Abstract
Tryptophan-rich antigens (TRAgs) are an antigen family that has been identified in human and rodent malaria parasites. TRAgs have been proposed as candidate antigens for potential vaccines. The Plasmodium vivax TRAg (PvTRAg) family includes 36 members. Each PvTRAg contains a tryptophan-rich (TR) domain in the C-terminal region. In this study, we recombinantly expressed all 36 PvTRAgs using a cell-free expression system, and, for the first time, profiled the IgG antibody responses against all PvTRAgs in the sera from 96 vivax malaria patients and 40 healthy individuals using protein microarray technology. The mean seropositive rate for all PvTRAgs was 60.3%. Among them, nine PvTRAgs were newly identified in this study and showed a seropositive rate of >50%. Five of them, PvTRAg_13, PvTRAg_15, PvTRAg_16, PvTRAg_26, and PvTRAg_29, produced higher levels of IgG antibody, even in low-endemicity countries. In addition, the results of an immunofluorescence analysis suggest that PvTRAgs are, at least in part, associated with caveola-vesicle complexes, a unique structure of P. vivax-infected erythrocytes. The mechanism of formation and the function of these abundant membrane structures are not known. Further investigation aimed at determining the functions of these proteins would lead to a better understanding of the blood-stage biology of P. vivax.
INTRODUCTION
In 2012, the global burden of malaria was estimated at 207 million cases and 0.63 million deaths, with an estimated 3.4 billion people being at risk of infection (1). Even though Plasmodium falciparum remains the leading cause of malaria in Africa, vivax malaria is the most geographically widespread of the human malarias (2, 3). It is prevalent throughout the tropics and subtropics, in the Middle East, Asia, the Western Pacific, and Latin America (2–4).
One of the control measures for malaria is vaccination; however, the development of a Plasmodium vivax-specific vaccine has been limited because of funding shortages and technical difficulties, such as the lack of efficient and continuous in vitro culture systems for parasites. Therefore, the majority of studies reported to date are on preerythrocytic and asexual blood-stage vaccines that are based on the P. falciparum ortholog antigens circumsporozoite protein, merozoite suface protein 1 (MSP1), and Duffy-binding protein. However, they remain the subject of preclinical studies (5). Extensive efforts, therefore, are required to identify new P. vivax antigens as vaccine candidates.
Tryptophan-rich antigens (TRAgs), which contain positionally conserved tryptophan residues in a tryptophan-rich (TR) domain, have been identified in murine and human malaria parasites (6–11). The tryptophan-rich proteins PypAg-1 and PypAg-3 were first characterized from Plasmodium yoelii, which showed binding to mouse erythrocytes (6, 7). PypAg-1 and PypAg-3, which are reportedly exported to the membranes of P. yoelii-infected erythrocytes, induce immune responses, and mice immunized with recombinant proteins were protected against parasite challenge infection with P. yoelii (7). Homologs of PypAg-1 and PypAg-3 have been identified in P. falciparum and named tryptophan-threonine-rich antigens (PfTryThrA) and P. falciparum merozoite-associated tryptophan-rich antigen (PfMaTrA), respectively (8, 12). Synthetic peptides derived from PfTryThrA inhibit the invasion of erythrocytes by merozoites (13). Remarkably, more tryptophan-rich protein-coding genes have been found in the P. vivax genome (36 genes) than in any other malaria parasite species (9). Fifteen of the tryptophan-rich antigens of P. vivax (PvTRAgs) that induced significant cellular and humoral responses in humans have been immunologically characterized and have shown few genetic polymorphisms in the parasite population (11). Recently, 10 of the 36 PvTRAgs have been shown to bind to human erythrocytes (14) and might be exploited to develop therapeutic agents against vivax malaria.
To evaluate the humoral immune response of the PvTRAg family as a whole, we characterized the immunoprofiling of PvTRAgs using wheat germ cell-free expression (WGCF) and protein array technologies and evaluated the correlation between antibody reactivities to paired recombinant PvTRAg proteins and PvMSP1-19. We also investigated the diversity of antibody responses against the five most seropositive PvTRAgs using sera from Korean, Myanmar, and Chinese patients, as well as the longevity of antibodies against the five PvTRAgs with archival serum samples. Moreover, we identified the subcellular localization of the PvTRAgs in blood-stage parasites of P. vivax.
MATERIALS AND METHODS
Sample and serum information.
We collected blood samples after the approval of protocols that were reviewed by the ethics committees of the Kangwon National University Hospital in the Republic of Korea; the Department of Medical Research (Lower Myanmar) in Myanmar; the Faculty of Tropical Medicine, Mahidol University, in Thailand; and the Jiangsu Institute of Parasitic Diseases (JIPD) in China.
The samples were divided into three types: patients (acute P. vivax infection), subjects exhibiting recovery (healthy individuals with vivax malaria history), and healthy subjects (healthy individuals without malaria history) (Table 1). The patients' samples from acute P. vivax infection were collected from three countries: the Republic of Korea (ROK), Myanmar, and China. The Korean patients' samples (n = 96) were collected from patients with symptoms and positive P. vivax parasitemia, as assessed by microscopic examination at local health centers and clinics in Gangwon and Gyeonggi Provinces, which are located within areas of endemicity in the Republic of Korea. The samples from Myanmar (n = 40) were collected in 2012 from patients from the Shwe Kyin area of Myanmar who were confirmed to be positive for vivax malaria by a malaria antigen rapid test (FK80; Standard Diagnostics, Gyeonggi, South Korea) and microscopic examination. The samples from China were kindly supplied by the JIPD, Wuxi, Jiangsu Province, China. The serum samples from healthy individuals with vivax malaria history were collected from Chinese residents who had an episode of vivax malaria infection in malaria-endemic areas of Anhui Province, China, in 2012 (n = 20) and from individuals who did not have a reinfection episode of vivax malaria in the preceding 5 (n = 30) and 12 (n = 30) years. Samples were classified into two groups: subjects exhibiting a 5-year recovery and subjects exhibiting a 12-year recovery. Individuals were designated “5-year recovery” subjects if they had a documented episode of vivax malaria in 2007 and no record of a malaria episode between 2007 and 2012. Other sera were collected from individuals who had a documented episode of vivax malaria in 2000 and no record of malaria episodes in the past 12 years. They were designated “12-year recovery” subjects. Serum samples from healthy malaria-naive individuals living in areas of nonendemicity in the ROK were also collected and used as controls.
TABLE 1.
Characteristics of study serum samples from areas of endemicity in Korea, Myanmar, and China
| Characteristic | Value |
||||||
|---|---|---|---|---|---|---|---|
| Acutely infected subjects |
Recovery subjects |
Healthy subjects (Korea) | |||||
| Korea (group 1) | Korea (group 2) | Myanmar | China | 5 yra | 12 yrb | ||
| Total (n) | 56 | 40 | 40 | 20 | 30 | 30 | 40 |
| Sex ratio (male:female) | 13:1 | 12.3:1 | 4:1 | 1.5:1 | 1.3:1 | 2:1 | 1:1 |
| Age (yr) | |||||||
| Mean (SD) | 33.89 (13.20) | 27.73 (9.80) | 22.88 (7.49) | 35.65 (16.14) | 52.47 (22.52) | 49.27 (16.47) | 10.00 (2.44) |
| Range | 13–72 | 18–59 | 10–42 | 17–58 | 7–85 | 19–78 | 6–13 |
| Parasitemia (%) | |||||||
| Mean (SD) | 0.15 (0.16) | 0.14 (0.15) | 0.10 (0.13) | 0.13 (0.15) | |||
| Range | 0.02–0.75 | 0.01–0.74 | 0.01–0.80 | 0.01–0.51 | |||
5 yr, infected with vivax malaria in 2007 with no record of malaria infection in the past 5 years in China.
12 yr, infected with vivax malaria in 2000 with no record of malaria infection in the past 12 years in China.
Cell-free expression of the PvTRAg domains.
