Abstract
Rapid advancement of next-generation sequencing (NGS) technologies has facilitated the search for genetic susceptibility factors that influence disease risk in the field of human genetics. In particular whole genome sequencing (WGS) has been used to obtain the most comprehensive genetic variation of an individual and perform detailed evaluation of all genetic variation. To this end, sophisticated methods to accurately call high-quality variants and genotypes simultaneously on a cohort of individuals from raw sequence data are required. On chromosome 22 of 818 WGS data from the Alzheimer's Disease Neuroimaging Initiative (ADNI), which is the largest WGS related to a single disease, we compared two multi-sample variant calling methods for the detection of single nucleotide variants (SNVs) and short insertions and deletions (indels) in WGS: (1) reduce the analysis-ready reads (BAM) file to a manageable size by keeping only essential information for variant calling (“REDUCE”) and (2) call variants individually on each sample and then perform a joint genotyping analysis of the variant files produced for all samples in a cohort (“JOINT”). JOINT identified 515,210 SNVs and 60,042 indels, while REDUCE identified 358,303 SNVs and 52,855 indels. JOINT identified many more SNVs and indels compared to REDUCE. Both methods had concordance rate of 99.60% for SNVs and 99.06% for indels. For SNVs, evaluation with HumanOmni 2.5M genotyping arrays revealed a concordance rate of 99.68% for JOINT and 99.50% for REDUCE. REDUCE needed more computational time and memory compared to JOINT. Our findings indicate that the multi-sample variant calling method using the JOINT process is a promising strategy for the variant detection, which should facilitate our understanding of the underlying pathogenesis of human diseases.
Introduction
Recent large-scale genome-wide association studies (GWAS) have identified and confirmed many susceptibility genes associated with human diseases and traits.1-3 However, only a small portion of their heritability is accounted for by all of the known susceptibility genes leaving a substantial proportion of the heritability remaining to be identified.4, 5 Next-generation sequencing (NGS) may enable discovery of novel genetic underpinnings that account for some of the missing heritability.6, 7 Rapid advancement of next-generation sequencing (NGS) technologies has facilitated the search for genetic susceptibility factors that influence disease risk and become a key technique for detecting pathogenic variants in human diseases.8, 9 Several sequencing-based association studies could identify functional risk variants with large effects on human disease pathogenesis within genes.10 Accumulating evidence shows that common and rare risk variants are likely to co-exist at the same locus (known as pleomorphic risk loci).11
In particular, whole-genome sequencing (WGS) has been used to obtain the most comprehensive genetic variation of an individual and perform detailed evaluation of all genetic variation.12 To this end, sophisticated methods to accurately call high-quality variants and genotypes simultaneously on a cohort of individuals from raw sequence data are required. Therefore, numerous methods have been proposed for high-throughput short read alignment and variant calling.13 Still highly accurate variant calling is one of the most important challenges. The reduction in the cost of sequencing a human genome has led make possible to sequence many samples completely. As multi-sample variant callings can use additional information from multiple samples at a single site, multi-sample variant callings are thought to have advantages compared to single-sample variant calling.14 However, the file size is a major roadblock for data analysis scalability, and multi-sample variant callings can require considerable computing time and resources. Therefore multi-sample variant calling methods are under active development.
Here we compared two multi-sample variant calling methods for the detection of single nucleotide variants (SNVs) and short insertions and deletions (indels) in WGS on chromosome 22 of 818 WGS data from the Alzheimer's Disease Neuroimaging Initiative (ADNI). The first type of multi-sample variant caller is to reduce the analysis-ready reads (BAM) file to a manageable size by keeping only essential information for variant calling that allows greater performance and scalability for multi-sample variant callers. The second type of multi-sample variant caller is to first call variants individually on each sample to produce a comprehensive record of genotype likelihoods and annotations for each site in the genome and then perform a joint genotyping analysis of the variant files produced for all samples in a cohort (www.broadinstitute.org/gatk/).
