Abstract
Staphylococcus aureus bacteremia cases are complicated by bacterial persistence and treatment failure despite the confirmed in vitro susceptibility of the infecting strain to administered antibiotics. A high incidence of methicillin-resistant S. aureus (MRSA) bacteremia cases are classified as persistent and are associated with poorer patient outcomes. It is still unclear how S. aureus evades the host immune system and resists antibiotic treatment for the prolonged duration of a persistent infection. In this study, the genetic changes and associated phenotypic traits specific to S. aureus persistent bacteremia were identified by comparing temporally dispersed isolates from persistent infections (persistent isolates) originating from two independent persistent S. aureus bacteremia cases with the initial infection isolates and with three resolved S. aureus bacteremia isolates from the same genetic background. Several novel traits were associated specifically with both independent sets of persistent S. aureus isolates compared to both the initial isolates and the isolates from resolved infections (resolved isolates). These traits included (i) increased growth under nutrient-poor conditions; (ii) increased tolerance of iron toxicity; (iii) higher expression of cell surface proteins involved in immune evasion and stress responses; and (iv) attenuated virulence in a Galleria mellonella larva infection model that was not associated with small-colony variation or metabolic dormancy such as had been seen previously. Whole-genome sequence analysis identified different single nucleotide mutations within the mprF genes of all the isolates with the adaptive persistence traits from both independent cases. Overall, our data indicate a novel role for MprF function during development of S. aureus persistence by increasing bacterial fitness and immune evasion.
INTRODUCTION
Staphylococcus aureus is a commensal bacterium of human skin and mucosal membranes. Asymptomatic carriage is common, with as many as 80% of people colonized at any one time (1). However, this opportunistic pathogen is best known for causing a range of diseases from minor skin infections to bacteremia and septic shock. Infections can be classified as health care associated (HA) or community associated (CA), with the latter category being linked to the more virulent S. aureus strains (2, 3). Recorded incidences of methicillin-resistant S. aureus (MRSA) bacteremia in the United Kingdom have exhibited a decline in recent years, whereas the incidence of bacteremia caused by methicillin-susceptible S. aureus (MSSA) has remained relatively consistent (4, 5). However, both MRSA and MSSA infections remain a major problem worldwide (6). The problem is further exacerbated by the proliferation of strains with resistance or reduced susceptibility to last-line antimicrobial agents such as vancomycin (7, 8) and daptomycin and linezolid (9, 10).
Bloodstream infections and bacteremia caused by S. aureus are further complicated by the phenomenon of bacterial persistence and treatment failure despite the confirmed in vitro susceptibility of the infecting strain to the administered antibiotics. Persistence is traditionally defined as the “continuation or recurrence of the infection over a period in excess of 7 days” (11–14). Up to 38% of MRSA bacteremia cases can be classified as persistent and are associated with poorer patient outcomes (15–17). It is still unclear how S. aureus evades the host immune system for the prolonged duration of a persistent infection.
Previous studies have identified several characteristics associated with isolates from persistent infections (persistent isolates). For example, persistent isolates have shown elevated resistance to host defense peptides (HDP), enhanced biofilm formation, increased adhesion capabilities, and accessory gene regulator (agr) dysfunction (11–14). Furthermore, small-colony variants (SCV) are frequently implicated in persistence. These subpopulations can be metabolically dormant, elicit a reduced immune response during infections, and display heightened antibiotic resistance (18–22). However, our overall understanding of the mechanisms employed by S. aureus for the development of bacteremia persistence is still limited, and those mechanisms need further investigation.
In this study, our aim was to define the molecular mechanisms that lead to S. aureus persistence by investigating the genetic and phenotypic differences between temporally dispersed isolates originating from two persistent S. aureus bacteremia cases and by comparing these traits with those of three S. aureus bacteremia isolates from resolved infections (resolved isolates) from the same genetic background. This approach defines any genetic mutations and resultant phenotypic changes that are specific to S. aureus that cause persistent bacteremia and not bacteremia per se. Several novel traits were associated specifically with both independent sets of persistent S. aureus isolates compared to the initial and resolved isolates. These traits included (i) increased growth under nutrient-poor conditions; (ii) increased tolerance of iron toxicity; (iii) higher expression of cell surface proteins involved in immune evasion and stress responses; and (iv) attenuated virulence in a Galleria mellonella larva infection model that was not associated with small-colony variation or metabolic dormancy such as has been seen previously. Biofilm formation increased in persistent isolates during survival in vivo; however, total biofilm levels were equivalent to those seen with resolved isolates, indicating that biofilm formation is not a persistence-specific mechanism. Whole-genome sequence analysis identified different single nucleotide mutations within the mprF genes of persistent infection isolates from both the independent cases which correlated with the emergence of persistence-associated phenotypes. These data led to the implication of a specific gene, mprF, in persistence development in S. aureus bacteremia.
MATERIALS AND METHODS
Bacterial isolates.
MRSA isolates relating to staphylococcal bacteremia cases were identified and collected from the University Hospitals of Leicester (UHL) archive and stored in tryptic soy broth (TSB; BD Diagnostics Systems) with 20% (vol/vol) glycerol at −80°C. Unless otherwise stated, isolates were cultured on Luria agar (LA; Oxoid) at 37°C in air followed by Luria broth (LB; Oxoid) and were incubated with shaking at 37°C. For nutrient-restrictive conditions, 6% horse blood agar (HBA; Oxoid) was used and strains were cultured at 37°C in 5% CO2 followed by CRPMI medium (CRPMI medium is RPMI 1640 medium [Sigma-Aldrich] depleted of metal ions via treatment with 6% [wt/vol] Chelex 100 [Sigma-Aldrich] but with 10% [vol/vol] untreated RPMI 1640 medium reapplied to provide the minimum elements required for growth) (23) cultured statically at 37°C in 5% CO2.
Antibiotic susceptibility.
MIC Evaluator strips (Oxoid) were used in accordance with CLSI guidelines to assess antibiotic sensitivities (24, 25). 2% NaCl was added to the medium for the oxacillin experiments according to standard practice. All readings were recorded after 24 h and confirmed at 48 h. Antibiotic sensitivity measurements were repeated 3 times; therefore, a 2-fold dilution range has been reported as recommended by the CLSI.
Growth analysis.
Nutrient-restrictive growth analysis was conducted by subculturing isolates into 20 ml Chelex-treated RPMI medium (pH 7.0) to reach an optical density of 0.1 at 600 nm (OD600) from a fresh overnight culture and incubating statically at 37°C in 5% CO2 for 24 h. To test the effect that toxic iron levels had upon growth, 200 μM FeSO4 was added to duplicate CRPMI inocula prepared as described above. All cultures were inverted until equal suspensions were achieved, and 1-ml samples were analyzed at OD600 with a Jenway 6705 UV-light/visible-light (UV/Vis) spectrometer. Experiments were repeated independently three times, means and standard deviations (SD) were calculated, and a two-tailed Student t test determined significance.
Quantitative hemolysis activity.
Bacterial exoproteins, including hemolysins, were extracted from overnight cultures by centrifugation. The supernatants were sterilized and concentrated using Amicon Ultra Centrifugal Concentrators (Millipore). A log2 serial dilution of the concentrated supernatant was made using phosphate-buffered saline (PBS) in a round-bottomed 96-well plate (Nunc). The negative lysis control did not contain any bacterial supernatant. A 4% concentration of sheep and rabbit blood was prepared by resuspending 400 μl of concentrated sheep and rabbit red blood cells (Oxoid) in 10 ml PBS. A 50-μl volume of 4% sheep blood–rabbit blood was added to all wells, and the plate was incubated at 37°C for 30 min. The terminal dilution where complete hemolysis still occurred was determined by eye and recorded for each sample. Experiments were repeated independently in triplicate to confirm reproducibility.
DNase activity.
DNase activity was assessed by spotting 10 μl from each overnight culture onto DNase agar plates (Fluka BioChemika) followed by incubation at 37°C for 24 h. After incubation, DNase plates were flooded with 1 N HCl, precipitating polymerized DNA and leaving clear halos where the DNA was hydrolyzed by bacterial DNases. The radiuses of the clear halos surrounding the bacterial colonies were measured and recorded. Experiments were repeated independently in triplicate to confirm reproducibility.
Non-covalently bound cell surface extracts and iTRAQ LC-MS.
Cell surface-associated protein extracts were prepared as described previously (23), except 16-h CRPMI (pH 7.0) cultures were used. Cells were harvested by centrifugation and the bacterial pellet weights calculated to normalize protein loading and to avoid bias from potential growth differences. The bacterial cell pellet (10 mg) was resuspended in 100 μl of 125 mM MOPS (morpholinepropanesulfonic acid; pH 7.0)–2% sodium dodecyl sulfate and boiled for 3 min followed by centrifugation and supernatant recovery. Samples were equalized with respect to protein concentration as determined by A280 values (using a Thermo Scientific 2000c NanoDrop instrument) and sent to the Protein Nucleic Acid Chemistry Laboratory (PNACL) proteomic facility at the University of Leicester. iTRAQ (isobaric tag for relative and absolute quantifications) (AB Sciex UK Ltd.) analysis was followed by liquid chromatography-mass spectrometry (LC-MS) for a maximum of eight individual samples. Data were analyzed using Scaffold 4 software (Proteome Software Inc.), which calculated P values from individual analysis of variance (ANOVA) tests. P values of <0.001 and <0.0001 showed that the results presented were statistically and significantly different under the compared conditions (https://proteome-software.wikispaces.com/file/view/looking-at-iTRAQ-data-in-scaffold-qplus).
Biofilm formation assay.
Biofilm assays were conducted as previously documented (23). Briefly, this involved inoculating quadruplicate wells of a 96-well flat-bottomed microtiter plate (Nunc) with 200 μl of diluted bacterial cell suspension at an optical density at 600 nm (OD600) of 0.05 followed by static incubation for 24 h at 37°C. OD600 values were recorded at 0 and 24 h, using a BMG Labtech FLUOstar Omega plate reader to record potential growth differences. The wells were washed three times with sterile phosphate-buffered saline (PBS) and heat fixed at 60°C for 30 min. The wells were stained with 200 μl of 1% safranin for 30 min and washed twice with deionized water. Biofilms values were measured at OD490. Experiments were repeated independently three times, means and standard errors were calculated, and a two-tailed Student t test was performed.