All 36 PvTRAgs were expressed using a cell-free expression system and used for immunoscreening by protein microarray. The predicted signal peptide in the amino acid sequence was basically excluded from the expression constructs. The regions of the PvTRAg family proteins that were expressed are underlined in Fig. 1. Detailed information about the 36 PvTRAg constructs, including the GeneID, chromosomal location, size of the insert, geographical origin of the parasites used as the genomic DNA (gDNA) source, and polymorphisms compared with the reference strain of P. vivax, the Sal-I strain, are summarized in Table S1 in the supplemental material. Genomic DNAs were extracted from P. vivax isolates from the ROK and used as templates for PCR amplification of 16 PvTRAgs in this study, and the remaining 20 PvTRAg clones were available from our previous studies (15, 16). The In-fusion primers for PvTRAg amplification were designed based on PvTRAgs of the P. vivax Sal-1 strain sequence, with each gene-specific sense primer extending at the 5′ terminus, 5′-GGG CGG ATA TCT CGA G-3′, and each antisense primer extending at the 3′ terminus, 5′-GCG GTA CCC GGG ATC C-3′ (see Table S2 in the supplemental material). The PCR amplification and In-fusion cloning of PCR products (Clontech, Palo Alto, CA, USA) into a vector of the WGCF expression system, pEU-E01-His-Tev-N2 (CellFree Sciences, Matsuyama, Japan), was performed as described in previous studies (15–18). The inserted nucleotide sequence was confirmed by sequencing analysis (Genotech, Daejon, South Korea). Purified plasmid DNA was prepared using a Midiplus ultrapure plasmid extraction system (Viogene, Taipei, Taiwan). These proteins were expressed using a WGCF expression system (CellFree Sciences), as described in a previous study (19), and the expression levels and solubility of each protein were evaluated by Western blot analysis using an anti-His tag antibody (Qiagen, Hilden, Germany). PvMSP1-19 and PvAMA1 were also expressed using a WGCF expression system (CellFree Sciences) and were purified using Ni-nitrilotriacetic acid (NTA) affinity chromatography (Qiagen), as described previously (20).
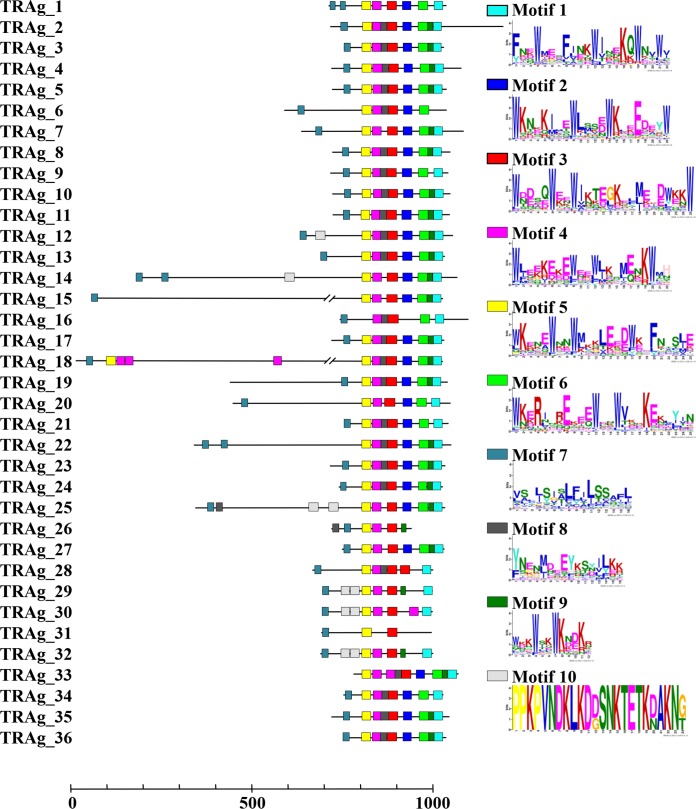
FIG 1.
Schematic depiction of the characteristics of all PvTRAg family members. Each PvTRAg protein is depicted with the size based on the number of amino acids. The regions of the proteins that were expressed are underlined.
Production and purification of E. coli-expressed recombinant proteins.
Among the 36 PvTRAg candidates, the five most antigenic PvTRAg proteins (PvTRAg_16, PvTRAG_29, PvTRAg_26, PvTRAg_13, and PvTRAg_15) were expressed as recombinant fusion proteins in E. coli, purified, and used for secondary screening by protein microarray and immune serum production. PvTRAg_16 was expressed using a pGEX-4T-2 (GE Healthcare, Uppsala, Sweden) expression vector with a glutathione S-transferase (GST) tag, and the other four PvTRAgs (PvTRAg_29, PvTRAg_26, PvTRAg_13, and PvTRAg_15) were expressed using a pEXP5-NT/TOPO expression vector with a His tag (Invitrogen, Carlsbad, CA, USA) (see Table S3 in the supplemental material). All five fragments were amplified from the plasmid clones that had been prepared for the WGCF expression system using high-fidelity DNA polymerase (Finnzymes, Espoo, Finland) and were cloned into the vector plasmid. Positive clones were confirmed by DNA sequencing analysis. The clones were then transformed into E. coli BL21 Star(DE3) cells (Invitrogen) for protein expression. Recombinant-protein expression was induced by isopropyl-β-d-thiogalactopyranoside (IPTG) under conditions that were optimized for each of the proteins. GST-tagged soluble proteins were purified using glutathione Sepharose 4B (GE Healthcare), and hexa-His-tagged proteins were purified using Ni-NTA (Qiagen), according to the manufacturer's protocol. The GST protein was expressed and used as a control. The purity of each recombinant protein was confirmed by SDS-PAGE and Western blot analysis according to protocols described previously (21). We also expressed the helical N terminus of P. vivax, interspersed with the subtelomeric/caveola–vesicle complexes-8195 (PHIST/CVC-8195) protein (PVX_093680), which was used as a control caveola-vesicle complex (CVC) marker (22), using a pGEX-4T-2 (GE Healthcare) expression vector with a GST tag (data not shown).
Protein microarrays.
Amine-coated slides were prepared as described previously (15). For the protein microarray primary screening, we analyzed the humoral immune response using well-type amine arrays of sera from 56 patients (group 1) with vivax malaria and 40 malaria-naive individuals. Briefly, 1 μl of an anti-penta-His antibody (Qiagen; 10 ng/μl in phosphate buffer, pH 7.4) was spotted into each well of the arrays and incubated for 2 h at 37°C. Then, 1 μl of the crude PvTRAg protein was spotted in duplicate into each well of the arrays and incubated for 1 h at 37°C, followed by blocking with 5% bovine serum albumin (BSA) in phosphate-buffered saline–Tween (0.1%) (PBS-T). The chips were then probed with sera from the malaria patients and malaria-naive individuals (1:10 dilution), which had been preabsorbed against a wheat germ lysate (1:100 dilution) to block anti-wheat germ antibodies. Alexa Fluor 546-conjugated goat anti-human IgG (10 ng/μl; Invitrogen) in PBS-T was used as the detection antibody, and the fluorescent signals were detected using a fluorescence scanner (ScanArray Express; PerkinElmer, Boston, MA, USA) and quantified as described previously (15). The cutoff value was defined as 2 standard deviations (SD) above the mean fluorescence intensity of the malaria-naive samples. To normalize the antibody array data, fluorescence intensities were divided by each cutoff value (see Table 3). After primary screening of PvTRAgs using human sera, the five PvTRAgs that exhibited the highest immunoreactivity were used for further evaluation of the humoral immune response among malaria patients. A serially diluted protein was used to optimize the coating antigen concentration (3 to 200 ng/μl) of each PvTRAg protein. The optimized concentrations of the purified recombinant proteins per well of the array were determined to be 50 to 100 ng/μl for the PvTRAg_16, PvTRAG_29, PvTRAg_26, PvTRAg_13, and PvTRAg_15 proteins. The IgG antibody responses against the five purified recombinant proteins were analyzed in vivax malaria patients from Korea (n = 40; group 2), Myanmar (n = 40), and China (n = 20) and from malaria-naive individuals (n = 32) from Korea.
TABLE 3.
