Abstract
Background
Intron sequences are common in 16S rRNA genes of specific thermophilic lineages of Archaea, specifically the Thermoproteales (phylum Crenarchaeota). Environmental sequencing (16S rRNA gene and metagenome) from geothermal habitats in Yellowstone National Park (YNP) has expanded the available datasets for investigating 16S rRNA gene introns. The objectives of this study were to characterize and curate archaeal 16S rRNA gene introns from high-temperature habitats, evaluate the conservation and distribution of archaeal 16S rRNA introns in geothermal systems, and determine which “universal” archaeal 16S rRNA gene primers are impacted by the presence of intron sequences.
Results
Several new introns were identified and their insertion loci were constrained to thirteen locations across the 16S rRNA gene. Many of these introns encode homing endonucleases, although some introns were short or partial sequences. Pyrobaculum, Thermoproteus, and Caldivirga 16S rRNA genes contained the most abundant and diverse intron sequences. Phylogenetic analysis of introns revealed that sequences within the same locus are distributed biogeographically. The most diverse set of introns were observed in a high-temperature, circumneutral (pH 6) sulfur sediment environment, which also contained the greatest diversity of different Thermoproteales phylotypes.
Conclusions
The widespread presence of introns in the Thermoproteales indicates a high probability of misalignments using different “universal” 16S rRNA primers employed in environmental microbial community analysis.
Reviewers
This article was reviewed by Dr. Eugene Koonin and Dr. W. Ford Doolittle.
Electronic supplementary material
The online version of this article (doi:10.1186/s13062-015-0065-6) contains supplementary material, which is available to authorized users.
Keywords: Archaea, Thermoproteales, Introns, 16S rRNA, Geothermal, Metagenomics
Background
The 16S rRNA is the central structural component of the bacterial and archaeal 30S ribosomal subunit and is required for the initiation of protein synthesis and the stabilization of correct codon-anticodon pairing in the A site of the ribosome during mRNA translation [1]. Due to the functional constancy and highly conserved nature of the 16S rRNA gene, it has been an important phylogenetic marker, and was used to define the three domains of Life [2–4]. Several lineages within the domain Archaea contain 16S rRNA gene introns, which are mobile genetic elements that do not appear to impact the host’s growth or metabolism. 16S rRNA gene introns have been identified in two genera of the order Desulfurococcales (Aeropyrum and Staphylothermus) [5–10] and four genera in the order Thermoproteales: Pyrobaculum (11 spp.), Thermoproteus (2 spp.), Caldivirga (1 sp.), and Vulcanisaeta (1 sp.) [10–19]. The only other archaea to contain 16S rRNA gene introns are the Aigarchaeota (i.e., Caldiarchaeum subterraneum) [20]. The orders Desulfurococcales and Thermoproteales also contain 23S rRNA and tRNA gene introns; however, the limited number of genomes in these orders prevents a robust analysis of the diversity and distribution of introns in these genes.
Currently, the Thermoproteales exhibit the most numerous and diverse rRNA and tRNA introns in the domain Archaea [21–23]. Although the life cycle of tRNA and rRNA intron sequences has been well characterized, little is known regarding the transmission and evolution of introns [23, 24]. A distinguishing characteristic of all archaeal rRNA and tRNA introns is the bulge-helix-bulge motif formed at the intron insertion site [25, 26]. This core motif is recognized by the tRNA splicing endoribonuclease, which is responsible for post-transcriptional excision of RNA introns [27–30] followed by ligation via the tRNA splicing ligase (RtcB; [31]) during the maturation process. Many rRNA introns encode homing endonuclease proteins with either one or two copies of the characteristic LAGLI-DADG motif [24, 32]. The two forms of the enzyme (a homodimer, which consists of two subunits each containing one motif copy, and a monomer, which is a single subunit containing two motif copies) each recognize long stretches (15 - 30 bp) of intron– loci and catalyze a double-strand break where the intron sequence is inserted via recombination and repair mechanisms [10, 33]. Frequent horizontal transmission within a population is required for intron persistence, while the remnant sequence is subjected to decreased selection pressure [10, 34]. A functional homing endonuclease gene or a mRNA transcript has been suggested for intercellular intron migration [25, 35].
Direct sequencing of 16S rRNA genes and/or metagenomes from environmental samples has become a standard and convenient method of assessing microbial population abundance, structure, and function in microbial communities (e.g. [36, 37]). Publically available 16S rRNA gene sequences from high-temperature geothermal environments in Yellowstone National Park (YNP) have increased dramatically, which provides a large dataset to determine the diversity and distribution of introns in (hyper)thermophilic archaeal communities. Consequently, the objectives of this study were to (i) characterize and curate all archaeal 16S rRNA gene introns found within currently available genomes and environmental sequence databases, (ii) perform a phylogenetic analysis to evaluate the conservation and distribution of archaeal 16S rRNA introns in geothermal systems, and (iii) determine which “universal” archaeal 16S rRNA gene primers are interrupted by the presence of intron sequences.
