Abstract
Usher syndrome type III is an autosomal recessive disorder characterized by progressive sensorineural hearing loss, vestibular dysfunction, and retinitis pigmentosa. The disease gene was localized to 3q25 and recently was identified by positional cloning. In the present study, we have revised the structure of the USH3 gene, including a new translation start site, 5′ untranslated region, and a transcript encoding a 232–amino acid protein. The mature form of the protein is predicted to contain three transmembrane domains and 204 residues. We have found four new disease-causing mutations, including one that appears to be relatively common in the Ashkenazi Jewish population. We have also identified mouse (chromosome 3) and rat (chromosome 2) orthologues, as well as two human paralogues on chromosomes 4 and 10.
Usher syndrome type III (USH3 [MIM #276902]) is unique among the clinical subtypes of Usher syndrome, in that it shows postlingual, progressive hearing loss and late onset of retinitis pigmentosa (RP), as well as a progressive loss of vestibular function (Kimberling et al. 2000). The disease locus was originally mapped to chromosome 3q25, between the markers WI-17533 and 486D12SP6, a region of ∼700 kb (Joensuu et al. 1996). In a recent publication (Joensuu et al. 2000), the USH3 locus was assigned to a region of 250 kb between 107G19CA7 and D3S3625, by means of haplotype and linkage disequilibrium analyses in Finnish carriers of the putative founder mutation.
A region near—but outside of—this partially sequenced (GenBank accession number AF388363) 250-kb critical region was subsequently reported to contain USH3A (GenBank accession number AF388366), on the basis of mutational analysis (Joensuu et al. 2001). This gene, which we refer to as “USH3Joensuu,” spanned 20,776 nucleotides and contained four exons that appeared to encode three differentially spliced mRNA species of 4.5, 1.5, and 1.0 kb. All three mRNA species were reported to code for a predicted protein of 120 amino acid residues, with two transmembrane (TM) domains and no homology to any known protein. Three mutations were reported: a nonsense mutation in exon 3 (Finnmajor); a missense mutation in exon 2 (Finnminor); and a 3-bp deletion in exon 3 in a large consanguineous Italian family (Joensuu et al. 2001). No mutations were detected in the first and fourth exons.
To corroborate the identity of the gene implicated in USH3, we analyzed the USH3Joensuu gene of 32 unrelated probands selected from a set of >1,300 families with Usher syndrome, of various genetic backgrounds, from Sweden and the United States. Fourteen families with the USH3 phenotype were compatible with linkage to 3q25 markers (data not shown); the remaining families were uninformative for linkage and were selected on the basis of clinical phenotype alone. The clinical diagnosis of USH3 was supported by audiometric measurement, a history of progressive hearing loss, and retinitis pigmentosa confirmed by electroretinography and funduscopic examination (data not shown).
The PCR primer pairs suggested by Joensuu et al. (2001) were constructed to amplify the four exons of USH3Joensuu (GenBank accession number AF388366) and exon 1b of isoform bJoensuu (GenBank accession number AF388368), using genomic DNA as template. The amplified products were screened for the presence of disease-causing mutations by heteroduplex analysis using denaturing high-performance liquid chromatography (DHPLC) on a Wave 2100 System (Transgenomic), as well as by direct sequencing, and the results are shown in table 1. Any mutations observed were confirmed by a second method, including restriction digest if possible, and by segregation with affected status within families. In three families of Finnish ancestry and in two families of Swedish ancestry, direct sequencing revealed the homozygous Finnmajor mutation. One patient of Scottish-Irish ancestry, residing in the United States, was found to have one copy of the Finnmajor mutation.
Table 1.