Adhesion, invasion, and intracellular persistence assay.
An H9C2 cell line was seeded and maintained in Dulbecco's modified Eagle medium (DMEM) GlutaMAX (Gibco) with 10% fetal bovine serum (FBS; PAA Laboratories), and the cells were grown to 100% confluence in 12-well tissue culture plates (Nunc) in preparation for invasion experiments. These were conducted using DMEM GlutaMAX with 1% FBS, and this was used to replace the previous growth media at least 30 min before the start of each experiment. For the bacterial inoculum, 10-ml LB overnight cultures were harvested and the bacterial pellets were washed three times in sterile PBS and resuspended in PBS to an OD600 of 1.0, equivalent to approximately 107 CFU per 10 μl. Inoculum CFU levels were confirmed during each experiment via serial dilution and plating. A 10-μl volume of each culture was used to inoculate triplicate wells in three separate 12-well plates containing confluent H9C2 cells. The plates were incubated at 37°C in 5% CO2 for 2 h. For two of the three plates, the medium was replaced with DMEM GlutaMAX with 1% FBS, including 200 μg/ml gentamicin (Sigma-Aldrich), and the reaction mixture was returned to the incubator for a further 3 h (invasion assay) and a further 72 h (intracellular persistence assay). The final plate was taken for the next step without further medium replacement or incubations (adhesion assay). At each of these time points, medium was aspirated and wells were washed twice with PBS. The H9C2 cells were then lysed in 1 ml of 1% Triton X-100 (Sigma-Aldrich)–PBS for 10 min at room temperature, serial dilutions were made, and suspensions were plated onto nonselective media to calculate bacterial CFU levels.
Galleria mellonella larva virulence model.
Galleria mellonella larvae from Live Foods Ltd. (United Kingdom) were stored at 4°C for up to 1 week. Prior to each experiment, larvae were left overnight at room temperature to acclimatize. Overnight bacterial cultures in 5 ml LB were centrifuged, and pellets were washed three times in sterile PBS and resuspended at an OD600 of 0.1, equivalent to 106 CFU per 20 μl. Groups of 10 Galleria larvae were used for each infecting isolate. Larvae were injected with 20 μl of bacterial inoculum via their last proleg using microfine 1-ml insulin syringes (BD) with a stepper repetitive pipette (Tridak) and were incubated at 37°C. Fatalities were recorded every 24 h for 96 h. Groups of 10 larvae were injected with 20 μl sterile PBS, and another 10 were not injected at all; these were used as negative controls. If a total of two larvae from either or both of the two control groups died, the experimental results were discarded. Experiments were repeated independently three times; results were plotted on Kaplan-Meier survival graphs, and log-rank statistical analysis was conducted with the aid of GraphPad Prism 5 software. Equal inoculum sizes were confirmed by serial dilutions and CFU plating from the inoculum at the start of each experiment.
CFU counts at 48 h postinfection and RNA isolation from Galleria mellonella larvae.
Relative bacterial burdens were investigated 48 h postinfection, and additional larvae were infected in parallel with each virulence experiment for this purpose. At 48 h postinfection, two living larvae per infecting strain per experiment were homogenized in 2 ml sterile PBS using zirconia/silica beads (Thistle Scientific, United Kingdom) (4 by 2.5 mm) in a minibead beater (Biospec Products, Inc.) at a high speed for 30 s. These were centrifuged at 2,500 rpm for 5 min at 4°C, and the liquid phase was retained and kept on ice. Serial dilutions were made and plated onto Staphylococcus-selective media (Mannitol salt agar; Oxoid), and CFU levels were measured after a minimum of 48 h of incubation at 37°C. The remaining liquid phase was centrifuged at full speed for 15 min, and the pellets were resuspended in 100 μl Tris-EDTA (TE) buffer containing 3 mg/ml lysozyme and 40 μg/ml lysostaphin (TELL; Ambi Products LLC, USA) and incubated at 37°C for 15 min. A 900-μl volume of TRI reagent (Sigma-Aldrich) was then added, and samples were incubated for a further 5 min at room temperature. After the addition of 200 μl of chloroform, samples were subjected to vortex mixing for 15 s followed by 15 min of incubation at room temperature. Samples were centrifuged for 15 min at full speed at 4°C, and the upper aqueous phase was transferred to a fresh tube for RNA isolation. A 0.5-ml volume of isopropanol (IPA) was added to the upper aqueous phase, and the reaction mixture was inverted for mixing and incubated at room temperature for 10 min. Samples were then centrifuged at full speed for 8 min at 4°C to pellet the RNA. The RNA pellet was then washed with 1 ml 75% ethanol and centrifuged again at 10,500 rpm for 5 min at 4°C. The pellet was briefly air dried before being resuspended in 50 μl diethyl pyrocarbonate (DEPC)-treated water and stored at −80°C.
Quantitative reverse transcription-PCR (qRT-PCR).
RNA samples were DNase treated using Turbo DNase (Ambion) according to the manufacturer's instructions, and the resulting samples were quantified using a Thermo Scientific 2000c NanoDrop instrument and normalized so that 12 μl contained 2 μg of RNA. Samples were stored at −80°C. Omniscript reverse transcription kits (Qiagen) were used to convert RNA to cDNA, using 12 μl (2 μg) of DNase-treated RNA; RNase inhibitor (Ambion) (10 U/μl) and random hexamers (Applied Biosystems) (10 μM) were also used as recommended but were not supplied within the kit. Samples were stored at −20°C for up to 1 week. Quantitative PCR (qPCR) was conducted using Fast SYBR green technology and an ABI 7500 machine. Briefly, this involved the use of a 20-μl reaction mixture containing 10 μl Fast SYBR green master mix (Applied Biosystems), 0.5 μl forward (ACGGTCTTGCTGTCACTTATT) and reverse (TACACATATGTTCTTCCCTAATAA) 16S rRNA 10 μM primer stocks (26), and the resulting cDNA (12 μl). Cycle conditions of 94°C for 20 s and 40 cycles of 95°C for 3 s and 60°C for 30 s were used, ending in a disassociation curve to confirm accurate primer binding and disassociation. Two technical repeats were completed per RNA sample. The resulting threshold cycle (CT) values were inverted and multiplied by 1,000 to make them manageable for comparisons.
Whole-genome sequencing.
Genomic DNA (gDNA) was prepared using a Wizard Genomic DNA purification kit (Promega) according to the manufacturer's instructions, with an added lysis step involving pellet resuspension in 100 μl of 50 mM EDTA containing 200 μg/ml lysostaphin and incubation at 37°C for 30 min. A further cleanup step was used involving genomic DNA clean and concentrator columns (Cambridge Bioscience) according to the manufacturer's instructions. A minimum of 4 μg purified gDNA (minimum concentration, 40 ng/μl) resuspended in distilled water (dH2O) was submitted to the Swansea Genome Centre (Swansea, United Kingdom) and subjected to paired-end high-throughput genome sequencing on an Illumina HiSeq 2500 machine. Bioinformatic analysis was conducted using SPECTRE (Special Computational Teaching and Research Environment) at the University of Leicester. Raw paired-end reads were quality checked and trimmed using a FASTX toolkit. Trimmed reads were aligned to a reference genome (HO 5096 0412) using Picard, BWA-MEM, and SAMtools. Genetic variations compared to the reference genome and between the sequence data of individual isolates were identified using VarScan.v2.3.6 and a variant threshold of >80%. Additional sequence analysis involved the assembly of short reads into contiguous sequence files using Velvet software (27). These were entered into the BIGS Staphylococcal database, hosted at Swansea University, for gene-by-gene analysis (28). Open reading frames were identified using a reference pan-genome approach (29), and a core genome of 2,918 genes found in 90% or more of the 12 isolates was defined. A neighbor-joining phylogenetic tree was constructed from a core genome alignment of 2,596,723 bp shared by all isolates using MEGA v6 (30).
RESULTS
Clinical and genetic backgrounds of isolates from persistent and resolved MRSA bacteremia cases.