Results and discussion
Intron sequences were confined to 13 loci across the 16S rRNA gene (Fig. 1, Table 1). The number of introns identified per locus ranged from 2 (locus 722) to 41 introns (locus 781). Phylogenetic analysis of host 16S rRNA genes revealed that the large majority of introns (>90 %) were observed within the order Thermoproteales, and less in the Desulfurococcales (Crenarchaeota) and Aigarchaeaota (Fig. 2). Pyrobaculum spp. contained the majority of known introns and these are found at diverse loci across the 16S rRNA gene (loci 374, 548, 781, 907, 908, 919, 1093, 1205, 1213, 1391). Other genera in the Thermoproteales also contain intron sequences at many of the same loci: Thermoproteus (loci 548, 722, 1093, 1205, 1213, 1391), Caldivirga (loci 374, 781, 901, 908, 919), and Vulcanisaeta (locus 1391). Desulfurococcales-like introns were identified in six loci (548, 901, 908, 919, 1093, 1205).
Fig. 1.
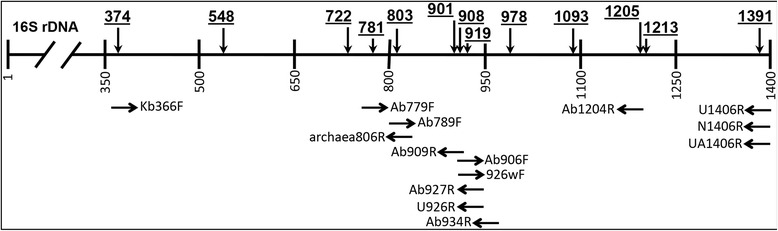
Position of introns identified in 16S rRNA genes within the Archaea. Vertical arrows indicate loci with positions underlined (E. coli 16S rRNA numbering). Horizontal arrows indicate “universal” archaeal primers interrupted by the presence of intron sequences (arrows not to scale)
Table 1.
Intron insertion loci identified in archaeal 16S rRNA genes
| Representative Taxa | Locusa | #CDSb,c | #HP2 | #PRUb,c | Interrupted “universal” primer(s) |
|---|---|---|---|---|---|
| Pyrobaculum, Caldivirga | 374 | 5 (5,0) | 10 | - | Kb366F |
| Pyrobaculum, Thermoproteus, Staphylothermus, Desulfurococcalesd | 548 | 2 (0,2) | 13 | - | -none- |
| Caldiarchaeum | 722 | - | - | 2 | -none- |
| Pyrobaculum, Thermoproteus, Vulcanisaeta, Thermoprotealesd, Caldivirga, Caldiarchaeum | 781 | 14 (2,4) | - | 28 | Ab779F |
| Desulfurococcales | 803 | - | - | 3 | Ab789F, Arc806R |
| Pyrobaculum, Caldivirga, Desulfurococcales | 901 | - | 8 | - | Ab909R |
| Pyrobaculum, Caldivirga, Aeropyrum, Caldiarchaeum | 908 | 4 (1,3) | - | 6 | Ab909R, Ab906F, 926wF, U926R, Ab927R |
| Pyrobaculum, Thermoproteus, Caldivirga, Vulcanisaeta, Caldiarchaeum, Desulfuroccocales | 919 | 6 (2,3) | - | 27 (1,0) | Ab906F, 926wF, U926R, Ab927R, A934R |
| Caldiarchaeum | 978 | - | 9 | - | -none- |
| Pyrobaculum, Thermoproteus, Desulfurococcales | 1093 | 15 (4,11) | 7 | 9 (2,1) | -none- |
| Pyrobaculum, Thermoproteus, Desulfurococcales | 1205 | 2 (0,2) | 17 | - | -none- |
| Pyrobaculum, Thermoproteus | 1213 | 18 (4,14) | - | 2 | -none- |
| Pyrobaculum, Thermoproteus, Vulcanisaeta | 1391 | 15 (7,2) | - | 5 | UA1406R, N1406R, U1406R |
aIntron insertion location in 16S rRNA gene (E. coli numbering, nucleotide position directly before insertion), b CDS Homing endonuclease Coding Sequence, HP small (<50 nt) RNA hairpin-forming introns, PRU Partial, Remnant, or Uncharacterized introns, cNumbers in parenthesis refer to the number of LAGLI-DADG motifs (1 or 2, respectively), dSpecifies an uncharacterized phyla within the Desulfurococcales and Thermoproteales, respectively
Fig. 2.
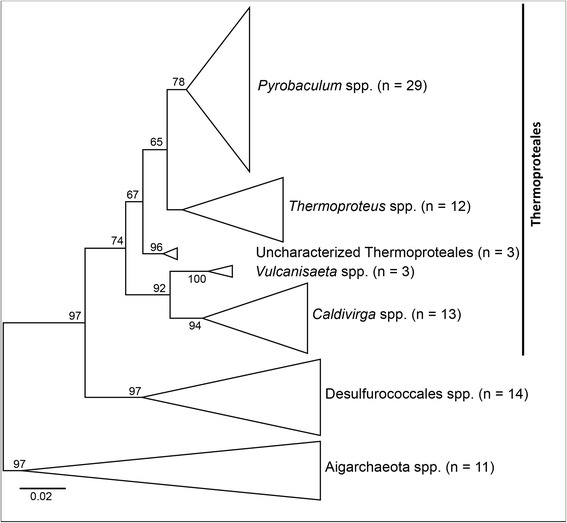
Phylogenetic tree of 16S rRNA genes that contain intron sequences. Intron sequences were not included in the alignments. The tree was constructed with Neighbor-Joining methods and bootstrap values were determined by resampling 1000 trees
Most (9/13) intron regions contained two or more predicted homing endonuclease genes, which code for either one or two of the canonical LAGLI-DADG motifs (Table 1). Short (<50 nt), hairpin-forming introns were identified in 7 intron loci, and intron loci 901, 978, and 1205 only contained short hairpins. Partial, remnant, or undetermined (PRU) intron sequences were identified in eight of the 13 loci. The activity of P. oguniense 16S rRNA homing endonuclease (Pog.S1213) promoted the homing of its own intron and guaranteed the co-conversion of both the downstream hairpin-forming intron (Pog.S1205) and the intervening eight nucleotides by cleaving at the intron− locus 1205 [38]. This co-conversion homing mechanism may be applicable to other 16S rRNA gene loci, specifically introns at loci 901 (HP-only) and 908 (homing endonuclease encoding), which are separated by seven nucleotides.
