Abstract
Sialic acids are found on all vertebrate cell surfaces and are part of a larger class of molecules known as nonulosonic acids. Many bacterial pathogens synthesize related nine-carbon backbone sugars; however, the role(s) of these non-sialic acid molecules in host-pathogen interactions is poorly understood. Vibrio vulnificus is the leading cause of seafood-related death in the United States due to its ability to quickly access the host bloodstream, which it can accomplish through gastrointestinal or wound infection. However, little is known about how this organism persists systemically. Here we demonstrate that sialic acid-like molecules are present on the lipopolysaccharide of V. vulnificus, are required for full motility and biofilm formation, and also contribute to the organism's natural resistance to polymyxin B. Further experiments in a murine model of intravenous V. vulnificus infection demonstrated that expression of nonulosonic acids had a striking benefit for bacterial survival during bloodstream infection and dissemination to other tissues in vivo. In fact, levels of bacterial persistence in the blood corresponded to the overall levels of these molecules expressed by V. vulnificus isolates. Taken together, these results suggest that molecules similar to sialic acids evolved to facilitate the aquatic lifestyle of V. vulnificus but that their emergence also resulted in a gain of function with life-threatening potential in the human host.
INTRODUCTION
Sialic acids are found on all vertebrate cells, positioned at the terminal end of glycan chains that modify proteins and lipids. The most common sialic acid in animals is N-acetylneuraminic acid (Neu5Ac). In mammals, sialic acids perform a wide range of functions, such as cell-cell interactions and immune modulation (1, 46). Sialic acids also appear to have an immunological status as “self-associated molecular patterns” in mammals (2, 47). In fact, many bacterial pathogens synthesize or acquire sialic acids (Neu5Ac) and incorporate these molecules into surface structures, such as lipopolysaccharides (LPS) and capsular polysaccharides (CPS) (reviewed in reference 3). Bacterial sialic acids have been shown to dampen host complement activity and neutrophil responses (2, 4) and to promote bacterial survival in the bloodstream and systemic dissemination to other tissues (5), especially when expressed at high levels. Sialic acid-modified surface structures are important for virulence in bloodstream and disseminated infections for well over a dozen pathogens (6–13). In contrast, many bacteria synthesize nine-carbon backbone sugars that do not meet the chemical definition of sialic acids, but the potential roles of these molecules in blood-borne diseases in mammals are not known.
Many bacteria synthesize nine-carbon backbone amino sugars, called legionaminic and pseudaminic acids, that are structurally similar to sialic acids (1). These molecules have been shown to play roles in motility and biofilm formation in vitro and in the chicken colonization and ferret diarrhea models in vivo (14–16). In fact, a growing body of evidence suggests that these acidic nine-carbon backbone amino sugars (collectively called nonulosonic acids, or NulOs) are widespread among prokaryotes, with ∼20% of sequenced prokaryotic genomes encoding a predicted nonulosonic acid biosynthetic (NAB) pathway (1). Collectively, these NulO molecules (including sialic acids) share an evolutionary origin, as evidenced by the fact that three enzymatic steps (Nab1, Nab2, and Nab3) of all NAB pathways are homologous across the three domains of life that synthesize them (1). Mammals do not synthesize legionaminic or pseudaminic acids.
Campylobacter jejuni is an interesting and well-studied example of a bacterium that synthesizes multiple types of NulOs—some strains can synthesize both sialic acids and sialic acid-like molecules (14, 17, 18, 20, 21). In some strains of C. jejuni, the LPS is capped with a sialic acid, while the unsheathed flagellum is modified with legionaminic and/or pseudaminic acid residues (20, 21). Mutations in key genes in the pseudaminic acid biosynthetic pathway of C. jejuni resulted in nonmotile phenotypes, defects in autoagglutination, and adherence to epithelial cells, as well as an attenuation of virulence, in a ferret diarrhea model (14). In contrast, mutations in the legionaminic acid biosynthetic gene cluster in C. jejuni did not have an impact on motility but led to decreased hydrophobicity, reduced biofilm formation, and a reduced ability to colonize the chicken intestine (15). Vibrio fischeri is another recent example in which legionaminic acid modifications present on LPS were shown to be important for motility and colonization of the bacterium's natural host, the Hawaiian bobtail squid (16).
Vibrio vulnificus is found in estuarine and coastal environments, both as a free-living bacterium and in association with a wide range of aquatic animals (22). Humans commonly come into contact with V. vulnificus via consumption of raw or undercooked shellfish, and this bacterium is the leading cause of seafood-related death in the United States; susceptible individuals have an alarming 50% case fatality rate (23). Vibrio vulnificus lethality appears to result from rapid progression of a localized infection (wound or gastroenteritis) to a systemic infection of the blood. However, little is known at the molecular level about virulence factors that allow V. vulnificus to survive in the bloodstream to cause this rapid-onset and commonly fatal disease.
Bioinformatic, phylogenetic, and biochemical analyses strongly suggest that V. vulnificus produces either the NulO legionaminic or pseudaminic acid but not the structurally similar sialic acid N-acetylneuraminic acid (1, 19, 24). Our previous study demonstrated that NulO molecules are expressed at some level by virtually all tested strains of V. vulnificus and suggested that clinical isolates are more likely to encode a NAB pathway genotype associated with higher levels of NulO expression (24). Here we sought to formally characterize the genetic basis for NulO biosynthesis in V. vulnificus, evaluate the significance of NulO production for motility and biofilm behaviors, and examine the hypothesis that these sialic acid-like molecules may also play a role in systemic pathogenesis. These studies implicate bacterial non-sialic acid NulO molecules in systemic mammalian pathogenesis for the first time and suggest that expression of these molecules in V. vulnificus also enhances its environmental fitness.
MATERIALS AND METHODS
Strains and culture conditions.
This study utilized the V. vulnificus clinical isolates CMCP6 and YJ016. References to wild-type (WT) and Δnab2 strains are for strains with the CMCP6 background unless stated otherwise. Strains and plasmids used are listed in Table 1. All strains were grown aerobically at 30°C in Luria-Bertani broth (LB) (Fisher Scientific, Fair Lawn, NJ) containing 2% NaCl or in marine broth 2216 (MB) (BD, Franklin Lakes, NJ), as noted. Stationary-phase cultures were prepared with a single colony and allowed to grow for 16 h. A 2% inoculum of stationary-phase culture was used to grow 4-h logarithmic-phase cultures. Growth curves of strains were examined by measuring the optical density at 595 nm (OD595) every hour for 24 h, using a Sunrise microplate reader and Magellan plate reader software (Tecan US, Durham, NC). There were no significant differences in growth between WT and nab deletion strains. Streptomycin-resistant strains (Table 1), used only in the animal experiments, were obtained as follows. Strains were grown aerobically from a single colony at 30°C in 5 ml of LB plus 2% NaCl for 16 h. The overnight culture was concentrated by centrifugation at 4,000 × g for 10 min, and the resulting pellet was resuspended in 100 μl of LB plus 2% NaCl, plated on LB plus 2% NaCl plus 1,000 μg/ml streptomycin, and incubated for 24 h at 30°C. Resulting growth was replated on LB plus 2% NaCl plus 200 μg/ml streptomycin to isolate resistant colonies. Careful characterization of the resistant strains demonstrated the same phenotypes seen for the mutant strains (e.g., motility and biofilm defects). We also verified that the Δnab2 resistant strain did not vary in its in vitro growth characteristics from the streptomycin-sensitive Δnab2 or WT strain.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| Vibrio vulnificus strains | ||
| CMCP6 | Clinical isolate | 54 |
| YJ016 | Clinical isolate | 55 |
| JBL0808 (Δnab2) | CMCP6 Δnab2 (VV1_0808) | This study |
| JBL0808 (Δnab2) Smr | CMCP6 Δnab2; spontaneously Sm-resistant isolate | This study |
| JBL0808C | CMCP6 Δnab2/pnab2 (pBB0808c) | This study |
| JBL0808C Smr | CMCP6 Δnab2/pnab2 (pBB0808c); spontaneously Sm-resistant isolate | This study |
| SAM0312 (Δnab2) | YJ016 Δnab2 (VV0312) | This study |
| SAM0312 (Δnab2) Smr | YJ016 Δnab2 (VV0312); Sm-resistant isolate | This study |
| JBL1950 (ΔflhF) Smr | CMCP6 ΔflhF (VV1_1950); Sm-resistant isolate | This study |
| JBL1950 (ΔflhF) | CMCP6 ΔflhF (VV1_1950) | This study |
| Escherichia coli strains | ||
| DH5α λ-pir | pir ϕ80dlacZΔM15 δ(lacZYA-argF)U169 recA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1 | |
| β2155 DAP | Donor for bacterial conjugation; thr-1004 pro thi strA hsdS lacZΔM15 (F lacZΔM15 lacTRQJ36 proAB) dapA Ermr pirRP4 (Kmr from SM10) | |
| Plasmids | ||
| pJet1.2/blunt | High-copy-number cloning vector | |
| pJet10808SOE | V. vulnificus VV1_0808 (nab2) SOE product | This study |
| pJet0312SOE | V. vulnificus VV0312 (nab1) SOE product | This study |
| pJet1950SOE | V. vulnificus VV1_1950 (flhF) SOE product | This study |
| pDS132 | Suicide plasmid; Cmr sacB | 30 |
| pDS0808 | V. vulnificus nab2 knockout vector (VV1_0808 SOE product) | This study |
| pDS0312 | V. vulnificus nab1 knockout vector (VV0312 SOE product) | This study |
| pDS1950 | V. vulnificus flhF knockout vector (VV1_1950 SOE product) | This study |
| pBBR1MCS | Expression plasmid; empty vector; Cmr | 56 |
| pBB0808c | nab2 (VV1_0808) expression plasmid (pnab2) | This study |
Allelic exchange mutagenesis and gene complementation in V. vulnificus.
