Abstract
The Chlamydiales are an order of obligate intracellular bacteria sharing a developmental cycle inside a cytosolic vacuole, with very diverse natural hosts, from amoebae to mammals. The clinically most important species is Chlamydia trachomatis. Many uncertainties remain as to how Chlamydia organizes its intracellular development and replication. The discovery of new Chlamydiales species from other families permits the comparative analysis of cell-biological events and may indicate events that are common to all or peculiar to some species and more or less tightly linked to “chlamydial” development. We used this approach in the infection of human cells with Waddlia chondrophila, a species from the family Waddliaceae whose natural host is uncertain. Compared to C. trachomatis, W. chondrophila had slightly different growth characteristics, including faster cytotoxicity. The embedding in cytoskeletal structures was not as pronounced as for the C. trachomatis inclusion. C. trachomatis infection generates proteolytic activity by the protease Chlamydia protease-like activity factor (CPAF), which degrades host substrates upon extraction; these substrates were not cleaved in the case of W. chondrophila. Unlike Chlamydia, W. chondrophila did not protect against staurosporine-induced apoptosis. C. trachomatis infection causes Golgi apparatus fragmentation and redirects post-Golgi sphingomyelin transport to the inclusion; both were absent from W. chondrophila-infected cells. When host cells were infected with both species, growth of both species was reduced. This study highlights differences between bacterial species that both depend on obligate intracellular replication inside an inclusion. Some features seem principally dispensable for intracellular development of Chlamydiales in vitro but may be linked to host adaptation of Chlamydia and the higher virulence of C. trachomatis.
INTRODUCTION
The Chlamydiales are an order of obligate intracellular bacteria in the phylum Chlamydiae. All tested members of the Chlamydiae share the unique developmental cycle of an actively dividing reticulate body and an almost inert elementary body and develop inside a cytosolic vacuole. Until about a decade ago, Chlamydia research was centered on well-known and highly prevalent human-pathogenic bacteria, especially on Chlamydia trachomatis and Chlamydia pneumoniae. C. trachomatis plays a significant role in human health as the most common bacterial agent of human sexually transmitted disease and the pathogen of trachoma, an often blinding infection of the eye (1, 2); C. pneumoniae is a very common pathogen, most often causing upper-airway infection (3).
More recently, an evolutionary relationship of human-pathogenic members of the genus Chlamydia (now in the family Chlamydiaceae) with a large and growing group of bacteria has been discovered (4). Numerous species have been described and placed into new families that are related to, for instance, C. trachomatis but that may have drastically different host preferences. Thus, the families Chlamydiaceae, Parachlamydiaceae, Simkaniaceae, and Waddliaceae have been formed within the Chlamydiales (4). An intriguing example is bacteria that replicate inside single-celled organisms, in particular, free-living amoebae (for instance, Parachlamydia and Protochlamydia, in the family Parachlamydiaceae), referred to as symbionts of these amoebae. Genetic studies illustrate that all chlamydiae evolved from a common ancestor about 700 million years ago; since at that time there were only single-celled organisms, this ancestor was associated with protozoa (5).
Presumably, the containment within the inclusion protects the bacteria against cellular defense systems and permits the generation of an environment specialized for bacterial replication. However, life within a cytoplasmic vacuole also requires the bacteria to solve a number of problems, such as the acquisition of nutrients for their growth; if the bacteria replicate, more inclusion membrane has to be synthesized or incorporated, and the bacteria have to organize their release from the cell or their transport to other cells. Numerous ways in which C. trachomatis affects its human host cell are known: inhibition of apoptosis and of immune signaling pathways, acquisition of cytoskeletal components, manipulation of vesicular transport linked to transport of sphingomyelin to the inclusion, fragmentation of the Golgi apparatus, and substantial transcriptional regulation are examples (6).
In coevolving with mammals, pathogenic chlamydiae had to cope with the developing, sophisticated defense systems of this lineage and will have evolved strategies for dealing with them. However, some of the above basic requirements of intracellular life probably stayed the same. Further, a substantial number of proteins proposed to be associated with chlamydial virulence in humans are also found in other bacteria that are not established human pathogens (such as Simkania, Waddlia, and members of the Parachlamydiaceae) (4). It is likely that a Chlamydia organism in a human cell has to contend with a situation that is in some ways different from that of amoeba-dwelling bacteria; however, some core features may overlap between the two situations. To understand core requirements of chlamydia-like life in an inclusion and to distinguish them from specialized host adaptations, it therefore seems a suitable approach to compare the cell biology of C. trachomatis with that of other, related species.
The natural host of Waddlia chondrophila is not known. It has been isolated from two aborted bovine fetuses (7, 8), and an association of antibodies reacting with W. chondrophila antigens and human miscarriage has been described (9). In a study of 387 patients with community-acquired pneumonia, DNA of W. chondrophila was found in one patient; in a similar investigation of another 561 samples in a different study, none was detected (10). Although it is too early to make a clear statement either way, W. chondrophila may have pathogenic potential for humans (11). However, W. chondrophila can also grow in amoebae (4), and that may be its natural niche; infections of humans may be an opportunistic accident.
W. chondrophila can grow in human cells (12–14), and experimental growth was recently also reported in an ovine trophoblast cell line (15). We here infected HeLa human cervical epithelial cells (the standard host cell line for the study of C. trachomatis) with W. chondrophila and tested for the morphology of the vacuole, association with the endolysosomal pathway, cytoskeletal recruitment, inhibition of apoptosis, cleavage of host cell proteins, the ability to compete with C. trachomatis in coinfection experiments, fragmentation of the Golgi apparatus, and transport of sphingomyelin to the Waddlia inclusion. Substantial differences relative to chlamydial behavior in most of these areas were recorded. The results provide evidence that a number of cell-biological alterations that occur during infection with C. trachomatis are not essentially linked to “chlamydial” growth (i.e., not required for all members of the Chlamydiales) but may be required only for growth of Chlamydia and perhaps be linked to bacterial virulence.
MATERIALS AND METHODS
Cell culture.
HeLa cells and the stable HeLa cell line YFP-Golgi-HeLa (16) were maintained in Dulbecco modified Eagle's minimal essential medium (DMEM) supplemented with 10% fetal calf serum (FCS; tetracycline negative; PAA Laboratories) and cultured at 37°C and 5% CO2.
Infection with W. chondrophila and C. trachomatis.
Chlamydia trachomatis LGV2 (L2) was obtained from the American Type Culture Collection (ATCC) and stored in SPG medium (0.2 M sucrose, 8.6 mM Na2HPO4, 3.8 mM KH2PO4, 5 mM glutamic acid [pH 7.4]) at −80°C. Waddlia chondrophila strain WSU 86-1044 (ATCC number VR-1470) was a kind gift of Gilbert Greub (Center for Research on Intracellular Bacteria, Institute of Microbiology, University Hospital Center and University of Lausanne, Lausanne, Switzerland) and stored in SPG medium at −80°C. One day prior to infection, cells were seeded in culture medium and incubated at 37°C with 5% CO2 overnight. Bacteria were added directly to the cells at the specified multiplicity of infection (MOI).
Immunofluorescence and 3D reconstruction.
For immunofluorescence, cells were seeded in 24-well plates on coverslips and infected as described above with an MOI of 1 to 3. For microscopy, cells were fixed in 4% PFA for 15 min, permeabilized for 10 min in 0.2% Triton X-100 (Sigma) in phosphate-buffered saline (PBS), and incubated in 5% bovine serum albumin (BSA; Sigma) in PBS. Antibodies used were rabbit anti-Chlamydia (1:3,000; Milan Analytica no. 20-698), rabbit anti-GPP130 (1:150; Covance no. PRB-144C), mouse anti-chlamydial Hsp60 (1:500; Enzo Life Sciences no. ALX-804-072), mouse anti-α-tubulin (1:400; Sigma no. T9026), rabbit anti-vimentin (1:100; Acris no. AP00289PU-N), rabbit anti-SEPT2 (1:200; Sigma no. HPA018481), goat anti-EEA1 (1:300; Santa Cruz no. sc-6415), goat anti-LAMP1 (1:300; Santa Cruz no. sc-8098), Alexa Fluor 647-conjugated donkey anti-rabbit IgG (1:500; Dianova no. 711-605-152), Alexa Fluor 488-conjugated donkey anti-rabbit IgG (1:300; Dianova no. 711-545-152), Alexa Fluor 488-conjugated goat anti-mouse IgG (1:300; Dianova no. 115-485-062), and Dylight 649-conjugated donkey anti-goat IgG (1:300; Dianova no. 705-495-147). Phalloidin Alexa Fluor 546 was used to detect F-actin (1:40; Life Technologies no. A22283). Subsequently, the samples were stained with Hoechst 33342 (1 μg/ml; Sigma) for 10 min before being mounted in Permafluor (Thermo Fisher). The samples were analyzed with a BZ 9000E microscope (Keyence) and processed using BZ II Analyzer software 1.42 (Keyence). Colocalization studies were performed by transiently transfecting HeLa cells with Rab5 and green fluorescent protein (GFP)-conjugated Rab7 constructs. Wild-type (WT) GFP-Rab7 was a gift from Richard Pagano (Addgene plasmid no. 12605) (17) and Rab5-peGFP-C1 was constructed by subcloning cDNA from DsRed-Rab5 (Addgene plasmid no. 13050) (18) via BglII and XhoI into peGFP-C1 (Clontech). Mitochondria were stained for 45 min with 250 nM MitoTracker orange CMTMRos (Life Technologies no. M7510) before fixation. For three-dimensional (3D) reconstruction of microtubules and actin, infected cells were fixed and permeabilized as described above in glass-bottom dishes (MatTek, Ashland, USA). Cells were transferred for 30 min to blocking solution (0.05% [vol/vol] Tween and 1% [wt/vol] BSA in PBS) and then incubated overnight at 4°C with mouse anti-α-tubulin (1:3,000; Sigma no. T9026) followed by washing (0.05% [vol/vol] Tween in PBS) and incubation for 1 h with Alexa Fluor 488-conjugated goat anti-mouse IgG (1:200; Life Technologies no. A-11029) and rhodamine phalloidin (250 ng/ml; Life Technologies no. P1951). Images were collected with an inverted Axiovert 200 M microscope (Zeiss) as described below. Images were assembled with Adobe Illustrator CS6 (Adobe).