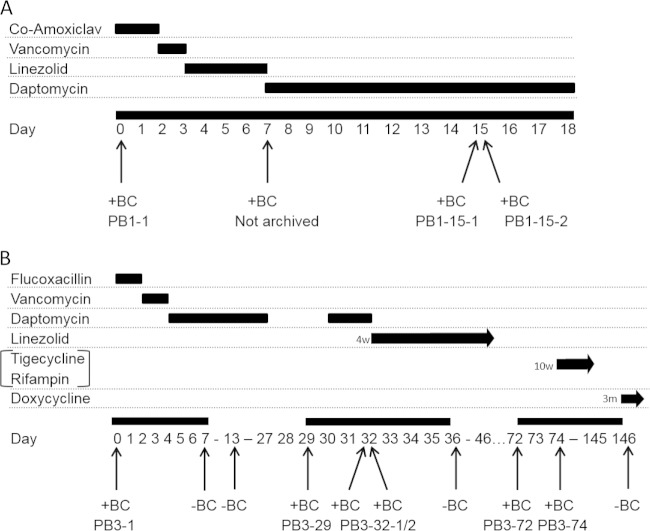
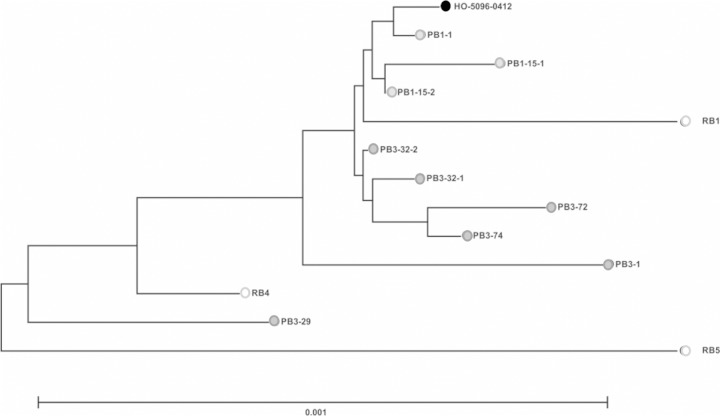
To investigate the genetic and phenotypic traits specifically associated with S. aureus persistence, multiple temporal isolates were sampled from two patients admitted to the University Hospitals of Leicester between 2009 and 2010 who had been diagnosed with persistent MRSA bacteremia (infection cases designated PB1 and PB3). Three additional patients were diagnosed with resolved MRSA bacteremia (RB1, RB4, and RB5); i.e., the infections were treated successfully. Individual infection timelines were produced (Fig. 1), and relevant clinical information was recorded (Table 1). Two individual positive blood cultures were collected on day 15 and day 32, respectively, for both PB1 and PB3; this allowed the confirmation of homogenous blood-borne bacterial populations in those cases at those time points. PB1 was diagnosed with infective endocarditis following transthoracic echocardiography (TTE) on day 6, which offers a potential source for the persistent and, ultimately, fatal infection in this case. In contrast, the MRSA bacteremia in PB3 was diagnosed as a secondary infection associated with an ulcer in an infected foot, which was amputated on day 20, thereby eliminating the only identified focus of infection prior to the subsequent positive blood cultures (day 29 onward). On day 76 following the first positive blood culture, a spinal abscess and osteomyelitis were suspected, providing alternative possible infective foci in PB3. All MRSA bacteremia isolates, independently of infection type (persistent or resolved bacteremia), belonged to sequence type 22 (ST22) and agr type I and exhibited a staphylococcal cassette chromosome mec (SCCmec) type IVh element (31–34), indicating that these isolates were derivatives of Epidemic MRSA-15 (EMRSA-15). Whole-genome analysis revealed clustering of isolates as determined on the basis of homologous sequence variation at shared core genes (Fig. 2).
FIG 1.
A diagrammatical representation of PB1 (A) and PB3 (B) treatment timelines. Time points for each blood culture collection and periods of antibiotic prescription are indicated relative to the initial positive blood culture for each bacteremia case. +BC, positive blood culture; −BC, negative blood culture; Co-Amoxiclav, amoxicillin-clavulanic acid.
TABLE 1.
Clinical details of MRSA persistent- and resolved-bacteremia cases used in this studya
| Patient | Bacteremia classification | Antibiotic(s) prescribed (specific to MRSA BSI) | Positive MRSA BC(s)b | Days of negative BC collection post-initial culture | Suspected bacteremia focus and source seen at indicated time | Bacteremia outcome |
|---|---|---|---|---|---|---|
| PB1 | Persistent | Vancomycin, linezolid, daptomycin | PB1-1, PB1-15-1, PB1-15-2 | None | Day 6, TTE, mitral valve endocarditis | Patient died (day 18) |
| PB3 | Persistent | Vancomycin, daptomycin, linezolid, tigecycline/rifampin, doxycycline | PB3-1, PB3-29, PB3-32-1, PB3-32-2, PB3-72, PB3-74 | 7, 13, 36, 37, 40, 42, 44, 47, 50, 57, 61, 146 | Day 20, left foot ulcer (amputated); day 76, paraspinal abscess and right foot osteomyelitis | Patient discharged (day 146) |
| RB1 | Resolved | Linezolid | RB1 | NA | Infected right hemithorax and diaphragmatic patch | Resolved; patient discharged |
| RB4 | Resolved | Vancomycin | RB4 | NA | Long-term catheter (pyelonephritis) and JJ stents | Resolved; patient discharged |
| RB5 | Resolved | Vancomycin | RB5 | NA | Infected right prosthetic hip joint | Resolved; patient discharged |
BSI, bloodstream infection; BC, blood culture; PB, persistent bacteremia: RB, resolved bacteremia; JJ stent, ureteric stent; NA, not applicable.
1st number, bacteremia case number; 2nd number, day of BC collection; 3rd number, identification number of isolate from multiple BCs collected on the same day.
FIG 2.
Genetic relatedness of bacteremia isolates. A neighbor-joining phylogenetic tree from a core genome alignment of 2,596,723 bp shared by all isolates constructed using MEGA v6 (30) is shown. The scale represents the number of substitutions per site. The reference EMRSA-15 genome (HO-5096-0412) is represented by a filled black circle, and resolved bacteremia isolates RB1, RB4, and RB5 are represented by open black circles.
Persistent isolates exhibited daptomycin nonsusceptibility.
All isolates were resistant to oxacillin and other β-lactams as confirmed by the oxacillin MICs being above the CLSI-recommended breakpoint (>2 μg/ml; Table 2). Data also showed that all EMRSA-15 PB3-derived isolates and all resolved bacteremia isolates were susceptible to vancomycin and linezolid (sensitivity, MIC ≤ 2 and ≤ 4 μg/ml, respectively). PB1 isolates showed intermediate resistance to vancomycin. Interestingly, daptomycin nonsusceptibility was judged to have developed during the PB1 and PB3 infections because the sequential isolates (PB1-15-1 and -15-2 and PB3-29, -32-1, and -32-2) showed a daptomycin resistance MIC (>1 μg/ml) whereas the cognate initial isolates (PB1-1 and PB3-1) showed susceptibility. Interestingly, PB3-72 and PB3-74 exhibited daptomycin susceptibility MIC values identical to that of the initial isolate (PB3-1).
TABLE 2.
MICs for oxacillin, vancomycin, linezolid, and daptomycina
| Isolate | MIC (μg/ml) |
|||
|---|---|---|---|---|
| Oxacillin | Vancomycin | Linezolid | Daptomycin | |
| PB1-1 | 128 | 3–4 | 2 | 1 |
| PB1-15-1 | 128 | 4 | 2 | 2–4 |
| PB1-15-2 | 128 | 4 | 2 | 2–4 |
| PB3-1 | 64 | 1–2 | 1–2 | 1 |
| PB3-29 | 128 | 2 | 2 | 2–4 |
| PB3-32-1 | 128 | 2 | 1–2 | 2 |
| PB3-32-2 | 128 | 2 | 2 | 2–4 |
| PB3-72 | 64–128 | 2 | 2 | 1 |
| PB3-74 | 64 | 2 | 1–2 | 1 |
| RB1 | 64 | 2 | 2 | 0.5 |
| RB4 | >256 | 2 | 2–4 | 0.5 |
| RB5 | 64–128 | 2 | 1–2 | 0.5 |
Experiments were repeated independently three times, and MIC data were recorded at up to a log2 dilution; a 2-fold dilution range is reported as recommended by the CLSI. MICs deemed to represent intermediate susceptibility or resistance according to published breakpoints are underlined. Resistance breakpoints (μg/ml): oxacillin, >2; vancomycin, >2; linezolid, >4; daptomycin, >1.
Enhanced growth of persistent MRSA isolates in a nutrient-deprived growth environment and during iron stress.
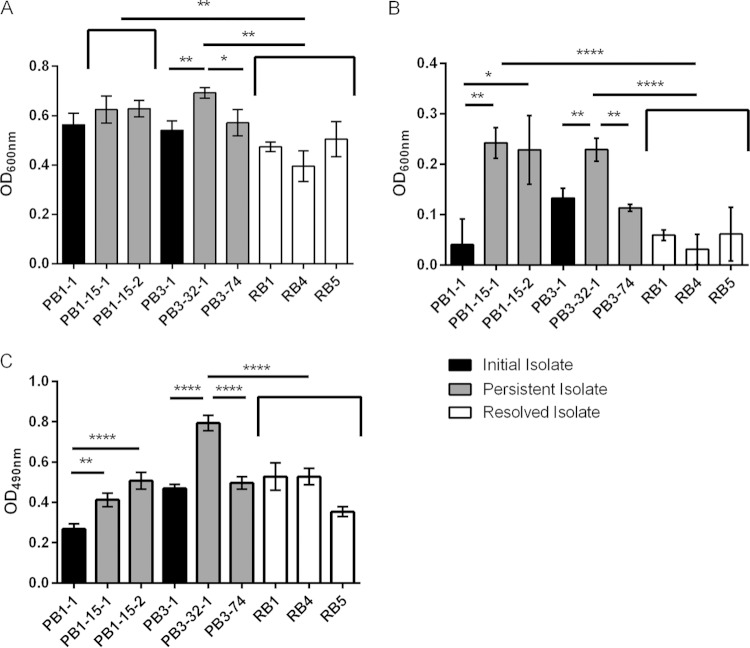
The formation of SCV has repeatedly been implicated in S. aureus persistence (18, 19, 21, 22); however, no significant differences in colony sizes between the initial, persistent, and resolved isolates examined in this study were observed, therefore indicating that none of the MRSA bacteremia isolates exhibited an SCV phenotype. Moreover, all MRSA bacteremia isolates showed no growth differences when cultured in a nutrient-replete medium (data not shown). In contrast, the EMRSA-15 PB1- and PB3-derived persistent isolates, PB1-15-1, PB1-15-2, and PB3-32-1, showed significantly increased growth compared to the control resolved EMRSA-15 bacteremia isolates when grown in nutrient-deprived, metal ion-restricted medium (Fig. 3A; P < 0.0001), a growth environment which is reflective of conditions in vivo (35). These data also showed that the persistent isolates had a growth advantage in CRPMI medium compared to the respective initial isolates (PB1-1 and PB3-1), although this difference was significant only for the PB3-1-versus-PB3-32-1 comparison (P < 0.01). Viability assays confirmed that persistent isolates showed an increased growth phenotype under nutrient-deprived conditions compared to both the initial infection and the control resolved isolates (data not shown). Interestingly, the measured growth of PB3-74 was equivalent to that of the initial PB3-1 isolate and was significantly lower than that of PB3-32-1 (P < 0.01), which is in agreement with the antibiotic resistance phenotypes.