V. vulnificus CMCP6 and YJ016 genome sequences were used for primer design, and oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA) (Table 2). An in-frame deletion mutant of the V. vulnificus CMCP6 nab2 (VV1_0808) gene was constructed using splicing by overlap extension (SOE) PCR and allelic exchange (29), using the primers labeled SOEVV1_0808. A 411-bp deletion within the full-length, 1,002-bp nab2 gene was constructed in the CMCP6 strain background as follows. Primers were designed (SOEVV1_0808A plus SOEVV1_0808B and SOEVV1_0808C plus SOEVV1_0808D) to PCR amplify two gene fragments that flank the targeted 411-bp deletion region within nab2. The fragments were annealed, ligated, and amplified using primers SOEVV1_0808A and SOEVV1_0808D to construct the 591-bp truncated nab2 gene fragment. The truncated gene was inserted into the high-copy-number cloning vector pJET 1.2 AR (Thermo Fisher Scientific, Waltham, MA) to generate pJet10808SOE, which was then transformed into the Escherichia coli cloning strain DH5α λ-pir. The cloned fragment was then subcloned from pJet10808SOE into the suicide vector pDS132 Cmr (30) to generate the nab2 knockout vector pDS0808. The knockout vector was then transformed into the diaminopimelic acid (DAP)-auxotrophic E. coli β2155 donor strain, which was conjugated to V. vulnificus by cross-streaking onto LB plates containing 0.3 mM DAP (Sigma-Aldrich, St. Louis, MO). Subsequent growth was plated on LB plus 2% NaCl plus 25 μg/ml chloramphenicol to isolate single-crossover recombinants of V. vulnificus containing pDS0808. V. vulnificus conjugants were cultured overnight without antibiotics to allow a second round of homologous recombination to occur, eliminating pDS0808 and producing an in-frame deletion mutant. The resulting overnight culture was plated onto LB plus 2% NaCl supplemented with 10% sucrose, which was lethal to members of the population still harboring a sacB-containing plasmid. A 396-bp deletion in the YJ016 nab2 (VV0312) allele, a 420-bp deletion in CMCP6 nab1, and an 837-bp deletion in the CMCP6 flhF (VV1_1950) gene were also constructed as described above. Primers for these mutants are provided in Table 2 and use the same naming convention as that shown for CMCP6 nab2. Deletion mutants were confirmed by PCR using SOE flanking primers and by sequencing. CMCP6 Δnab2 was complemented by amplifying the VV1_0808 gene, using primers VV1_0808Fc and VV1_0808Rc (Table 2). The gene was cloned into pBBR1MCS Cmr to generate the complementation plasmid pBB0808c (pnab2). Selection for the complemented strain was achieved by plating on LB plus 2% NaCl plus 25 μg/ml chloramphenicol. It should be noted that the complementation strains displayed a moderate growth defect compared to the wild type, which could not be attributed to expression of the empty vector or to overexpression of Nab2, as the same effect was not observed for CMCP6 plus pnab2, CMCP6 plus empty vector or CMCP6 Δnab2 plus empty vector. Despite this growth defect, the complemented strain restored the in vitro phenotypes of the Δnab2 mutant and was able to outcompete the Δnab2 mutant in vivo.
TABLE 2.
Primers used for this study
| Primer name | Sequence (5′-3′) | Tm (°C) | Product size (bp) |
|---|---|---|---|
| SOEVV1_0808A | TCTAGACGCCAAGCACCAAAGTTATT | 60 | |
| SOEVV1 0808B | CTCTTCGTCTGGCGTTGACA | 60 | 356 |
| SOEVV1_0808C | TGTCAACGCCAGACGAAGAGGCTAGCCTCGAGCCACAAGA | 60 | |
| SOEVV1_0808D | GCATGCTCAAGTTGCGGATCATTTTG | 60 | 373 |
| SOEVV1_0808FLF | CCAGAGAAGAGGCACCAGTT | ||
| SOEVV1_0808FLR | CCAGAGAAGAGGCACCAGTT | 60 | 1,237 |
| SOEVV0312A | CTTATGGGCCAAGGTGATAGCAG | 61 | |
| SOEVV0312B | GGTCGAAAGGAACTCAATACCC | 61 | 402 |
| SOEVV0312C | GGGTATTGAGTTCCTTTCGACCGATTTACCTGCGGGTTACTCTG | 60 | |
| SOEVV0312D | CTCGAGCATCTCCCAATACTG | 60 | 369 |
| SOEVV0312FLF | GTTACCTTGGGTGAAGAAGCAC | 60 | |
| SOEVV0312FLR | CACTTCAGCAAAGGAAGAGACC | 60 | 1,936 |
| SOEVV1_1950A | TCTAGATCGAATTGTACAGGCTGTGG | 60 | |
| SOEVV1_1950B | CAAAGCGTTTGGTCATGGAT | 61 | 399 |
| SOEVV1_1950C | ATCCATGACCAAACGCTTTGTCATGCAAGAGAGTGG GGAA | 61 | |
| SOEVV1_1950D | GAGCTCGCTTGATCGTGTATCATATTCTCA | 59 | 377 |
| SOEVV1_1950FLF | AGGCGAACCAGCAGTCTTAC | 59 | |
| SOEVV1_1950FLR | GCCAAACCTAGGGTCACATT | 59 | 1,838 |
| SOEVV1_0803A | TCTAGAAAGCGTGCACAAGTGGATATTCTCA | 60 | |
| SOEVV1_0803B | AACGGCACCTCTGCCCCCGCT | 60 | 241 |
| SOEVV1_0803C | AGCGGGGGCAGAGGTGCCGTTAAGCTCGGTTGATATTGACCATGAG | 56 | |
| SOEVV1_0803D | GAGCTCGCCCCACTCCACTTGCTGA | 56 | 227 |
| SOEVV1_0803FLF | GATTACTTCGGTGGCGAAAG | 60 | |
| SOEVV1_0803LFR | GAATCAGGTAACGTGGTGCT | 60 | 1,063 |
| VV1_0808Fc | AAGCTTCGCCAAGCACCAAAGTTATT | 60 | |
| VV1_0808Rc | TCTAGAGTGGCGGTGACTACACAGATT | 60 | 1,079 |
HPLC analysis of V. vulnificus NulOs.