Ceramide transport and live-cell imaging.
The experimental setup for ceramide transport has been described before (16). Briefly, HeLa cells were seeded in glass-bottom dishes (MatTek, Ashland, MA, USA) and infected with W. chondrophila or C. trachomatis L2 for 24 h. The medium was replaced with fresh medium containing 100 nM BODIPY (boron-dipyrromethene) FL C5 ceramide (Invitrogen no. B22650). After 30 min, cells were subjected to time-lapse microscopy at 37°C in a chamber with a humidified atmosphere (6.5% CO2 and 9% O2). Images were collected with an inverted Axiovert 200 M microscope (Zeiss) equipped with a Yokogawa CSU-X1 spinning-disc confocal head (Tokyo, Japan). Fluorescence intensities were measured inside the inclusion in a 22.88-μm2 area (C. trachomatis, 38 inclusions; W. chondrophila, 35 inclusions) at various time points using MetaMorph imaging software version 7.7.11.0 (Universal Imaging). Averages and standard errors of the means (SEM) from two independent experiments were calculated using Microsoft Excel 2010. For live-cell microscopy, cells were seeded as described above and infected with W. chondrophila for 22 h, and time-lapse microscopy was performed for 4 h as described above. Pictures were taken every minute and assembled into a movie using the MetaMorph imaging software.
Quantitative real-time PCR.
A total of 150,000 HeLa cells were seeded in 6-well plates and infected at an MOI of 1 with W. chondrophila or C. trachomatis L2 or coinfected with both species at the same time (both at an MOI of 1). At 24 and 30 h postinfection (p.i.), DNA was extracted using the DNeasy blood and tissue kit (Qiagen no. 69504) according to the manufacturer's instructions. The DNA was quantified with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). For quantitative reverse transcription-PCR (qRT-PCR) analysis, 50 ng DNA of all samples was used in a total volume of 10 μl with SYBR select master mix (Applied Biosystems no. 4472908). Primers targeting the 16S DNA of W. chondrophila have been described before (forward, 5′-GGCCCTTGGGTCGTAAAGTTCT; reverse, 5′-CGGAGTTAGCCGGTGCTTCT [19]). Primers targeting the 16S DNA of C. trachomatis were 5′-CGGTAATACGGAGGGTGCTA (forward) and 5′-CTACGCATTTCACCGCTACA (reverse) (20). The cycle conditions were 10 min at 95°C followed by 45 cycles of 15 s at 95°C and 1 min at 60°C with an ABI Prism 7900HT system (Applied Biosystems). Quantification was achieved using a standard curve derived of a dilution series of DNA extracted from purified W. chondrophila or C. trachomatis, respectively. The DNA amount was calculated with the formula (real-time PCR applications guide; Bio-Rad).
Western blotting.
For Western blot analysis, 200,000 HeLa cells were seeded in 6-well plates and infected as described above. Extracts using radioimmunoprecipitation assay (RIPA) buffer or 8 M urea dissolved in water were prepared as described before (16). Ten micrograms of protein was loaded onto 10% SDS gels and transferred to nitrocellulose membranes. After blocking of the membranes (5% milk in Tris-buffered saline–Tween [TBS-T]), they were incubated with the respective antibody at 4°C overnight. Primary antibodies were directed against β-actin (mouse, 1:10,000; Sigma no. A5441), Bim C34C5 (rabbit, 1:5,000; Cell Signaling no. 2933), vimentin (rabbit, 1:1,000; Acris no. AP00289PU-N), NF-κB p65 (rabbit, 1:5,000; Cell Signaling no. 4764), cytokeratin-8 (1:20,.000; Acris no. BM5045P), RFX5 (rabbit, 1:1,000; Rockland no. 200-401-194), USF-1 (rabbit, 1:2,000; Santa Cruz no. sc-8983), Cyclin B1 (mouse, 1:2,000; Cell Signaling no. 4135), GAPDH (mouse, 1:10,000; Millipore no. MAB374) and α-tubulin (mouse, 1:10,000; Sigma no. T9026). Secondary antibodies used were goat anti-rabbit IgG (1:5,000 to 1:20,000; Sigma no. A6667) and goat anti-mouse IgG (1:3,000 to 1:30,000; Jackson ImmunoResearch no. 115-035-166).
Cytotoxicity assay.
Cytotoxicity of W. chondrophila and C. trachomatis L2 for HeLa cells was measured by colorimetric quantification of released activity of the cytosolic lactate dehydrogenase (LDH) using the cytotoxicity detection kit (Roche no. 11644793001). A total of 30,000 HeLa cells were seeded in 24-well plates and infected with C. trachomatis L2 or W. chondrophila (MOI of 1.5). At various time points after infection, 1 ml 2% Triton X-100 (Sigma) was added to the positive control, and 1 ml medium was added to all other samples. The supernatants were sterile filtrated and centrifuged at 8,000 rpm for 5 min. LDH release was measured according to the manufacturer's protocol (Roche). All experiments were repeated three times independently, and samples were measured in triplicate at 490 nm. LDH release was calculated, and the positive control (uninfected cells treated with Triton X-100) was set to 100% at each time point.
Detection of apoptosis.
A total of 200,000 HeLa cells were seeded in 6-well plates and infected with W. chondrophila or C. trachomatis L2 (MOI of 3) for 24 and 36 h. Subsequently, cells were treated for 7 h with staurosporine (1 μM; Sigma). Active caspase-3 staining was performed as described before (21). Briefly, cells were washed, fixed, and incubated for 30 min in permeabilization buffer (0.5% [wt/vol] BSA and 0.5% [wt/vol] saponin in PBS) with monoclonal antibody against active caspase-3 (1:500; BD Pharmingen). After washing, cells were resuspended in permeabilization buffer with Cy5-conjugated secondary antibody (1:500; Dianova), washed, and analyzed by flow cytometry using a FACSCalibur flow cytometer (Becton-Dickinson).
RESULTS
Growth and cytotoxicity of W. chondrophila in HeLa cells.
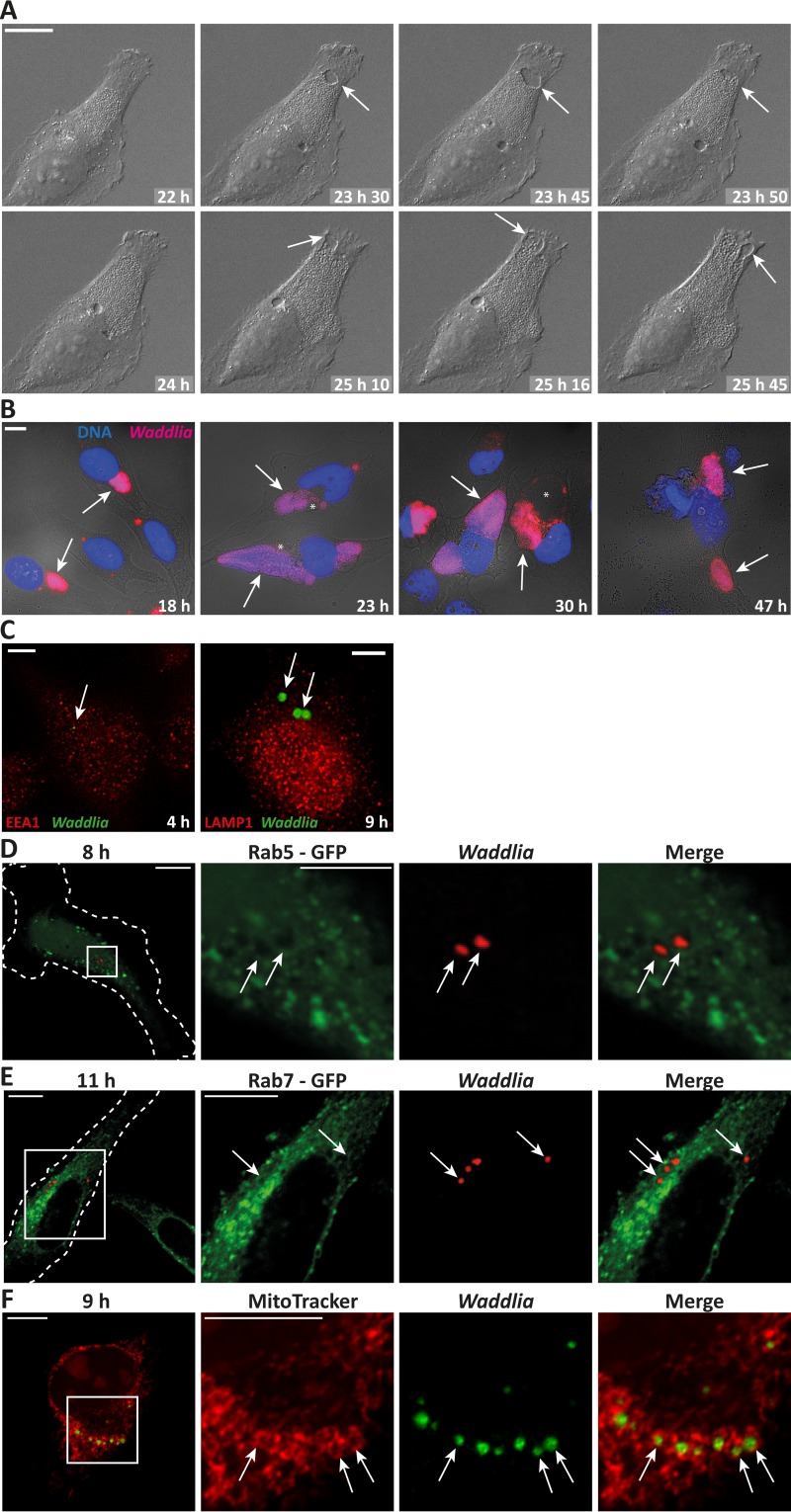
The ability of W. chondrophila to grow in mammalian cells has been established. We found that W. chondrophila readily infected HeLa cells (see below) as well as various other human cell lines (data not shown) and formed big inclusions. W. chondrophila growth was visible after about 12 h; at later stages, growth was very rapid and the inclusions appeared irregular, similar to the patterns reported in other human cells (13) or an ovine (15) cell line. On top of the main inclusion formed in an infected HeLa cell, growth of the bacteria involved the appearance of cytosolic “holes” that increased in size over about 1 to 4 h before gaining access to the inclusion and being filled up by the bacteria (Fig. 1A and B; also, see Video S1 in the supplemental material). These structures are very likely membranous, empty early inclusions. Growth, inclusion morphology, and appearance of these membrane-surrounded cytosolic structures were similar to those in HeLa cells in a number of additional human cells lines tested (we tested two melanoma lines [1205Lu and WM35], two lung cancer cell lines [H1650 and HCC827], HaCaT human keratinocytes, and 293 embryonic kidney cells [data not shown]).