Secondary structures of intron sequences
Analysis of the intron insertion loci in the modeled secondary structure of an archaeal 16S rRNA molecule [39] revealed that nearly all of the intron loci (loci 374, 722, 781, 803, 901, 908, 919, 978, 1093, 1213, 1391) were either located in a bulge motif or in a helix structure very near a bulge motif (548, 1205; Fig. 3). These locations may provide the necessary flexibility in secondary structure for the insertion of intron sequences, which are often > 700 nt. There are many other bulge motifs in the secondary structure of the 16S rRNA molecule that do not contain intron sequences. This might be attributed to secondary or tertiary structure restrictions (e.g., steric hindrance), sequence specificity of insertion, or an inability to detect introns at these locations in the environment. Alternatively, these loci may not provide sufficient access to the tRNA splicing endoribonuclease. All V-regions (V1 - V9; [40]) of the 16S rRNA gene were free of introns, indicating that many introns are confined to the most highly-conserved loci in the 16S rRNA gene. Intron loci 374 and 803 flank variable regions V4 and V8, respectively, and could interrupt primers designed around these regions (see below).
Fig. 3.
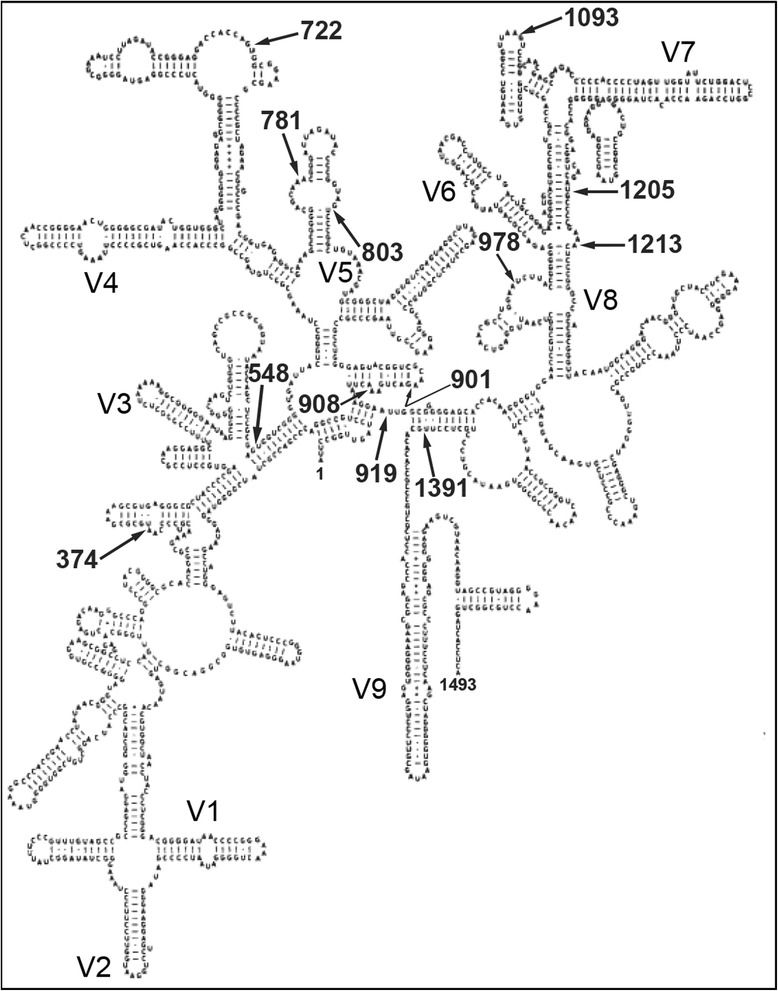
The location (E. coli numbering) of intron sequences (identified in the current study) within the transcribed secondary structure of the 16S rRNA gene. Variable regions (V1 - V9) are shown for reference
Transcribed intron sequences also had predictable, thermodynamically favorable secondary structures (e.g., Fig. 4a), similar to observations of 23S rRNA gene introns [25]. Intron sequences sharing high nucleotide identity within the same insertion locus maintained similar secondary structure (data not shown). However, the predicted secondary structures were not generally conserved within insertion loci. The predicted folding of each transcribed intron is thermodynamically favorable (at 37 °C; Fig. 4), but considering the predominance of introns in high-temperature habitats, the themostability of secondary structures may play a yet uncharacterized role in intron distribution and propagation. The average G + C content of the intron sequences was 57 ± 11 %, indicating that some thermostability could be attributed to strong G + C bonding; however, this value is still lower than the average % G + C content of the host 16S rRNA genes (67 ± 2 %).
Fig. 4.