Bacterial cultures were washed with phosphate-buffered saline (PBS) and subjected to mild acid hydrolysis in 2 N acetic acid at 80°C for 3 h, as previously described (31). After mild acid treatment to release NulO residues, cellular debris was removed by centrifugation, and a low-molecular-weight fraction was isolated by centrifugation through a 10,000-molecular-weight-cutoff filter (Vivaspin 500; Vivaproducts, Littleton, MA). The filtrate was derivatized with 1,2-diamino-4,5-methylene dioxybenzene (DMB), which is specific to the keto acid portion of NulO residues. Reaction mixtures consisted of 7 mM DMB, 18 mM sodium hydrosulfite, 1.4 M acetic acid, and 0.7 M 2-mercaptoethanol (Sigma-Aldrich) and were incubated in the dark at 50°C for 2 h. Derivatized NulO residues were resolved by high-pressure liquid chromatography (HPLC) using a C18 column (Tosoh Bioscience, King of Prussia, PA) and eluted isocratically using 85% Milli-Q ultrapure water, 8% acetonitrile, and 7% methanol at a rate of 0.9 ml/min for a period of 50 min, as previously described (31). Detection of fluorescent NulO derivatives was achieved using an excitation wavelength of 373 nm and an emission wavelength of 448 nm. 3-Deoxy-d-manno-octulosonic acid (Kdo) is an eight-carbon-backbone monosaccharide related to the nonulosonic acids that is a conserved component of the lipopolysaccharide core. Kdo is also released by mild acid hydrolysis and derivatized by DMB and thus served as a convenient internal positive control in the analyses and could also be used to calculate relative levels of NulO production.
Lipopolysaccharide analysis.
Lipopolysaccharide was analyzed by the method detailed by Amaro et al. (32), with minor modifications. Briefly, 1.5-ml log cultures grown in marine broth were adjusted to an OD600 of 0.8, centrifuged at 400 × g for 5 min, and resuspended in 50 μl of Laemmli buffer. Samples were boiled for 10 min, and 25 μg of proteinase K in 10 μl of Laemmli buffer was added and incubated at 60°C for 1 h. The extracted LPS was separated by SDS-PAGE on a 4 to 15% acrylamide gradient gel (Bio-Rad, Hercules, CA) and visualized by periodic acid oxidation, fluorescence labeling with Pro-Q Emerald 300 stain (Molecular Probes/Invitrogen, Carlsbad, CA), and imaging under UV light.
Antibiotic sensitivity.
Sensitivity to polymyxin B was assessed by assays of the zone of clearance. One-hundred-microgram aliquots of polymyxin B (Sigma) were applied to 1/4-in. sterile blank cellulose antibiotic susceptibility discs. The following antibiotic discs (BD) were used to determine the general sensitivity of the strains: tetracycline, 30 μg; ciprofloxacin, 5 μg; nalidixic acid, 30 μg; erythromycin, 15 μg; and gentamicin, 10 μg. Log-phase cultures were spread plated onto LB plus 2% NaCl and allowed to dry for 10 min, antibiotic discs were applied, and the plates were incubated at 30°C overnight. The assay was performed in duplicate or triplicate, followed by measurement of the zones of clearance by use of calipers.
V. vulnificus motility.
Motility was assessed by inoculating 1 μl of a log-phase culture (adjusted to an OD600 of 1.0) into the center of a 10-cm MB soft agar plate (0.3% agar). After incubation for 16 h at room temperature, the diameter of bacterial movement during growth was measured in millimeters by using calipers. Measurements were performed in triplicate, with at least two biological replicates for each.
Visualization and counting of V. vulnificus flagella.
Transmission electron microscopy was used to visualize flagella and quantify the population with flagellar defects. V. vulnificus was cultured overnight at 30°C in MB, washed 3 times by centrifugation at 4,000 × g, and resuspended in Dulbecco's phosphate-buffered saline. Samples were allowed to adsorb to Formvar/carbon-coated copper grids for 10 min. Grids were washed in distilled H2O and stained with 1% aqueous uranyl acetate (Ted Pella Inc., Redding, CA) for 1 min. Excess liquid was gently wicked off, and grids were allowed to air dry. Samples were viewed on a JEOL 1200EX transmission electron microscope (JEOL USA, Peabody, MA) at an accelerating voltage of 80 kV. Flagellar phenotypes of 100 bacteria were counted for each strain in two independent experiments.
Biofilm assay.
The ability to form biofilms was assessed by inoculating 96-well plates with 1:100 dilutions of stationary cultures in MB, followed by incubation periods of up to 24 h at room temperature. The culture medium was removed, and the wells were washed three times with sterile PBS. One hundred microliters of 0.1% crystal violet was added to each well and incubated for 30 min at room temperature. Unbound crystal violet was then removed, and wells were washed three times with 100 μl of sterile PBS. Crystal violet was extracted using 100 μl of dimethyl sulfoxide (DMSO), and the OD595 was recorded. Biofilm assays were performed in triplicate, with at least two biological replicates for each.
Mouse infection model.
Animal infection studies were performed in accordance with approved protocols from the Washington University Division of Comparative Medicine. Bacterial strains to be used in the competitive infections were grown overnight aerobically at 30°C in LB plus 2% NaCl. Overnight cultures were diluted 1:50 in fresh medium to grow samples to log phase. V. vulnificus strains used in each competition were washed in sterile PBS and normalized to an OD600 of 0.3 prior to preparation of the mixed strain inoculum. Male 8- to 10-week-old CD1 mice (Charles River, Wilmington, MA) were infected with 5 × 106 to 1 × 107 total CFU in 100 μl PBS by tail vein injection. Blood was collected from the submandibular vein at 30 and/or 90 min postinfection. The animals were sacrificed at 90 min postinfection, and in some experiments, the liver was harvested and homogenized in sterile PBS. Spontaneously streptomycin-resistant isolates of the CMCP6 Δnab2 mutant and Δnab2/pnab2 complemented strain were generated specifically for use in the in vivo competition experiments. The collected samples were serially diluted and plated on LB agar, with and without streptomycin (Sm) to distinguish WT from Δnab2 Smr mutant or Δnab2/pnab2 Smr colonies. The combination of chloramphenicol and streptomycin proved too toxic for the slow-growing Δnab2/pnab2 complemented strain (refer to “Strains and culture conditions,” above); therefore, in competitions with the CMCP6 Δnab2 mutant, the Δnab2/pnab2 complemented strain was distinguished by its small colony size. The competitive index (CI) was determined as follows: CI = ratio out (WT/mutant)/ratio in (WT/mutant). A CI of >1 indicates that the WT outcompeted the mutant.
RESULTS
Establishment of the genetic basis for nonulosonic acid biosynthesis in V. vulnificus.
V. vulnificus CMCP6 and YJ016 contain phylogenetically distinct clusters predicted to be involved in the synthesis of NulO molecules. Three common steps of NAB pathways across the three domains of life are catalyzed by homologs of CMP-N-acetylneuraminic acid synthetase (Nab1), N-acetylneuraminic acid synthase (Nab2), and UDP-N-acetylglucosamine 2-epimerase (Nab3). HPLC analysis previously revealed that strains with a CMCP6-like genotype produced much larger amounts of NulO than those with a YJ016-like genotype (24). In fact, the nab clusters in these strains are highly divergent from one another, with no synteny and limited sequence identity (Fig. 1A). In YJ016, the nab genes are on the positive strand, and in CMCP6, they are on the negative strand. Although the nab genes in V. vulnificus are related to genes encoding a sialic acid (N-acetylneuraminic acid), there is limited sequence identity between NAB pathway components in V. vulnificus and the more well-described enzymes involved in sialic acid synthesis in E. coli K1. For example, CMCP6 Nab1 shares only 38% amino acid identity (E value, 5e−34) with E. coli NeuA, Nab2 shares 44% identity (E value, 3e−91) with E. coli NeuB, and Nab3 shares 40% identity (E value, 9e−84) with E. coli NeuC.
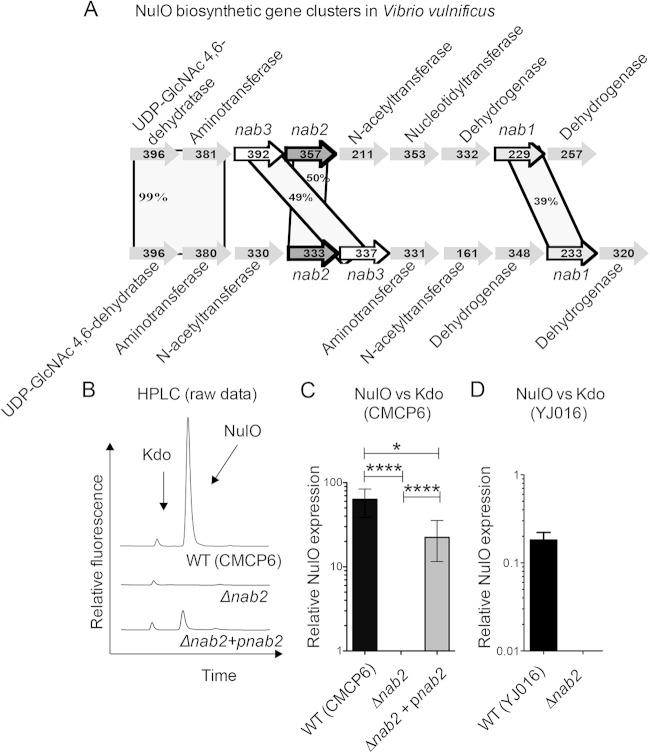
FIG 1.