FIG 1.
Waddlia chondrophila growth and association with the endolysosomal pathway in HeLa cells. (A) HeLa cells were seeded for live-cell imaging, infected with W. chondrophila at an MOI of 1, and subjected to time-lapse microscopy at 22 h p.i. Shown are selected pictures of the indicated time points after infection. Scale bar, 20 μm. HeLa cells were infected with W. chondrophila at an MOI of 1, incubated for the indicated times, and processed for immunofluorescence. (B) Blue, Hoechst DNA stain; pink, Waddlia (anti-Chlamydia stain). Scale bar, 10 μm. (C) HeLa cells were infected with W. chondrophila at an MOI of 2, centrifuged for 60 min at 550 × g, incubated for the indicated times, and processed for immunofluorescence. Green, Waddlia; red, EEA1 and LAMP1. Scale bar, 5 μm. (D and E) HeLa cells were transiently transfected with Rab5-GFP (D) or Rab7-GFP (E). Four hours later, they were infected for the indicated times as described above (for panel C). Green, Rab5-GFP or Rab7-GFP; red, Waddlia. Scale bar, 10 μm (D and E) or 5 μm (Rab5 magnification). (F) HeLa cells were infected as described above (for panel C) and incubated with MitoTracker, showing a clear and complete association of all inclusions with mitochondria. Scale bar, 10 μm. Arrows point to inclusions; asterisks mark holes (membrane-surrounded spaces).
Although an initial association of W. chondrophila with early endosomal compartments has been shown in macrophages by colocalization with EEA1 (early endosomal antigen 1) (22), no such association could be detected in HeLa cells at 10, 20, 30, 60, 120, or 240 min after infection (Fig. 1C and data not shown). Likewise, no association of W. chondrophila with the late endosomal marker LAMP1 was detected (Fig. 1C). We further tested for recruitment of the early endosomal marker proteins Rab5 and Rab7 to the Waddlia inclusion at early time points (8 to 11 h p.i.) (Fig. 1D and E) as well as later during infection (16 to 24 h p.i.) (data not shown). Colocalization of neither marker was seen.
Close proximity of W. chondrophila inclusions with mitochondria has been shown in bovine, monkey, and mouse cells as well as in human macrophages and Ishikawa epithelial cells (13, 22). This close association with mitochondria was also seen in HeLa cells (Fig. 1F). Even as early as 9 h p.i., every inclusion analyzed was surrounded by mitochondria.
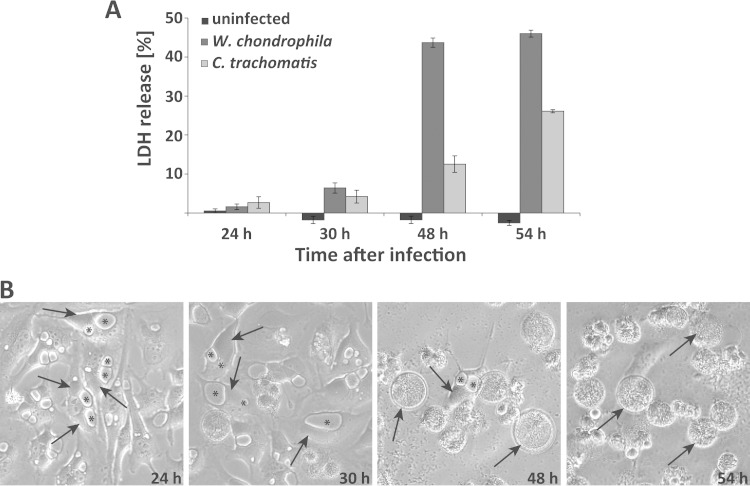
Cytotoxicity of W. chondrophila has been observed in human (13) as well as ovine (15) cells. In HeLa cells, there was little cytotoxicity for the first 30 h of infection with an MOI of about 1 to 3 but very substantial cell death (by morphology and as measured by release of the enzyme activity of the cytosolic protein LDH) between 30 and 48 h (Fig. 2A). Around this time, numerous cells seemed to rupture (Fig. 2B), suggesting that cytotoxicity was linked to expansive growth of the bacteria inside the cell.
FIG 2.
Waddlia chondrophila growth and cytotoxicity in HeLa cells. (A) Cytotoxicity was determined by measuring LDH release. At the indicated time points, uninfected cells were lysed as positive controls, the measured LDH release was set to 100%, and all LDH values were calculated relative to this positive control. Shown are results of three independent experiments and SEM. (B) Phase contrast pictures of HeLa cells infected with W. chondrophila at an MOI of 1.5 incubated for 24, 30, 48, or 54 h along with uninfected control cells. Arrows point to inclusions and asterisks mark the holes (membrane-surrounded spaces) in three randomly chosen cells.
Cytoskeletal embedding of W. chondrophila in HeLa cells.
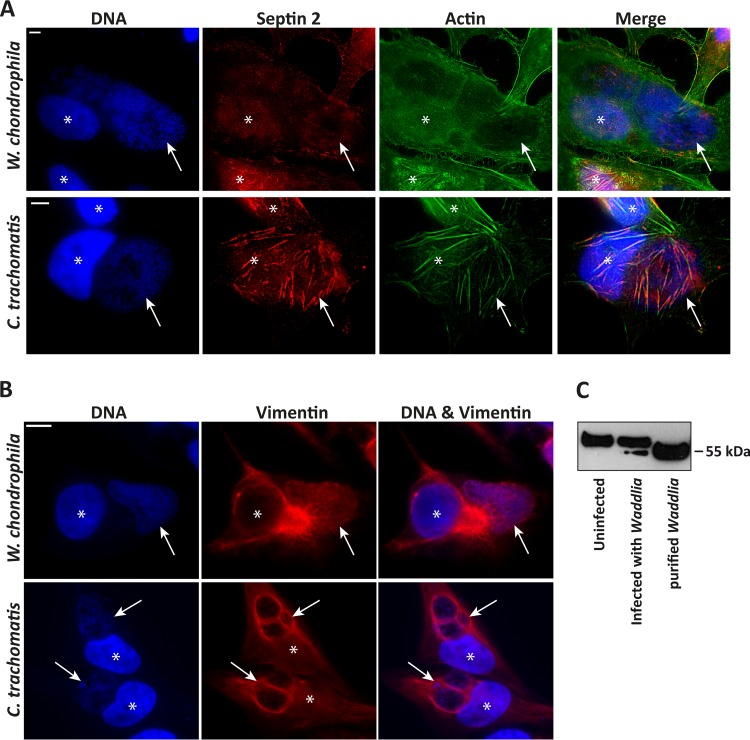
The arrangement of cytoskeletal structures around the inclusion is a feature of C. trachomatis infection. The major cytoskeletal components, microtubules and actin, were arranged around the W. chondrophila inclusion (see Fig. S1 in the supplemental material), as is known to be the case for C. trachomatis (23). Although clearly detectable, this association was less pronounced than during chlamydial infection (for instance, see reference 24). Confocal 3D reconstruction illustrated a tubulin structure around the Waddlia inclusion (see Fig. S1 in the supplemental material); the cytosolic membranous structures (referred to as holes) were also encased in microtubular structure but were only very thinly, if at all, coated on the top of the cell grown on a solid support (see Fig. S1 in the supplemental material). The intermediate filament vimentin formed the reported “cage” around the C. trachomatis inclusion. However, this was only very discrete in case of the W. chondrophila inclusion (Fig. 3A). (The faint staining of the inclusion probably shows the cross-reactivity of the vimentin antibody with Waddlia antigens, as purified W. chondrophila also gave a signal with this antibody in Western blotting [Fig. 3C].) We recently reported that septins are arranged around the chlamydial inclusion and play a role in the arrangement of F-actin fibers (24). In contrast, no such coating with septins was visible on Waddlia inclusions (Fig. 3A).