Summary of Mutations and SNPs in Disequilibrium (by Family)
| Family | Ancestry (Proband'sCountry of Origin) | First PathologicalMutation | SecondPathologicalMutation | SNPHaplotypea | Linkage toChromosome 3b |
| 486 | Finnish (Sweden) | 528T→G | 528T→G | A-T/A-T | Compatible |
| 540 | Finnish (Sweden) | 528T→G | 528T→G | A-T/A-T | NI |
| 635 | Scottish-Irish (United States) | 149delCAGG/insTGTCCAAT | 528T→G | G-A/A-T |
Compatible |
| 638 | Dutch (United States) | 165delC | Not found | A-A/G-A |
Compatible |
| 803 | Jewish (United States) | 144T→G | 144T→G | A-A/A-A | NI |
| 805 | Swedish (Sweden) | 528T→G | 528T→G | A-T/A-T | Compatible |
| 885 | Jewish (United States) | 144T→G | 144T→G | A-A/A-A | NI |
| 947 | Jewish (United States) | 144T→G | 449T→C | A-A/A-A |
NI |
| 1053 | Jewish (United States) | 144T→G | 144T→G | A-A/A-A | Compatible |
| 1059 | Finnish (Sweden) | 528T→G | 528T→G | A-T/A-T | Compatible |
| 2417 | Jewish (United States) | 144T→G | 144T→G | A-A/A-A | NI |
| 2539 | Jewish (United States) | 144T→G | 144T→G | A-A/A-A | NI |
| 2714 | Swedish (Sweden) | 528T→G | 528T→G | A-T/A-T | NI |
| 3087 | Other northern European(United States) | 149delCAGG/insTGTCCAAT | 149delCAGG/insTGTCCAAT | G-A/G-A | NI |
In those families in which segregation can be determined, −71A→G and 57A→T link with pathological mutations. SNP haplotypes are listed in gene orientation order: −71A→G, then 57A→T, with a slash (/) dividing the alleles. The −71A→G and 57A→T SNPs appeared to be nonrandomly associated with the two most common USH3 mutations. The [−71A]-[57A] was always found in cis with the Jewish 144T→G, and we always found [−71A]-[57T] in cis with the Finnish 528T→G. In addition, two of the SNPs, 1012T→C and 1069T→C, occurred solely in cis orientation, with 1012T-1069T and 1012C-1069C the only alleles observed. Underlined alleles have been confirmed to be in cis phase with pathological mutations in these families.
NI = not informative; compatible = compatible with linkage to 3q25-26.
Several of our patients whose clinical and/or linkage data made them very likely to have USH3 did not exhibit mutations in the USH3Joensuu gene-coding region. We therefore suspected that the USH3 gene might be incompletely characterized. We first searched for putative missing exons, by performing PCR using human retina Marathon Ready cDNA (Clontech) and performing RT-PCR using mRNA obtained from a Y79 retinoblastoma cell line (American Type Culture Collection [ATCC] #HTB-18) and total RNA from human retina.
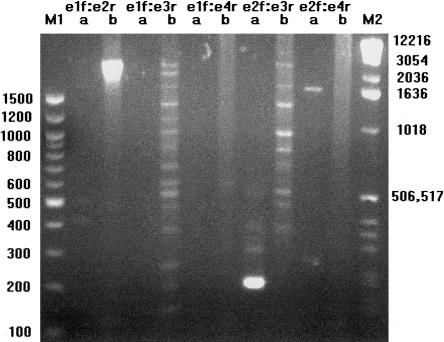
On the basis of the USH3Joensuu sequence, we designed primer pairs to amplify products between the first and second (e1f:e2r), first and third (e1f:e3r), first and fourth (e1f:e4r), second and third (e2f:e3r), and second and fourth (e2f:e4r) published exons. Only the primer pair designed to amplify across the splice between the second and third exons showed the expected product as a strong band; other combinations of the primers produced extremely faint bands (fig. 1), even when we used two times the recommended concentration of Marathon Ready cDNA as template for the reactions, as shown in figure 1. These same primer pairs yielded no detectable products of the expected size, with the exception of e2f:e3r, when used for RT-PCR on poly(A) RNA from Y79 retinoblastoma cells or for RT-PCR on total RNA from human retina (data not shown). Since these PCR and RT-PCR experiments could not convincingly connect the first exon to the second, third, or fourth exons, or the second exon to the fourth exon, we concluded that the first and fourth exons of USH3Joensuu were not part of the primary USH3 transcript.
Figure 1.
PCR analysis of the USH3Joensuu transcript. Primers based on the published sequence of USH3Joensuu were used to amplify DNA fragments between the first and second (e1f:e2r), first and third (e1f:e3r), first and fourth (e1f:e4r), second and third (e2f:e3r), and second and fourth (e2f:e4r) exons, with 35 cycles of 30 s at 96°C, 30 s at 58°C, and 4 min at 72°C. Marathon Ready human retina cDNA at two times the recommended concentration from Clontech (lane a) and human genomic DNA at 5 ng/μL (lane b) were used as template. Only the primer pair e2f:e3r produced a robust product of the expected size on retina-specific cDNA. Faint bands are visible in the cDNA amplifications for e1f:e2r and e1f:e4r, at approximately the expected sizes but at orders of magnitude below the intensity of e2f:e3r. Expected sizes for USH3Joensuu are 412, 574, 780, 186, and 392 bp for lane a, with an additional 87 nucleotides between exons 1 and 2 in the case of isoform bJoensuu. Evidence for an additional splice variant was observed in lane a, for e2f:e4r, and reamplification followed by sequencing showed that the band at about 165 bp represents a direct splice between exons 2 and 4, skipping exon 3.