V. vulnificus nab genetic organization and role of nab2 in NulO biosynthesis. (A) Schematic of the V. vulnificus NulO biosynthetic clusters in YJ016 and CMCP6. Open reading frames and the direction of transcription are designated by arrows. nab1 (encoding a CMP-neuraminic acid synthase homolog), nab2 (encoding an N-acetylneuraminic acid synthase homolog), and nab3 (encoding a UDP-N-acetylglucosamine 2-epimerase homolog) are highlighted. Percentages in shaded areas indicate amino acid identities of translated genes. (B) DMB-HPLC analysis of the CMCP6 WT and isogenic Δnab2 and complemented (Δnab2+pnab2) strains confirmed that nab2 encodes the V. vulnificus NulO synthase. (C and D) NulO expression was normalized to that of 3-deoxy-d-manno-octulosonic acid (Kdo) for quantitative comparisons of NulO expression between strains. Error bars represent standard deviations for three experiments. One-way analysis of variance (ANOVA) with the Bonferroni multiple-comparison posttest (P < 0.05) was used to examine the statistical significance of differences in NulO production. *, P < 0.05; ****, P < 0.0001.
To definitively determine the genetic basis for NulO production in V. vulnificus, we performed an in-frame deletion of the VV1_0808 synthase (nab2) in the V. vulnificus CMCP6 wild-type (WT) strain (Fig. 1A). The Δnab2 mutant strain (JBL0808) did not exhibit any significant defects in growth in vitro. To determine the effect of deletion of nab2 on NulO biosynthesis, NulO residues in the WT and Δnab2 strains were released by mild acid hydrolysis and derivatized with the fluorescent molecule DMB, which reacts with the alpha-keto acid of NulOs. HPLC analysis of total cell-associated NulOs revealed a complete absence of NulO expression in the Δnab2 strain (Fig. 1B and C). Complementation of the Δnab2 strain was accomplished by plasmid-based overexpression from the nab2 plasmid to create strain JBL0808C, here referred to as the Δnab2/pnab2 strain (Table 1). Deletion of nab2 in the YJ016 background also resulted in the elimination of NulO biosynthesis (Fig. 1D). We note that NulO synthesis was restored in the Δnab2/pnab2 strain but that NulO levels were lower than the WT levels (Fig. 1C). These analyses formally demonstrate the genetic basis for NulO synthesis in V. vulnificus, showing that the N-acetylneuraminic acid synthase homolog encoded by nab2 is required for NulO biosynthesis.
V. vulnificus LPS is modified with NulO residues.
The LPS O antigen is a frequent site of NulO modification in Gram-negative organisms (16, 29–31). To investigate whether V. vulnificus LPS contains NulO residues, LPS was isolated from the WT and Δnab2 strains. Polyacrylamide gel electrophoresis followed by glycan visualization using the Pro-Q Emerald stain demonstrated a lower-molecular-weight band in the Δnab2 strains than in the WT strain (Fig. 2A), suggesting a truncation of LPS. The higher-molecular-weight band seen with WT LPS was partially restored in the Δnab2/pnab2 strain (Fig. 2A). This finding is consistent with the intermediate levels of NulO measured in the Δnab2/pnab2 complemented strain (Fig. 1C). The material used for electrophoresis was also subjected to fluorescence derivatization and HPLC analysis, verifying that NulOs were present in the LPS preparations (data not shown). In contrast, HPLC of flagellum subunits from isolated SDS-PAGE bands did not yield evidence of the presence of NulOs, despite the fact that positive controls analyzed in parallel (bacterial species known to produce glycosylated flagella) gave the expected result (data not shown). We note that a more sensitive detection method may be required to completely rule out the possibility of direct glycosylation of flagellar proteins in V. vulnificus.
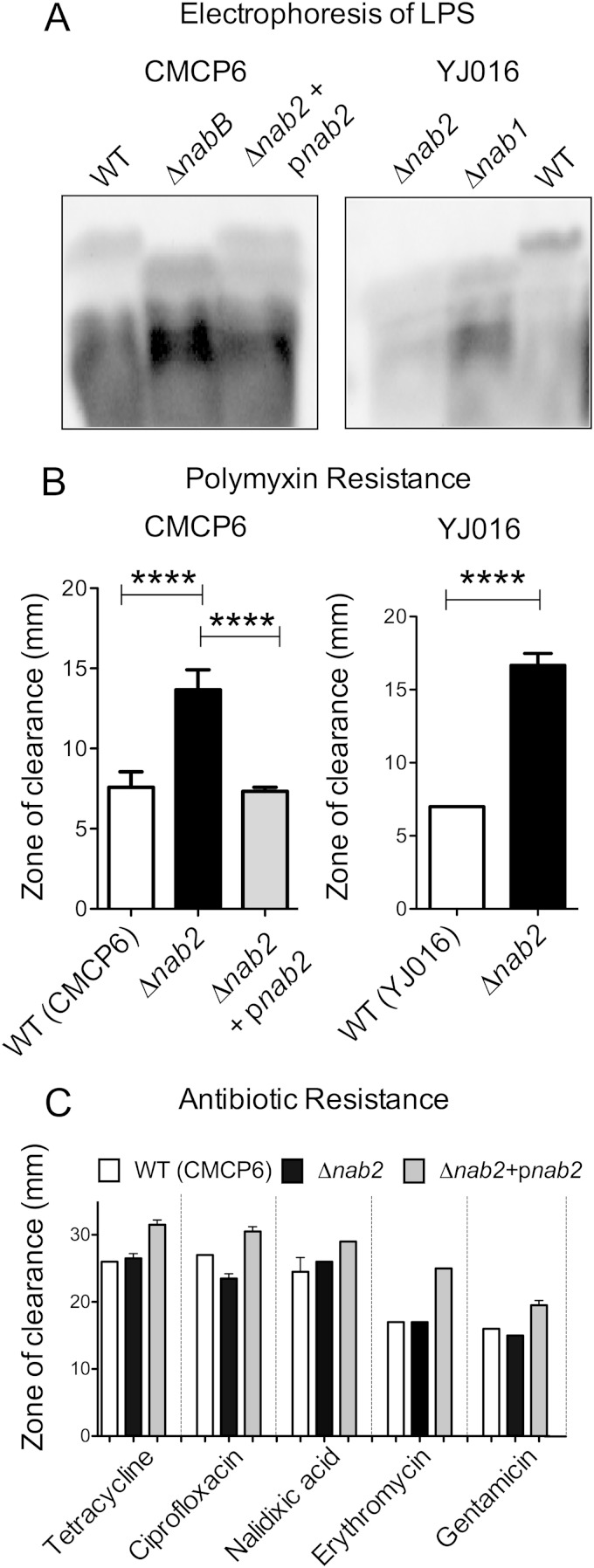
FIG 2.
V. vulnificus LPS is modified with NulO, which contributes to polymyxin B resistance. (A) V. vulnificus LPS was analyzed by SDS-PAGE with staining of fluorescent glycans (Pro-Q Emerald 300). (B) Sensitivity to the cationic antimicrobial peptide polymyxin B was determined by measuring zones of clearance following overnight growth on plates with discs containing 100 μg of polymyxin B. ****, P < 0.0001. (C) Resistance of V. vulnificus to other antibiotics.
NulO modification of LPS contributes to the natural resistance of V. vulnificus to polymyxin B.
Polymyxins are a class of cationic antibiotics produced by Gram-positive bacteria that target LPS found in Gram-negative organisms. Polymyxins are considered one of the “last resort” antibiotics that can be used for infections with multidrug-resistant pathogens. Vibrio vulnificus is naturally resistant to polymyxin B; however, the basis for this resistance is not known (36). Based on the NulO modification of LPS, we investigated whether NulO residues on V. vulnificus influence the organism's sensitivity to polymyxin B by measuring the zones of clearance after bacterial growth around discs impregnated with the antibiotic. These experiments revealed that the Δnab2 strains (in both the CMCP6 and YJ016 backgrounds) had more pronounced zones of clearance than the WT strains (P < 0.001) (Fig. 2B), indicating an enhanced sensitivity to polymyxin B. Despite only partial restoration of NulO LPS modification, complementation (Δnab2/pnab2 strain) completely restored polymyxin B resistance. To determine whether the increased sensitivity of the Δnab2 strain to polymyxin B was specific or whether the mutation may have weakened the bacterial surface architecture, possibly allowing greater overall permeability, we examined the bacterial sensitivity to a broader range of antibiotics. These experiments demonstrated that the Δnab2 strain did not have increased susceptibility to tetracycline, ciprofloxacin, nalidixic acid, erythromycin, or gentamicin (Fig. 2C). Together, these results strongly suggest that NulO modification of LPS shields V. vulnificus LPS from interaction with polymyxin B, thereby contributing to the natural resistance of V. vulnificus to this cationic antimicrobial peptide.