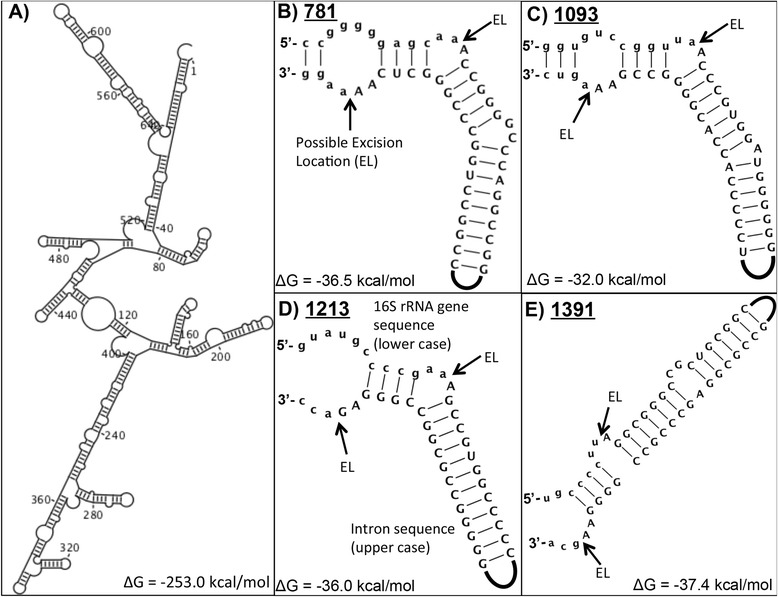
Predicted secondary structures of 16S rRNA gene introns. a Predicted secondary structure of the Pyrobaculum yellowstonensis strain WP30 16S rRNA gene intron at locus 1391, illustrating the highly-structured nature of transcribed intron sequences. Numbers denote nucleotide position along the intron sequence. b-e Predicted secondary structures based on consensus sequences (weblogo) of four different 16S rRNA gene introns. Lower and upper case nucleotides denote 16S rRNA gene sequence and intron sequence, respectively. Arrows indicate hypothesized excision locations (EL) within the bulge-helix-bulge motif
Comparison of intron sequences within the same loci identified highly-conserved nucleotide residues near the intron-exon junctions, which represent conserved intron cores [14, 25]. These cores form a bulge-helix-bulge (BHB) motif (Fig. 4 b-e) post-transcription that is very similar to the characterized BHB motifs of intron-containing archaeal tRNAs [41] and 23S rRNAs [23], both of which are excised by the same splicing endoribonuclease [23, 42–44]. The order Thermoproteales, and especially the genus Pyrobaculum, contain the majority of intron-containing tRNAs and 23S rRNA gene introns in the domain Archaea. The flanking 16S rRNA gene sequences of each intron locus were also highly conserved across entries in the domain Archaea (also one of the reasons “universal” primers have been designed around these loci). The nucleotide sequences comprising the DNA insertion sites and the BHB motif vary among loci and it is unclear how sequence specificity of the homing endoribonuclease or the splicing ligase contributes to intron propagation and distribution. Several intron loci were identified in both the Crenarchaeota and Aigarchaeota (e.g., loci 908, 919, 1093, and 1205), which suggests that these loci have been sites of intron insertions since the divergence of these two phyla. Based on extant insertion loci, intron sequences radiated throughout the Thermoproteales, specifically in the Pyrobaculum and Thermoproteus spp.
Phylogenetic analysis of 16S rRNA gene introns
Intron sequences within the same loci were successfully aligned at the nucleotide level and subsequent phylogenetic analysis revealed both intra-locus and geographic separation (Fig. 5). Intron sequences within loci tended to clade with other introns from similar geographic locations, rather than with related genera. For example, the Thermoproteales-like homing endonuclease encoding genes at loci 781, 1093, and 1213 (shown in Fig. 5a) formed clades corresponding to sequences from Japan, YNP, Kamchatka, and/or Iceland and not by genus, although Kamchatka and Iceland were only represented by solitary isolates Pyrobaculum sp. 1860 [12] and P. neutrophilum [45], respectively. Intron sequences grouped first by geographic location (Fig. 5); for example, intron sequences from Japan grouped together compared to entries from YNP (i.e., loci 781 and 1213). Locus 1391 contained several sequences from YNP, including a novel intron identified in the YNP isolate P. yellowstonensis strain WP30 [46] that is highly related to intron sequences obtained from the Bison Pool metagenome (Fig. 5b). The first, and only, previously described intron at this location was a Vulcanisaeta distributa IC-065 clone from Japan [18]; however this sequence is significantly divergent from the YNP entries. These entries clade based on the phylogeny of the host 16S rRNA gene (i.e. Pyrobaculum and Thermoproteus). Introns found at different 16S rRNA gene loci were too different for phylogenetic comparison (<20 % nt/aa identity).
Fig. 5.