FIG 3.
Growth analysis and biofilm assessment. (A and B) S. aureus strains were grown in iron-restricted nutrient-deprived CRPMI medium (A) and under conditions of iron stress in CRPMI medium with 200 μM FeSO4 (B). Growth was assessed by OD600 measurement after 24 h of static growth at 37°C in 5% CO2. (C) Biofilm formation was assessed by OD490 measurement after incubation in brain heart infusion (BHI)–1% glucose for 24 h. Data represent means and standard deviations of the results of at least three independent experiments. Significance is indicated with asterisks. *, P < 0.05; **, P < 0.01; ****, P ≤ 0.0001.
Comprehensive assessment of the isolates' sensitivities was performed for a range of environmental stimuli. Interestingly, the EMRSA-15 PB1 and PB3 persistent isolates demonstrated significantly increased growth in the presence of toxic levels of iron (200 μM FeSO4) compared to the combined average of the growth measurements of the resolved isolates used as a control (Fig. 3B; P < 0.0001). In addition, persistent isolates PB1-15-1, PB1-15-2, and PB3-32-1 (Fig. 3B; P < 0.01 to 0.05) showed increased growth during iron stress compared to the respective initial isolates (PB1-1 and PB3-1). PB3-74 again displayed growth similar to that of the initial isolate (PB3-1), and the abundance of growth was significantly lower than that of PB3-32-1 (P < 0.01). These data suggest that the persistent isolates had acquired stable adaptations giving them a growth advantage under nutrient-deprived conditions and toxic iron stress conditions, both of which are environments that can be encountered during long-term survival in vivo; neither has been previously associated with S. aureus persistence.
Persistent isolates show increased levels of cell surface-associated proteins involved in immune system evasion, host cell adhesion, and bacterial stress response.
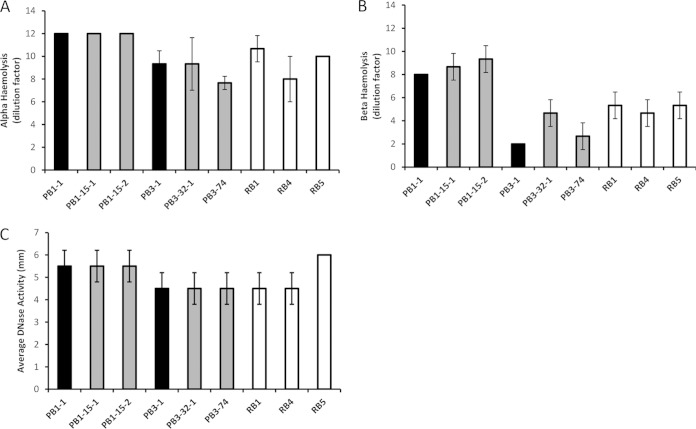
Persistent S. aureus bacteremia isolates have previously been associated with decreased toxin expression and activity, particularly of alpha-hemolysin, usually in combination with a recognized SCV phenotype and/or agr dysfunction (11, 13, 18, 19). No differences were found between the initial, persistent, and resolved MRSA bacteremia isolates with regard to alpha-hemolysin, beta-hemolysin, and DNase activity or cell wall covalently bound protein profiles in this study (Fig. 4 and unpublished data). However, several reproducible differences between the persistent, initial, and resolved EMRSA-15 bacteremia isolates in the non-covalently bound cell surface-associated protein fraction after growth in nutrient- and metal ion-restricted CRPMI medium were recorded. To identify individual proteins, quantitative proteomic profiling was conducted using iTRAQ (isobaric tag for relative and absolute quantification) coupled with LC-MS (liquid chromatography-mass spectrometry). The iTRAQ–LC-MS analysis identified 214 and 178 individual proteins in each biological repeat across the full set of isolates tested, and mean relative protein values were calculated. Those proteins which exhibited >0.5 log2-fold differences (P > 0.05) in abundance between each persistent or resolved MRSA bacteremia isolate and the respective initial isolate are listed in Tables 3 and 4, and the data are summarized in Fig. 5.
FIG 4.
DNase and hemolysin activity. (A and B) Alpha (A) and beta (B) hemolytic activities were assessed as the final dilution (DF, dilution factor) of concentrated bacterial supernatant where complete hemolysis still occurred. Dilution factor means were calculated and plotted from the results of three independent experiments ± 1 standard deviation. (C) DNase activity was assessed by measuring the size (in millimeters) of the halo around the test isolates indicating hydrolyzed DNA. Means were calculated and plotted from the results of three independent experiments ± 1 standard deviation.
TABLE 3.
Quantitative proteomic profiling of surface-associated proteins in PB1 persistent-bacteremia isolates and EMRSA-15 resolved-bacteremia isolates compared to the PB1 initial isolatea
| Accession no. | Protein name | Gene | 0.5 log2 ratio |
|||
|---|---|---|---|---|---|---|
| PB1-15-1 | PB1-15-2 | RB1 | RB5 | |||
| Q5HEI2 | Map adherence protein | eap | 0.80 | 0.7 | −0.75 | −1.05 |
| Q2FIK4 | Extracellular matrixbinding protein | emp | 0.05 | 0.35 | −0.65 | −1.15 |
| Q2YVZ4 | Immunoglobulin binding protein | sbi | 0.05 | 0.30 | −0.65 | −2.15 |
| Q2FH36 | Cold shock protein | cspA | 0.40 | 0.30 | −0.85 | −0.95 |
| Q5HJW3 | Penicillin binding protein 2 | pbp2′ | 0.70 | 0.85 | −0.70 | −1.65 |
| A7WZT2 | Enolase | eno | 1.75 | 1.75 | −0.15 | 0.10 |
| A7X1J8 | Acyl carrier protein | 0.45 | 0.25 | −0.60 | −0.80 | |
| Q2YUM5 | RNA polymerase | rpoE | 3.15 | 2.7 | 0.85 | 1.1 |
| Q2FHT6 | Thioredoxin | trx | 2.10 | 0.95 | −0.25 | −0.15 |
Bold values represent significant relative changes of 0.5 log2 ratio differences between the initial PB1-1 isolate and persistent PB1 infection isolates or resolved EMRSA-15 infection isolates. Values without a minus sign represent increases; values with a minus sign represent decreases. Relative protein change data represent averages of the results of two biological repeats (inclusive of two technical repeats). Proteins identified showed P values of <0.001 and <0.0001, calculated using the Scaffold Q+ proteomic software from individual ANOVA tests, showing the significance of the difference in protein abundance between two conditions.
TABLE 4.
Quantitative proteomic profiling of surface-associated proteins in PB3 persistent bacteremia isolates and resolved bacteremia (RB) EMRSA-15 isolates compared to the PB3 initial isolatea
| Accession no. | Protein name | Gene | 0.5 log2 ratio |
|||
|---|---|---|---|---|---|---|
| PB3-32-1 | PB3-74 | RB1 | RB5 | |||
| Q5HEI2 | Map adherence protein | eap | 1.1 | −0.20 | 0.0 | −0.3 |
| Q2FIK4 | Extracelllar matrix binding protein | emp | 1.4 | −0.15 | −0.2 | −0.8 |
| Q2YVZ4 | Immunoglobin binding protein | sbi | 0.7 | −0.10 | 0.55 | −0.95 |
| Q2FH36 | Cold shock protein | cspA | 0.75 | 0.10 | −0.4 | −0.55 |
| Q5HJW3 | Penicillin binding protein 2 | pbp2′ | 1.1 | 0.05 | −0.1 | −1.15 |
| A7WZT2 | Enolase | eno | 0.1 | −0.15 | −0.70 | −0.5 |
| A7X1J8 | Acyl carrier protein | 0.8 | 0.00 | 0.0 | −0.25 | |
| Q2YUM5 | RNA polymerase | rpoE | 0.95 | 0.30 | 0.1 | 0.35 |
| Q2FHT6 | Thioredoxin | trx | 1.1 | 0.00 | −0.45 | −0.45 |
Bold values represent significant relative changes of 0.5 log2 ratio differences between the initial PB1-1 isolate and persistent PB1 infection isolates or resolved EMRSA-15 infection isolates. Values without a minus sign represent increases; values with a minus sign represent decreases. Relative protein change data represent averages of the results of two biological repeats (inclusive of two technical repeats). Proteins identified showed P values of <0.001 and <0.0001, calculated using the Scaffold Q+ proteomic software from individual ANOVA tests, showing the significance of the difference in protein abundance between two conditions.
FIG 5.
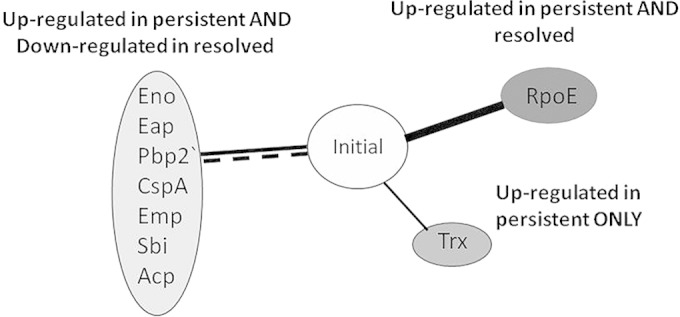
Summary of quantitative proteomic profiling of surface-associated proteins in EMRSA-15 persistent isolates and resolved-bacteremia EMRSA-15 isolates compared to the initial bacteremia isolates. Solid black lines indicate upregulation of proteins compared to the initial isolates; dashed lines indicate downregulation. The data representing the indicated proteins show changes relative to the initial isolates in one or more persistent or resolved isolate samples.
Interestingly, the proteomic profiling showed that several non-covalently associated surface proteins displayed increased levels in persistent isolates and decreased levels in the resolved isolates compared to the initial persistent bacteremia isolates, suggesting that these proteins play important roles in S. aureus persistence. Proteins with increased levels in the persistent isolates compared to the initial isolates in both the PB1 and PB3 infections included extracellular adhesion protein (Eap), penicillin binding protein 2a (PBP2A′), and acyl carrier protein. Enolase (Eno) levels showed a substantial increase only in the PB1 persistent isolates, whereas extracellular matrix binding protein (Emp), bifunctional autolysin (Atl), immunoglobulin binding protein (Sbi), and cold shock protein A (CspA) showed increased levels in the PB3 infection.