Prevalences, 95% confidence intervals, and normalized mean fluorescence intensities of IgG responses to five PvTRAgs, MSP1-19, and AMA1 in serum samples from human patients and healthy individuals
| Antigen | Sample | No. positive | No. negative | Sensitivity [total (%)] | Specificity [total (%)] | 95% CIa | Normalized MFIb (cutoff value) | P valuec |
|---|---|---|---|---|---|---|---|---|
| MSP1-19 | Patients | 90 | 10 | 100 (90.0) | 82.6–94.5 | 3.078 (6,679) | <0.0001 | |
| 5 yr | 6 | 24 | 30 (20.0) | 9.5–37.3 | 0.781 (6,679) | 0.002 | ||
| 12 yr | 2 | 28 | 30 (6.6) | 1.9–21.3 | 0.481 (6,679) | 0.910 | ||
| Healthy | 3 | 29 | 32 (96.8) | 75.8–96.8 | 0.456 (6,679) | |||
| AMA1 | Patients | 52 | 48 | 100 (52.0) | 42.3–61.5 | 1.259 (3,921) | <0.0001 | |
| 5 yr | 5 | 25 | 30 (16.7) | 7.3–33.6 | 0.799 (3,921) | 0.261 | ||
| 12 yr | 1 | 29 | 30 (3.9) | 5.6–12.7 | 0.558 (3,921) | 0.231 | ||
| Healthy | 2 | 30 | 32 (93.8) | 79.9–98.3 | 0.585 (3,921) | |||
| TRAg_16 | Patients | 75 | 25 | 100 (75.0) | 65.7–82.5 | 1.554 (4,796) | <0.0001 | |
| 5 yr | 4 | 26 | 30 (13.3) | 5.3–29.7 | 0.749 (4,796) | 0.257 | ||
| 12 yr | 0 | 30 | 30 (0) | 0–11.35 | 0.597 (4,796) | 0.588 | ||
| Healthy | 1 | 31 | 32 (96.9) | 84.3–99.5 | 0.622 (4,796) | |||
| TRAg_29 | Patients | 61 | 39 | 100 (61.0) | 51.2–70.0 | 1.384 (5,389) | <0.0001 | |
| 5 yr | 5 | 25 | 30 (16.7) | 7.3–33.6 | 0.734 (5,389) | 0.006 | ||
| 12 yr | 4 | 26 | 30 (13.3) | 5.3–29.7 | 0.640 (5,389) | 0.110 | ||
| Healthy | 1 | 31 | 32 (96.9) | 84.3–99.5 | 0.535 (5,389) | |||
| TRAg_26 | Patients | 54 | 46 | 100 (54.0) | 44.3–63.4 | 1.197 (3,079) | <0.0001 | |
| 5 yr | 3 | 27 | 30 (10.0) | 3.5–25.6 | 0.726 (3,079) | 0.662 | ||
| 12 yr | 0 | 30 | 30 (0) | 0.584 (3,079) | 0.731 | |||
| Healthy | 1 | 31 | 32 (96.9) | 84.3–99.5 | 0.618 (3,079) | |||
| TRAg_13 | Patients | 51 | 49 | 100 (51.0) | 41.4–60.6 | 1.157 (3,351) | <0.0001 | |
| 5 yr | 3 | 27 | 30 (10.0) | 3.5–25.6 | 0.767 (3,351) | 0.035 | ||
| 12 yr | 2 | 28 | 30 (6.6) | 1.9–21.3 | 0.639 (3,351) | 0.564 | ||
| Healthy | 1 | 31 | 32 (96.9) | 84.3–99.5 | 0.602 (3,351) | |||
| TRAg_15 | Patients | 60 | 40 | 100 (60.0) | 50.2–69.1 | 1.183 (3,238) | <0.0001 | |
| 5 yr | 18 | 12 | 30 (60.0) | 42.3–75.4 | 1.085 (3,238) | <0.0001 | ||
| 12 yr | 6 | 24 | 30 (20.0) | 9.5–37.3 | 0.714 (3,238) | 0.002 | ||
| Healthy | 1 | 31 | 32 (96.9) | 84.3–99.5 | 0.565 (3,238) | |||
CI, confidence interval.
Normalized MFI, fluorescence intensities divided by each cutoff value (2 standard deviations above the mean fluorescence intensity of the malaria-naive samples).
Differences in the total IgG level for each antigen between vivax malaria patients and healthy individuals were calculated with the Mann-Whitney U test. A P value of <0.05 was considered statistically significant.
Production of animal immune sera.
For rabbit antiserum production, two rabbits were immunized subcutaneously with 250 μg of five purified recombinant PvTRAgs (PvTRAg_16, PvTRAG_29, PvTRAg_26, PvTRAg_13, and PvTRAg_15) in Freund's complete adjuvant (Sigma-Aldrich, St. Louis, MO, USA), followed by 250 μg of each protein in Freund's incomplete adjuvant (Sigma-Aldrich). All immunizations were administered three times at 3-week intervals. The antisera were collected 2 weeks after the final boost. To generate mouse antibodies against PvPHIST/CVC-8195, two 5-week-old female BALB/c mice (Daehan Biolink Co., Eumsung, ROK) were immunized subcutaneously with 20 μg of purified proteins plus Freund's complete adjuvant, followed by 20 μg with Freund's incomplete adjuvant. All immunizations were performed three times at 3-week intervals. The antisera were collected 2 weeks after the final boost. All animal experimental protocols were approved by the Kangwon National University Hospital ethics committee and followed the Ethical Guidelines for Animal Experiments of Kangwon National University.
Immunofluorescence assays (IFAs).
Intraerythrocytic-stage P. vivax parasites were collected from a malaria patient in Thailand. Slides smeared with parasite-infected blood were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature, dried, and stored at −80°C. Before use, the slides were thawed on blue silica gel (Samchun Chemical, Pyeongtaek, South Korea) and blocked for 30 min with 5% nonfat milk in PBS at 37°C. The slides were then incubated with 1:100 dilutions of rabbit anti-PvTRAg immune sera and mouse anti-PvPHIST/CVC-8195 immune sera and washed three times with cold PBS. The slides were then stained with Alexa Fluor 488-conjugated goat anti-mouse IgG or Alexa Fluor 568-conjugated goat anti-rabbit IgG secondary antibodies (Invitrogen) and the nuclear stain 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen) at 37°C for 30 min. Subsequently, the slides were mounted in Prolong Gold antifade reagent (Invitrogen), and fluorescence was visualized under oil immersion using a confocal laser-scanning microscope (FV1000; Olympus, Tokyo, Japan). Images were captured using the FV10-ASW 3.0 Viewer software (Olympus) and processed using Adobe Photoshop CS5 (Adobe Systems, San Jose, CA, USA).
Data analysis.
The putative signal peptide (SP) cleavage site was predicted using SignalP version 3.0 (http://www.cbs.dtu.dk/services/SignalP/). The TMHMM Server version 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) was used to search for transmembrane (TM) domains. Conserved TR motifs were evaluated using the MEME package (http://meme.sdsc.edu/meme4/cgibin/meme.cgi). The correlation between duplicate spots of protein arrays and the antibody reactivities of different concentrations of the recombinant proteins were determined using the GraphPad Prism software, version 5.0 (GraphPad, San Diego, CA, USA) and PASW Statistics 18.0 (SPSS Inc., Chicago, IL, USA). Sensitivity was measured based on the percentage of patients who had a positive test result, and specificity was evaluated based on the percentage of healthy individuals who had a negative test result. The Mann-Whitney U test was used to test the significance of differences in mean fluorescence intensity (MFI) values between every two groups. Hierarchical clustering of PvTRAgs from antibody reactivity profiles was calculated and drawn using the TIGR multiarray experiment viewer (MeV) software with 1,000 bootstrap iterations (23).
RESULTS
Identification of PvTRAg proteins.
The P. vivax genome database shows that 36 members of the tryptophan-rich protein family (9) are located in chromosomes 2, 4, 5, 8, 9, 10, 11, 12, and 14 and that 9 of them (PvTRAg_25 to PvTRAg_36) were not yet assigned to chromosomes in the PlasmoDB database (see Table S1 in the supplemental material). Here, we renamed the tryptophan-rich proteins PvTRAg_1 to PvTRAg_36 based on the localization of genes on chromosomes (PlasmoDB [http://plasmodb.org/plasmo]). Most PvTRAg proteins were predicted to consist of two exons, with the exception of four proteins (PvTRAg_8, PvTRAg_17, PvTRAg_18, and PvTRAg_33) that were encoded by a single exon. A schematic depiction of the features of all PvTRAg family members is shown in Fig. 1. Each PvTRAg protein contained a tryptophan-rich domain in the C-terminal region. An SP and/or TM domain was predicted to be present in the N-terminal regions of 29 of the 36 PvTRAg proteins, with the exception of PvTRAg_19, which contained a TM domain in the central part of its second exon. The Multiple EM for Motif Elicitation (MEME) (24) algorithm was used to search for conserved motifs in PvTRAg proteins. Ten motifs were identified among the 36 PvTRAg proteins, seven of which were tryptophan-rich motifs (Fig. 2; see Table S4 in the supplemental material). It is of special interest that almost all of the PvTRAg proteins contained those seven motifs in the order M5, M4, M3, M2, M6, M9, and M1 from the N terminus, whereas pairs of tryptophan-rich motifs showed low correlation (<0.60) (data not shown). Motif 8 was found in 25 PvTRAg proteins and existed mainly between motifs 3 and 4. Motif 10 had only a lysine-rich motif that was identified in three PvTRAg proteins (PvTRAg_29, PvTRAg_30, and PvTRAg_32) (Fig. 2). Moreover, the sequence alignment of the TR domain also showed the presence of conserved tryptophan residues in the TR domain. Thus, members of the TRAg family share several conserved motifs in the TR domain. Overall, these bioinformatics analyses proved that all 36 molecules have conserved characteristics that are specific to TRAgs (see Fig. S1 in the supplemental material).