Materials and Methods
Subjects
All individuals used in this report were participants of the Alzheimer's Disease Neuroimaging Initiative Phase 1 (ADNI-1) and/or its subsequent extension (ADNI-GO/2). The initial phase (ADNI-1) was launched in 2003 to test whether serial magnetic resonance imaging (MRI), position emission tomography (PET), other biological markers, and clinical and neuropsychological assessment could be combined to measure the progression of MCI and early AD. The ADNI-1 participants were recruited from 59 sites across the U.S. and Canada and include approximately 200 cognitively normal older individuals (healthy controls (HC)), 400 patients diagnosed with MCI, and 200 patients diagnosed with early probable AD aged 55-90 years. ADNI-1 has been extended to its subsequent phases (ADNI-GO and ADNI-2) for follow-up for existing participants and additional new enrollments. Inclusion and exclusion criteria, clinical and neuroimaging protocols, and other information about ADNI have been published previously and can be found at www.adni-info.org. Demographic information, raw scan data, APOE and whole genome sequencing data, neuropsychological test scores, and diagnostic information are available from the ADNI data repository (http://www.loni.usc.edu/ADNI/). Written informed consent was obtained at the time of enrollment for imaging and genetic sample collection and protocols of consent forms were approved by each participating sites’ Institutional Review Board (IRB).
Whole genome sequencing (WGS) analysis
WGS was performed on blood-derived genomic DNA samples obtained from 818 ADNI participants. Samples were sequenced on the Illumina HiSeq2000 using paired-end read chemistry and read lengths of 100bp (www.illumina.com). The resulting Illumina qseq files were converted into fastq files, a text-based format for storing both sequence reads and their corresponding quality information in Phred format. Short-read sequences were mapped to the NCBI reference human genome (build 37) using BWA, allowing for up to two mismatches in each read. During the alignment, we use only bases with Phred Quality > 15 in each read to include soft clipping of low-quality bases, retain only uniquely mapped pair-end reads, and remove potential PCR duplicates. After completing initial alignment, the alignment is further refined by locally realigning any suspicious reads. The reported base calling quality scores obtained from the sequencer are re-calibrated to account for covariates of base errors such as sequencing technology and machine cycle. Finally, the realigned reads are written to a BAM file for further analysis (see Figure 1). Variant Discovery: The analysis-ready BAM files are analyzed to identify all variants with statistical evidence for an alternate allele present among samples using the HaplotypeCaller module of GATK for multi-sample variant callings. The first type of multi-sample variant caller is to reduce the analysis-ready reads (BAM) file to a manageable size by keeping only essential information for variant calling that allows greater performance and scalability for multi-sample variant callers (“REDUCE”). The second type of multi-sample variant caller is to first call variants individually on each sample to produce a comprehensive record of genotype likelihoods and annotations for each site in the genome and then perform a joint genotyping analysis of the variant files produced for all samples in a cohort (“JOINT”). The HaplotypeCaller module of GATK calls SNVs and indels simultaneously via local de-novo assembly of haplotypes in an active region. The quality of the variant calls was assessed by comparing sequencing-derived SNVs with those obtained from the Illumina Omni 2.5M genotyping array in order to estimate the concordance rate. Among 818 subjects, two subjects had concordance rates less than 99% and had been removed from our analysis.
Fig 1. Whole Genome Sequencing Analysis Pipeline.