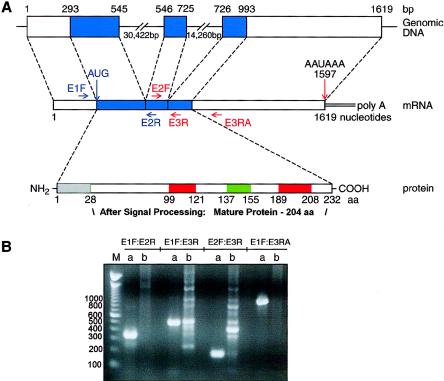
To search for the missing portions of the primary transcript, we performed 5′ and 3′ rapid amplification of cDNA ends (RACE) reactions (Clontech protocols) using primers based on the second and third exons. We selected, cloned, and sequenced a 5′ RACE product (789 bp) and a 3′ RACE product (1,080 bp) and assembled the overlapping cDNA fragments into a 1,642-bp cDNA contig (fig. 2). This contig contained the published second and third exons, with novel sequences to the 5′ and 3′ of these two exons. We aligned the 1,642-bp-long sequence with that of BAC clone RP11-251C9 (GenBank accession number AC020636) and used this alignment to identify intron/exon junctions, which, in this case, adhered to the AG/GT rule for predicted splice signals. We then verified the exon/intron boundaries by sequencing PCR products from genomic DNA, and we concluded that the transcript contained three exons. We then designed new PCR primer pairs for cDNA amplification, through use of the USH3revised sequence (table 2).
Figure 2.
Structure of the USH3revised gene (AF482697) and protein. A, Schematic structure of the USH3revised gene and its mRNA and protein products. The TMPred program predicted two inside-outside (green) and two outside-inside (red) TM domains, arranged in tandems in the USH3 protein (232 aa). There may also be a signal peptide (SignalP Server prediction; data not shown), which, if removed, could result in a mature protein with three TM domains, 204 aa in length. This signal peptide is grey in this figure, including the first inside-outside TM domain. No other previously characterized conserved domains were identified. B, PCR analysis of the transcript, performed using primers based on the sequence of USH3revised cDNA. The expected sizes of the amplified products were E1F/E2R, 337 bp; E1F/E3R, 499 bp; E2F/E3R, 187 bp; and E1F/ER3, 912 bp. M = molecular weight marker (Invitrogen catalogue number 15628019).
Table 2.
Primers (Listed 5′→3′) Used to Screen USH3 Gene for Mutations
| Primer Pair | Sequence | Length(bp) |
| 5′ UTR-Exl-U1 | CTCCTGCATTTTCATATTTCTGTA | |
| 5′ UTR-Exl-L1 | CTGCCTTCAAGTATCTCCTCTGT | 485 |
| Exl-5′-U1 | AGACAAAAGGCTGAGGAAGG | |
| Exl-5′-L1 | CCCGTTTTGCAGAGGACAGT | 507 |
| Exl-3′-U1 | CCGTCGATGGTGAAGTTG | |
| Exl-3′-L1 | CTGGGAAGAGTCTGCCTAAAG | 447 |
| Exl-3′-U3 | TGCCAAGCCAACAGAAGAAAATCA | |
| Exl-3′-L3 | TCCCAACCCACACTGCCTCAC | 230 |
| Exl-IVS1-U3 | GGCAGTCCCTTCCCATTG | |
| Exl-IVS1-U3 | TAAAAAGTCCTGCAGTAAACACG | 430 |
| Exl-IVS1-U4 | GGAGAGGGTGTGAGGCAGTGT | |
| Exl-IVS1-U4 | GCAGGAATAATGGGAGGGAGTG | 508 |
| Exl-IVS1-U5 | TTTGAGAATTTTGCCGTGTTTAC | |
| Exl-IVS1-U5 | CATCCATTTCTTTCCCAGTTAGC | 508 |
| IVS1-Ex2-U1 | AATAGATTTGGCGTGTTT | |
| IVS1-Ex2-L1 | TAGGGTTAGAAGAAGTTT | 575 |
| IVS1-Ex2-U3 | ACCCTAGTTTTGTCTTATCT | |
| IVS1-Ex2-L3 | AGCGTTTATCCTCTTGA | 465 |
| P2F2a | TCCCAGTGAGCATCCACGTC | |
| P2R2a | TGAAAAGCACATTTGTCTTCAGAGG | 198 |
| IVS2-Ex3-U1 | GCCTAGCAATTCAGCCTTCAC | |
| IVS2-Ex3-L1 | TTTTCACTTTGCGTTTTGTAGAC | 446 |
| IVS2-Ex3-U2 | CTCCTGTGGCTGTCTTGTCA | |
| IVS2-Ex3-L2 | CTTTCCAGCCTGTATCCTTAGTA | 424 |