FIG 3.
Cytoskeleton rearrangement in W. chondrophila-infected cells. (A) HeLa cells were infected with W. chondrophila or C. trachomatis L2 at an MOI of 1 for 30 h, processed for immunofluorescence and stained for DNA (blue), septin-2 (red), and actin (green). (B) HeLa cells were infected with W. chondrophila or C. trachomatis L2 at an MOI of 1 for 24 h, processed for immunofluorescence and stained for DNA (blue) and vimentin (red). While there is clearly a vimentin cage around the chlamydial inclusions, no such structure is clearly discernible in Waddlia infection. Asterisks indicate nuclei; arrows point to inclusions. Scale bar, 5 μm. The faint stain of the inclusion very likely indicates cross-reactivity of W. chondrophila with the antibody, as suggested by a vimentin Western blot in panel C. (C) Whole-cell lysates of HeLa cells either uninfected or infected with W. chondrophila (MOI of 1) for 30 h were prepared with urea extraction buffer. Purified W. chondrophila was lysed for 10 min at 95°C in 2× Laemmli buffer and loaded as a control. Eight micrograms of protein was loaded onto each lane. The upper band corresponds to vimentin, and the lower band shows cross-reaction of the antibody with a W. chondrophila protein.
In the case of C. trachomatis infection, it is believed that the cytoskeleton contributes to the acceptance of the inclusion by the cell, by providing physical support (especially vimentin and actin fibers [23]) and by organizing support pathways. Our results show that the W. chondrophila inclusion is similarly well accepted and that its growth is supported by the cell, although it is not as tightly embedded in cytoskeletal elements. This may be the reason for the irregular shape of the Waddlia inclusions.
Degradation and loss of host cell proteins.
One feature of chlamydial infection, which has recently generated a heated debate, is the degradation of host cell proteins in the course of the infection. Many degradation events have been reported in the literature (25), but the ones tested have been found, as far as can be tested by simple methods, to be extraction artifacts (26). Nevertheless, although most of the cleavage probably does not occur prior to the rupture of the inclusion, this cleavage may still have a function during rupture, for instance by the degradation of cytoskeletal structures for release of the bacteria (postulated for vimentin [27]). In C. trachomatis infection, at least most of this activity is exerted by the chlamydial protease CPAF (Chlamydia protease-like activity factor) (26). W. chondrophila has the CPAF gene (in analogy to the Chlamydia protein, the product could be called Waddlia protease-like activity factor [WPAF]; we prefer the term Waddlia CPAF). Whether similar host cell proteolysis is seen either in intact cells or upon cell lysis is not known.
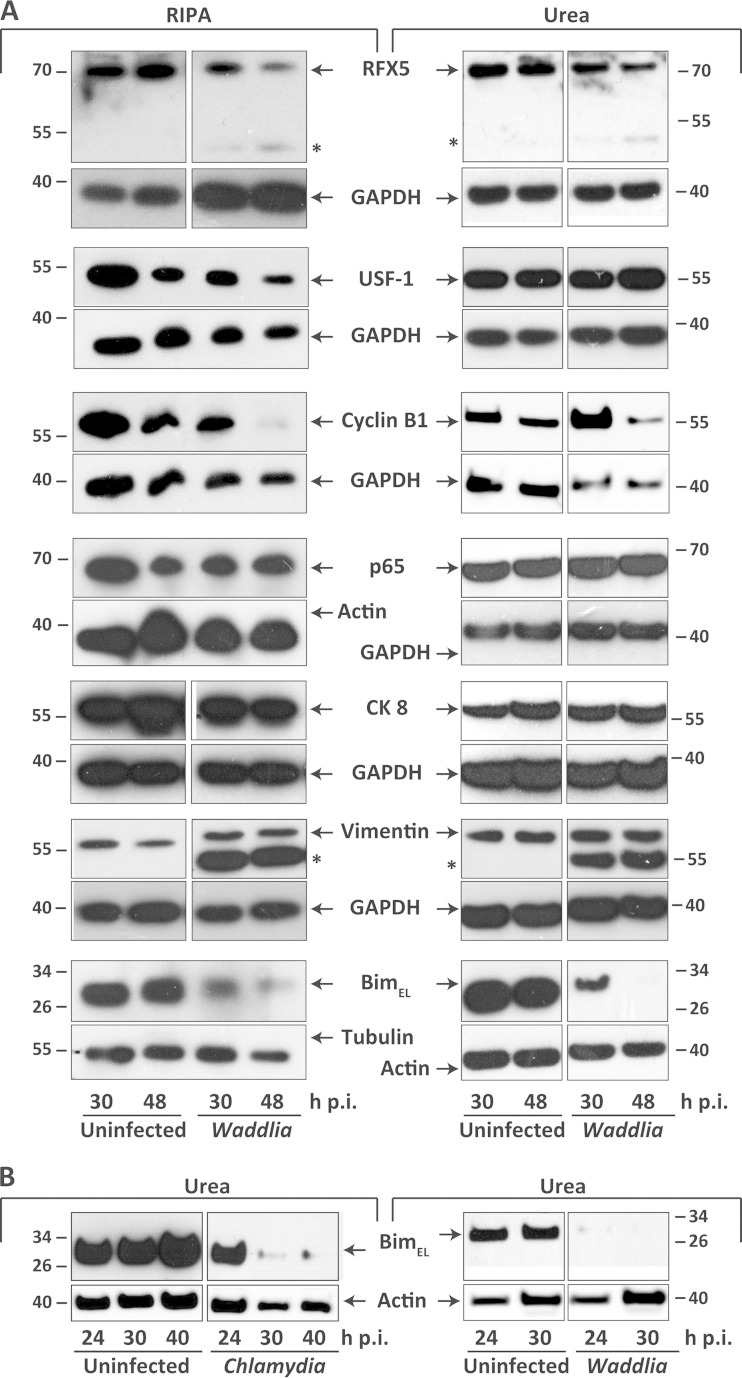
We therefore tested for such cleavage events. A number of proteins were selected that are known to be potential CPAF substrates (25): RFX5, USF-1 (both transcription factors), cyclin B1, p65/RelA (a member of the NF-κB-family), the intermediate filament proteins vimentin and cytokeratin-8, and the proapoptotic Bcl-2 family protein Bim. We performed the analysis either on samples extracted with RIPA buffer (permitting proteolysis during extraction) or on samples extracted with 8 M urea (which has been found to inhibit extraction-associated protein degradation during extraction of C. trachomatis-infected cultures [26]).
As shown in Fig. 4A, there was no cleavage detectable for most of the proteins selected in either detergent or urea buffer. Figure 4A shows the results up to 48 h postinfection, when substantial cytotoxicity was already observed (Fig. 2). In some experiments we analyzed up to 72 h; at this time, the results are difficult to interpret, since most proteins, including the controls, start disappearing, most likely due to lysis of the cells (not shown). However, even at that point, we never saw a cleavage product corresponding to what has often been observed in C. trachomatis-infected cells for most of the proteins tested.
FIG 4.
Impact of Waddlia chondrophila infection on integrity of host cell proteins. (A) HeLa cells were infected with W. chondrophila at an MOI of 1 and whole-cell lysates were prepared with either RIPA (left) or urea (right) extraction buffer (see Materials and Methods) at 30 and 48 h p.i. Ten micrograms of protein was loaded onto each lane. Shown are representative Western blots of uninfected or W. chondrophila-infected cells. The depicted results were reproduced independently. RFX5, n = 3 (urea) and 5 (RIPA); cyclin B1, n = 2 (urea) and 4 (RIPA); BimEL, n = 4 (urea) and 6 (RIPA); USF-1, n = 3 (urea) and 3 (RIPA); p65, n = 3 (urea) and 4 (RIPA); vimentin, n = 3 (urea) and 6 (RIPA); cytokeratin 8 (CK 8), n = 3 (urea) and 5 (RIPA). Actin, tubulin, and GAPDH were used as loading controls. (B) Representative Western blots of uninfected, W. chondrophila-infected, and C. trachomatis-infected cells. Urea lysates were prepared at 24, 30, or 40 h p.i.
In the vimentin blots, an additional smaller band was seen, but there was no decrease of the band corresponding to intact vimentin. The lower band corresponded to a W. chondrophila protein, as a band of the same size also appeared when W. chondrophila lysates were probed (Fig. 3C and 4A). The same pattern was observed in detergent- and in urea-extracted samples. Vimentin therefore is also not cleaved detectably during W. chondrophila infection.
However, it was evident that two of the investigated proteins, cyclin B1 and Bim, were detectably reduced during infection with W. chondrophila. This effect was less clear for cyclin B1 (and we cannot exclude the possibility that at 48 h, lysis of the cells is responsible) but prominent for Bim. Loss of Bim appeared to be the same in urea and in RIPA buffer, indicating that it was not an extraction artifact. Bim protein levels were substantially reduced as early as 24 h p.i. (Fig. 4B), at which time infection-induced cell death and release of proteins were still negligible (Fig. 2B); loss of Bim therefore seems not to be a result of infection-associated cell lysis. We also reproduced the loss of Bim during C. trachomatis infection here, as this still appears to be a contentious issue. For Fig. 4B, we used 8 M urea to extract the infected cells to prevent artificial lysis of proteins during preparation, as previously suggested (26). The result was similar to the one seen when cells were extracted with buffer containing 2% SDS, as we reported previously (28) (a recent report suggests that 1% SDS is sufficient to prevent extraction-associated degradation of proteins [27]). Although we can only speculate as to its mechanism, this loss of Bim was thus not inhibited by preventing lysis-associated artifacts (28) and may be caused by protein turnover effects.