Proteins showing a decreased level in the control resolved isolates compared to the PB1 initial isolate included Eap, Emp, Atl, Sbi, CspA, PBP2A′, and acyl carrier protein. Enolase was the only protein to show a significant level decrease in resolved isolates compared to the PB3 initial isolate (PB3-1). RNA polymerase RpoE showed increased levels in both persistent and resolved isolates compared to the initial isolates, whereas the thioredoxin (Trx) stress response protein level was increased in persistent isolates only. The PB3-74 isolate did not show any significant differences from the PB3 initial isolate. Overall, these data suggest the PB1- and PB3-infecting strains evolved during their individual infections, leading to similar changes in their cell surface protein profiles (Fig. 5). Additionally, the levels of the majority of these altered proteins were decreased in the resolved isolates, indicating the importance of these proteins for S. aureus persistence during bacteremia.
Persistent isolates exhibited enhanced glucose-mediated biofilm formation.
Bacterial biofilms have been previously implicated as general mechanisms for immune system modulation and antibiotic tolerance (36–38) and therefore could be involved in persistence development. Biofilm formation was detected in the isolates after incubation in the presence of glucose (Fig. 3C). Interestingly, the EMRSA-15 persistent isolates PB1-15-1 (P < 0.01), PB1-15-2 (P < 0.0001), and PB3-32-1 (P < 0.0001) exhibited glucose-mediated biofilm levels that were significantly higher than those produced by the respective EMRSA-15 initial isolates. However, only PB3-32-1 showed a biofilm level significantly higher than that seen with the control EMRSA-15 resolved bacteremia isolates (P < 0.0001). Therefore, these data show that, although the persistent isolates had evolved during the infection to increase biofilm formation, there was no significant difference from the control resolved bacteremia isolates with respect to the level of biofilm, suggesting that biofilm formation is not an important aspect of persistence in these cases.
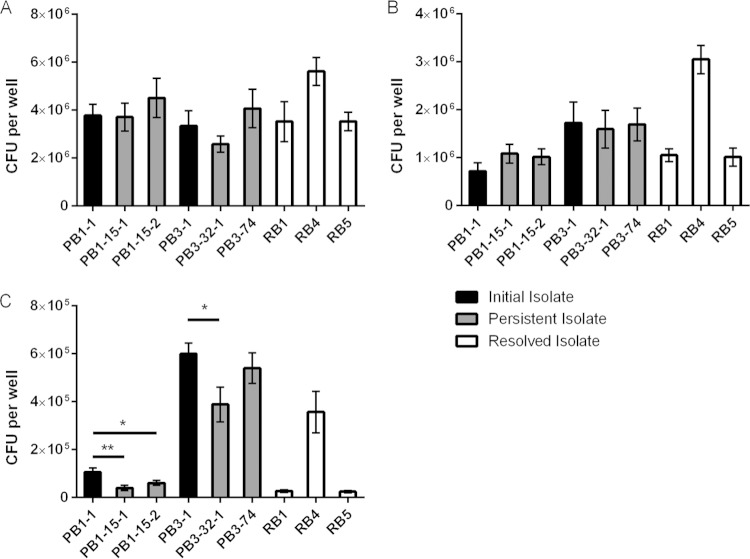
Persistent isolates did not exhibit enhanced adhesion, invasion, or intracellular persistence.
Host cell invasion and intracellular persistence have both been implicated as mechanisms of persistence (39, 40); therefore, we assessed these phenotypes using an H9C2 rat myocardium cell line infection assay (Fig. 6). No differences in adhesion or invasion rates were observed between the initial, persistent, and control resolved EMRSA-15 bacteremia isolates; in fact, resolved isolate RB4-1 showed a significantly higher invasion level than PB3-1, which was the isolate with the next highest invasion rate (P < 0.05). Furthermore, in contrast to previous studies, both the PB1- and PB3-associated persistent isolates actually showed reduced levels of intracellular persistence compared with the respective initial isolates (PB1-15-1, P < 0.01; PB1-15-2, P < 0.05; PB3-32-1, P < 0.05). These data show that there was no association between adhesion, invasion, or intracellular persistence capabilities and persistence in this study.
FIG 6.
Adhesion, invasion, and intracellular eukaryotic cell persistence assay. Confluent H9C2 cells were cultured and infected with 107 bacterial CFU, and the numbers of bacterial cells able to adhere (A), invade (B), and persist intracellularly (C) were counted. Significance is indicated with asterisks. *, P < 0.05; **, P < 0.01.
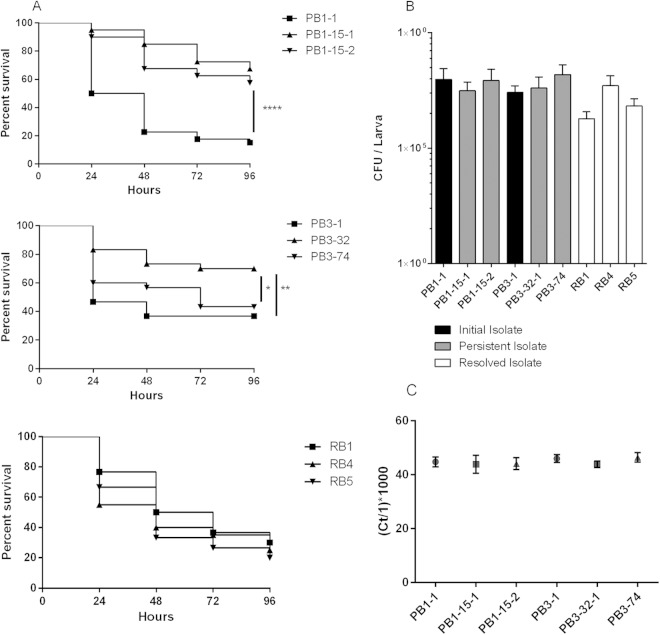
Persistent isolates displayed attenuated virulence in a Galleria mellonella larva infection model.
The general virulence behavior of the initial or persistent EMRSA-15 isolates was compared to that of the control resolved EMRSA-15 bacteremia isolates using a Galleria mellonella larva infection model where larvae mortalities are recorded every 24 h over a 96-h period (Fig. 7A). Persistent isolates associated with both PB1 and PB3 showed significantly lower virulence than the respective initial isolates (PB1-15-1, P < 0.001; PB1-15-2, P < 0.001; PB3-32, P < 0.01). There was also a significant difference between PB3-32-1 and PB3-74 (P < 0.05), with the latter exhibiting an enhanced Galleria killing phenotype equivalent to that seen with the PB3-1 initial isolate.
FIG 7.
Galleria mellonella larva virulence assay. (A) Groups of 10 Galleria larvae were injected with ∼106 CFU of bacteria and fatalities recorded every 24 h over a 96-h period. Data were plotted on Kaplan-Meier survival plots and log-rank tests used to determine significance. Significance is indicated with asterisks. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. (B) Bacterial CFU levels were calculated from individual homogenized larva using selective media. (C) Total RNA was extracted from homogenized larvae, and 16S rRNA was quantified via qRT-PCR. CT values were inverted and multiplied by 1,000 to give manageable relative values for comparisons.
Intra-Galleria bacterial burdens were measured 48 h postinfection to assess the relative contribution of bacterial load to the virulence data previously observed (Fig. 7B). Importantly, none of the S. aureus colonies obtained following retrieval of bacteria from homogenized infected Galleria larvae exhibited a SCV phenotype. One-way analysis of variance (ANOVA) showed no significant differences in CFU per larva (CFU/larva) at 48 h between the Galleria larvae infected with initial or cognate persistent MRSA bacteremia isolates (PB1 isolates, P = 0.21; PB3 isolates, P = 0.27). However, the CFU/larva data differed substantially even between individual larvae infected with the same isolate within the same experiment (coefficient of variation, 95.72% to 228.69%). Therefore, an alternative, molecular quantification method was employed involving 16S rRNA quantitative reverse transcription-PCR (qRT-PCR) and RNA extracted from homogenized Galleria specimens (Fig. 7C). According to the data determined by this non-culture-based molecular method, intra-Galleria variation was substantially reduced but there were still no significant differences observed between the PB1- or PB3-associated initial isolates and the persistent isolates, as shown by one-way ANOVA (PB1-associated isolates, P = 0.14; PB3-associated isolates, P = 0.08). These data demonstrated that the attenuated virulence exhibited by persistent EMRSA-15 isolates derived from two individual bacteremia cases was not a result of differences in bacterial loads.
All persistent isolates showing altered phenotypes had single nucleotide mutations in the mprF gene.
The reproducibility of the phenotypic traits associated with the persistent isolates in both cases of persistent EMRSA-15 bacteremia indicated that a stable, genetic basis for persistence had evolved during the two independent infections. Therefore, Illumina genome sequencing and whole-genome comparisons were conducted between the persistent and initial isolates within the individual infections to identify novel variants associated with persistence development.
FastQ files were quality checked, trimmed, and mapped to the closest available relative, EMRSA-15 (available from the Wellcome Trust Sanger Institute website; accession number HE681097 [HO 5096 0412]) (41), and variations present at >80% were recorded. Totals of 137 and 295 single nucleotide polymorphisms (SNPs) and insertions or deletions (indels) were identified in the initial PB1 and PB3 isolates, respectively, compared to the EMRSA-15 reference strain (Table 5). Comparisons of the genome sequences of the persistent isolates and the respective initial isolates showed that a series of genetic mutations (between 2 and 18 per isolate) had occurred in the persistent isolates during the course of the infection (Table 6).
TABLE 5.