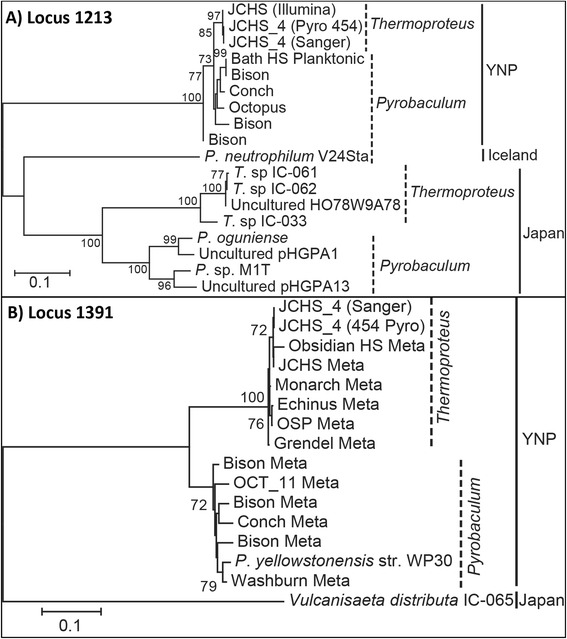
Phylogenetic analysis of intron sequences (homing endonuclease encoding) at loci 1213 (a) and 1392 (b). Unrooted trees were constructed with Neighbor-Joining methods and bootstrap values were determined by resampling 1000 trees
The phylogenetic distribution of intron sequences by geographic location is consistent with the hypothesis that once a homing endonuclease becomes fixed in a genome, selection pressure for the gene significantly decreases resulting in degeneration (mutation) and enforcing the requirement of frequent horizontal transmissions between populations to maintain intron propagation [34]. Populations related to the genus Pyrobaculum contain the most abundant 16S rRNA introns in the order Thermoproteales and although they predominate in high-temperature, near-neutral to alkaline (pH > 6) geothermal systems, populations of the other Thermoproteales genera (Caldivirga, Vulcanisaeta and Thermoproteus) are found with Pyrobaculum in > 65 °C, pH 5 - 7 hot springs [47, 48]. Therefore, the optimal conditions for intron transmission among the Thermoproteales may lie within this very defined temperature and pH range. Considering that rRNA introns are likely ancient [49], these environmental conditions may reflect constraints on the origin of RNA gene introns. Introns may be perpetuated in extant Thermoproteales species and result in genetic variation, analogous to the extensive mobile elements identified in some Sulfolobales genomes [50]. Very little is known about the dispersal of thermophilic archaea, however, many intron sequences were highly similar among YNP habitats (e.g., locus 1391). Additional archaeal 16S rRNA introns obtained from other geothermal systems will help resolve observed patterns of intron distribution and diversity.
“Universal” 16S rRNA gene primers
Primer sequences designed for archaeal 16S rRNA genes (n = 51) were manually aligned to intron-containing 16S rRNA gene sequences to determine which primers were interrupted by, or spanned intron sequences (Fig. 1). Fourteen primers spanned at least one intron insertion locus, including five primers that spanned two insertion loci (Ab909R, Ab906F, 926wF, U926R, Ab927R) and are located near the center of the 16S rRNA gene. “Universal” primers designed to anneal near the middle of the 16S rRNA gene have the greatest potential of being interrupted by an intron sequence. The remaining archaeal primers evaluated here did not directly overlap with currently known introns; however, depending on the chosen complementary primer for PCR, the resulting amplicons may contain complete or partial intervening intron sequences. These non-16S rRNA gene fragments are then often included in long-fragment 16S rRNA gene sequences, and have a high probability of being misinterpreted (either by human or computer) as non-specific amplification, chimeras, sequencing error, and/or assembly error, which may result in their exclusion from public databases. The failure to detect intron-containing 16S rRNA genes in thermal habitats using many universal primers would potentially underestimate several abundant Thermoproteales organisms. Consequently, specific efforts to quantify members of the Thermoproteales using short or long-fragment sequencing technologies should be aware of intron-containing 16S rRNA genes, and avoid primer sets that are centered on common sites of intron sequence, or intron insertion loci.
Conclusions
Intron sequences were confined to 13 loci across the 16S rRNA gene and were most abundant in the order Thermoproteales (phylum Crenarchaeota). All transcribed introns form predictable secondary structure including the characteristic bulge-helix-bulge motif. Many intron sequences encoded homing endonucleases, although other introns were short and/or non-coding sequences. Phylogenetic analysis revealed that intron sequences grouped together by geographic location and then by host taxa. The presence of introns in highly conserved regions of 16S rRNA introns directly interferes with use of “universal” primers often used in environmental gene surveys.
Methods
Intron sequences were identified by querying (blastn/blastx) the NCBI nr and the DOE-Joint Genome Institute (DOE-JGI) Integrated Microbial Genomes (with Microbiome Samples (IMG/M) databases) with previously identified 16S gene introns (Additional file 1: Table S1). Approximately 100 16S rRNA genes were identified that contained over 180 intron sequences (Additional file 2: Table S2) resulting in a total dataset of ~230 intron sequences (in ~115 16S rRNA genes) distributed at 13 different loci across the entire length of the 16S rRNA gene (Fig. 1, Table 1). Homing endonucleases were identified and secondary structures were predicted using CLC Main Workbench (v6.9.1; Qiagen). Longer intron sequences (> ~100 nt) were translated to amino acid sequence (if possible) and searched against the Pfam database (v. 27.0; [51]) to identify LAGLI-DADG motif(s) that are indicative of homing endonucleases [33]. Intron sequences were grouped into the following categories: (i) introns encoding a homing endonuclease, (ii) introns without an obvious open reading frame (remnant), (iii) introns forming small (< ~ 50 nt) predictable hairpin structures that maintain the bulge-helix-bulge motif, or (iv) partial, truncated, or uncharacterized intron sequences.