FIG 2.
Conserved motifs in PvTRAg proteins. Ten conserved motifs in PvTRAg proteins were identified using MEME. Seven motifs contain 3 to 5 tryptophan (W) residues. Motifs 7, 8, and 10 do not contain tryptophan residues.
To analyze the transcription pattern across the intraerythrocytic cell cycle, we summarized the PvTRAg transcriptome data from a previous study (25). Among the 36 PvTRAg genes, 33 genes with transcriptome data were selected for analysis (see Fig. S2 in the supplemental material). We found that almost all of these proteins showed an expression pattern with a peak in the ring (TP1 to TP3) or schizont (TP6 to TP9) stages of the parasites. Moreover, the genes were clustered into three types after hierarchical-clustering analysis. Type 1 genes were highly transcribed in the ring and trophozoite stages, whereas type 3 genes were highly transcribed in the schizont stage. Type 2 genes showed high transcription patterns in both the ring and late schizont stages. These results suggest that the PvTRAg family members do not act at a specific developmental stage; rather, they seem to act throughout the blood-stage cycle.
Expression of recombinant PvTRAg proteins and Western blot analysis.
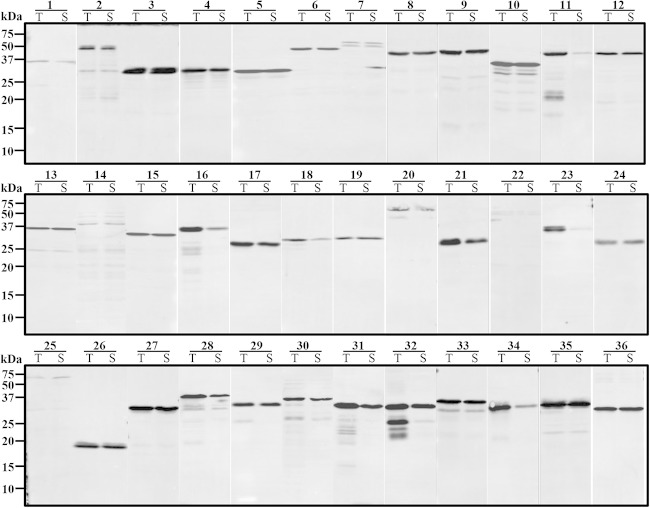
A total of 36 TR fragments of PvTRAg proteins were expressed for primary screening using the WGCF system. The recombinant N-terminallyHis-tagged PvTRAg proteins were detected by Western blot analysis using an anti-His tag antibody (Fig. 3). The results showed that the PvTRAg proteins (30/36; 83.3%) were highly expressed as a mean of 74.0% of soluble fractions (range, 20.0% to 98.6%), with an average concentration of 36.5 ng/μl (range, 5.0 to 97.0 ng/μl), with the exception of five proteins (TRAg_7, TRAg _14, TRAg _18, TRAg _22, and TRAg _25) that had lower expression and one protein that was expressed mainly in the insoluble fraction (TRAg_23) (Table 2 and Fig. 3). After the primary immunoscreening, as shown in Table 2, the five top-ranked immunoreactive proteins, PvTRAg_16, PvTRAg_29, PvTRAg_26, PvTRAg_13, and PvTRAg_15, were selected for secondary screening, and their recombinant proteins were successfully expressed by E. coli systems in good agreement with their theoretical molecular masses at 66, 31, 20, 37, and 35 kDa, respectively (Table 2 and Fig. 4). SDS-PAGE analysis showed that all the recombinant proteins were recovered as a soluble fraction in the supernatant (Fig. 4A). The PvTRAg_16 protein was expressed as a GST fusion protein, whereas the remaining four proteins, PvTRAg_29, PvTRAg_26, PvTRAg_13, and PvTRAg_15, were expressed as His-tagged proteins. All five proteins were used to immunize rabbits to produce polyclonal antibodies. Western blotting data showed that recombinant PvTRAg proteins were recognized by anti-His/GST-tag antibodies, pooled sera from vivax malaria patients from Korea, and rabbit immune sera (Fig. 4B). Western blot analysis of the parasite lysate probed with anti-PvTRAg_16, -PvTRAg_29, -PvTRAg_26, and -PvTRAg_13 rabbit antisera showed that the proteins were detected at 42, 36, 26, and 38 kDa, respectively, which was in good agreement with the expected molecular mass of the full-length target (Fig. 4C, arrowheads). The observation that the anti-PvTRAg_15 antibody recognized a band of approximately 70 kDa, and not of the expected size of 157 kDa, from the parasite antigen suggests that proteolytic processing occurs at approximately 70 kDa from the C terminus of the protein. The specificity of the rabbit antibodies was confirmed based on these results, because no band was detected at the identical position in the erythrocyte lysates (Fig. 4C, lanes E).
FIG 3.
Western blot analysis of the recombinant PvTRAg proteins expressed using the wheat germ cell-free system and probed with an anti-His tag antibody. The numbers represent PvTRAg proteins. T, total fraction; S, soluble fraction.
TABLE 2.
Summary of PvTRAgs and cell-free expressiona
| Name | SP | TM | Insert size (position) (aa) | Mass (kDa) | Concn.b | Solubilityc (%) | No. (%) positive | Normalized MFI (cutoff value)d | Functione |
|---|---|---|---|---|---|---|---|---|---|
| TRAg_16 | Yf | N | 335 (24–359) | 39.7 | 19.1 | 25.3 | 47 (83.9) | 2.294 (3,573.2) | |
| TRAg_29 | Y | Y | 281 (33–313) | 31.0 | 41.6 | 87.2 | 44 (78.6) | 1.922 (2,944.0) | PvTRAg36, EBA |
| TRAg_26 | N | N | 165 (60–210) | 20.0 | 64.1 | 92.2 | 42 (75.0) | 1.621 (4,054.2) | PvTRAg26.3, EBA |
| TRAg_13 | Y | N | 317 (23–340) | 36.5 | 25.2 | 94 | 42 (75.0) | 1.623 (5,392.0) | |
| TRAg_15 | Y | Y | 290 (1125–1415) | 35.1 | 38.6 | 97.3 | 41 (73.2) | 1.636 (5,168.8) | |
| TRAg_34 | Y | N | 251 (30–280) | 31.7 | 21.0 | 31.1 | 39 (69.6) | 1.255 (3,187.0) | |
| TRAg_5 | N | Y | 259 (58–317) | 31.9 | 45.5 | 94.1 | 38 (67.9) | 1.381 (6,761.4) | PvTRAg38, EBA |
| TRAg_9 | N | Y | 326 (2–327) | 39.8 | 52.0 | 93.2 | 38 (67.9) | 1.362 (3,277.6) | |
| TRAg_10 | N | Y | 287 (61–1041) | 35.9 | 49.6 | 98.6 | 38 (67.9) | 1.500 (5,343.6) | |
| TRAg_21 | Y | N | 253 (23–276) | 31.1 | 40.6 | 58.3 | 38 (67.9) | 1.398 (2,838.6) | PvTRAg33.5, EBA |
| TRAg_22 | N | Y | 646 (2–646) | 74.0 | 5.0 | 69 | 38 (67.9) | 1.389 (3,408.4) | PvTRAg74, EBA |
| TRAg_25 | N | N | 635 (59–693) | 74.2 | 5.0 | 47.3 | 38 (67.9) | 1.460 (3,602.4) | |
| TRAg_3 | Y | N | 254 (23–277) | 31.3 | 97.0 | 94.9 | 37 (66.1) | 1.417 (4,749.2) | |
| TRAg_6 | N | Y | 387 (59–445) | 47.1 | 20.7 | 88.5 | 37 (66.1) | 1.319 (6,591.8) | |
| TRAg_11 | N | Y | 326 (2–327) | 40.0 | 9.0 | 25.5 | 37 (66.1) | 1.270 (3,479.0) | PvTRAg, EBA |
| TRAg_30 | Y | N | 282 (33–314) | 33.5 | 40.0 | 90 | 37 (66.1) | 1.515 (3,091.2) | |