NulO residues play a role in the generation of V. vulnificus biofilms.
Given the common involvement of bacterial surface carbohydrates in biofilm formation (15, 33, 37–39), we next evaluated the potential role of NulO biosynthesis in the ability of V. vulnificus to generate biofilms on abiotic surfaces. The results showed that in the CMCP6 background, nab2 deletion resulted in a defect in biofilm formation compared to that of the WT (Fig. 3A). Moreover, complementation (Δnab2/pnab2 strain) completely restored the ability to form biofilms (Fig. 3A). These data implicate NulO molecules in the generation of V. vulnificus biofilms. However, in contrast to CMCP6, YJ016 (which expresses lower overall levels of NulOs) exhibited minimal, if any, nab2-dependent biofilm formation (not shown).
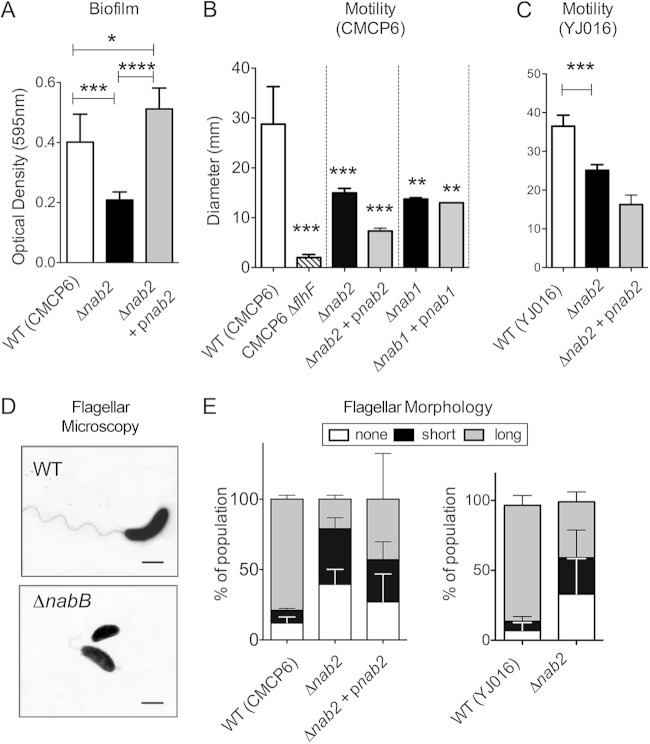
FIG 3.
NulO contributes to biofilm formation and motility. (A) Abiotic biofilm assays were conducted in 96-well polystyrene plates as described in Materials and Methods. At least 2 biological replicates were performed for each experimental condition. The data from the 6-h time point are shown. Error bars represent standard deviations. Statistical evaluation employed one-way ANOVA with the Bonferroni multiple-comparison posttest (P < 0.05). (B and C) Swimming motility was assessed by placing aliquots of WT and isogenic Δnab2 strains onto marine agar plates (0.3%) and measuring zones of motility following 16 h of growth at room temperature. The ΔflhF strain is a mutant of a known flagellar regulator that leads to a nonmotile, aflagellar strain. (D) Transmission electron micrographs of WT CMCP6 and its isogenic Δnab2 strain. (E) Flagellar morphology was assessed by electron microscopy. Several hundred individual bacteria were examined and showed that larger proportions of the Δnab2 population exhibited shortened or missing flagella. Error bars represent standard deviations. At least 2 biological replicates were tested for each experimental condition. One-way ANOVA with Dunnet's posttest was used to examine whether differences between the WT strain and isogenic derivatives were statistically significant. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01 ; *, P < 0.05.
Motility is reduced in V. vulnificus nab mutants.
Assessments of swimming motility revealed that mutations of NulO biosynthesis—in multiple strains and at multiple steps of the pathway—resulted in measurable reductions in motility (Fig. 3B and C). Notably, these strains are not nonmotile but rather exhibit a reduction of normal motility (see the comparison of the aflagellate ΔflhF strain to the Δnab strains in Fig. 3B). Visualization of flagellar filaments by transmission electron microscopy revealed that the deletion of nab2 resulted in a missing or shortened flagellum (Fig. 3D and E). Although the vast majority (∼80%) of individual bacteria in the WT population produced a single long polar flagellum characteristic of this organism, the CMCP6 Δnab2 population contained a much larger proportion of individual bacteria with shortened or missing flagella, with only ∼23% of the population exhibiting a long polar flagellum (Fig. 3D). Independent deletion of nab2 in the YJ016 strain background had the same effect on flagellum morphology and motility (Fig. 3D). Complementation of the motility phenotype was not possible by plasmid-based expression of the mutated genes. These results are consistent with the incomplete restoration of full NulO levels in the complemented versus WT strains (Fig. 1C) and a corresponding intermediate phenotype of the LPS, with both lower- and higher-molecular-weight forms (Fig. 2A). These data suggest a complex relationship between LPS structure and proper sheathing of the V. vulnificus flagellum, which will require further investigation. The fact that the Δnab2/pnab2 strain exhibited a complete restoration of polymyxin B resistance (Fig. 2B) and biofilm formation (Fig. 3A) despite its reduced motility clearly indicates that (i) NulO expression has biological effects that are independent of motility and (ii) we can use the mutant strains to distinguish motility-independent phenotypes.
NulO expression promotes bacterial survival in blood during systemic infection.
To determine whether NulO molecules play a role in V. vulnificus systemic virulence, a tail vein injection model was used to directly administer bacteria to the bloodstream in CD1 mice. A spontaneously streptomycin-resistant isolate of the CMCP6 Δnab2 strain (Δnab2 Smr) was generated specifically for use in these experiments to distinguish mutant from WT bacteria during competition in vivo. This strain retained the phenotypes observed for the Δnab2 strain and did not vary from the parental strain in its growth characteristics in vitro (not shown). Strains were set up to compete against each other in a pairwise fashion, using a 1:1 mixed inoculum, to examine relative fitness levels. Blood was drawn at early time points postinfection, and the numbers of CFU of the strains were determined based on differential resistance to streptomycin. These experiments revealed that the nab2 gene confers a strong survival advantage to the WT strain in the bloodstream in vivo. The WT strain was present at approximately 5,000 CFU/ml of blood at 90 min postinfection, whereas the Δnab2 Smr strain was completely cleared by most animals at this time point (Fig. 4A).
FIG 4.
Elaboration of NulO as a virulence factor during V. vulnificus bloodstream infection. (A to C) Animals were infected with approximately 107 bacteria via the tail vein, using a 1:1 mixture of the strain pair shown in each panel (a spontaneously streptomycin-resistant isolate of the Δnab2 strain was utilized). Blood was drawn from the submandibular vein 90 min after infection, and serial dilutions were plated on selective media containing streptomycin or chloramphenicol to distinguish between competing strains, as described in Materials and Methods. (D) The competitive index was calculated as described in Materials and Methods and takes into account both the inoculum dose and 90-min blood titers for both strains. Data shown are for two independent experiments comparing the stated strain competitions in parallel, using 4 mice per group. The Mann-Whitney U test was used to examine statistical significance; *, P < 0.03, ***, P < 0.001; ns, not statistically significant.