Sequence alignments were performed (manually and/or with ClustalW) before phylogenetic analysis and/or Weblogo3 analysis [52]. Phylogenetic trees of 16S rRNA genes and intron sequences were constructed by employing Neighbor-Joining or Maximum likelihood methods (Mega 5.2.2; [53]). Over 50 “universal” archaeal primers were manually aligned to 16S rRNA genes to identify those interrupted by, or that spanned intron loci (Additional file 3: Table S3).
Reviewers’ comments
Reviewer 1: Dr. Eugene Koonin
Report form: Jay and Inskeep report the distribution of Group I introns in 16S RNA genes from hyperthermophilic archaea, and in particular Thermoproteles, where these introns are most common. The analysis is carefully performed, and there is an important conclusion on the evolution of self-splicing introns, namely that they group by geographic location, i.e. spread primarily horizontally. Also, this analysis is of practical value because the authors carefully assess the effect of introns on the use of universal primers.
Authors’ response: We thank the reviewer for these comments.
Quality of written English: Acceptable.
Reviewer 2: Dr. W. Ford Doolittle
Report form: This is a perfectly fine and straightforward report and summary of the distribution of introns in the rRNA genes of members of the Thermoproteales. It will provide a basis for future experimental and comparative studies, and raises some interesting questions, such as the means by which introns are transferred between different lineages in a given environment and the structural adaptations of the intron RNA to high temperature, but does not attempt to answer them. It also points out how intron presence might result in failure to amplify specific regions of rRNA genes by PCR. Members of Thermoproteales might thus be underestimated in environmental sampling, and one wonders whether there are abundant major groups that go undetected because they always have such introns. Indeed, is there anything to keep introns from becoming colonized by genes essential for cellular survival, so that pieces of the rRNA gene become permanently separated, and the cells that bear them become undetectable with standard molecular methods?
Authors response: We thank the reviewer for these comments, and for the very interesting question regarding possible ‘colonization’ of introns by genes coding for essential function, and subsequent separation of rRNA gene fragments (i.e., within the genome). The question regarding intron colonization by other genes is difficult to answer due to the complexities of intron propagation. As we currently understand 16S rRNA gene introns in the Thermoproteales, a homing endonuclease (gene or transcript) is required for intron insertion into a functional rRNA. Consequently, any disruption to the homing endonuclease would not allow for insertion and propagation of the intron sequence. Any cell that may bear intron fragments by definition will still have a functioning rRNA somewhere in the genome and therefore would be detectable with standard methods.
Quality of written English: Acceptable.
Acknowledgements
We appreciate support from the DOE-Pacific Northwest National Laboratory (subcontract no. 112443), the Department of Energy (DOE)-Joint Genome Institute Community Sequencing Program (CSP 787081), and the NSF IGERT Program (DGE 0654336) for support to ZJJ. The work conducted by the Joint Genome Institute (DOE-AC02-05CH11231) and the Pacific Northwest National Laboratory (Foundational Scientific Focus Area) is supported by the Genomic Science Program, Office of Biological and Environmental Research, U.S. DOE. We also thank C. Hendrix, S. Gunther, and D. Hallac (Center for Resources, Yellowstone National Park) for research permitting (permits YELL-SCI-5068 and -5686). The open access fees were generously provided by the Library’s Author Fund at Montana State University.
Abbreviations
- BHB
Bulge-helix-bulge
- YNP
Yellowstone National Park
- RtcB
tRNA splicing ligase
Additional files
Archaea with known 16S rRNA gene introns.
Environmental metadata of sites in Yellowstone National Park where 16S rRNA gene introns were identified.
“Universal” archaeal 16S rRNA gene primers interrupted by introns common in members of the Thermoproteales (phylum Crenarchaeota).
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ZJJ Designed the study, conducted the experiments, collected and analyzed output data, and wrote the manuscript. WPI Obtained funding, assisted with experimental design, discussed the results, and edited the manuscript at all stages. Both authors read and approved the manuscript.
Contributor Information
Zackary J. Jay, Email: zackary.jay@montana.edu
William P. Inskeep, Email: binskeep@montana.edu
References
- 1.Wimberly BT, Brodersen DE, Clemons WM, Morgan-Warren RJ, Carter AP, Vonrhein C, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 2.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woese CR. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sako Y, Nomura N, Uchida A, Ishida Y, Morii H, Koga Y, et al. Aeropyrum pernix gen. nov., sp. nov., a novel aerobic hyperthermophilic archaeon growing at temperatures up to 100 °C. Int J Syst Bacteriol. 1996;46:1070–1077. doi: 10.1099/00207713-46-4-1070. [DOI] [PubMed] [Google Scholar]