Taken together, these results indicate that the proteolytic activity that is generated in W. chondrophila-infected cells appears either to be substantially lower than during infection with C. trachomatis or to have substantially different cleavage specificity.
Apoptosis inhibition in cells infected with W. chondrophila.
The inhibition of apoptosis, first reported in 1998 (29), is a prominent feature of infection with C. trachomatis. The mechanism of apoptosis inhibition is still uncertain, and it has variously been proposed to be linked to high expression of Mcl-1 (30), to inhibitor-of-apoptosis proteins (IAPs) (31), and to the loss of BH3-only proteins such as Bim (28). Although resistance to other stimuli is also clear (31–33), most studies have used the strong proapoptotic chemical staurosporine to test for this. Staurosporine induces apoptosis in most cells via the mitochondrial (Bcl-2-regulated) apoptotic pathway, although the molecular details of its action remain uncertain.
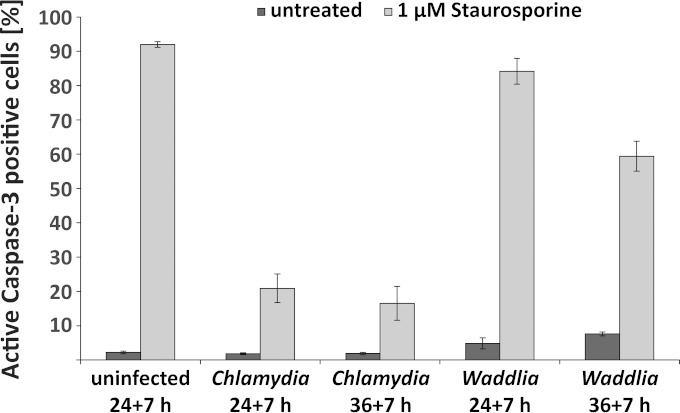
We used staurosporine-induced apoptosis to test whether W. chondrophila shares the antiapoptotic activity of C. trachomatis. We tested this by treating infected cells with staurosporine and measuring the activation of caspase-3, which is a very robust and specific feature of apoptosis. The anti-apoptotic effect of C. trachomatis was impressively reproduced, even when cells were treated with staurosporine for the period of 36 to 43 h postinfection (Fig. 5). However, there was very little if any such effect in W. chondrophila-infected cells. We tested two time points of infection, 24 h and 36 h (this was the time point when staurosporine was added for 7 h). The cytotoxic effect of W. chondrophila infection is detectable from about 30 h postinfection (see Fig. 2). When staurosporine was added for the time period between 24 and 31 h postinfection, there was no or very little protection (Fig. 5). When staurosporine was added to cells infected with W. chondrophila for the time between 36 and 43 h postinfection, there was less active caspase-3 detected in infected cells than in control cells. However, at this stage, cell lysis is very substantial (Fig. 2). It therefore seems likely that cells were unable to undergo apoptosis because they were in the process of being lysed by the infection at this late stage. We conclude that W. chondrophila inhibits apoptosis, if at all, to a much lower level than C. trachomatis. It should further be noted that, unlike C. trachomatis, W. chondrophila had a low level of proapoptotic activity, with 5 to 10% caspase-3-positive cells in infected cultures at 31 and 43 h postinfection (Fig. 5).
FIG 5.
Waddlia chondrophila is unable to inhibit apoptosis in infected cells. HeLa cells were infected with W. chondrophila or C. trachomatis L2 at an MOI of 3 and incubated for 24 or 36 h prior to incubation with 1 μM staurosporine for 7 h. Cells were then fixed, stained for active caspase-3, and analyzed by flow cytometry. Results are the means ± SEM from three independent experiments measured in triplicate.
W. chondrophila does not depend on Golgi apparatus fragmentation during infection.
An effect that has received much attention is the fragmentation of the Golgi apparatus during infection with C. trachomatis. The Golgi apparatus is arranged around the inclusion, and since protease inhibitors that inhibit Golgi apparatus fragmentation also inhibit chlamydial growth, it has been proposed that Golgi apparatus fragmentation is important for chlamydial replication (34). The mechanism of Golgi apparatus fragmentation during chlamydial infection is in dispute (for a recent discussion, see reference 35).
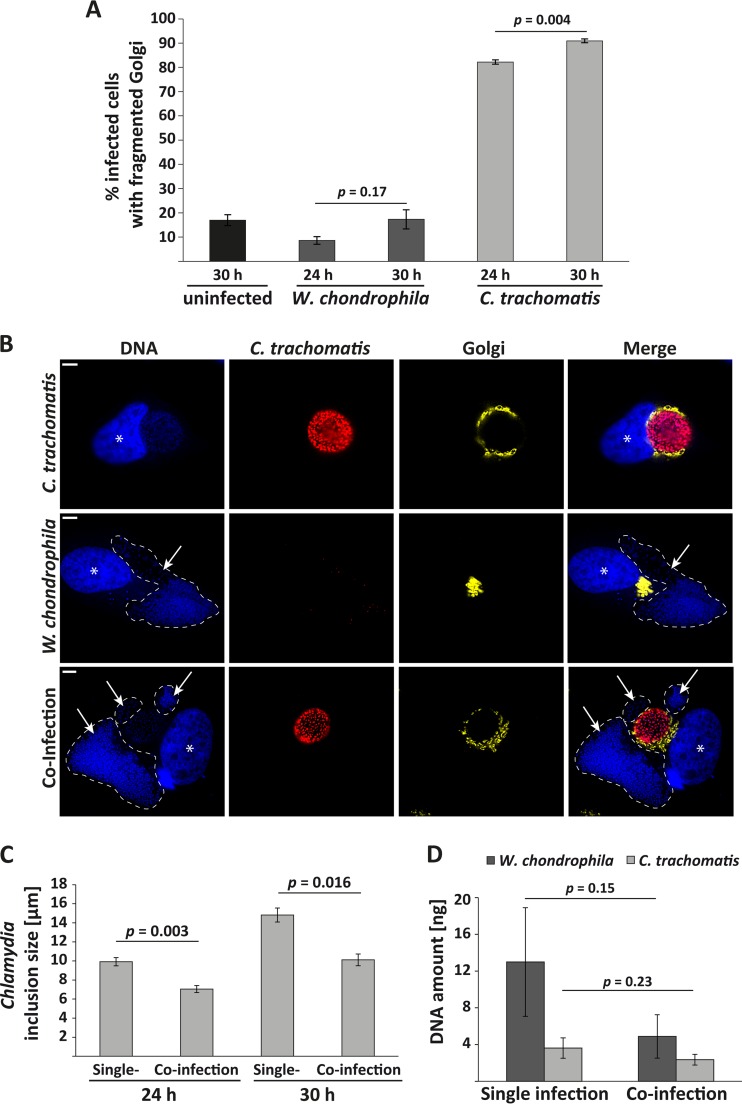
Golgi apparatus fragmentation is a very common event that can occur during numerous instances of cell-biological perturbations (for example, see references 36 to 41). We tested whether Golgi apparatus fragmentation also occurs during infection with W. chondrophila and therefore may be an essential feature of replication in a chlamydial vacuole. However, W. chondrophila was able to grow in the absence of detectable Golgi apparatus fragmentation (Fig. 6A and B). Even at 30 h p.i., when Waddlia inclusions were massive, there was no Golgi apparatus fragmentation above background (around 15% of cells), while 91% of the C. trachomatis-infected cells displayed a fragmented Golgi apparatus (Fig. 6A).
FIG 6.
Appearance of the Golgi apparatus during W. chondrophila infection. (A) Quantification of GPP130-stained HeLa cells or YFP-Golgi-HeLa cells showing a fragmented Golgi apparatus. All infected cells were assessed for Golgi apparatus fragmentation, and the ratio of fragmentation-positive cells was calculated. As a control, Golgi apparatus fragmentation was determined in uninfected samples (numbers of cells counted: 401, 305, 326, 327, and 309 [left to right]). The Golgi apparatus was considered fragmented when its normal, dense organization was disrupted and when individual cisternae were identifiable around the inclusion. The results are the means ± SEM from three independent experiments. (B) YFP-Golgi-HeLa cells were infected with W. chondrophila or C. trachomatis L2 at an MOI of 1, incubated for 24 h, processed for immunofluorescence and stained for DNA (blue) and C. trachomatis (red); Golgi apparatus is shown in yellow. For better visualization, Waddlia inclusions are marked with dashed lines. The Golgi apparatus is clearly fragmented and arranged around the chlamydial inclusion but not the Waddlia inclusion. Asterisks indicate nuclei, and arrows point to Waddlia inclusions. Scale bar, 5 μm. (C) Measurement of inclusion size in HeLa cells either infected with C. trachomatis L2 alone (MOI of 1) or coinfected with W. chondrophila and C. trachomatis L2 (both at an MOI of 1). Numbers of inclusions counted: 442, 398, 364, and 305 (left to right). Shown are the results of four (24 h) and three (30 h) independent experiments. Error bars represent the standard errors of the mean, and statistical significance was calculated by two-tailed Student's t test. (D) Quantification of DNA amounts of W. chondrophila and C. trachomatis in individual-infection and in coinfection experiments by qRT-PCR. Shown are the results of three independent experiments, each measured in duplicate, the error bars represent the standard deviation. The reduction in DNA amount in coinfection experiments is not statistically significant as calculated by two-tailed Student's t test.