| P3Fa | ATGTCAATGGGGATGATGGT | |
| P3Ra | GGAGCCCATTCAGAAAATGA | 291 |
| Ex3-ORF-U1 | TTATGTCTACAAAACGCAAAGT | |
| Ex3-ORF-L1 | TTCCCACCAGATAAAACAA | 383 |
| Ex3-3′ UTR-U1b | CAGGACCCTTCGTGACAATG | |
| Ex3-3′ UTR-L1 | CACGCCTGGCCTAAGAGTAT | 419 |
| Ex3-3′ UTR-U2 | GCTGCAATCGCTTTCCTA | |
| Ex3-3′ UTR-L2 | GATTCCTCAGTGGTCCTAACA | 424 |
Designed by Joensuu et al. (2001).
Fragment also contains a VNTR.
We prepared cDNA from Y79 mRNA and amplified the USH3revised cDNA to verify the composite cDNA sequence derived from RT-PCR and RACE assays. In parallel with the Y79 cDNA, we used human retina Marathon Ready cDNA at suggested concentrations as a template in PCRs. The resulting PCR products were of the predicted size and amplified consistently across the predicted transcript (fig. 2). The sequence of the PCR products was identical to the USH3revised cDNA (GenBank accession number AF482697). In addition, we were able to amplify a single product, 1,376 bp in length, containing the entire coding region of USH3revised from human retina Marathon Ready cDNA. Through use of the same primer pair for RT-PCR, we amplified a fragment of the same size from mRNA extracted from the Epstein-Barr virus–transformed lymphoblasts of an unaffected control individual. Direct sequencing confirmed the presence of all of the coding region and portions of the UTRs in the same message.
The 1,642-bp USH3revised cDNA is 198 bp longer than that of USH3Joensuu, with an ORF of 699 bp (vs. 363 bp for USH3Joensuu). The USH3revised gene has a first exon of 545 bp, with an ORF of 252 nucleotides (vs. 24 bp in exon 1 of USH3Joensuu). The second exon (180 bp) is common to all transcripts so far described. The USH3revised gene also has a longer third exon of 576 bp, with 267 bp in the continued ORF (USH3Joensuu shares 135 bp of that ORF in its exon 3, with an additional 22 bp in the continued ORF in its exon 4). No portion of exon1, exon 1b, or exon 4 of USH3Joensuu is present in the USH3revised cDNA. The predicted peptides of USH3revised (233 amino acids) and USH3Joensuu (120 amino acids) share 104 amino acids—namely, 9–112 of the USH3Joensuu peptide.
Reamplification (35 cycles) of aliquots of the products shown in figure 1, which had already been amplified (35 cycles) from twice the recommended concentrations of Marathon Ready cDNA, was necessary to obtain sufficient template for sequencing reactions to confirm the presence of exons 1 and 4 of the USH3Joensuu transcript. They can be present only in a very few transcripts, perhaps 1/100 as frequent as USH3revised. We also were able to confirm that, in some cases, exon 1b is spliced between exons 1 and 2 of USH3Joensuu. In addition, transcripts were observed where exons 2 and 4 of USH3Joensuu were directly spliced, without exon 3, at perhaps half the frequency of the USH3Joensuu transcripts. These results suggest that the USH3Joensuu transcripts are real but minor components that are barely detectable when our methods are used. Technological differences may be responsible for the different results between the two studies.