Summary of single nucleotide polymorphisms and insertions and deletions in infection isolates compared to the EMRSA-15 genome (HO 5096 0412)a
| Isolate | No. of SNPsb | No. of indels | Total no. of SNPs and indelsb | No. of SNPs and indels shared with respective initial isolatec |
|---|---|---|---|---|
| PB1 | 122 | 15 | 137 | NA |
| PB1-15-1 | 124 | 15 | 139 | 136 |
| PB1-15-2 | 119 | 14 | 133 | 132 |
| PB3-1 | 273 | 23 | 296 | NA |
| PB3-32-1 | 270 | 22 | 292 | 285 |
| PB3-32-2 | 273 | 20 | 293 | 286 |
| PB3-72 | 276 | 22 | 298 | 295 |
| PB3-74 | 277 | 23 | 300 | 295 |
SNPs, single nucleotide polymorphisms; indels, insertions and deletions.
Data represent the number of SNPs and indels in each bacteremia isolate compared to the EMRSA-15 reference genome strain (accession number HE681097 [HO 5096 0412]) (42); variations present at >80% were recorded.
Data represent the number of SNPs and indels identified from the EMRSA-15 reference strain that are shared between each of the persistent-bacteremia isolates and the corresponding initial isolate.
TABLE 6.
Genetic changes associated with unique open reading frames in persistent-bacteremia isolates compared to the respective initial isolates
| Isolate | Genea | Positionb | Reference nucleotide | Nucleotide variation(s)c | Function | Amino acid change or function |
|---|---|---|---|---|---|---|
| PB1-1 | SAEMRSA15_16780 | 1860581 | G | T | NAD binding | A271D |
| PB1-15-1 | mprF | 1343017 | C | T | Membrane modification | P314L |
| pyrG | 2206316 | G | A | CTP synthase | P412S | |
| 2550433 | T | C | Intergenic | |||
| PB1-15-2 | mprF | 1344552 | C | T | Membrane modification | L826F |
| PB3-29 | spa | 98739 | G | A | IgG binding protein | D339E |
| 401923 | T | −TTTAC | Intergenic | |||
| 938796 | G | T | Intergenic | |||
| mprF | 1342960 | C | T | Membrane modification | S295L | |
| PB3-32-1 | SAEMRSA15_00120* | 16087 | G | +T | Putative hydrolase | Promoter |
| spa | 98739 | G | A | IgG binding protein | D339E | |
| 401923 | T | −TTTAC | Intergenic | |||
| SAEMRSA15_03710* | 434768 | T | G | Hypothetical | W1072L | |
| 938796 | G | T | Intergenic | |||
| mprF | 1342960 | C | T | Membrane modification | S295L | |
| PB3-32-2 | SAEMRSA15_00120* | 16087 | G | +T | Putative hydrolase | Promoter |
| spa | 98690 | T | C | IgG binding protein | N356D | |
| 938796 | G | T | Intergenic | |||
| mprF | 1344552 | C | T | Membrane modification | L826F | |
| arsR2* | 1861879 | A | −ATAT | Arsenic resistance repressor | Promoter | |
| SAEMRSA15_18700* | 2057502 | A | T | Membrane protein | G2018G | |
| fnbA | 2591495 | T | C | Fibronectin binding | P892P | |
| PB3-72 | spa | 98739 | G | A | IgG binding protein | D339E |
| SAEMRSA15_03710* | 434757 | T | +GGAAAAAATTG | Hypothetical | ||
| PB3-74 | spa | 98739 | G | A | IgG binding protein | D339E |
| 401923 | T | −TTTAC | Intergenic |
*, reversions to EMRSA-15 sequence from PB3-1 preexisting SNP.
Data represent base positions in the EMRSA-15 reference genome sequence (accession number HE681097).
Data represent variant bases relative to EMRSA-15 present at >80%.
The extensive pattern of shared SNPs seen in the genome analysis shows clearly that the persistent isolates had evolved from the respective initial isolates during the course of the infections. Interestingly, the mutation in the initial PB1 isolate in gene SAEMRSA-15_16780 (which encodes a predicted NAD binding protein) that causes a nonpolar-to-acidic amino acid substitution (A271D) was repaired in both persistent PB1 isolates. Multiple other SNPs were also identified among the persistent isolates (Table 5); however, either these genotypic traits were found in only one of the persistent isolates from the infection case or the mutation was found to cause a conservative amino acid change. Two distinct mutations were identified in the spa gene in PB3 persistent isolates compared to the initial isolate; however, only one of the two mutations leads to a nonsynonymous amino acid change, and in all strains the spa gene appears to retain the frameshift mutation first identified in the EMRSA-15 genome sequence. The two different spa mutations do, however, indicate that PB3-29, PB3-32-1, PB3-72, and PB3-74 diverged from PB3-32-2 at an early stage of the infection, with PB3-29 and PB3-32-1 then gaining further mutations independently of PB3-72 and PB3-74.
Interestingly, the whole-genome sequencing revealed several different mutations in the mprF gene in the persistent isolates of PB1 and PB3 that were not observed in resolved EMRSA-15 isolates RB1, RB4, and RB5. The presence of different mprF SNPs, in separate divergent isolates taken from blood cultures on the same day, in two independent infections provides good evidence that there was a strong selective pressure for mprF mutation in vivo in these cases.
The terminal isolates of the PB3 lineage, PB3-72 and PB3-74, have the same synonymous SNP-containing spa alleles as PB3-29 and PB3-32-1; however, both PB3-72 and PB3-74 have the mprF allele of the initial isolate/wild type. This suggests that the spa mutation arose prior to the mprF mutation and that PB3-74 arose from a divergent bacterial population originating from the PB3 initial isolate prior to the emergence of PB3-29 and -32-1. In addition, the PB3-72 and PB3-74 isolates share 294 of the 295 SNPs and indels detected in PB3-1 (compared to the EMRSA-15 reference strain) whereas the PB3-32 isolates have only 285 of these original SNPs and indels, suggesting that the PB3-72 and PB3-74 isolates diverged directly from PB3-1. Therefore, the genome data support the phenotypic data showing that PB3-74 is more phenotypically similar to the initial PB3-1 isolate than to the other PB3 persistent isolates, such as PB3-32.
The sequence alignments of the initial and persistent isolates of both PB1 and PB3 were also examined using the Tablet viewer to look for large genomic insertions or deletions (43). Compared to the EMRSA-15 genome sequence, several large deletions or potential insertions were present in rRNA, phage, or transposon-related sequences of the bacteremia isolates; however, these mutations were found in all of the isolates, indicating that they likely had no role in the emergence of persistence (data not shown).
In summary, the mprF gene, in the two independent cases of persistent bacteremia, gained different mutations in the divergent persistent isolates that are strongly associated with the distinct novel phenotypic adaptations. These data therefore indicate that there is a strong association of in vivo mprF allelic variation with S. aureus persistence.
DISCUSSION
In this study, we directly compared multiple temporal isolates from two independent cases of persistent EMRSA-15 bacteremia to contemporary isolates from resolved EMRSA-15 bacteremia infections. We identified several novel phenotypic traits which were associated exclusively with the persistent isolates from the two independent bacteremia cases and not with the respective initial isolate or the control resolved isolates. These novel persistence-associated traits include the following: (i) enhanced growth in a nutrient-deprived environment; (ii) increased tolerance of toxic iron conditions; (iii) increased abundance of cell surface-associated proteins involved in immune evasion, host cell adhesion, and bacterial stress response systems; and (iv) attenuated virulence in an insect larva infection model.
Whole-genome sequence comparisons identified several different SNPs in the mprF gene of the phenotypically adapted persistent isolates of both PB1 and PB3 bacteremia. The mprF SNPs were not found in non-phenotypically adapted persistent or resolved isolates, indicating that the mprF SNPs are associated with the genetic evolution of persistence. The three mprF SNPs identified in this study (S295L, P314L, and L826F) have previously been reported to be gain-of-function mutations and associated with the emergence of daptomycin nonsusceptibility and increased resistance to HDP (43–47). MprF is responsible for synthesis and translocation of lysyl-phosphotidylglycerol (L-PG) into the cell membrane, resulting in a reduced negative charge exhibited by the bacterial cell surface. It is proposed that enhanced MprF activity leads to greater abundance of L-PG in the cell membrane and an altered cell surface charge that leads to an increase in resistance to cationic antimicrobial agents such as HDP and calcium-coupled daptomycin (43–47). This is consistent with our observation of increased daptomycin resistance in the persistent isolates with mprF gain-of-function SNPs. However, the precise mechanism by which these mprF mutations lead to subsequent daptomycin nonsusceptibility remains unidentified. The fact that our study identified different causal mprF SNPs in separate isolates, taken from independent blood cultures collected on the same day, in two independent cases of daptomycin-treated persistent bacteremia provides significant evidence of a strong selective pressure for mprF mutation in vivo.
Indeed, our data suggest that there are in fact multiple selective pressures in vivo because novel conserved phenotypic changes were identified in the isolates with mprF gain-of-function mutations. These in vitro phenotypic traits (detailed above) increase bacterial fitness in vivo by increasing growth and colonization under the nutrient-poor conditions found in a host and by promoting evasion of the immune system (Fig. 8). None of these phenotypic changes have been associated with persistence previously and so represent novel mechanisms of persistence.
FIG 8.
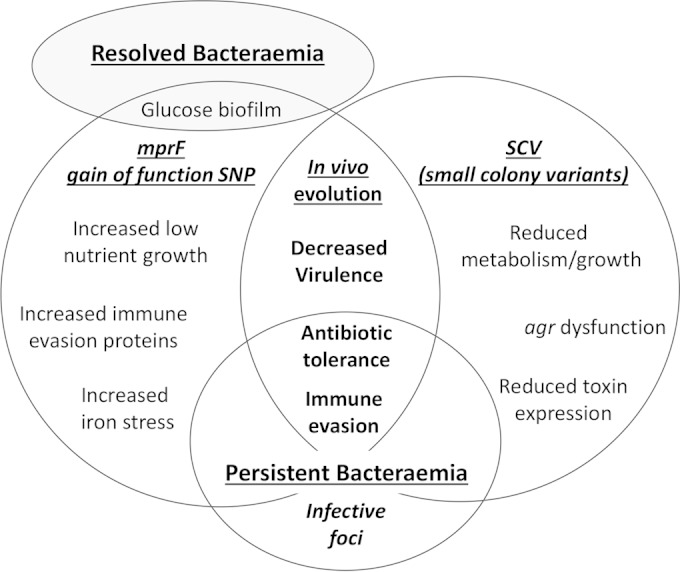
Model showing conserved mechanisms of S. aureus bacteremia persistence. Major mechanisms of persistence include in vivo genome evolution resulting in altered phenotypes and survival in a protected infective focus, both of which enable S. aureus to evade the immune system and tolerate treatment antibiotics.