Results
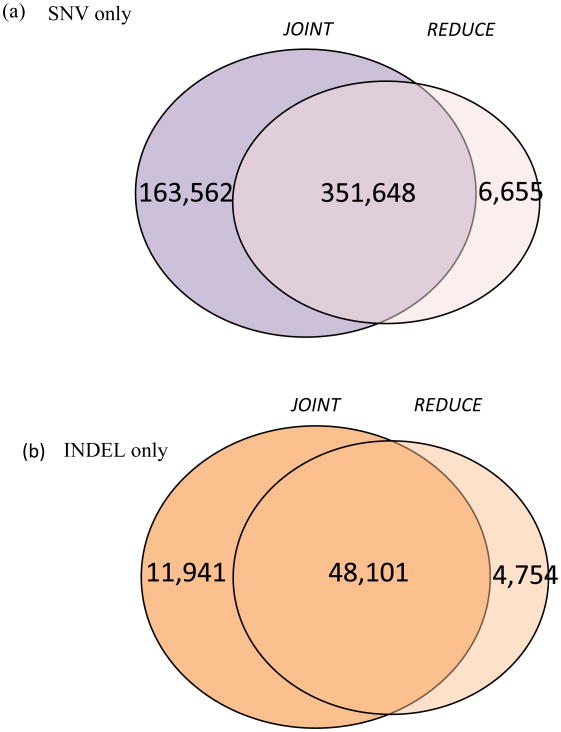
We used a same pre-calling procedure and two different multi-sample variant calling methods to identify SNVs and indels from 818 ADNI WGS data. First we compared the numbers of SNVs and indels across two multi-sample variant callers. Figure 2 and Table 1 summarized the distribution of the number of SNVs and indels identified using two different callers.
Fig 2. Variants (SNVs and indels) identified on chromosome 22 of 816 genomes by two multi-sample variant calling methods.
Table 1. Numbers of identified SNVs and indels on chromosome 22 of 816 genomes.
| SNV only | INDEL only | |||
|---|---|---|---|---|
| JOINT | REDUCE | JOINT | REDUCE | |
| Exonic | 8,594 | 5,458 | 184 | 177 |
| Intergenic | 233,991 | 164,549 | 27,170 | 23,430 |
| Intronic | 226,289 | 156,430 | 27,156 | 24,195 |
| Splicing | 57 | 35 | 8 | 8 |
| UTR 3′ | 7,984 | 5,508 | 944 | 893 |
| UTR 5′ | 1,834 | 1,142 | 167 | 156 |
The final variant file (VCF) indicated that the mean depth of mapped unique reads (after removing reads with more than two mismatches in each read) at all identified variants on chromosome 22 are 24.6X for JOINT. JOINT identified 515,210 SNVs and 60,042 indels, while REDUCE identified 358,303 SNVs and 52,853 indels. For the JOINT SNVs, 8,594 exonic SNVs, of which 4,650 SNVs (54.1%) are non-synonymous, were found in the protein-coding regions. For the REDUCE SNVs, 5,458 SNVs, of which 2,908 SNVs (53.3%) are non-synonymous, were found in the protein-coding regions. JOINT increased the proportion of called variants, i.e., identified 43% and 14% more SNVs and indels compared to REDUCE. 98.1% (351,648 SNVs) and 91.0% (48,101 indels) of the REDUCE SNV and indel calls, respectively, are also present in the JOINT set. The concordance ratios of the common SNVs and indels from two caller methods are 99.60% and 99.06%, respectively. The observed transition-to-transversion ratios for the SNV sets on chromosome 22 for JOINT and REDUCE are 2.39 and 2.36, respectively. In order to assess the quality of the variant calls, we compared sequencing-derived SNVs with those obtained from the Illumina Omni 2.5M genotyping array and overall genotype consistency rates are 99.7% for the JOINT SNV set and 99.5% for the REDUCE SNV set.
Discussion
Our understanding of the association of the genetic variation with human disease has been greatly advanced using high-throughput NGS technologies. NGS has become a powerful tool for explaining the missing heritability of human diseases through rare and de novo variants. One of the most important challenges in NGS analysis is to accurately call high-quality variants (SNVs and indels) and genotypes simultaneously on a cohort of individuals from raw sequence data and is still under an active research topic. Multi-sample variant callings have been shown to have more advantages than the corresponding single-sample variant callings. However, under current computing resources, it is not possible to call multi-sample variants using all mapped reads simultaneously from 818 WGS. Here we compared two multi-sample variant calling methods for SNVs and indels on chromosome 22 of 818 WGS data from ADNI, which is the largest WGS related to a single disease.