| TRAg_35 | N | N | 272 (60–331) | 34.1 | 89.2 | 85.5 | 36 (64.3) | 1.418 (4,875.0) | |
| TRAg_2 | N | Y | 421 (61–481) | 48.2 | 23.7 | 73.7 | 35 (62.5) | 1.414 (4,662.8) | |
| TRAg_24 | Y | N | 262 (27–288) | 32.8 | 20.1 | 98.6 | 35 (62.5) | 1.207 (4,484.8) | |
| TRAg_19 | N | Y | 268 (321–589) | 34.5 | 16.1 | 88.6 | 33 (58.9) | 1.256 (5,757.0) | PvTRAg69.4, EBA |
| TRAg_36 | Y | N | 258 (27–284) | 32.4 | 70.8 | 98.3 | 33 (58.9) | 1.264 (2,732.6) | PvTRAg35.2, EBA |
| TRAg_8 | N | N | 323 (2–323) | 40.0 | 41.0 | 94 | 32 (57.1) | 1.265 (3,787.8) | |
| TRAg_7 | N | Y | 397 (62–458) | 49.5 | 5.7 | 98 | 31 (55.4) | 1.149 (6,787.4) | |
| TRAg_14 | Y | N | 313 (558–870) | 38.2 | 5.0 | 95 | 31 (55.4) | 1.273 (3,865.8) | |
| TRAg_17 | Y | Y | 256 (61–316) | 32.1 | 49.2 | 74.3 | 31 (55.4) | 1.150 (4,474.0) | |
| TRAg_20 | N | N | 550 (63–613) | 66.3 | 11.9 | 65.2 | 30 (53.6) | 1.209 (3,820.2) | |
| TRAg_18 | Y | Y | 280 (2383–2663) | 35.6 | 7.3 | 20.9 | 29 (51.8) | 1.200 (5,517.0) | |
| TRAg_28 | Y | N | 303 (34–336) | 35.3 | 33.9 | 46.2 | 29 (51.8) | 1.179 (3,564.0) | |
| TRAg_12 | Y | N | 357 (2–357) | 43.0 | 32.6 | 85.9 | 27 (48.2) | 1.160 (4,229.8) | |
| TRAg_23 | N | Y | 321 (23–321) | 39.8 | 5.0 | 20 | 27 (48.2) | 1.070 (5,142.4) | |
| TRAg_32 | Y | Y | 285 (30–314) | 33.5 | 70.1 | 80.4 | 27 (48.2) | 1.059 (4,837.2) | PvTRAg36.6, EBA |
| TRAg_33 | N | N | 295 (2–295) | 37.4 | 71.7 | 85.9 | 27 (48.2) | 1.071 (4,092.2) | |
| TRAg_31 | Y | N | 287 (17–303) | 32.4 | 63.9 | 67.7 | 26 (46.4) | 1.062 (3,702.2) | PvTRAg34, EBA |
| TRAg_27 | Y | N | 260 (30–289) | 32.3 | 73.7 | 87 | 25 (44.6) | 1.056 (4,852.0) | |
| TRAg_1 | Y | N | 298 (30–328) | 36.8 | 10.0 | 44.8 | 22 (39.3) | 1.005 (3,965.2) | |
| TRAg_4 | N | N | 263 (2–295) | 32.3 | 49.1 | 68 | 19 (33.9) | 0.997 (3,919.0) |
The P. vivax GeneID, mass, SP, and TM data were obtained from the PlasmoDB database (http://www.plasmodb.org).
Concn, the concentration of expressed target protein estimated by using Image J.
Solubility, the intensity ratio of a specific protein band versus the total intensity calculated from the intensity of the bands from Western blot analysis.
Normalized MFI, fluorescence intensities divided by each cutoff value (2 standard deviations above the mean fluorescence intensity of the malaria-naive samples).
Data from reference 14. EBA, erythrocyte binding activity.
Y, yes; N, no.
FIG 4.

SDS-PAGE and Western blot analysis of five recombinant PvTRAg proteins expressed by E. coli. (A) Affinity-purified PvTRAg_16 (65.7 kDa; GST tag), PvTRAg_29 (31.1 kDa; His tag), PvTRAg_26 (20.0 kDa; His), PvTRAg_13 (36.5 kDa; His), and PvTRAg_15 (35.1 kDa; His) expressed in E. coli soluble fractions. (B) Recombinant PvTRAg proteins recognized by an anti-His/GST tag antibody and by pooled sera of vivax malaria patients from Korea. H, His tag; G, GST tag; Pt, pooled patient sera; R, immune rabbit sera. (C) Native PvTRAg proteins are detected by immune rabbit sera. E, erythrocyte lysates; P, parasite lysates.
Immunoprofiling of PvTRAgs.
The immunoreactivity of proteins of the PvTRAg family was screened with group 1 sera from 56 patients with vivax malaria and 40 malaria-naive individuals from the ROK. Among the 36 PvTRAg recombinant proteins, the mean seropositive rate was 60.3%. Five proteins (PvTRAg_16, PvTRAg_29, PvTRAg_26, PvTRAg_13, and PvTRAg_15) yielded a positive rate of >71.4% (40/56), 23 proteins displayed a positive rate between 69.6% (39/56) and 51.8% (29/56), and the remaining 8 proteins showed a positive rate between 48.2% (27/56) and 33.9% (19/56) (Table 2). Among them, nine TRAg proteins (PvTRAg_3, PvTRAg_7, PvTRAg_13, PvTRAg_14, PvTRAg_15, PvTRAg_18, PvTRAg_20, PvTRAg_26, and PvTRAg_35) with a high IgG prevalence (≥51.8%) were newly identified in this study. On closer inspection (Table 2), we found that some antigens had high IgG titers but did not show a high positive rate in patients. This may be caused by their high level of polymorphism; further investigation is needed to address this issue. Moreover, these top five high-prevalence PvTRAgs also showed high IgG titers in the sera of individual patients and are potential vaccine targets.
To analyze the cross-reaction among the PvTRAgs based on this study, we observed the reactivity profiles of PvTRAgs with individual sera by hierarchical-clustering analysis using the MeV software with 1,000 bootstrap iterations (see Fig. S3 in the supplemental material).
The clustering analysis showed that they could be significantly (bootstrap value, >95) clustered in only three combinations consisting of two antigens, i.e., TRAg_22 and TRAg_25, TRAg_14 and TRAg_30, and TRAg_23 and TRAg_32. This result suggests the existence of cross-reactions only among the combinations. Moreover, we compared the mean transcript abundances of the genes and normalized the MFI of the protein microarray. However, we did not find any antigens with a high MFI and a low transcriptional level (data not shown). This finding suggests the absence of antigens that cross-react with more abundant paralogs.
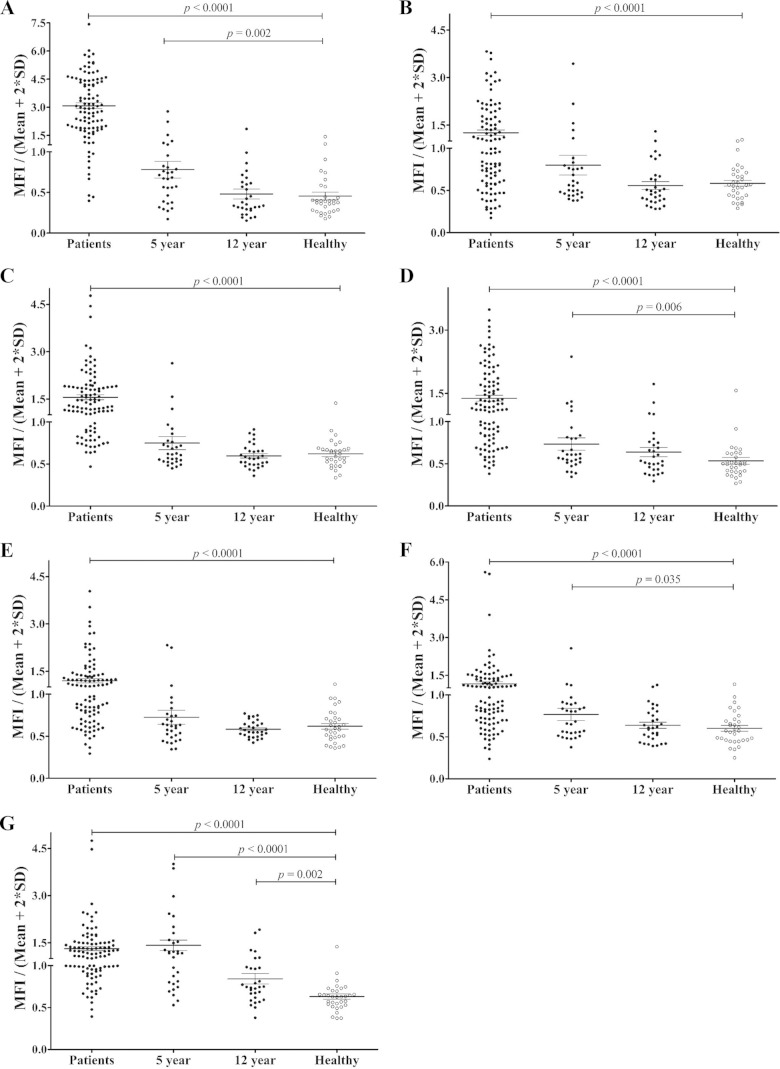
To evaluate further the humoral immune response against the five highly antigenic proteins PvTRAg_16, PvTRAg_29, PvTRAg_26, PvTRAg_13, and PvTRAg_15, we evaluated serum antibody responses against the five antigens in vivax malaria patients from Korea (n = 40; group 2), Myanmar (n = 40), and China (n = 20), as well as in malaria-naive individuals (n = 32) from Korea. The prevalences of total IgG for PvTRAg_16, PvTRAg_29, PvTRAg_26, PvTRAg_13, and PvTRAg_15 were greater than 50% (75%, 61%, 54%, 51%, and 60%, respectively). The specificities for all PvTRAgs were >96.9% (Fig. 5 and Table 3).