Our data suggest that, in addition to in vivo genome evolution, another mechanism of persistence involves S. aureus “hiding” in a protected focus and then reemerging into the bloodstream. In the PB3 infection, the PB3-74 isolate showed antibiotic resistance, growth characteristics, and phenotypic traits identical to those seen with the initial PB3 isolate. In addition, the genome sequence analysis indicated that PB3-72 and PB3-74 share 100% of the EMRSA-15 reference genome SNPs and indels with the initial PB3-1 isolate, whereas PB3-32-1 and PB3-32-2 have only 96% identity, suggesting that the PB3-32 isolates have diverged further from PB3-1 than the PB3-72 and -74 isolates. Therefore, it is likely that the PB3-72 and -74 isolates reemerged into the bloodstream from a focus such as an abscess where the bacteria had been protected from the immune system and treatment antibiotics; they are unlikely to be growing at a high rate, hence the very low mutation rate.
Several persistence-associated traits have been proposed in previous studies. In this study, however, no evidence was found to support a correlation between SCV formation or eukaryotic intracellular persistence and persistent isolates. Indeed, SCV were not identified in any of the MRSA isolates used in this study, either during the initial clinical diagnosis at the University Hospitals of Leicester (UHL) or during subsequent in vitro analysis. This finding is significant because the formation of SCV is generally considered the main mechanism for persistence (18, 19, 21, 22). In addition, our study did not find any evidence of altered bacterial virulence, due to decreased exoprotein expression and/or reduced in vivo mortality, in combination with agr dysfunction and/or SCV formation as previously observed (11, 13, 18, 19). The persistent isolates in this study did, however, show attenuated virulence in an insect model of infection compared to the respective initial isolates or the resolved bacteremia isolates. Therefore, these data provide evidence for a role for reduced virulence in persistence that is not dependent on agr dysfunction, reductions in levels of secreted exoproteins or toxins, or SCV formation. Instead, our proteomic profiling found, for the first time, persistent isolate-specific increases in the levels of several non-covalently bound cell surface proteins.
The proteins with increased expression in persistent isolates included Emp and Eap, known mediators of low-iron biofilm formation, host protein adhesion enabling eukaryotic cell invasion (23, 48), and T-cell response disruption (49). The level of immune evasion protein Sbi, an immunoglobulin G binding protein, was also increased in persistent isolates (50, 51). The thioredoxin (Trx) and cold shock protein A (CspA) proteins and oxidative and cold shock pressure stress proteins also showed increased expression in persistent isolates (52, 53). Other proteins which showed differential abundance levels included PBP2′, enolase, a laminin adhesion protein (52), and two general cell maintenance proteins (acyl carrier protein and RNA polymerase subunit E [RpoE]).
Increased biofilm formation has previously been suggested to be a mechanism of persistence (14, 38). Consistent with this, our data suggest improved glucose-mediated biofilm formation in both the PB1- and PB3-infecting strains; however, the control resolved isolates demonstrated equally high levels of biofilms, suggesting that biofilm formation alone is not a specific persistence mechanism. This finding in particular emphasizes the importance of comparing initial infecting isolates with multiple persistent isolates from the same infection and contemporary resolved infection isolates to eliminate the possibility of bias introduced by any infection-specific traits.
Overall, our data suggest that the PB1- and PB3-infecting strains adapted to long-term survival within a host environment by upregulating surface protein-associated immune system defenses and stress response systems in combination with altered metabolism and virulence capabilities. Moreover, genomic evidence from independent infections shows a novel association between mprF mutation and the emergence of these novel persistence-specific virulence phenotypes during the progression of complex S. aureus bacteremia and the development of persistence in vivo. In summary, S. aureus has multiple mechanisms of persistence, including in vivo genome evolution and protection within a focus, which result in immune system evasion and tolerance of treatment antibiotics (Fig. 8).
ACKNOWLEDGMENTS
This work was supported by a Wellcome Trust grant (097828Z11Z) to J.A.M. and K.R.
We thank Andrew R. Bottrill of the PNACL Proteomics Facility, University of Leicester (http://www2.le.ac.uk/colleges/medbiopsych/facilities-and-services/cbs), for the proteomic analyses.
REFERENCES
- 1.van Belkum A, Verkaik NJ, de Vogel CP, Boelens HA, Verveer J, Nouwen JL, Verbrugh HA, Wertheim HFL. 2009. Reclassification of Staphylococcus aureus nasal carriage types. J Infect Dis 199:1820–1826. doi: 10.1086/599119. [DOI] [PubMed] [Google Scholar]
- 2.Stevens DL. 2003. Community-acquired Staphylococcus aureus infections: increasing virulence and emerging methicillin resistance in the new millennium. Curr Opin Infect Dis 16:189–191. doi: 10.1097/00001432-200306000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827. doi: 10.1016/S0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 4.Public Health England (PHE). 2012. Voluntary reporting of Staphylococcus aureus bacteraemia in England, Wales and Northern Ireland, 2012. https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/342872/S__aureus_bacteraemia_in_England_Wales_and_Northern_Ireland_2012_FINAL.pdf. [Google Scholar]
- 5.PHE. 2013. Quarterly epidemiological commentary: mandatory MRSA, MSSA and E. coli bacteraemia, and C. difficile infection data (up to July–September 2013). https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/346075/QEC_12_December__Jul_to_Sep_2013.pdf. [Google Scholar]
- 6.WHO. 2014. Antimicrobial resistance—global report on surveillance. WHO, Geneva, Switzerland. [Google Scholar]
- 7.Howden BP, Davies JK, Johnson PDR, Stinear TP, Grayson ML. 2010. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev 23:99–139. doi: 10.1128/CMR.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khatib R, Jose J, Musta A, Sharma M, Fakih MG, Johnson LB, Riederer K, Shemes S. 2011. Relevance of vancomycin-intermediate susceptibility and heteroresistance in methicillin-resistant Staphylococcus aureus bacteraemia. J Antimicrob Chemother 66:1594–1599. doi: 10.1093/jac/dkr169. [DOI] [PubMed] [Google Scholar]
- 9.Fischer A, Yang SJ, Bayer AS, Vaezzadeh AR, Herzig S, Stenz L, Girard M, Sakoulas G, Scherl A, Yeaman MR, Proctor RA, Schrenzel J, François P. 2011. Daptomycin resistance mechanisms in clinically derived Staphylococcus aureus strains assessed by a combined transcriptomics and proteomics approach. J Antimicrob Chemother 66:1696–1711. doi: 10.1093/jac/dkr195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hentschke M, Saager B, Horstkotte MA, Scherpe S, Wolters M, Kabisch H, Grosse R, Heisig P, Aepfelbacher M, Rohde H. 2008. Emergence of linezolid resistance in a methicillin resistant Staphylococcus aureus strain. Infection 36:85–87. doi: 10.1007/s15010-007-7220-7. [DOI] [PubMed] [Google Scholar]
- 11.Fowler VG, Sakoulas G, McIntyre LM, Meka VG, Arbeit RD, Cabell CH, Stryjewski ME, Eliopoulos GM, Reller LB, Corey GR, Jones T, Lucindo N, Yeaman MR, Bayer AS. 2004. Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J Infect Dis 190:1140–1149. doi: 10.1086/423145. [DOI] [PubMed] [Google Scholar]
- 12.Xiong YQ, Fowler VG Jr, Yeaman MR, Perdreau-Remington F, Kreiswirth BN, Bayer AS. 2009. Phenotypic and genotypic characteristics of persistent methicillin-resistant Staphylococcus aureus bacteremia in vitro and in an experimental endocarditis model. J Infect Dis 199:201–208. doi: 10.1086/595738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park S-Y, Chong YP, Park HJ, Park K-H, Moon SM, Jeong J-Y, Kim M-N, Kim S-H, Lee S-O, Choi S-H, Woo JH, Kim YS. 12 October 2012, posting date. agr dysfunction and persistent methicillin-resistant Staphylococcus aureus bacteremia in patients with removed eradicable foci. Infection doi: 10.1007/s15010-012-0348-0. [DOI] [PubMed] [Google Scholar]
- 14.Seidl K, Bayer AS, Fowler VG, McKinnell JA, Abdel Hady W, Sakoulas G, Yeaman MR, Xiong YQ. 2011. Combinatorial phenotypic signatures distinguish persistent from resolving methicillin-resistant Staphylococcus aureus bacteremia isolates. Antimicrob Agents Chemother 55:575–582. doi: 10.1128/AAC.01028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khatib R, Johnson LB, Sharma M, Fakih MG, Ganga R, Riederer K. 2009. Persistent Staphylococcus aureus bacteremia: incidence and outcome trends over time. Scand J Infect Dis 41:4–9. doi: 10.1080/00365540802441711. [DOI] [PubMed] [Google Scholar]
- 16.Khatib R, Johnson LB, Fakih MG, Riederer K, Khosrovaneh A, Shamse Tabriz M, Sharma M, Saeed S. 2006. Persistence in Staphylococcus aureus bacteremia: incidence, characteristics of patients and outcome. Scand J Infect Dis 38:7–14. doi: 10.1080/00365540500372846. [DOI] [PubMed] [Google Scholar]