Golgi apparatus fragmentation, a salient feature of C. trachomatis infection, is therefore principally dispensable for intracellular growth of W. chondrophila. Interestingly, when HeLa cells were simultaneously infected with C. trachomatis and W. chondrophila, the Golgi apparatus was fragmented and arranged around the chlamydial but not the Waddlia inclusion (Fig. 6B). During these coinfection experiments, a considerable difference in chlamydial inclusion size was observed (Fig. 6C). In terms of genome equivalents, coinfection with W. chondrophila reduced growth of C. trachomatis to about 65% at 24 h (compared to HeLa infection with C. trachomatis alone), while coinfection with C. trachomatis reduced the growth of W. chondrophila to about 40% (Fig. 6D).
Sphingomyelin transport to the inclusion.
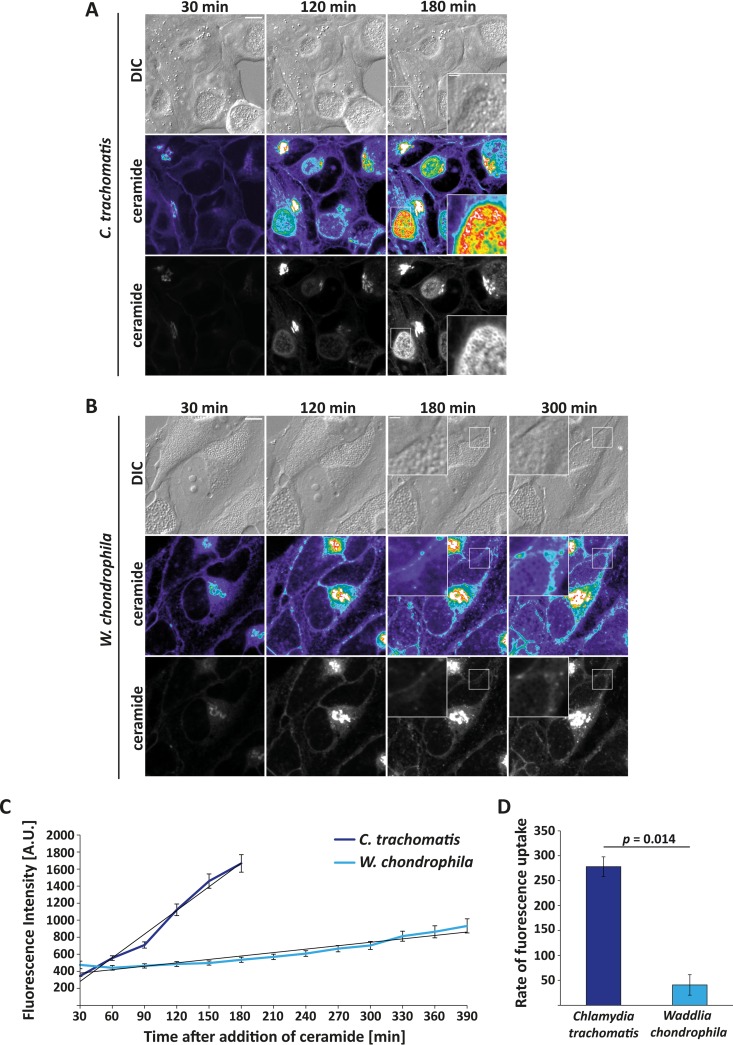
Almost 20 years ago, it was reported that Chlamydia can redirect post-Golgi transport and by this mechanism acquires sphingolipids from the host (42, 43). This transport can be measured by following the transport of fluorescent ceramide to the infected cells, which accumulates in the Golgi apparatus before being transported as sphingomyelin to the C. trachomatis inclusion membrane and ultimately into the bacteria (reproduced in Fig. 7A, C, and D). Intriguingly, there was little or no such transport to W. chondrophila (Fig. 7B to D). Like Golgi apparatus fragmentation, sphingomyelin acquisition by redirecting post-Golgi transport therefore appears to be dispensable for W. chondrophila and is thus not a universal feature of members of the Chlamydiae.
FIG 7.
Sphingomyelin transport into Waddlia chondrophila and Chlamydia trachomatis inclusions. HeLa cells were infected with C. trachomatis L2 (A) or W. chondrophila (B) at an MOI of 1 for 24 h. Confocal microscopic pictures were taken every 30 min. Representative pictures taken 30, 120, 180, and 300 min after addition of BODIPY-FL C5 ceramide are shown. The magnified insets show the fluorescence uptake into the bacterial cell walls. The heat map ranges from purple (low intensity) to white (high intensity). (C) Quantification of fluorescence intensity. Averages of two independent experiments (38 inclusions with C. trachomatis and 35 inclusions with W. chondrophila infection) and the trend line are given; error bars represent the standard errors of the means. (D) Rate of fluorescence uptake by C. trachomatis and W. chondrophila, compared by depicting the slope of the trend line of the two experiments whose results are shown in panel C. Error bars represent standard deviations, and P values were calculated using a two-tailed Student's t test.
DISCUSSION
Our study identifies a number of cell-biological events during infection of human cells with W. chondrophila. The comparison with the infection with C. trachomatis gives us the opportunity to identify events that are not indispensably linked with the intracellular life style of the Chlamydiales but may represent adaptation features of the family Chlamydiaceae or of the genus Chlamydia to their vertebrate hosts. The inclusions of both C. trachomatis and W. chondrophila are embedded by the cytoskeleton into the cell, but there are no comparable cage-like structures formed by actin, septin-2, and vimentin around Waddlia inclusions. As observed before, the inclusion is less regular in W. chondrophila infection, and growth entails the curious feature of partly propagating through the generation of initially apparently empty, presumably membrane-surrounded spaces (holes), which later gain access to the inclusion. These membrane-surrounded structures may indeed have origins similar to those of structures that have been described as secondary inclusions during infection with some strains of C. trachomatis: initially empty membranous vesicles that later on fill with chlamydial reticulate bodies (RBs) and that are connected to the primary inclusion by IncA-laden fibers (44).
W. chondrophila was found to be more cytotoxic than C. trachomatis but unable to inhibit staurosporine-induced apoptosis. We also observed less proteolytic activity toward C. trachomatis CPAF substrates. Unlike during infection with C. trachomatis, there was no detectable fragmentation of the Golgi apparatus or sphingomyelin transport to the bacteria. Thus, although W. chondrophila can grow readily in all human cell lines tested, W. chondrophila infection lacks a number of features associated with C. trachomatis infection. These features may reflect the adaptation of Chlamydia to their hosts, and the lack of these abilities may be a barrier to W. chondrophila to establish itself as a human pathogen.
At the same time, the different cell biology may be interpreted to mean that the two organisms and their growth characteristics are not as closely associated as is suggested by the “chlamydial” developmental cycle inside a cytosolic inclusion. It may certainly be argued that the different bacteria use different cell biologies, which are characterized by different requirements. However, since all members of the Chlamydiales appear to have originated from a common progenitor (5), all life styles very likely built on one original set of cell-biological features. Differences between the two species, as observed here, therefore permit the distinction into features that are probably essential and features that reflect adaptation to particular hosts or conditions.
Cytoskeletal interactions of C. trachomatis are understood to some extent. Microtubules are used for initial trafficking of the nascent inclusion (45), and defined microdomains on the inclusion appear to be responsible for this interaction (46). Actin recruitment has been investigated with small-molecule inhibitors, and a contribution of bacterial activities and a number of host cell networks involved have been identified (47). Intermediate filaments have been suggested to convey stability to the inclusion (23). We found tubulin rearrangements around the Waddlia inclusion but neither actin/septin networks nor the vimentin cage. The function of actin/septin arrangements is in part the release of the intact C. trachomatis inclusion by “extrusion” (48); the large amount of cell lysis by W. chondrophila may be a correlate of the lack of extrusion. The intermediate filament/vimentin structures have been suggested to convey stability. The lack of these structures in W. condrophila infection may be the reason for the irregular shape of the inclusions.
The subject of proteolysis during chlamydial infection has attracted controversy. C. trachomatis expresses CPAF, which is likely to translocate in part from the inclusion to the cytosol (16, 49, 50), although it is difficult to be absolutely certain of this for technical reasons and although the bulk of it is probably released only if the inclusion ruptures toward the end of the developmental cycle (27). Most of the reported massive degradation events due to CPAF activity are probably extraction artifacts (26), although small amounts of degradation occurring in intact cells cannot be excluded at present (see discussion in reference 35). Against this background, it is interesting that the reported CPAF substrates that we have investigated are not (or at least not strongly) degraded by Waddlia CPAF, providing additional evidence that their cleavage is not essentially linked to the chlamydial life style. We cannot at this stage make the distinction whether Waddlia CPAF shows less activity or has a different activity toward host cell proteins.
We included the host protein Bim in this analysis, since it has been shown that Bim can be degraded by an activity in detergent extracts from C. trachomatis-infected cells, which in turn can be inhibited by lactacystin (which blocks CPAF activity) (51). It is likely that this activity is released during detergent lysis (26). However, the loss of Bim during chlamydial infection is also seen when cells are lysed in harsher conditions probably precluding artifacts (using direct lysis in 2% SDS [28] or 8 M urea [our unpublished data]), and although CPAF can have the effect of reducing Bim expression, it is very likely not through direct cleavage (52). A very similar effect appears to operate in W. chondrophila infection, where Bim also disappeared, also under conditions where extraction-associated lysis was prevented.