FIG 5.
IgG antibody responses to five recombinant PvTRAg proteins in the sera of malaria patients (Patients), archival vivax malaria patients (5-year and 12-year recovery), and malaria-naive individuals (Healthy). (A) PvMSP1-19. (B) PvAMA1. (C) PvTRAg_16. (D) PvTRAg_29. (E) PvTRAg_26. (F) PvTRAg_13. (G) PvTRAg_15. The vertical axis (MFI/Mean + 2SD) represents the mean fluorescence intensity divided by the mean fluorescence intensity plus 2 standard deviations of the malaria-naive samples. The horizontal bars indicate the means ± standard deviations. The P values were calculated using the Mann-Whitney U test.
We also analyzed the antigenicity of PvMSP1-19 and PvAMA1 using the same sera, and our data reconfirmed the high IgG prevalence against PvMSP1-19 and PvAMA1; in addition, PvAMA1 displayed an IgG reactivity that was similar to those of PvTRAgs (Table 3). We also observed that the five PvTRAgs produced similar IgG titers among isolates from the three countries (data not shown). Taken together, these results suggest that most of the PvTRAgs are antigenic proteins in humans, even among the acute cases from low-endemicity countries, such as the ROK. Moreover, five highly antigenic proteins displayed an IgG response that was similar to that of PvAMA1, and their IgG reactivity was conserved across malaria areas from three Asian countries.
Evaluation of the longevity of IgG antibodies to PvTRAgs.
To determine the longevity of anti-PvTRAg antibody responses, we analyzed the IgG antibody reactivities of five PvTRAg proteins using long-term archival vivax samples (5-year recovery and 12-year recovery samples) from China. Three of the PvTRAgs (PvTRAg_29, PvTRAg_13, and PvTRAg_15) showed significantly higher IgG MFI (0.734, 0.767, and 1.085, respectively; P < 0.05) in 5-year recovery sera than in those of malaria-naive individuals (0.535, 0.602, and 0.565, respectively) (Fig. 5 and Table 3). Simultaneously, the IgG prevalence and MFI against PvMSP1-19 in 5-year recovery patients dropped from 90% to 20% and from 3.078 to 0.781, respectively (P < 0.05) (Fig. 5). Moreover, in most cases, IgG antibody levels in individuals who had malaria 12 years before had decreased to baseline levels. However, PvTRAg_15 showed a significantly higher IgG antibody response in 12-year recovery sera (P = 0.002) (Fig. 5). These analyses suggest that IgG antibody responses to some PvTRAgs, especially PvTRAg_15, are stably sustained in this population.
Subcellular localization of PvTRAgs.
To understand the high antigenicity of the five PvTRAgs in humans, the subcellular localization of the five proteins was investigated. An immunofluorescence assay was performed using unpurified parasites. We found that antibodies against the five PvTRAgs specifically recognized parasites as a multiple-dot pattern in the infected erythrocytes of ring/trophozoite-stage parasites. This feature was similar to the appearance of Schüffner's dots in Giemsa-stained slides. To examine their specific subcellular localization, we purified parasites and performed dual labeling using mouse polyclonal antibodies against PvPHIST/CVC-8195 (a CVC marker) (22) as a control (Fig. 6A). All five PvTRAgs showed similar immunofluorescence patterns: small dots across the infected red blood cell cytoplasm at the ring/early trophozoite stage, with the abundance and intensity of the dots increasing greatly in the later trophozoite and schizont stages; in addition, a strong signal was observed around the parasitophorous vacuolar space (Fig. 6B). Most of the dot-like structures were closely localized and/or partially merged with PvPHIST/CVC-8195, which was localized in the CVC tubular extensions (Fig. 6).
FIG 6.
Subcellular localization of five PvTRAg proteins in asexual blood-stage parasites of P. vivax. The 4% paraformaldehyde-fixed ring-stage (A) and trophozoite/schizont-stage (B) P. vivax parasites were dually labeled with a mouse anti-PvPHIST/CVC-8195 polyclonal antibody (green; caveola-vesicle complex marker) and a rabbit immune serum against each PvTRAg protein (red). DIC, differential interference contrast; Merge, merged images of PvTRAgs and PvPHIST/CVC-8195. Scale bars, 5 μm.
DISCUSSION
The members of the PvTRAg protein family mainly contained a C-terminal TR domain, which consisted of seven kinds of TR motifs, as assessed using MEME analysis. According to the transcriptome profiles of PvTRAgs in erythrocytic-stage parasites (25) (see Fig. S2 in the supplemental material), all 33 proteins were differentially transcribed in the ring, trophozoite, and/or schizont stage, respectively, indicating that these proteins may play specific roles in blood-stage development. We expressed all the recombinant PvTRAgs using a WGCF system and produced more than 80% of the targets with 74.0% solubility. The humoral immune response to these PvTRAg proteins was evaluated in the sera of vivax malaria patients and malaria-naive individuals using protein array methods; the mean antibody-positive rate was 60.3% (range, 33.9% to 83.9%). Moreover, antibodies against some of the PvTRAgs were detected in individuals who had had a vivax malaria episode 5 or 12 years before. In addition, the subcellular localization of PvTRAgs was partially associated with the CVC of P. vivax parasites.
Antibodies are crucial for naturally acquired protective immunity against the intraerythrocytic stage of malaria parasites. These antibodies induced from malaria parasite infection were found to play important roles in several mechanisms, including inhibition of parasite invasion, blockage of parasite development, and enhancement of the phagocytic activity of monocytes and macrophages (26, 27). In the current study, we analyzed IgG antibody responses against all of the PvTRAg family proteins in a larger number of serum samples from vivax malaria patients and malaria-naive individuals than in previous PvTRAg studies (11, 16). Nine novel TRAg proteins (PvTRAg_3, PvTRAg_7, PvTRAg_13, PvTRAg_14, PvTRAg_15, PvTRAg_18, PvTRAg_20, PvTRAg_26, and PvTRAg_35) showed higher IgG positivity (≥51.8%) in patients' sera. Further analysis of the five most seropositive PvTRAgs showed conserved IgG reactivity in all three areas of Asian countries with low malaria endemicity, indicating the universal antigenicity of these PvTRAgs compared with those of two known blood-stage antigens, PvMSP1-19 and PvAMA1. Moreover, we found that four out of the five highly immunogenic proteins showed a high transcription profile in schizont-stage parasites, which suggests their involvement in the invasion process. Among them, TRAg_26 exhibited a high expression profile only in the ring stage (see Fig. S2 in the supplemental material). Thus, TRAg_26 may be expressed on the CVC of ring-stage parasite-infected erythrocytes and may induce host immune response after the disruption of the infected erythrocytes. However, this warrants confirmation by further study.
It is widely believed that periodic reinfection is required to evoke protective immunity to malaria and that malaria antibodies are usually short-lived in the absence of reinfection (28). These short-lived antibody responses against P. falciparum MSP1 (PfMSP1), P. falciparum AMA1 (PfAMA1), and the P. falciparum erythrocyte membrane protein 1 (PfEMP1) have been observed in previous studies (29, 30), especially in young children (31, 32). However, several studies have demonstrated stable antibody responses to PfMSP1-19, P. falciparum merozoite surface protein 2 (PfMSP2), and PfAMA1 antigens (31, 33, 34). Moreover, long-lived antibody and memory B-cell responses to P. falciparum (PfMSP1-19, PfMSP2, PfCSP, and PvAMA1) and P. vivax (PvMSP1-19, PvAMA1, and PvDBP) were identified recently (35, 36). Here, we also observed that antibodies against several PvTRAgs (TRAg_13, TRAg_15, and TRAg_29) and PvMSP1-19 were detected in individuals from Jiangsu Province, China (where malaria is not recently endemic), who had had a vivax malaria episode 5 or 12 years before. The pharmacological half-life of human IgG molecules is around 21 days (37), and the long-term maintenance of IgG titers might result from a larger proportion of plasmablasts that differentiated into long-lived plasma cells in circumstances in which malaria infection is infrequent (35). Boosting the response or memory response of anti-PvTRAg antibodies may induce memory T cells and/or B cells. The stability of anti-PvTRAg antibodies in 5- or 10-year recovery subjects could determine the antigenicity of PvTRAgs in the induction of memory Th2 cell response in a previous study (11). However, it is difficult to verify whether anti-PvTRAg is stable in the P. vivax population based on incident measurement of antibody levels among different individuals. In the current study, we evaluated antibody responses against all PvTRAgs and found different sensitivities, whereas a previous report described a 75% to 100% seropositive rate (11). This may be due to the different origins of the serum samples, as we used samples from low-endemicity area, whereas the previous study used samples from areas of high endemicity, where reinfection was common (11).