Our observation that the Δnab2 strain displayed reduced motility compared to the WT strain required us not to rely solely on comparisons between the WT and Δnab2 strains to test the hypothesis that NulO expression may affect pathogenesis (since motility could also conceivably be a factor). To rule out the possibility that effects on motility contributed to the bloodstream survival phenotype in vivo, we also performed experiments in which the Δnab2 strain competed against the complemented Δnab2/pnab2 strain, which expresses NulO but retains the motility defect of the Δnab2 strain (see above). The results of this experiment demonstrated that plasmid-based complementation of nab2 restored the ability of V. vulnificus to survive in the mammalian bloodstream (Fig. 4B). This finding was echoed by the results of competition between the Δnab2 and Δnab2/pnab2 strains, which demonstrated that the complemented strain was significantly better able to survive in the bloodstream, exhibiting approximately 10-fold higher titers in blood than those of the Δnab2 strain 90 min after infection. To account for both input and output numbers of CFU of both strains and in each animal, a competitive index (CI) was calculated: values of >1 indicate a relative fold increase in fitness of the WT versus the competing strain. In competition experiments between the WT and Δnab2 strains, the median CI at 90 min postinfection was 28.3 (range, 5.7 to 480.5) (Fig. 4D). In contrast, in the Δnab2 complemented strain (Δnab2/pnab2), there was an increase in the ability to compete against the WT strain, thus resulting in a significantly lower median CI (4.3; range, 0.484 to 49.2) than that for the WT and Δnab2 strain competition (Fig. 4D). The median CI for animals infected with the Δnab2 and Δnab2/pnab2 strains was ∼10.7 (range, 0.667 to 77.4). Taken together, these data show that sialic acid-like NulO expression (independent of motility) contributes to the survival of V. vulnificus during systemic infection.
Higher levels of NulOs correspond to a greater ability to cause bloodstream infection and dissemination.
We previously observed that V. vulnificus strains with a CMCP6-like nab genotype had approximately 100-fold higher levels of NulO expression than strains with a YJ016-like genotype (24) (Fig. 5A). We next set out to measure the extent to which these two nab genotypes and their corresponding NulO phenotypes contribute to pathogenesis. To test the hypothesis that the CMCP6 strain may display a greater extent of nab2-dependent bloodstream survival and dissemination than YJ016, each of the WT strains were competed in parallel against their respective isogenic Δnab2 strains in the murine bacteremia model. Deletion of nab2 in the YJ016 background significantly reduced bacterial survival at 90 min postinfection when this strain competed against the WT YJ016 strain. Head-to-head comparison of nab2-dependent bloodstream survival revealed a pronounced, ∼50-fold difference in the competitive index between the two strain backgrounds (Fig. 5B). Whereas the CMCP6 strain outcompeted its Δnab2 derivative in the blood at 90 min postinfection, by about 500-fold (median, 508.3; n = 15), the YJ016 strain outcompeted its Δnab2 derivative by only about 10-fold (median, 11.22; n = 14). This finding strongly supports the interpretation that higher levels of NulO synthesis in connection with the CMCP6 nab genotype contribute to a greater capacity for bloodstream survival. These differences in nab2-dependent survival in blood in vivo also translate into differences in bacterial dissemination to other tissues, as evidenced by bacterial titers in the liver at 90 min postinfection (P < 0.001) (Fig. 5B).
FIG 5.

Relationship between CMCP6 and YJ016 NulO expression levels and nab2-dependent bacteremia with organ dissemination. (A) NulO expression levels were measured among environmental and clinical V. vulnificus isolates and expressed relative to the eight-carbon backbone monosaccharide Kdo. PCR was utilized to determine CMCP6-type or YJ016-type alleles of nab2. These data were previously published (24), and the results of a new analysis are shown here for comparison to panel B. (B) Animals were infected with approximately 107 bacteria via the tail vein, using 1:1 mixtures of the WT reference strains YJ016 and CMCP6, each in competition with the Δnab2 mutant generated in the corresponding background. As described in the legend to Fig. 4, these experiments utilized a spontaneously streptomycin-resistant isolate of the Δnab2 strain to distinguish between the strains. The Mann-Whitney U test was used to determine statistical significance. **, P < 0.01; ***, P < 0.001. Data from four independent experiments were combined.
DISCUSSION
Vibrio vulnificus is a leading cause of food-related death in the United States due to its ability to quickly access the host bloodstream. However, virtually nothing is known about how this organism persists in the systemic vasculature during life-threatening systemic disease. Here we (i) formally elucidate the genetic basis of NulO biosynthesis in V. vulnificus, (ii) show that sialic acid-like modifications are present on bacterial LPS, (iii) demonstrate key roles for these molecules in behaviors and phenotypes likely to benefit the organism in its aquatic niche (e.g., antimicrobial resistance, biofilm formation, and motility), and (iv) demonstrate sialic acid-like molecules as the first virulence factor that promotes V. vulnificus survival during bloodstream infection in vivo.
Gene deletions at different steps of the NulO biosynthesis pathway in different V. vulnificus backgrounds led to measurable reductions in motility that could not be restored upon plasmid-based complementation. These motility defects may be due to alteration of the composition of the polar flagellum sheath, which has been shown in both V. cholerae and V. parahaemolyticus to contain LPS residues (40). Multiple lineages of Vibrionaceae express flagella that are sheathed with LPS (40–42). Moreover, a reduction or elimination of motility has been demonstrated upon alteration of the LPS O-antigen structure (via genetic manipulation) (16, 43) or through host antibody responses directed against the LPS O antigen (44, 45). For V. vulnificus, we demonstrated that NulO biosynthesis returns upon plasmid-based gene complementation. However, consistent with reduced NulO levels in the complemented versus WT strains, there was a significant amount of truncated LPS that remained. Unfortunately, the role of the sheath in flagellar function is largely unknown; some species contain a sheathed flagellum, whereas others do not. The apparent requirement of LPS O-antigen glycosylation for bacterial motility in several bacterial species with sheathed flagella requires additional study.
The mechanism by which NulO molecules promote survival of V. vulnificus in vivo is unknown at present. Sialic acid-like NulO molecules are structurally very similar to the canonical sialic acids and may act through similar mechanisms. Molecular mimicry of N-acetylneuraminic acid (the most common mammalian sialic acid) by bacterial pathogens has been studied and reviewed extensively (3, 46–50); thus, we provide only a few brief examples to illustrate how sialic acid-like molecules of V. vulnificus could result in enhanced bloodstream survival and dissemination. For example, sialic acid modifications of the LPS O antigens of Campylobacter jejuni, Neisseria meningitidis, and Haemophilus influenzae, the K1 (capsular) antigen of Escherichia coli, and the capsular polysaccharide of Streptococcus agalactiae have been shown to confer serum resistance and/or resistance to complement-mediated opsonization (25, 26, 51–53). Bacterial sialic acids have also been shown to interact with a family of sialic acid-binding, Ig-like lectin receptors known as Siglecs (recently reviewed in reference 27). For example, terminal sialic acids of the capsular polysaccharide of Streptococcus agalactiae dampen immune responses by coopting Siglecs expressed on the surfaces of neutrophils and monocytes, resulting in a reduced bactericidal capacity (2, 27, 28). Future efforts will examine whether V. vulnificus sialic acid-like molecules protect against host immune responses by mechanisms similar to those of the canonical sialic acid-expressing pathogens.
Taken together, our results demonstrate that NulO residues modify LPS in V. vulnificus and contribute to the bacterium's motility, biofilm formation, and resistance to the cationic antimicrobial peptide polymyxin B. Moreover, our studies of systemic virulence in a mouse intravenous infection model provide the first demonstration that NulO molecules resembling host sialic acids are an important virulence factor promoting survival of V. vulnificus in the bloodstream. These studies highlight the importance of NulO molecules in the biology of V. vulnificus in its aquatic niche and suggest that the enhanced virulence brought about by these molecules in mammals may be purely accidental. Nevertheless, the results of these studies strongly suggest that strains possessing high levels of NulOs may be more dangerous to susceptible individuals. Thus, technologies able to identify strains with high-level NulO expression could prove useful for safety monitoring of oysters and other shellfish or the waters from which they are harvested.
ACKNOWLEDGMENTS
We gratefully acknowledge Wandy Beatty for assistance with transmission electron microscopy and Deborah Frank for constructive feedback on the manuscript.
This research was supported in part by a National Science Foundation Career award (award DEB-0844409) to E.F.B. and by departmental funds to A.L.L. J.-B.L. was supported in part by the Chemistry-Biology Interface Graduate Program at the University of Delaware. N.M.G. was supported in part by a postdoctoral fellowship from the American Heart Association.