The JOINT method identified much more SNVs and indels, and required considerably less computation time and resources. The JOINT method identified 43% more SNVs, although the JOINT method identified 14% more indels. In particular, 98.1% and 91.0% of SNVs and indels identified by the REDUCE method were also called by the JOINT method with more than 99% concordance. Both methods showed very high concordance with both each other and the Illumina Omni 2.5M genotyping array. The concordance analysis indicated that the JOINT method performed considerably better than the REDUCE method.
In conclusion, our data indicate that the multi-sample variant calling method to first call variants individually on each sample in order to produce a comprehensive record of genotype likelihoods and annotations for each site in the genome and then perform a joint genotyping analysis of the variant files produced for all samples in a cohort is a promising strategy for the variant detection. As the development of multi-sample variant calling methods is a rapidly evolving target, these methods will require frequent re-evaluation for further improvement.
Acknowledgments
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer's Association; Alzheimer's Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Samples from the National Cell Repository for AD (NCRAD), which receives government support under a cooperative agreement grant (U24 AG21886) awarded by the National Institute on Aging (AIG), were used in this study. Additional support for data analysis was provided by NLM R00 LM011384-02, NIA R01 AG19771, and NIA P30 AG10133. This research was supported in part by Lilly Endowment, Inc., through its support for the Indiana University Pervasive Technology Institute, and in part by the Indiana METACyt Initiative. The Indiana METACyt Initiative at IU is also supported in part by Lilly Endowment, Inc. This material is based upon work supported by the National Science Foundation under Grant No. CNS-0521433.
References
- 1.Naj AC, Jun G, Beecham GW, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nature genetics. 2011 May;43(5):436–41. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stein JL, Medland SE, Vasquez AA, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nature genetics. 2012 May;44(5):552–61. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rietveld CA, Medland SE, Derringer J, et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013 Jun 21;340(6139):1467–71. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eichler EE, Flint J, Gibson G, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nature reviews Genetics. 2010 Jun;11(6):446–50. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. American journal of human genetics. 2011 Mar 11;88(3):294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guerreiro R, Wojtas A, Bras J, et al. TREM2 variants in Alzheimer's disease. The New England journal of medicine. 2013 Jan 10;368(2):117–27. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nho K, Corneveaux JJ, Kim S, et al. Whole-exome sequencing and imaging genetics identify functional variants for rate of change in hippocampal volume in mild cognitive impairment. Molecular psychiatry. 2013 Jul;18(7):781–7. doi: 10.1038/mp.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Metzker ML. Sequencing technologies - the next generation. Nature reviews Genetics. 2010 Jan;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 9.Goldstein DB, Allen A, Keebler J, et al. Sequencing studies in human genetics: design and interpretation. Nature reviews Genetics. 2013 Jul;14(7):460–70. doi: 10.1038/nrg3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruchaga C, Karch CM, Jin SC, et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer's disease. Nature. 2014 Jan 23;505(7484):550–4. doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singleton A, Hardy J. A generalizable hypothesis for the genetic architecture of disease: pleomorphic risk loci. Human molecular genetics. 2011 Oct 15;20(R2):R158–62. doi: 10.1093/hmg/ddr358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dewey FE, Grove ME, Pan C, et al. Clinical interpretation and implications of whole-genome sequencing. JAMA : the journal of the American Medical Association. 2014 Mar 12;311(10):1035–45. doi: 10.1001/jama.2014.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011 May;43(5):491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Han S, Wang Z, Gelernter J, Yang BZ. Variant callers for next-generation sequencing data: a comparison study. PloS one. 2013;8(9):e75619. doi: 10.1371/journal.pone.0075619. [DOI] [PMC free article] [PubMed] [Google Scholar]