During the invasion and maturation of the intracellular parasite, a series of dramatic and extensive changes occur in the structural and functional properties of the infected erythrocytes (38). In contrast to the development of protruding knob structures in P. falciparum, P. vivax induces unique CVC structures, which were first identified by transmission electron microscopy in 1975 (39). The function of the CVCs remains largely unknown. However, it was hypothesized that they have an endocytic function (39). Recently, a member of the plasmodium helical interspersed subtelomeric (PHIST) superfamily, which is named PHIST/CVC-8195, was identified as a 95-kDa CVC protein and localized on the cytoplasmic side of the CVC tubular extensions. This protein may be essential for the survival of vivax malaria parasites (22). In the current study, we observed that five PvTRAg proteins were partially merged with PvPHIST/CVC-8195, indicating the CVC-associated localization of these proteins. Similarly, the CVC-associated localization of another PvTRAg protein, PvTRAg_11, was reported in a previous study (40). These combined effects beg the question of whether the localization of other members of the PvTRAg family is also associated with CVCs. Of these five PvTRAg proteins, PvTRAg_26 and PvTRAg_29 showed binding activity to host erythrocytes in a recent study (14). However, the detailed functions of the proteins remain unknown, and further investigations are required.
Our data analysis showed that the positive rate varied over a wide range (33.9% to 83.9%) among each PvTRAg antigen, even if, here, we expressed larger fragments than the TR domain for reactivity profiles of serum samples. If cross-reactivity exists in PvTRAg proteins, they may show similar positive rates. Furthermore, we applied a cluster analysis of the reactivity profiles of the individual sera to address whether antigens can be grouped according to reactivity profiles. Clustering analysis results showed that the reactivity profiles of PvTRAgs in individual serum samples were very variable: only three clusters, each consisting of two antigens, were clustered significantly. The pairs were also highly correlated using correlation analysis (data not shown), suggesting that cross-reactions occur only between two antigens in the same cluster. Moreover, the comparison between the mean MFI and transcriptional levels confirmed that rare antigens reacted with antibodies against major antigens. Although PvTRAgs share common motifs, these analyses clearly showed that most of our findings regarding the reactivity of PvTRAgs were antigen specific.
In summary, 36 PvTRAg family proteins were characterized using a proteomics microarray, and we found a mean positive rate of 60.3% (range, 33.9% to 83.9%). Moreover, conserved TR motifs were observed in most PvTRAgs. Five PvTRAg proteins exhibited high antigenicity against vivax malaria parasite infection, and three proteins (TRAg_13, TRAg_15, and TRAg_29) may produce universal antibody responses among the residents of low-endemicity areas of three Asian countries. A total of 10 clusters were produced, most of which included 1 to 9 PvTRAg proteins. The cross-reactivity among antigens in the same cluster may warrant further study. Moreover, the results of an immunofluorescence analysis suggest that PvTRAgs are, at least in part, associated with caveola-vesicle complexes, which are unique structures of P. vivax-infected erythrocytes. Further investigation aimed at determining the functions of these proteins will be needed for a better understanding of the blood-stage biology of P. vivax.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the staff at JIPD, China, and DMR for serum collection. We thank Deok-Hoon Kong at Kwon-Soo Ha's laboratory for technical assistance.
This work was supported by a grant from the Korea Health Technology R&D Project, Ministry of Health and Welfare, Republic of Korea (A121180). This work was also supported in part by MEXT KAKENHI (23117008) and JSPS KAKENHI (26670202, 26253026), Japan.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.03067-14.
REFERENCES
- 1.WHO. 2013. World malaria report 2013. WHO, Geneva, Switzerland. [Google Scholar]
- 2.Gething PW, Elyazar IR, Moyes CL, Smith DL, Battle KE, Guerra CA, Patil AP, Tatem AJ, Howes RE, Myers MF, George DB, Horby P, Wertheim HF, Price RN, Mueller I, Baird JK, Hay SI. 2012. A long neglected world malaria map: Plasmodium vivax endemicity in 2010. PLoS Negl Trop Dis 6:e1814. doi: 10.1371/journal.pntd.0001814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arevalo-Herrera M, Chitnis C, Herrera S. 2010. Current status of Plasmodium vivax vaccine. Hum Vaccin 6:124–132. doi: 10.4161/hv.6.1.9931. [DOI] [PubMed] [Google Scholar]
- 4.Mendis K, Sina BJ, Marchesini P, Carter R. 2001. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg 64:97–106. [DOI] [PubMed] [Google Scholar]
- 5.Arevalo-Herrera M, Herrera S. 2001. Plasmodium vivax malaria vaccine development. Mol Immunol 38:443–455. doi: 10.1016/S0161-5890(01)00080-3. [DOI] [PubMed] [Google Scholar]
- 6.Salzwedel K, West JT, Hunter E. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J Virol 73:2469–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns JM, Adeeku EK, Belk CC, Dunn PD. 2000. An unusual tryptophan-rich domain characterizes two secreted antigens of Plasmodium yoelii-infected erythrocytes. Mol Biochem Parasitol 110:11–21. doi: 10.1016/S0166-6851(00)00252-8. [DOI] [PubMed] [Google Scholar]
- 8.Ntumngia FB, Bouyou-Akotet MK, Uhlemann AC, Mordmuller B, Kremsner PG, Kun JF. 2004. Characterisation of a tryptophan-rich Plasmodium falciparum antigen associated with merozoites. Mol Biochem Parasitol 137:349–353. doi: 10.1016/j.molbiopara.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, Cheng Q, Coulson RM, Crabb BS, Del Portillo HA, Essien K, Feldblyum TV, Fernandez-Becerra C, Gilson PR, Gueye AH, Guo X, Kang'a S, Kooij TW, Korsinczky M, Meyer EV, Nene V, Paulsen I, White O, Ralph SA, Ren Q, Sargeant TJ, Salzberg SL, Stoeckert CJ, Sullivan SA, Yamamoto MM, Hoffman SL, Wortman JR, Gardner MJ, Galinski MR, Barnwell JW, Fraser-Liggett CM. 2008. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyagi RK, Sharma YD. 2012. Erythrocyte binding activity displayed by a selective group of Plasmodium vivax tryptophan rich antigens is inhibited by patients' antibodies. PLoS One 7:e50754. doi: 10.1371/journal.pone.0050754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeeshan M, Bora H, Sharma YD. 2013. Presence of memory T cells and naturally acquired antibodies in Plasmodium vivax malaria exposed individuals against a group of tryptophan-rich antigens with conserved sequences. J Infect Dis 207:175–185. doi: 10.1093/infdis/jis650. [DOI] [PubMed] [Google Scholar]
- 12.Uhlemann AC, Oguariri RM, McColl DJ, Coppel RL, Kremsner PG, Anders RF, Kun JF. 2001. Properties of the Plasmodium falciparum homologue of a protective vaccine candidate of Plasmodium yoelii. Mol Biochem Parasitol 118:41–48. doi: 10.1016/S0166-6851(01)00370-X. [DOI] [PubMed] [Google Scholar]
- 13.Curtidor H, Ocampo M, Rodriguez LE, Lopez R, Garcia JE, Valbuena J, Vera R, Puentes A, Leiton J, Cortes LJ, Lopez Y, Patarroyo MA, Patarroyo ME. 2006. Plasmodium falciparum TryThrA antigen synthetic peptides block in vitro merozoite invasion to erythrocytes. Biochem Biophys Res Commun 339:888–896. doi: 10.1016/j.bbrc.2005.11.089. [DOI] [PubMed] [Google Scholar]