To further validate the predicted structure and sequence of the USH3revised cDNA, we searched for its orthologues in rodents. RT-PCR was performed on mRNA obtained from rat retina through use of the PCR primer pairs E1F/E2R and E2F/E3R, and the products were cloned and sequenced; two clones showed homology to the USH3 revised cDNA sequence. We then extended the 0.5-kb sequence by 3′ RACE, using primer E3F and rat retina mRNA as the template; cloned the products; and sequenced them. The resultant rat cDNA contig of 1,075 bp contains the entire coding region but lacks some of the 5′ and 3′ UTRs. The rat cDNA (GenBank accession number AF482698) has a high degree of homology to the human sequence, and the rat gene has an intron/exon structure identical to that of the human gene. Both human and rat cDNA probes identify a message of ∼1.6 kb in northern blotting (data not shown) within species.
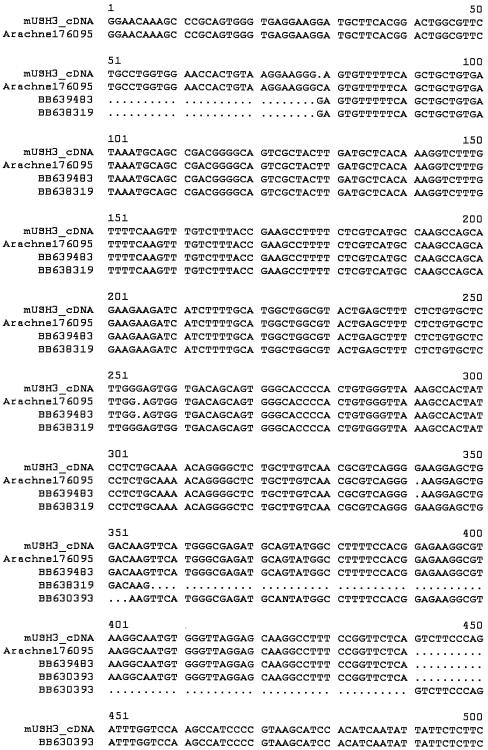
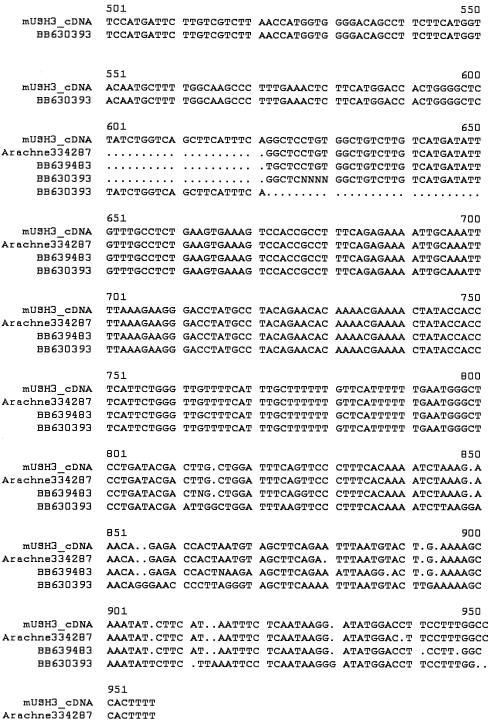
Homology searches, using the USH3revised predicted peptide sequence against the translated mouse EST database and Arachne Whole Genome Shotgun assembly (see the Blast the Mouse Genome, Genome Survey Sequences Database, High-Throughput Genomic Sequences, MouseBLAST, NCBI BLAST, NCBI Expressed Sequence Tags Database, and UCSC Human Genome Working Draft Web sites), identified three overlapping ESTs (GenBank accession numbers AF630393, BB638319, and BB639483) and two genomic segments (Ensembl-Arachne contigs 176095 and 334287) that were used to form a cDNA contig of 955 bp (fig. 3). Because this contig is a derived sequence and has not yet been experimentally verified, it has no unique accession number.
Figure 3.
Alignment of mouse EST and Ensembl-Arachne data used to predict the mouse USH3revised orthologue. The original alignment was accomplished using DNASTAR's Seqman program in the Lasergene version 5.0 software package, and formatting was provided by Multalin (Corpet et al. 1988), with default settings and manual adjustment.
Using radiation hybrid panels from Invitrogen, we mapped the rat and mouse USH3 genes to rat chromosome 2, between d2mit9 (proximal) and d2mgh15 (distal), and to mouse chromosome 3, between D3Mit173 (proximal) and D3Mit228 (distal) (data not shown); these regions are orthologous (Virtual Comparative Mapping Web site) to the human 3q region containing the USH3 gene.