- 17.Chong YP, Park S-J, Kim HS, Kim ES, Kim M-N, Park K-H, Kim S-H, Lee S-O, Choi S-H, Jeong J-Y, Woo JH, Kim YS. 2013. Persistent Staphylococcus aureus bacteremia: a prospective analysis of risk factors, outcomes, and microbiologic and genotypic characteristics of isolates. Medicine (Baltimore) 92:98–108. doi: 10.1097/MD.0b013e318289ff1e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao W, Chua K, Davies JK, Newton HJ, Seemann T, Harrison PF, Holmes NE, Rhee H-W, Hong J-I, Hartland EL, Stinear TP, Howden BP. 2010. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog 6:e1000944. doi: 10.1371/journal.ppat.1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao W, Cameron DR, Davies JK, Kostoulias X, Stepnell J, Tuck KL, Yeaman MR, Peleg AY, Stinear TP, Howden BP. 2013. The RpoB H481Y rifampicin resistance mutation and an active stringent response reduce virulence and increase resistance to innate immune responses in Staphylococcus aureus. J Infect Dis 207:929–939. doi: 10.1093/infdis/jis772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pascoe B, Dams L, Wilkinson TS, Harris LG, Bodger O, Mack D, Davies AP. 2014. Dormant cells of Staphylococcus aureus are resuscitated by spent culture supernatant. PLoS One 9:e85998. doi: 10.1371/journal.pone.0085998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol 4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- 22.von Eiff C, Peters G, Becker K. 2006. The small colony variant (SCV) concept—the role of staphylococcal SCVs in persistent infections. Injury 37:S26–S33. doi: 10.1016/j.injury.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Johnson M, Cockayne A, Morrissey JA. 2008. Iron-regulated biofilm formation in Staphylococcus aureus Newman requires ica and the secreted protein Emp. Infect Immun 76:1756–1765. doi: 10.1128/IAI.01635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.CLSI. 2007. Performance standards for antimicrobial susceptibility testing; 17th informational supplement Vol 27; document M100-S17. CLSI, Wayne, PA. [Google Scholar]
- 25.CLSI. 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement Vol 31; document M100-S21. CLSI, Wayne, PA. [Google Scholar]
- 26.Wada M, Lkhagvadorj E, Bian L, Wang C, Chiba Y, Nagata S, Shimizu T, Yamashiro Y, Asahara T, Nomoto K. 2010. Quantitative reverse transcription-PCR assay for the rapid detection of methicillin-resistant Staphylococcus aureus. J Appl Microbiol 108:779–788. doi: 10.1111/j.1365-2672.2009.04476.x. [DOI] [PubMed] [Google Scholar]
- 27.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheppard SK, Jolley KA, Maiden MCJ. 2012. A gene-by-gene approach to bacterial population genomics: whole genome MLST of Campylobacter. Genes (Basel) 3:261–277. doi: 10.3390/genes3020261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Méric G, Yahara K, Mageiros L, Pascoe B, Maiden MCJ, Jolley KA, Sheppard SK. 2014. A reference pan-genome approach to comparative bacterial genomics: identification of novel epidemiological markers in pathogenic Campylobacter. PLoS One 9:e92798. doi: 10.1371/journal.pone.0092798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boye K, Bartels MD, Andersen IS, Meller JA, Westh H. 2007. A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I–V. Clin Microbiol Infect Dis 13:725–727. doi: 10.1111/j.1469-0691.2007.01720.x. [DOI] [PubMed] [Google Scholar]
- 33.Milheiriço C, Oliveira DC, de Lencastre H. 2007. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex'. J Antimicrob Chemother 60:42–48. doi: 10.1093/jac/dkm112. [DOI] [PubMed] [Google Scholar]
- 34.Campbell SJ, Deshmukh HS, Nelson CL, Bae I-G, Stryjewski ME, Federspiel JJ, Tonthat GT, Rude TH, Barriere SL, Corey R, Fowler VG. 2008. Genotypic characteristics of Staphylococcus aureus isolates from a multinational trial of complicated skin and skin structure infections. J Clin Microbiol 46:678–684. doi: 10.1128/JCM.01822-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrissey JA, Cockayne A, Hammacott J, Denman-Johnson A, Hill PJ, Bishop K, Williams P. 2002. Conservation, surface exposure, and in vivo expression of the Frp family of iron-regulated cell wall proteins in Staphylococcus aureus. Infect Immun 70:2399–2407. doi: 10.1128/IAI.70.5.2399-2407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thurlow LR, Hanke ML, Fritz T, Angle A, Aldrich A, Williams SH, Engebretsen IL, Bayles KW, Horswill AR, Kielian T. 2011. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J Immunol 186:6585–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh R, Ray P, Das A, Sharma M. 2010. Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. J Antimicrob Chemother 65:1955–1958. doi: 10.1093/jac/dkq257. [DOI] [PubMed] [Google Scholar]
- 38.Abdelhady W, Bayer AS, Seidl K, Moormeier DE, Bayles KW, Cheung A, Yeaman MR, Xiong YQ. 2014. Impact of vancomycin on sarA-mediated biofilm formation: role in persistent endovascular infections due to methicillin-resistant Staphylococcus aureus. J Infect Dis 209:1231–1240. doi: 10.1093/infdis/jiu007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clement S, Vaudaux P, Francois P, Schrenzel J, Huggler E, Kampf S, Chaponnier C, Lew D, Lacroix J-S. 2005. Evidence of an intracellular reservoir in the nasal mucosa of patients with recurrent Staphylococcus aureus rhinosinusitis. J Infect Dis 192:1023–1028. doi: 10.1086/432735. [DOI] [PubMed] [Google Scholar]
- 40.Garzoni C, Kelley WL. 2009. Staphylococcus aureus: new evidence for intracellular persistence. Trends Microbiol 17:59–65. doi: 10.1016/j.tim.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 41.Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform 14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- 42.Holden MTG, Hsu L-Y, Kurt K, Weinert LA, Mather AE, Harris SR, Strommenger B, Layer F, Witte W, de Lencastre H, Skov R, Westh H, Zemlicková H, Coombs G, Kearns AM, Hill RLR, Edgeworth J, Gould I, Gant V, Cooke J, Edwards GF, McAdam PR, Templeton KE, McCann A, Zhou Z, Castillo-Ramírez S, Feil EJ, Hudson LO, Enright MC, Balloux F, Aanensen DM, Spratt BG, Fitzgerald JR, Parkhill J, Achtman M, Bentley SD, Nübel U. 2013. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res 23:653–664. doi: 10.1101/gr.147710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mishra NN, Yang S-J, Chen L, Muller C, Saleh-Mghir A, Kuhn S, Peschel A, Yeaman MR, Nast CC, Kreiswirth BN, Crémieux A-C, Bayer AS. 2013. Emergence of daptomycin resistance in daptomycin-naïve rabbits with methicillin-resistant Staphylococcus aureus prosthetic joint infection is associated with resistance to host defense cationic peptides and mprF polymorphisms. PLoS One 8:e71151. doi: 10.1371/journal.pone.0071151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peleg AY, Miyakis S, Ward DV, Earl AM, Rubio A, Cameron DR, Pillai S, Moellering RC Jr, Eliopoulos GM. 2012. Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS One 7:e28316. doi: 10.1371/journal.pone.0028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rubio A, Moore J, Varoglu M, Conrad M, Chu M, Shaw W, Silverman JA. 2012. LC-MS/MS characterization of phospholipid content in daptomycin-susceptible and -resistant isolates of Staphylococcus aureus with mutations in mprF. Mol Membr Biol 29:1–8. doi: 10.3109/09687688.2011.640948. [DOI] [PubMed] [Google Scholar]
- 46.Yang S-J, Mishra NN, Rubio A, Bayer AS. 2013. Causal role of single nucleotide polymorphisms within the mprF gene of Staphylococcus aureus in daptomycin resistance. Antimicrob Agents Chemother 57:5658–5664. doi: 10.1128/AAC.01184-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mishra NN, Bayer AS, Weidenmaier C, Grau T, Wanner S, Stefani S, Cafiso V, Bertuccio T, Yeaman MR, Nast CC, Yang S-J. 2014. Phenotypic and genotypic characterization of daptomycin-resistant methicillin-resistant Staphylococcus aureus strains: relative roles of mprF and dlt operons. PLoS One 9:e107426. doi: 10.1371/journal.pone.0107426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hussain M, Haggar A, Peters G, Chhatwal GS, Herrmann M, Flock J-I, Sinha B. 2008. More than one tandem repeat domain of the extracellular adherence protein of Staphylococcus aureus is required for aggregation, adherence, and host cell invasion but not for leukocyte activation. Infect Immun 76:5615–5623. doi: 10.1128/IAI.00480-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee LY, Miyamoto YJ, Mcintyre BW, Höök M, Mccrea KW, Mcdevitt D, Brown EL. 2002. The Staphylococcus aureus Map protein is an immunomodulator that interferes with T cell-mediated responses. J Clin Invest 110:1461–1471. doi: 10.1172/JCI16318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang L, Jacobson K, Vasi J, Lindberg M, Frykberg L. 1998. A second IgG-binding protein in Staphylococcus aureus. Microbiology 144:985–991. doi: 10.1099/00221287-144-4-985. [DOI] [PubMed] [Google Scholar]
- 51.Smith EJ, Visai L, Kerrigan SW, Speziale P, Foster TJ. 2011. The Sbi protein is a multifunctional immune evasion factor of Staphylococcus aureus. Infect Immun 79:3801–3809. doi: 10.1128/IAI.05075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uziel O, Borovok I, Schreiber R, Cohen G, Aharonowitz Y. 2004. Transcriptional regulation of the Staphylococcus aureus thioredoxin and thioredoxin reductase genes in response to oxygen and disulfide stress. J Bacteriol 186:326–334. doi: 10.1128/JB.186.2.326-334.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katzif S, Danavall D, Bowers S, Balthazar JT, Shafer WM. 2003. The major cold shock gene, cspA, is involved in the susceptibility of Staphylococcus aureus to an antimicrobial peptide of human cathepsin G. Infect Immun 71:4304–4312. doi: 10.1128/IAI.71.8.4304-4312.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]