- 14.Zeeshan M, Tyagi RK, Tyagi K, Alam MS, Sharma YD. 2015. Host-parasite interaction: selective numbers of ‘Pv-fam-a’ family proteins of Plasmodium vivax bind to a restricted number of human erythrocyte receptors. J Infect Dis 211:1111–1120. doi: 10.1093/infdis/jiu558. [DOI] [PubMed] [Google Scholar]
- 15.Chen JH, Jung JW, Wang Y, Ha KS, Lu F, Lim CS, Takeo S, Tsuboi T, Han ET. 2010. Immunoproteomics profiling of blood stage Plasmodium vivax infection by high-throughput screening assays. J Proteome Res 9:6479–6489. doi: 10.1021/pr100705g. [DOI] [PubMed] [Google Scholar]
- 16.Lu F, Li J, Wang B, Cheng Y, Kong DH, Cui L, Ha KS, Sattabongkot J, Tsuboi T, Han ET. 2014. Profiling the humoral immune responses to Plasmodium vivax infection and identification of candidate immunogenic rhoptry-associated membrane antigen (RAMA). J Proteomics 102:66–82. doi: 10.1016/j.jprot.2014.02.029. [DOI] [PubMed] [Google Scholar]
- 17.Cheng Y, Wang Y, Ito D, Kong DH, Ha KS, Chen JH, Lu F, Li J, Wang B, Takashima E, Sattabongkot J, Tsuboi T, Han ET. 2013. PvMSP1P, merozoite surface protein 1 paralog, is a novel erythrocyte-binding ligand of Plasmodium vivax. Infect Immun 81:1585–1595. doi: 10.1128/IAI.01117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang B, Lu F, Cheng Y, Li J, Ito D, Sattabongkot J, Tsuboi T, Han ET. 2013. Identification and characterization of the Plasmodium falciparum RhopH2 ortholog in Plasmodium vivax. Parasitol Res 112:585–593. doi: 10.1007/s00436-012-3170-9. [DOI] [PubMed] [Google Scholar]
- 19.Tsuboi T, Takeo S, Iriko H, Jin L, Tsuchimochi M, Matsuda S, Han ET, Otsuki H, Kaneko O, Sattabongkot J, Udomsangpetch R, Sawasaki T, Torii M, Endo Y. 2008. Wheat germ cell-free system-based production of malaria proteins for discovery of novel vaccine candidates. Infect Immun 76:1702–1708. doi: 10.1128/IAI.01539-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen JH, Wang Y, Ha KS, Lu F, Suh IB, Lim CS, Park JH, Takeo S, Tsuboi T, Han ET. 2011. Measurement of naturally acquired humoral immune responses against the C-terminal region of the Plasmodium vivax MSP1 protein using protein arrays. Parasitol Res 109:1259–1266. doi: 10.1007/s00436-011-2370-z. [DOI] [PubMed] [Google Scholar]
- 21.Cheng Y, Wang B, Sattabongkot J, Lim CS, Tsuboi T, Han ET. 2014. Immunogenicity and antigenicity of Plasmodium vivax merozoite surface protein 10. Parasitol Res 113:2559–2568. doi: 10.1007/s00436-014-3907-8. [DOI] [PubMed] [Google Scholar]
- 22.Akinyi S, Hanssen E, Meyer EV, Jiang J, Korir CC, Singh B, Lapp S, Barnwell JW, Tilley L, Galinski MR. 2012. A 95 kDa protein of Plasmodium vivax and P. cynomolgi visualized by three-dimensional tomography in the caveola-vesicle complexes (Schuffner's dots) of infected erythrocytes is a member of the PHIST family. Mol Microbiol 84:816–831. doi: 10.1111/j.1365-2958.2012.08060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378. [DOI] [PubMed] [Google Scholar]
- 24.Bailey TL, Williams N, Misleh C, Li WW. 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bozdech Z, Mok S, Hu G, Imwong M, Jaidee A, Russell B, Ginsburg H, Nosten F, Day NP, White NJ, Carlton JM, Preiser PR. 2008. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc Natl Acad Sci U S A 105:16290–16295. doi: 10.1073/pnas.0807404105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wipasa J, Elliott S, Xu H, Good MF. 2002. Immunity to asexual blood stage malaria and vaccine approaches. Immunol Cell Biol 80:401–414. doi: 10.1046/j.1440-1711.2002.01107.x. [DOI] [PubMed] [Google Scholar]
- 27.Beeson JG, Osier FH, Engwerda CR. 2008. Recent insights into humoral and cellular immune responses against malaria. Trends Parasitol 24:578–584. doi: 10.1016/j.pt.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 28.Langhorne J, Ndungu FM, Sponaas AM, Marsh K. 2008. Immunity to malaria: more questions than answers. Nat Immunol 9:725–732. doi: 10.1038/ni.f.205. [DOI] [PubMed] [Google Scholar]
- 29.Cavanagh DR, Elhassan IM, Roper C, Robinson VJ, Giha H, Holder AA, Hviid L, Theander TG, Arnot DE, McBride JS. 1998. A longitudinal study of type-specific antibody responses to Plasmodium falciparum merozoite surface protein-1 in an area of unstable malaria in Sudan. J Immunol 161:347–359. [PubMed] [Google Scholar]
- 30.Dorfman JR, Bejon P, Ndungu FM, Langhorne J, Kortok MM, Lowe BS, Mwangi TW, Williams TN, Marsh K. 2005. B cell memory to 3 Plasmodium falciparum blood-stage antigens in a malaria-endemic area. J Infect Dis 191:1623–1630. doi: 10.1086/429671. [DOI] [PubMed] [Google Scholar]
- 31.Taylor RR, Egan A, McGuinness D, Jepson A, Adair R, Drakely C, Riley E. 1996. Selective recognition of malaria antigens by human serum antibodies is not genetically determined but demonstrates some features of clonal imprinting. Int Immunol 8:905–915. doi: 10.1093/intimm/8.6.905. [DOI] [PubMed] [Google Scholar]
- 32.Akpogheneta OJ, Duah NO, Tetteh KK, Dunyo S, Lanar DE, Pinder M, Conway DJ. 2008. Duration of naturally acquired antibody responses to blood-stage Plasmodium falciparum is age dependent and antigen specific. Infect Immun 76:1748–1755. doi: 10.1128/IAI.01333-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Udhayakumar V, Kariuki S, Kolczack M, Girma M, Roberts JM, Oloo AJ, Nahlen BL, Lal AA. 2001. Longitudinal study of natural immune responses to the Plasmodium falciparum apical membrane antigen (AMA-1) in a holoendemic region of malaria in western Kenya: Asembo Bay Cohort Project VIII. Am J Trop Med Hyg 65:100–107. [DOI] [PubMed] [Google Scholar]
- 34.Drakeley CJ, Corran PH, Coleman PG, Tongren JE, McDonald SL, Carneiro I, Malima R, Lusingu J, Manjurano A, Nkya WM, Lemnge MM, Cox J, Reyburn H, Riley EM. 2005. Estimating medium- and long-term trends in malaria transmission by using serological markers of malaria exposure. Proc Natl Acad Sci U S A 102:5108–5113. doi: 10.1073/pnas.0408725102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wipasa J, Suphavilai C, Okell LC, Cook J, Corran PH, Thaikla K, Liewsaree W, Riley EM, Hafalla JC. 2010. Long-lived antibody and B cell memory responses to the human malaria parasites, Plasmodium falciparum and Plasmodium vivax. PLoS Pathog 6:e1000770. doi: 10.1371/journal.ppat.1000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ndungu FM, Lundblom K, Rono J, Illingworth J, Eriksson S, Farnert A. 2013. Long-lived Plasmodium falciparum specific memory B cells in naturally exposed Swedish travelers. Eur J Immunol 43:2919–2929. doi: 10.1002/eji.201343630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hopkins RJ, Kramer WG, Blackwelder WC, Ashtekar M, Hague L, Winker-La Roche SD, Berezuk G, Smith D, Leese PT. 2004. Safety and pharmacokinetic evaluation of intravenous vaccinia immune globulin in healthy volunteers. Clin Infect Dis 39:759–766. doi: 10.1086/422998. [DOI] [PubMed] [Google Scholar]
- 38.Cooke BM, Mohandas N, Coppel RL. 2001. The malaria-infected red blood cell: structural and functional changes. Adv Parasitol 50:1–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aikawa M, Miller LH, Rabbege J. 1975. Caveola-vesicle complexes in the plasmalemma of erythrocytes infected by Plasmodium vivax and P. cynomolgi. Unique structures related to Schuffner's dots. Am J Pathol 79:285–300. [PMC free article] [PubMed] [Google Scholar]
- 40.Jalah R, Sarin R, Sud N, Alam MT, Parikh N, Das TK, Sharma YD. 2005. Identification, expression, localization and serological characterization of a tryptophan-rich antigen from the human malaria parasite Plasmodium vivax. Mol Biochem Parasitol 142:158–169. doi: 10.1016/j.molbiopara.2005.01.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.