389/fcimb.2013.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Roman J., Leon-Sicairos N., Flores-Villasenor H., Villafana-Rauda S., Canizalez-Roman A. (2012). Association of pandemic Vibrio parahaemolyticus O3:K6 present in the coastal environment of Northwest Mexico with cases of recurrent diarrhea between 2004 and 2010. Appl. Environ. Microbiol. 78 1794–1803. 10.1128/AEM.06953-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongxay K., Pan Z., Zhang X., Wang S., Cheng S., Mei L., et al. (2008). Occurrence of pandemic clones of Vibrio parahaemolyticus isolates from seafood and clinical samples in a Chinese coastal province. Foodborne Pathog. Dis. 5 127–134. 10.1089/fpd.2007.0045 [DOI] [PubMed] [Google Scholar]
- Wandersman C., Delepelaire P. (2004). Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58 611–647. 10.1146/annurev.micro.58.030603.123811 [DOI] [PubMed] [Google Scholar]
- Wandersman C., Delepelaire P. (2012). Haemophore functions revisited. Mol. Microbiol. 85 618–631. 10.1111/j.1365-2958.2012.08136.x [DOI] [PubMed] [Google Scholar]
- Weaver E. A., Wyckoff E. E., Mey A. R., Morrison R., Payne S. M. (2013). FeoA and FeoC are essential components of the Vibrio cholerae ferrous iron uptake system, and FeoC interacts with FeoB. J. Bacteriol. 195 4826–4835. 10.1128/JB.00738-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg E. D. (1974). Iron and susceptibility to infectious disease. Science 184 952–956. 10.1126/science.184.4140.952 [DOI] [PubMed] [Google Scholar]
- Wen Y., Kim I. H., Son J. S., Lee B. H., Kim K. S. (2012). Iron and quorum sensing coordinately regulate the expression of vulnibactin biosynthesis in Vibrio vulnificus. J. Biol. Chem. 287 26727–26739. 10.1074/jbc.M112.374165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H. C., Lee Y. S. (1994). Regulation of iron on bacterial growth and production of thermostable direct hemolysin by Vibrio parahaemolyticus in intraperitoneal infected mice. Microbiol. Immunol. 38 367–371. 10.1111/j.1348-0421.1994.tb01792.x [DOI] [PubMed] [Google Scholar]
- Wong H. C., Liu C. C., Yu C. M., Lee Y. S. (1996). Utilization of iron sources and its possible roles in the pathogenesis of Vibrio parahaemolyticus. Microbiol. Immunol. 40 791–798. 10.1111/j.1348-0421.1996.tb01144.x [DOI] [PubMed] [Google Scholar]
- Wong H. C., Liu S. H., Ku L. W., Lee I. Y., Wang T. K., Lee Y. S., et al. (2000). Characterization of Vibrio parahaemolyticus isolates obtained from foodborne illness outbreaks during 1992 through 1995 in Taiwan. J. Food Prot. 63 900–906. [DOI] [PubMed] [Google Scholar]
- Wong H. C., Shyu J. T. (1994). Identification and characterization of a protease produced by Vibrio parahaemolyticus in iron-limited medium. Zhonghua Min Guo Wei Sheng Wu Ji Mian Yi Xue Za Zhi. 27 173–185. [PubMed] [Google Scholar]
- Wright A. C., Simpson L. M., Oliver J. D. (1981). Role of iron in the pathogenesis of Vibrio vulnificus infections. Infect. Immun. 34 503–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff E. E., Boulette M. L., Payne S. M. (2009). Genetics and environmental regulation of Shigella iron transport systems. Biometals 22 43–51. 10.1007/s10534-008-9188-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff E. E., Mey A. R., Payne S. M. (2007). Iron acquisition in Vibrio cholerae. Biometals 20 405–416. 10.1007/s10534-006-9073-4 [DOI] [PubMed] [Google Scholar]
- Wyckoff E. E., Schmitt M., Wilks A., Payne S. M. (2004). HutZ is required for efficient heme utilization in Vibrio cholerae. J. Bacteriol. 186 4142–4151. 10.1128/JB.186.13.4142-4151.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Funahashi T., Ikai H., Shinoda S. (1997). Cloning and sequencing of the Vibrio parahaemolyticus fur gene. Microbiol. Immunol. 41 737–740. 10.1111/j.1348-0421.1997.tb01919.x [DOI] [PubMed] [Google Scholar]
- Yamamoto S., Okujo N., Matsuura S., Fujiwara I., Fujita Y., Shinoda S. (1994). Siderophore-mediated utilization of iron bound to transferrin by Vibrio parahaemolyticus. Microbiol. Immunol. 38 687–693. 10.1111/j.1348-0421.1994.tb01843.x [DOI] [PubMed] [Google Scholar]
- Yano Y., Hamano K., Satomi M., Tsutsui I., Ban M., Aue-Umneoy D. (2014). Prevalence and antimicrobial susceptibility of Vibrio species related to food safety isolated from shrimp cultured at inland ponds in Thailand. Food control 38 30–36. 10.1016/j.foodcont.2013.09.019 [DOI] [Google Scholar]
- Zhang L., Orth K. (2013). Virulence determinants for Vibrio parahaemolyticus infection. Curr. Opin. Microbiol. 16 70–77. 10.1016/j.mib.2013.02.002 [DOI] [PubMed] [Google Scholar]
- Zhu J., Mekalanos J. J. (2003). Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5 647–656. 10.1016/S1534-5807(03)00295-8 [DOI] [PubMed] [Google Scholar]