J.-B.L., W.G.L., N.M.G., E.F.B., and A.L.L. designed experiments. J.-B.L. performed genetic manipulation of V. vulnificus in the Boyd lab and HPLC in the Lewis lab. S.A.-M. made the YJ016 nab1 mutant in the Boyd lab. J.-B.L. performed growth curve assays, biofilm assays, and some of the motility studies in the Boyd lab. J.-B.L. performed LPS analysis, antibiotic susceptibility assays, transmission electron microscopy experiments, and some of the motility studies in the Lewis lab, with the guidance of W.G.L. N.M.G., J.-B.L., C.M.W., W.G.L., and A.L.L. contributed to the completion of the animal experiments, all performed in the Lewis lab. J.-B.L., W.G.L., N.M.G., E.F.B., and A.L.L. analyzed data. J.-B.L., W.G.L., N.M.G., E.F.B., and A.L.L. wrote the paper, and all coauthors read and approved the written document.
REFERENCES
- 1.Lewis AL, Desa N, Hansen EE, Knirel YA, Gordon JI, Gagneux P, Nizet V, Varki A. 2009. Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. Proc Natl Acad Sci U S A 106:13552–13557. doi: 10.1073/pnas.0902431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlin AF, Lewis AL, Varki A, Nizet V. 2007. Group B streptococcal capsular sialic acids interact with siglecs (immunoglobulin-like lectins) on human leukocytes. J Bacteriol 189:1231–1237. doi: 10.1128/JB.01155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vimr E, Lichtensteiger C. 2002. To sialylate, or not to sialylate: that is the question. Trends Microbiol 10:254–257. doi: 10.1016/S0966-842X(02)02361-2. [DOI] [PubMed] [Google Scholar]
- 4.Parsons NJ, Patel PV, Tan EL, Andrade JR, Nairn CA, Goldner M, Cole JA, Smith H. 1988. Cytidine 5′-monophospho-N-acetyl neuraminic acid and a low molecular weight factor from human blood cells induce lipopolysaccharide alteration in gonococci when conferring resistance to killing by human serum. Microb Pathog 5:303–309. doi: 10.1016/0882-4010(88)90103-9. [DOI] [PubMed] [Google Scholar]
- 5.Zelmer A, Bowen M, Jokilammi A, Finne J, Luzio JP, Taylor PW. 2008. Differential expression of the polysialyl capsule during blood-to-brain transit of neuropathogenic Escherichia coli K1. Microbiology 154:2522–2532. doi: 10.1099/mic.0.2008/017988-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inzana TJ, Balyan R, Howard MD. 2012. Decoration of Histophilus somni lipooligosaccharide with N-acetyl-5-neuraminic acid enhances bacterial binding of complement factor H and resistance to killing by serum and polymorphonuclear leukocytes. Vet Microbiol 161:113–121. doi: 10.1016/j.vetmic.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Feng Y, Cao M, Shi J, Zhang H, Hu D, Zhu J, Zhang X, Geng M, Zheng F, Pan X, Li X, Hu F, Tang J, Wang C. 2012. Attenuation of Streptococcus suis virulence by the alteration of bacterial surface architecture. Sci Rep 2:710. doi: 10.1038/srep00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costa-Hurtado M, Olvera A, Martinez-Moliner V, Galofré-Milà N, Martínez P, Dominguez J, Aragon V. 2013. Changes in macrophage phenotype after infection of pigs with Haemophilus parasuis strains with different levels of virulence. Infect Immun 81:2327–2333. doi: 10.1128/IAI.00056-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ram S, Sharma AK, Simpson SD, Gulati S, McQuillen DP, Pangburn MK, Rice PA. 1998. A novel sialic acid binding site on factor H mediates serum resistance of sialylated Neisseria gonorrhoeae. J Exp Med 187:743–752. doi: 10.1084/jem.187.5.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim KS, Itabashi H, Gemski P, Sadoff J, Warren RL, Cross AS. 1992. The K1 capsule is the critical determinant in the development of Escherichia coli meningitis in the rat. J Clin Invest 90:897–905. doi: 10.1172/JCI115965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johswich KO, Zhou J, Law DKS, St Michael F, McCaw SE, Jamieson FB, Cox AD, Tsang RSW, Gray-Owen SD. 2012. Invasive potential of nonencapsulated disease isolates of Neisseria meningitidis. Infect Immun 80:2346–2353. doi: 10.1128/IAI.00293-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diallo IS, Frost AJ. 2000. Survival of avian strains of Pasteurella multocida in chicken serum. Vet Microbiol 72:153–161. doi: 10.1016/S0378-1135(99)00195-9. [DOI] [PubMed] [Google Scholar]
- 13.Bax M, Kuijf ML, Heikema AP, van Rijs W, Bruijns SCM, García-Vallejo JJ, Crocker PR, Jacobs BC, van Vliet SJ, van Kooyk Y. 2011. Campylobacter jejuni lipooligosaccharides modulate dendritic cell-mediated T cell polarization in a sialic acid linkage-dependent manner. Infect Immun 79:2681–2689. doi: 10.1128/IAI.00009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerry P, Ewing CP, Schirm M, Lorenzo M, Kelly J, Pattarini D, Majam G, Thibault P, Logan S. 2006. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol Microbiol 60:299–311. doi: 10.1111/j.1365-2958.2006.05100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howard SL, Jagannathan A, Soo EC, Hui JPM, Aubry AJ, Ahmed I, Karlyshev A, Kelly JF, Jones MA, Stevens MP, Logan SM, Wren BW. 2009. Campylobacter jejuni glycosylation island important in cell charge, legionaminic acid biosynthesis, and colonization of chickens. Infect Immun 77:2544–2556. doi: 10.1128/IAI.01425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Post DMB, Yu L, Krasity BC, Choudhury B, Mandel MJ, Brennan CA, Ruby EG, McFall-Ngai MJ, Gibson BW, Apicella MA. 2012. O-antigen and core carbohydrate of Vibrio fischeri lipopolysaccharide: composition and analysis of their role in Euprymna scolopes light organ colonization. J Biol Chem 287:8515–8530. doi: 10.1074/jbc.M111.324012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.St Michael F, Szymanski CM, Li J, Chan KH, Khieu NH, Larocque S, Wakarchuk WW, Brisson J-R, Monteiro MA. 2002. The structures of the lipooligosaccharide and capsule polysaccharide of Campylobacter jejuni genome sequenced strain NCTC 11168. Eur J Biochem 269:5119–5136. doi: 10.1046/j.1432-1033.2002.03201.x. [DOI] [PubMed] [Google Scholar]
- 18.Schoenhofen IC, Vinogradov E, Whitfield DM, Brisson J-R, Logan SM. 2009. The CMP-legionaminic acid pathway in Campylobacter: biosynthesis involving novel GDP-linked precursors. Glycobiology 19:715–725. doi: 10.1093/glycob/cwp039. [DOI] [PubMed] [Google Scholar]
- 19.Vinogradov E, Wilde C, Anderson EM, Nakhamchik A, Lam JS, Rowe-Magnus DA. 2009. Structure of the lipopolysaccharide core of Vibrio vulnificus type strain 27562. Carbohydr Res 344:484–490. doi: 10.1016/j.carres.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 20.Thibault P, Logan SM, Kelly JF, Brisson J-R, Ewing CP, Trust TJ, Guerry P. 2001. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J Biol Chem 276:34862–34870. doi: 10.1074/jbc.M104529200. [DOI] [PubMed] [Google Scholar]
- 21.Logan SM, Kelly JF, Thibault P, Ewing CP, Guerry P. 2002. Structural heterogeneity of carbohydrate modifications affects serospecificity of Campylobacter flagellins. Mol Microbiol 46:587–597. doi: 10.1046/j.1365-2958.2002.03185.x. [DOI] [PubMed] [Google Scholar]
- 22.Blackwell KD, Oliver JD. 2008. The ecology of Vibrio vulnificus, Vibrio cholerae, and Vibrio parahaemolyticus in North Carolina estuaries. J Microbiol 46:146–153. doi: 10.1007/s12275-007-0216-2. [DOI] [PubMed] [Google Scholar]
- 23.Bross MH, Soch K, Morales R, Mitchell RB. 2007. Vibrio vulnificus infection: diagnosis and treatment. Am Fam Physician 76:539–544. [PubMed] [Google Scholar]