- 6.Kawarabayasi Y, Hino Y, Horikawa H, Yamazaki S, Haikawa Y, Jin-no K, et al. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res Int J Rapid Publ Rep Genes Genomes. 1999;6:83–101. doi: 10.1093/dnares/6.2.83. [DOI] [PubMed] [Google Scholar]
- 7.Nomura N, Sako Y, Uchida A. Molecular characterization and postsplicing fate of three introns within the single rRNA operon of the hyperthermophilic archaeon Aeropyrum pernix K1. J Bacteriol. 1998;180:3635–3643. doi: 10.1128/jb.180.14.3635-3643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiala G, Stetter KO, Jannasch HW, Langworthy TA, Madon J. Staphylothermus marinus sp. nov. Represents a novel genus of extremely thermophilic submarine heterotrophic archaebacteria growing up to 98 °C. Syst Appl Microbiol. 1986;8:106–113. doi: 10.1016/S0723-2020(86)80157-6. [DOI] [Google Scholar]
- 9.Anderson IJ, Dharmarajan L, Rodriguez J, Hooper S, Porat I, Ulrich LE, et al. The complete genome sequence of Staphylothermus marinus reveals differences in sulfur metabolism among heterotrophic Crenarchaeota. BMC Genomics. 2009;10:145–158. doi: 10.1186/1471-2164-10-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morinaga Y, Nomura N, Sako Y. Population dynamics of archaeal mobile introns in natural environments: a shrewd invasion strategy of the latent parasitic DNA. Microbes Environ. 2002;17:153–163. doi: 10.1264/jsme2.17.153. [DOI] [Google Scholar]
- 11.Burggraf S, Larsen N, Woese CR, Stetter KO. An intron within the 16S ribosomal RNA gene of the archaeon Pyrobaculum aerophilum. Proc Natl Acad Sci. 1993;90:2547–2550. doi: 10.1073/pnas.90.6.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mardanov AV, Gumerov VM, Slobodkina GB, Beletsky AV, Bonch-Osmolovskaya EA, Ravin NV, et al. Complete genome sequence of strain 1860, a crenarchaeon of the genus Pyrobaculum able to grow with various electron acceptors. J Bacteriol. 2012;194:727–728. doi: 10.1128/JB.06465-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takai K, Horikoshi K. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics. 1999;152:1285–1297. doi: 10.1093/genetics/152.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itoh T, Suzuki K, Nakase T. Occurrence of introns in the 16S rRNA genes of members of the genus Thermoproteus. Arch Microbiol. 1998;170:155–161. doi: 10.1007/s002030050628. [DOI] [PubMed] [Google Scholar]
- 15.Fischer F, Zillig W, Stetter KO, Schreiber G. Chemolithoautotrophic metabolism of anaerobic extremely thermophilic archaebacteria. Nature. 1983;301:511–513. doi: 10.1038/301511a0. [DOI] [PubMed] [Google Scholar]
- 16.Itoh T, Suzuki K, Nakase T. Vulcanisaeta distributa gen. nov., sp. nov., and Vulcanisaeta souniana sp. nov., novel hyperthermophilic, rod-shaped crenarchaeotes isolated from hot springs in Japan. Int J Syst Evol Microbiol. 2002;52:1097–1104. doi: 10.1099/ijs.0.02152-0. [DOI] [PubMed] [Google Scholar]
- 17.Sako Y, Nunoura T, Uchida A. Pyrobaculum oguniense sp. nov., a novel facultatively aerobic and hyperthermophilic archaeon growing at up to 97° C. Int J Syst Evol Microbiol. 2001;51(Pt 2):303–309. doi: 10.1099/00207713-51-2-303. [DOI] [PubMed] [Google Scholar]
- 18.Itoh T, Nomura N, Sako Y. Distribution of 16S rRNA introns among the family Thermoproteaceae and their evolutionary implications. Extremophiles. 2003;7:229–233. doi: 10.1007/s00792-003-0314-y. [DOI] [PubMed] [Google Scholar]
- 19.Itoh T, Suzuki K, Sanchez PC, Nakase T. Caldivirga maquilingensis gen. nov., sp. nov., a new genus of rod-shaped crenarchaeote isolated from a hot spring in the Philippines. Int J Syst Bacteriol. 1999;49 Pt 3:1157–1163. doi: 10.1099/00207713-49-3-1157. [DOI] [PubMed] [Google Scholar]
- 20.Nunoura T, Takaki Y, Kakuta J, Nishi S, Sugahara J, Kazama H, et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2011;39:3204–3223. doi: 10.1093/nar/gkq1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugahara J, Kikuta K, Fujishima K, Yachie N, Tomita M, Kanai A. Comprehensive analysis of archaeal tRNA genes reveals rapid increase of tRNA introns in the Order Thermoproteales. Mol Biol Evol. 2008;25:2709–2716. doi: 10.1093/molbev/msn216. [DOI] [PubMed] [Google Scholar]
- 22.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37(Database issue):D93–97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP. Evolution of introns in the archaeal world. Proc Natl Acad Sci U S A. 2011;108:4782–4787. doi: 10.1073/pnas.1100862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoddard BL. Homing endonuclease structure and function. Q Rev Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 25.Lykke-Andersen J, Garrett RA. Structural characteristics of the stable RNA introns of archaeal hyperthermophiles and their splicing junctions. J Mol Biol. 1994;243:846–855. doi: 10.1006/jmbi.1994.1687. [DOI] [PubMed] [Google Scholar]
- 26.Diener JL, Moore PB. Solution Structure of a Substrate for the Archaeal Pre-tRNA Splicing Endonucleases: The Bulge-Helix-Bulge Motif. Mol Cell. 1998;1:883–894. doi: 10.1016/S1097-2765(00)80087-8. [DOI] [PubMed] [Google Scholar]
- 27.Lykke-Andersen J, Garrett RA. RNA-protein interactions of an archaeal homotetrameric splicing endoribonuclease with an exceptional evolutionary history. EMBO J. 1997;16:6290–6300. doi: 10.1093/emboj/16.20.6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fabbri S, Fruscoloni P, Bufardeci E, Negri EDN, Baldi MI, Attardi DG, et al. Conservation of substrate recognition mechanisms by tRNA splicing endonucleases. Science. 1998;280:284–286. doi: 10.1126/science.280.5361.284. [DOI] [PubMed] [Google Scholar]