Antiapoptotic activity is a prominent feature of C. trachomatis infection. It was therefore surprising that staurosporine-induced apoptosis was not inhibited by W. chondrophila. It should be noted that we do not know why C. trachomatis needs this activity. One possibility is that C. trachomatis infection induces apoptosis and therefore also has to have the ability to block it. We observed some low-level proapoptotic activity during Waddlia infection; that may be seen as supporting this hypothesis.
It is also possible that the activity is required during infection in vivo, where the immune system may induce apoptosis in infected cells. Indeed, it is conceivable that W. chondrophila, although it can grow well in human cells in vitro, is not a highly prevalent human pathogen because it cannot in some way counter the immune system in the same manner as Chlamydia, and this could be linked to the ability to prevent apoptosis.
How chlamydial antiapoptosis works is still not clear. We have proposed that it is linked to the loss of Bim and other proapoptotic molecules (28, 33). This loss is indeed likely to have an effect, since these BH3-only proteins, and most prominently Bim, are important triggers of apoptosis, and the loss of Bim has strong effects on apoptosis in a number of cells (53, 54). However, this effect is not sufficient for inhibition of staurosporine-induced apoptosis in HeLa cells, since W. chondrophila-infected cells have very little Bim left yet are almost fully sensitive to staurosporine. Thus, while it is likely that the loss of Bim has an antiapoptotic effect, the prominent antiapoptotic effect of C. trachomatis is probably independent of Bim.
Perhaps the greatest surprise was the lack of Golgi apparatus fragmentation and of post-Golgi sphingomyelin acquisition by W. chondrophila. This pathway is very well established in chlamydial infection. The mechanism of Golgi apparatus fragmentation by C. trachomatis is not known. The only molecular explanation that had been put forward (i.e., that it occurs through degradation of the Golgi apparatus matrix protein golgin-84 [34]) is probably not correct, since it is doubtful that golgin-84 is degraded in intact infected cells (26). However, Golgi apparatus fragmentation during chlamydial infection has been found to require Rab proteins (55) and is therefore likely linked to vesicular transport; Golgi apparatus fragmentation may even be a consequence of the redirection of post-Golgi vesicular transport to the inclusion. If that is the case, it is not surprising that both events (Golgi fragmentation and post-Golgi sphingomyelin transport to the bacteria) can be seen in Chlamydia infection but that neither is seen in Waddlia infection.
Post-Golgi-sphingomyelin transport is not the only known means of lipid acquisition by Chlamydia. Ceramide and sphingomyelin can be transported to the inclusion by the cytosolic lipid transporter CERT; lipids may be acquired through multivesicular bodies or through lipid droplets; glycerophospholipids as well as sphingomyelin may be acquired through host cell signaling processes (for a review, see reference 56). W. chondrophila may therefore use options other than these to satisfy its need for host lipids. Alternatively, or in addition to this, W. chondrophila may have means of synthesizing lipids through additional pathways and thus use other precursors. The W. chondrophila genome contains 2,028 coding sequences (CDSs) (4), which is more than twice the number in C. trachomatis, and may therefore also encode additional ways of acquiring versus synthesizing nutrients, as has been suggested based on genomic analyses (11).
In summary, although the infection biology of C. trachomatis and W. chondrophila is generally similar, there are substantial differences. A number of features of C. trachomatis infection are not indispensable for the biphasic developmental cycle and growth inside the cytosolic inclusion. The differences are likely adaptations of C. trachomatis to vertebrate hosts and the abilities to inhibit apoptosis, to be accepted by cytoskeletal structures, and to redirect sphingomyelin transport into the inclusion may all be features that are necessary for successful establishment of C. trachomatis in humans in vivo. Although it has been proposed that W. chondrophila is an emerging pathogen, the number of cases where these bacteria have been isolated from human patients is very small, and a causative role of W. chondrophila has not been demonstrated for any clinical condition. Understanding these different features may therefore help us understand how C. trachomatis has come to be such a successful pathogen.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the German Research Foundation (grants SPP1580 and HA2128/16-1 to G.H.). We thank G. Greub for providing the Waddlia chondrophila strain, Arlena Metz for expert technical assistance, and Carsten Schwan (Institute of Experimental and Clinical Pharmacology and Toxicology, University of Freiburg, Freiburg, Germany) for his help with the confocal microscopy.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00322-15.
REFERENCES
- 1.Pascolini D, Mariotti SP. 2012. Global estimates of visual impairment: 2010. Br J Ophthalmol 96:614–618. doi: 10.1136/bjophthalmol-2011-300539. [DOI] [PubMed] [Google Scholar]
- 2.Low N, Cassell JA, Spencer B, Bender N, Hilber AM, van Bergen J, Andersen B, Herrmann B, Dubois-Arber F, Hamers FF, van de Laar M, Stephenson JM. 2012. Chlamydia control activities in Europe: cross-sectional survey. Eur J Public Health 22:556–561. doi: 10.1093/eurpub/ckr046. [DOI] [PubMed] [Google Scholar]
- 3.Hahn DL, Azenabor AA, Beatty WL, Byrne GI. 2002. Chlamydia pneumoniae as a respiratory pathogen. Front Biosci 7:e66–76. doi: 10.2741/hahn. [DOI] [PubMed] [Google Scholar]
- 4.Collingro A, Tischler P, Weinmaier T, Penz T, Heinz E, Brunham RC, Read TD, Bavoil PM, Sachse K, Kahane S, Friedman MG, Rattei T, Myers GS, Horn M. 2011. Unity in variety—the pan-genome of the Chlamydiae. Mol Biol Evol 28:3253–3270. doi: 10.1093/molbev/msr161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horn M, Collingro A, Schmitz-Esser S, Beier CL, Purkhold U, Fartmann B, Brandt P, Nyakatura GJ, Droege M, Frishman D, Rattei T, Mewes HW, Wagner M. 2004. Illuminating the evolutionary history of chlamydiae. Science 304:728–730. doi: 10.1126/science.1096330. [DOI] [PubMed] [Google Scholar]
- 6.Valdivia RH. 2008. Chlamydia effector proteins and new insights into chlamydial cellular microbiology. Curr Opin Microbiol 11:53–59. doi: 10.1016/j.mib.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Dilbeck PM, Evermann JF, Crawford TB, Ward AC, Leathers CW, Holland CJ, Mebus CA, Logan LL, Rurangirwa FR, McGuire TC. 1990. Isolation of a previously undescribed rickettsia from an aborted bovine fetus. J Clin Microbiol 28:814–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henning K, Schares G, Granzow H, Polster U, Hartmann M, Hotzel H, Sachse K, Peters M, Rauser M. 2002. Neospora caninum and Waddlia chondrophila strain 2032/99 in a septic stillborn calf. Vet Microbiol 85:285–292. doi: 10.1016/S0378-1135(01)00510-7. [DOI] [PubMed] [Google Scholar]
- 9.Baud D, Thomas V, Arafa A, Regan L, Greub G. 2007. Waddlia chondrophila, a potential agent of human fetal death. Emerg Infect Dis 13:1239–1243. doi: 10.3201/eid1308.070315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niemi S, Greub G, Puolakkainen M. 2011. Chlamydia-related bacteria in respiratory samples in Finland. Microbes Infect 13:824–827. doi: 10.1016/j.micinf.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 11.de Barsy M, Greub G. 2013. Waddlia chondrophila: from biology to pathogenicity. Microbes Infect 15:1033–1041. doi: 10.1016/j.micinf.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Goy G, Croxatto A, Greub G. 2008. Waddlia chondrophila enters and multiplies within human macrophages. Microbes Infect 10:556–562. doi: 10.1016/j.micinf.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Kebbi-Beghdadi C, Cisse O, Greub G. 2011. Permissivity of Vero cells, human pneumocytes and human endometrial cells to Waddlia chondrophila. Microbes Infect 13:566–574. doi: 10.1016/j.micinf.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 14.Jacquier N, Frandi A, Pillonel T, Viollier P, Greub G. 2014. Cell wall precursors are required to organize the chlamydial division septum. Nat Commun 5:3578. doi: 10.1038/ncomms4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wheelhouse N, Coyle C, Barlow PG, Mitchell S, Greub G, Baszler T, Rae MT, Longbottom D. 2014. Waddlia chondrophila infects and multiplies in ovine trophoblast cells stimulating an inflammatory immune response. PLoS One 9:e102386. doi: 10.1371/journal.pone.0102386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dille S, Herbst K, Volceanov L, Nolke T, Kretz O, Hacker G. 2014. Golgi fragmentation and sphingomyelin transport to Chlamydia trachomatis during penicillin-induced persistence do not depend on the cytosolic presence of the chlamydial protease CPAF. PLoS One 9:e103220. doi: 10.1371/journal.pone.0103220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choudhury A, Dominguez M, Puri V, Sharma DK, Narita K, Wheatley CL, Marks DL, Pagano RE. 2002. Rab proteins mediate Golgi transport of caveola-internalized glycosphingolipids and correct lipid trafficking in Niemann-Pick C cells. J Clin Invest 109:1541–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma DK, Choudhury A, Singh RD, Wheatley CL, Marks DL, Pagano RE. 2003. Glycosphingolipids internalized via caveolar-related endocytosis rapidly merge with the clathrin pathway in early endosomes and form microdomains for recycling. J Biol Chem 278:7564–7572. doi: 10.1074/jbc.M210457200. [DOI] [PubMed] [Google Scholar]