- 24.Lewis AL, Lubin JB, Argade S, Naidu N, Choudhury B, Boyd EF. 2011. Genomic and metabolic profiling of nonulosonic acids in Vibrionaceae reveal biochemical phenotypes of allelic divergence in Vibrio vulnificus. Appl Environ Microbiol 77:5782–5793. doi: 10.1128/AEM.00712-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mushtaq N, Redpath MB, Luzio JP, Taylor PW. 2004. Prevention and cure of systemic Escherichia coli K1 infection by modification of the bacterial phenotype. Antimicrob Agents Chemother 48:1503–1508. doi: 10.1128/AAC.48.5.1503-1508.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogel U, Weinberger A, Frank R, Muller A, Kohl J, Atkinson JP, Frosch M. 1997. Complement factor C3 deposition and serum resistance in isogenic capsule and lipooligosaccharide sialic acid mutants of serogroup B Neisseria meningitidis. Infect Immun 65:4022–4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang YC, Nizet V. 2014. The interplay between Siglecs and sialylated pathogens. Glycobiology 24:818–825. doi: 10.1093/glycob/cwu067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang YC, Olson J, Beasley FC, Tung C, Zhang J, Crocker PR, Varki A, Nizet V. 2014. Group B Streptococcus engages an inhibitory Siglec through sialic acid mimicry to blunt innate immune and inflammatory responses in vivo. PLoS Pathog 10:e1003846. doi: 10.1371/journal.ppat.1003846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 30.Philippe N, Alcaraz JP, Coursange E, Geiselmann J, Schneider D. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51:246–255. doi: 10.1016/j.plasmid.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Lewis AL, Nizet V, Varki A. 2004. Discovery and characterization of sialic acid O-acetylation in group B Streptococcus. Proc Natl Acad Sci U S A 101:11123–11128. doi: 10.1073/pnas.0403010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amaro C, Biosca E, Fouz B, Garay E. 1992. Electrophoretic analysis of heterogeneous lipopolysaccharides from various strains of Vibrio vulnificus biotypes 1 and 2 by silver staining and immunoblotting. Curr Microbiol 25:99–104. doi: 10.1007/BF01570967. [DOI] [PubMed] [Google Scholar]
- 33.Anderson GG, Goller CC, Justice S, Hultgren SJ, Seed PC. 2010. Polysaccharide capsule and sialic acid-mediated regulation promote biofilm-like intracellular bacterial communities during cystitis. Infect Immun 78:963–975. doi: 10.1128/IAI.00925-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knirel YA, Moll H, Helbig JH, Zahringer U. 1997. Chemical characterization of a new 5,7-diamino-3,5,7,9-tetradeoxynonulosonic acid released by mild acid hydrolysis of the Legionella pneumophila serogroup 1 lipopolysaccharide. Carbohydr Res 304:77–79. doi: 10.1016/S0008-6215(97)00211-5. [DOI] [PubMed] [Google Scholar]
- 35.Knirel YA, Vinogradov EV, L'Vov VL, Kocharova NA, Shashkov AS, Dmitriev BA, Kochetkov NK. 1984. Sialic acids of a new type from the lipopolysaccharides of Pseudomonas aeruginosa and Shigella boydii. Carbohydr Res 133:C5–C8. doi: 10.1016/0008-6215(84)85213-1. [DOI] [PubMed] [Google Scholar]
- 36.Oliver JD, Guthrie K, Preyer J, Wright A, Simpson LM, Siebeling R, Morris JG Jr. 1992. Use of colistin-polymyxin B-cellobiose agar for isolation of Vibrio vulnificus from the environment. Appl Environ Microbiol 58:737–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jurcisek J, Greiner L, Watanabe H, Zaleski A, Apicella MA, Bakaletz LO. 2005. Role of sialic acid and complex carbohydrate biosynthesis in biofilm formation by nontypeable Haemophilus influenzae in the chinchilla middle ear. Infect Immun 73:3210–3218. doi: 10.1128/IAI.73.6.3210-3218.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kierek K, Watnick PI. 2003. The Vibrio cholerae O139 O-antigen polysaccharide is essential for Ca2+-dependent biofilm development in sea water. Proc Natl Acad Sci U S A 100:14357–14362. doi: 10.1073/pnas.2334614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malcova M, Karasova D, Rychlik I. 2009. aroA and aroD mutations influence biofilm formation in Salmonella Enteritidis. FEMS Microbiol Lett 291:44–49. doi: 10.1111/j.1574-6968.2008.01433.x. [DOI] [PubMed] [Google Scholar]
- 40.Fuerst JA, Perry JW. 1988. Demonstration of lipopolysaccharide on sheathed flagella of Vibrio cholerae O:1 by protein A-gold immunoelectron microscopy. J Bacteriol 170:1488–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brennan CA, Hunt JR, Kremer N, Krasity BC, Apicella MA, McFall-Ngai MJ, Ruby EG. 2014. A model symbiosis reveals a role for sheathed-flagellum rotation in the release of immunogenic lipopolysaccharide. eLife 3:e01579. doi: 10.7554/eLife.01579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Norqvist A, Wolf-Watz H. 1993. Characterization of a novel chromosomal virulence locus involved in expression of a major surface flagellar sheath antigen of the fish pathogen Vibrio anguillarum. Infect Immun 61:2434–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valiente E, Jimenez N, Merino S, Tomas JM, Amaro C. 2008. Vibrio vulnificus biotype 2 serovar E gne but not galE is essential for lipopolysaccharide biosynthesis and virulence. Infect Immun 76:1628–1638. doi: 10.1128/IAI.01393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leitner DR, Feichter S, Schild-Prufert K, Rechberger GN, Reidl J, Schild S. 2013. Lipopolysaccharide modifications of a cholera vaccine candidate based on outer membrane vesicles reduce endotoxicity and reveal the major protective antigen. Infect Immun 81:2379–2393. doi: 10.1128/IAI.01382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bishop AL, Schild S, Patimalla B, Klein B, Camilli A. 2010. Mucosal immunization with Vibrio cholerae outer membrane vesicles provides maternal protection mediated by antilipopolysaccharide antibodies that inhibit bacterial motility. Infect Immun 78:4402–4420. doi: 10.1128/IAI.00398-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varki A, Gagneux P. 2012. Multifarious roles of sialic acids in immunity. Ann N Y Acad Sci 1253:16–36. doi: 10.1111/j.1749-6632.2012.06517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varki A. 2011. Since there are PAMPs and DAMPs, there must be SAMPs? Glycan “self-associated molecular patterns” dampen innate immunity, but pathogens can mimic them. Glycobiology 21:1121–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. 2004. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev 68:132–153. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moran AP, Prendergast MM, Appelmelk BJ. 1996. Molecular mimicry of host structures by bacterial lipopolysaccharides and its contribution to disease. FEMS Immunol Med Microbiol 16:105–115. doi: 10.1111/j.1574-695X.1996.tb00127.x. [DOI] [PubMed] [Google Scholar]
- 50.Mandrell RE, Apicella MA. 1993. Lipo-oligosaccharides (LOS) of mucosal pathogens: molecular mimicry and host-modification of LOS. Immunobiology 187:382–402. doi: 10.1016/S0171-2985(11)80352-9. [DOI] [PubMed] [Google Scholar]
- 51.Edwards MS, Kasper DL, Jennings HJ, Baker CJ, Nicholson-Weller A. 1982. Capsular sialic acid prevents activation of the alternative complement pathway by type III, group B streptococci. J Immunol 128:1278–1283. [PubMed] [Google Scholar]
- 52.Guerry P, Ewing CP, Hickey TE, Prendergast MM, Moran AP. 2000. Sialylation of lipooligosaccharide cores affects immunogenicity and serum resistance of Campylobacter jejuni. Infect Immun 68:6656–6662. doi: 10.1128/IAI.68.12.6656-6662.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hood DW, Makepeace K, Deadman ME, Rest RF, Thibault P, Martin A, Richards JC, Moxon ER. 1999. Sialic acid in the lipopolysaccharide of Haemophilus influenzae: strain distribution, influence on serum resistance and structural characterization. Mol Microbiol 33:679–692. doi: 10.1046/j.1365-2958.1999.01509.x. [DOI] [PubMed] [Google Scholar]
- 54.Kim YR, Lee SE, Kim CM, Kim SY, Shin EK, Shin DH, Chung SS, Choy HE, Progulske-Fox A, Hillman JD, Handfield M, Rhee JH. 2003. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect Immun 71:5461–5471. doi: 10.1128/IAI.71.10.5461-5471.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen CY, Wu KM, Chang YC, Chang CH, Tsai HC, Liao TL, Liu YM, Chen HJ, Shen AB, Li JC, Su TL, Shao CP, Lee CT, Hor LI, Tsai SF. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res 13:2577–2587. doi: 10.1101/gr.1295503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kovach ME, Phillips RW, Elzer PH, Roop RM II, Peterson KM. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800–802. [PubMed] [Google Scholar]