- 29.Xue S, Calvin K, Li H. RNA recognition and cleavage by a splicing endonuclease. Science. 2006;312:906–910. doi: 10.1126/science.1126629. [DOI] [PubMed] [Google Scholar]
- 30.Hirata A, Fujishima K, Yamagami R, Kawamura T, Banfield JF, Kanai A, et al. X-ray structure of the fourth type of archaeal tRNA splicing endonuclease: insights into the evolution of a novel three-unit composition and a unique loop involved in broad substrate specificity. Nucleic Acids Res. 2012;40:10554–10566. doi: 10.1093/nar/gks826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Englert M, Sheppard K, Aslanian A, Yates JR, Söll D. Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc Natl Acad Sci. 2011;108:1290–1295. doi: 10.1073/pnas.1018307108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001;29:3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jurica MS, Monnat RJ, Jr, Stoddard BL. DNA Recognition and cleavage by the LAGLIDADG homing endonuclease I-Cre I. Mol Cell. 1998;2:469–476. doi: 10.1016/S1097-2765(00)80146-X. [DOI] [PubMed] [Google Scholar]
- 34.Burt A, Koufopanou V. Homing endonuclease genes: the rise and fall and rise again of a selfish element. Curr Opin Genet Dev. 2004;14:609–615. doi: 10.1016/j.gde.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Aagaard C, Dalgaard JZ, Garrett RA. Intercellular mobility and homing of an archaeal rDNA intron confers a selective advantage over intron- cells of Sulfolobus acidocaldarius. Proc Natl Acad Sci. 1995;92:12285–12289. doi: 10.1073/pnas.92.26.12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere.”. Proc Natl Acad Sci. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tazi L, Breakwell DP, Harker AR, Crandall KA. Life in extreme environments: microbial diversity in Great Salt Lake, Utah. Extrem Life Extreme Cond. 2014;18:525–535. doi: 10.1007/s00792-014-0637-x. [DOI] [PubMed] [Google Scholar]
- 38.Nakayama H, Morinaga Y, Nomura N, Nunoura T, Sako Y, Uchida A. An archaeal homing endonuclease I-PogI cleaves at the insertion site of the neighboring intron, which has no nested open reading frame. FEBS Lett. 2003;544:165–170. doi: 10.1016/S0014-5793(03)00497-6. [DOI] [PubMed] [Google Scholar]
- 39.Gutell RR, Weiser B, Woese CR, Noller HF. Comparative anatomy of 16-S-like ribosomal RNA. In PROG NUCLEIC ACID RES&MOLECULAR BIO. Volume 32. Edited by Cohn WE, Moldave K. Orlando, FL: Academic Press; 1985:155–216. [DOI] [PubMed]
- 40.Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. At Least 1 in 20 16S rRNA Sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol. 2005;71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marck C, Grosjean H. Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: evolutionary implications. RNA. 2003;9:1516–1531. doi: 10.1261/rna.5132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biniszkiewicz D, Cesnaviciene E, Shub DA. Self-splicing group I intron in cyanobacterial initiator methionine tRNA: evidence for lateral transfer of introns in bacteria. EMBO J. 1994;13:4629–4635. doi: 10.1002/j.1460-2075.1994.tb06785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abelson J, Trotta CR, Li H. tRNA Splicing. J Biol Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- 44.Li H, Trotta CR, Abelson J. Crystal structure and evolution of a transfer RNA splicing enzyme. Science. 1998;280:279–284. doi: 10.1126/science.280.5361.279. [DOI] [PubMed] [Google Scholar]
- 45.Chan PP, Cozen AE, Lowe TM. Reclassification of Thermoproteus neutrophilus stetter and zillig 1989 as Pyrobaculum neutrophilum comb. nov. based on phylogenetic analysis. Int J Syst Evol Microbiol. 2012;63(Pt 2):751–754. doi: 10.1099/ijs.0.043091-0. [DOI] [PubMed] [Google Scholar]
- 46.Jay ZJ, Beam JP, Dohnalkova A, Lohmayer R, Bodle B, Planer-Friedrich B, et al. Pyrobaculum yellowstonensis str. WP30 respires on elemental sulfur and/or arsenate in circumneutral sulfidic geothermal sediments of Yellowstone National Park. Appl Environ Microbiol. 2015, in press. [DOI] [PMC free article] [PubMed]
- 47.Inskeep WP, Rusch DB, Jay ZJ, Herrgard MJ, Kozubal MA, Richardson TH, et al. Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS ONE. 2010;5:e9773. doi: 10.1371/journal.pone.0009773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inskeep WP, Jay ZJ, Herrgard MJ, Kozubal MA, Rusch DB, Tringe SG, et al. Phylogenetic and functional analysis of metagenome sequence from high-temperature archaeal habitats demonstrate linkages between metabolic potential and geochemistry. Front Microb Physiol Metab. 2013;4:95–116. doi: 10.3389/fmicb.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dalgaard JZ, Garrett RA, Belfort M. A site-specific endonuclease encoded by a typical archaeal intron. Proc Natl Acad Sci U S A. 1993;90:5414–5417. doi: 10.1073/pnas.90.12.5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brügger K, Redder P, She Q, Confalonieri F, Zivanovic Y, Garrett RA. Mobile elements in archaeal genomes. FEMS Microbiol Lett. 2002;206:131–141. doi: 10.1016/S0378-1097(01)00504-3. [DOI] [PubMed] [Google Scholar]
- 51.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]