- 19.Goy G, Croxatto A, Posfay-Barbe KM, Gervaix A, Greub G. 2009. Development of a real-time PCR for the specific detection of Waddlia chondrophila in clinical samples. Eur J Clin Microbiol Infect Dis 28:1483–1486. doi: 10.1007/s10096-009-0804-7. [DOI] [PubMed] [Google Scholar]
- 20.Madico G, Quinn TC, Boman J, Gaydos CA. 2000. Touchdown enzyme time release-PCR for detection and identification of Chlamydia trachomatis, C. pneumoniae, and C. psittaci using the 16S and 16S-23S spacer rRNA genes. J Clin Microbiol 38:1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weber A, Auslander D, Hacker G. 2013. Mouse Noxa uses only the C-terminal BH3-domain to inactivate Mcl-1. Apoptosis 18:1093–1105. doi: 10.1007/s10495-013-0868-9. [DOI] [PubMed] [Google Scholar]
- 22.Croxatto A, Greub G. 2010. Early intracellular trafficking of Waddlia chondrophila in human macrophages. Microbiology 156:340–355. doi: 10.1099/mic.0.034546-0. [DOI] [PubMed] [Google Scholar]
- 23.Kumar Y, Valdivia RH. 2008. Actin and intermediate filaments stabilize the Chlamydia trachomatis vacuole by forming dynamic structural scaffolds. Cell Host Microbe 4:159–169. doi: 10.1016/j.chom.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Volceanov L, Herbst K, Biniossek M, Schilling O, Haller D, Nolke T, Subbarayal P, Rudel T, Zieger B, Hacker G. 2014. Septins arrange F-actin-containing fibers on the Chlamydia trachomatis inclusion and are required for normal release of the inclusion by extrusion. mBio 5:e01802–14. doi: 10.1128/mBio.01802-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong G. 2009. Killing me softly: chlamydial use of proteolysis for evading host defenses. Trends Microbiol 17:467–474. doi: 10.1016/j.tim.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen AL, Johnson KA, Lee JK, Sutterlin C, Tan M. 2012. CPAF: a chlamydial protease in search of an authentic substrate. PLoS Pathog 8:e1002842. doi: 10.1371/journal.ppat.1002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snavely EA, Kokes M, Dunn JD, Saka HA, Nguyen BD, Bastidas RJ, McCafferty DG, Valdivia RH. 2014. Reassessing the role of the secreted protease CPAF in Chlamydia trachomatis infection through genetic approaches. Pathog Dis doi: 10.1111/2049-632X.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer SF, Vier J, Kirschnek S, Klos A, Hess S, Ying S, Hacker G. 2004. Chlamydia inhibit host cell apoptosis by degradation of proapoptotic BH3-only proteins. J Exp Med 200:905–916. doi: 10.1084/jem.20040402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan T, Lu H, Hu H, Shi L, McClarty GA, Nance DM, Greenberg AH, Zhong G. 1998. Inhibition of apoptosis in Chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J Exp Med 187:487–496. doi: 10.1084/jem.187.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajalingam K, Sharma M, Lohmann C, Oswald M, Thieck O, Froelich CJ, Rudel T. 2008. Mcl-1 is a key regulator of apoptosis resistance in Chlamydia trachomatis-infected cells. PLoS One 3:e3102. doi: 10.1371/journal.pone.0003102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajalingam K, Sharma M, Paland N, Hurwitz R, Thieck O, Oswald M, Machuy N, Rudel T. 2006. IAP-IAP complexes required for apoptosis resistance of C. trachomatis-infected cells. PLoS Pathog 2:e114. doi: 10.1371/journal.ppat.0020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer SF, Harlander T, Vier J, Hacker G. 2004. Protection against CD95-induced apoptosis by chlamydial infection at a mitochondrial step. Infect Immun 72:1107–1115. doi: 10.1128/IAI.72.2.1107-1115.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ying S, Seiffert BM, Hacker G, Fischer SF. 2005. Broad degradation of proapoptotic proteins with the conserved Bcl-2 homology domain 3 during infection with Chlamydia trachomatis. Infect Immun 73:1399–1403. doi: 10.1128/IAI.73.3.1399-1403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heuer D, Lipinski AR, Machuy N, Karlas A, Wehrens A, Siedler F, Brinkmann V, Meyer TF. 2009. Chlamydia causes fragmentation of the Golgi compartment to ensure reproduction. Nature 457:731–735. doi: 10.1038/nature07578. [DOI] [PubMed] [Google Scholar]
- 35.Hacker G. 2014. The chlamydial protease CPAF: important or not, important for what? Microbes Infect 16:367–370. doi: 10.1016/j.micinf.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 36.Lucocq JM, Warren G. 1987. Fragmentation and partitioning of the Golgi apparatus during mitosis in HeLa cells. EMBO J 6:3239–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Acharya U, Mallabiabarrena A, Acharya JK, Malhotra V. 1998. Signaling via mitogen-activated protein kinase kinase (MEK1) is required for Golgi fragmentation during mitosis. Cell 92:183–192. doi: 10.1016/S0092-8674(00)80913-7. [DOI] [PubMed] [Google Scholar]
- 38.Welch WJ, Suhan JP. 1985. Morphological study of the mammalian stress response: characterization of changes in cytoplasmic organelles, cytoskeleton, and nucleoli, and appearance of intranuclear actin filaments in rat fibroblasts after heat-shock treatment. J Cell Biol 101:1198–1211. doi: 10.1083/jcb.101.4.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonatas NK, Stieber A, Gonatas JO. 2006. Fragmentation of the Golgi apparatus in neurodegenerative diseases and cell death. J Neurol Sci 246:21–30. doi: 10.1016/j.jns.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 40.Zhou Z, Mogensen MM, Powell PP, Curry S, Wileman T. 2013. Foot-and-mouth disease virus 3C protease induces fragmentation of the Golgi compartment and blocks intra-Golgi transport. J Virol 87:11721–11729. doi: 10.1128/JVI.01355-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mounier J, Boncompain G, Senerovic L, Lagache T, Chretien F, Perez F, Kolbe M, Olivo-Marin JC, Sansonetti PJ, Sauvonnet N. 2012. Shigella effector IpaB-induced cholesterol relocation disrupts the Golgi complex and recycling network to inhibit host cell secretion. Cell Host Microbe 12:381–389. doi: 10.1016/j.chom.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 42.Hackstadt T, Scidmore MA, Rockey DD. 1995. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc Natl Acad Sci U S A 92:4877–4881. doi: 10.1073/pnas.92.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scidmore MA, Fischer ER, Hackstadt T. 1996. Sphingolipids and glycoproteins are differentially trafficked to the Chlamydia trachomatis inclusion. J Cell Biol 134:363–374. doi: 10.1083/jcb.134.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suchland RJ, Rockey DD, Weeks SK, Alzhanov DT, Stamm WE. 2005. Development of secondary inclusions in cells infected by Chlamydia trachomatis. Infect Immun 73:3954–3962. doi: 10.1128/IAI.73.7.3954-3962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clausen JD, Christiansen G, Holst HU, Birkelund S. 1997. Chlamydia trachomatis utilizes the host cell microtubule network during early events of infection. Mol Microbiol 25:441–449. doi: 10.1046/j.1365-2958.1997.4591832.x. [DOI] [PubMed] [Google Scholar]
- 46.Mital J, Miller NJ, Fischer ER, Hackstadt T. 2010. Specific chlamydial inclusion membrane proteins associate with active Src family kinases in microdomains that interact with the host microtubule network. Cell Microbiol 12:1235–1249. doi: 10.1111/j.1462-5822.2010.01465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chin E, Kirker K, Zuck M, James G, Hybiske K. 2012. Actin recruitment to the Chlamydia inclusion is spatiotemporally regulated by a mechanism that requires host and bacterial factors. PLoS One 7:e46949. doi: 10.1371/journal.pone.0046949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hybiske K, Stephens RS. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc Natl Acad Sci U S A 104:11430–11435. doi: 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong G, Fan P, Ji H, Dong F, Huang Y. 2001. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J Exp Med 193:935–942. doi: 10.1084/jem.193.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bauler LD, Hackstadt T. 2014. Expression and targeting of secreted proteins from Chlamydia trachomatis. J Bacteriol 196:1325–1334. doi: 10.1128/JB.01290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dong F, Pirbhai M, Xiao Y, Zhong Y, Wu Y, Zhong G. 2005. Degradation of the proapoptotic proteins Bik, Puma, and Bim with Bcl-2 domain 3 homology in Chlamydia trachomatis-infected cells. Infect Immun 73:1861–1864. doi: 10.1128/IAI.73.3.1861-1864.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paschen SA, Christian JG, Vier J, Schmidt F, Walch A, Ojcius DM, Hacker G. 2008. Cytopathicity of Chlamydia is largely reproduced by expression of a single chlamydial protease. J Cell Biol 182:117–127. doi: 10.1083/jcb.200804023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, Adams JM, Strasser A. 1999. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286:1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- 54.Tan TT, Degenhardt K, Nelson DA, Beaudoin B, Nieves-Neira W, Bouillet P, Villunger A, Adams JM, White E. 2005. Key roles of BIM-driven apoptosis in epithelial tumors and rational chemotherapy. Cancer Cell 7:227–238. doi: 10.1016/j.ccr.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 55.Rejman Lipinski A, Heymann J, Meissner C, Karlas A, Brinkmann V, Meyer TF, Heuer D. 2009. Rab6 and Rab11 regulate Chlamydia trachomatis development and golgin-84-dependent Golgi fragmentation. PLoS Pathog 5:e1000615. doi: 10.1371/journal.ppat.1000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elwell CA, Engel JN. 2012. Lipid acquisition by intracellular Chlamydiae. Cell Microbiol 14:1010–1018. doi: 10.1111/j.1462-5822.2012.01794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.