In the longest ORF of the human USH3revised cDNA, the translation initiator ATG occurs at nucleotide position 292, with the TGA translation termination codon at position 988. This codes for a predicted peptide of 232 amino acid residues. TMPred predicted four TM domains (fig. 2): inside-out, outside-in, inside-out, and outside-in; however, the first TM domain might be lost with a signal peptide (Nielsen et al. 1997; SignalP Server prediction), leaving the amino terminus outside and the carboxy terminus inside the cell and resulting in a mature protein of 204 amino acid residues with a calculated molecular mass of 22.7 kD without other modifications. The rat and mouse peptide sequences are ∼88% and ∼84% identical to the human sequence, respectively; all three orthologous cDNAs code for a peptide of 232 amino acid residues, with a predicted 204 amino acids in the mature protein and three TM domains in the same positions (Nielsen et al. 1997; SignalP Server; TMPred Web site).
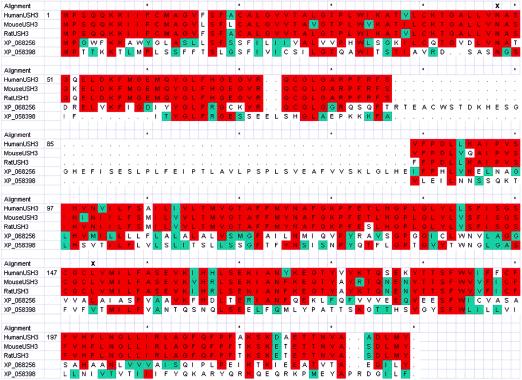
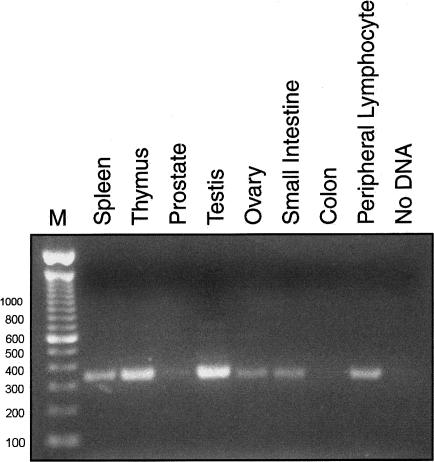
In addition, the protein predicted by the revised USH3 showed paralogy to two hypothetical proteins, with GenBank accession numbers XP_058398 and XP_068256, which also are likely to have three TM domains in their mature form (fig. 4; XP_068256 has an extra exon that we have not verified). The USH3revised mRNA was expressed in various human tissues, when probed by RT-PCR (fig. 5) and northern hybridization (data not shown), and there were weaker signals at ∼4.4 and ∼7.6 kb in northern blotting (data not shown). These additional bands were not detectable by RT-PCR and so may be from USH3 paralogues. We did not detect the USH3revised mRNA in rat retinal pigment epithelium–derived cells (RPE-J; available as ATCC #CRL-2240).
Figure 4.
Alignment of human, mouse, and rat USH3revised proteins and two human USH3revised protein paralogues. “X” signifies sites of missense mutations found in the present study; a red background indicates identity of 3/5 residues at that position; blue indicates similarity of 3/5. Alignment of a putative USH3-like gene from human chromosomes 4 and 10 and the putative USH3 orthologues from rat and mouse show that Leu150 can be replaced by isoleucine but is otherwise conserved. Secondary-structure predictions indicate that the L150P mutation would disrupt a helical region (DNAStar's Protean program in the Lasergene version 5.0 software package; data not shown). The alignment also shows that Asn48 is conserved, except in the putative USH3-like gene on human chromosome 4, where it is replaced by an aspartate. Lysine, with its longer alkyl chain and lack of a carboxyl moiety, would be a radical substitution (N48K) in this region, which is predicted to form an extended loop. The original alignment was accomplished using DNAStar's Megalign program in the Lasergene version 5.0 software package; formatting was provided by The Sequence Manipulation Suite: Multiple Align Show.
Figure 5.
Expression profile of USH3revised. Single-stranded cDNA from the indicated tissues—with primer pair E1F:E2R, as in figure 2—was used to amplify a 337-bp product corresponding to the first and second exons of USH3revised. The experiment was performed three times for the Clontech MTC panel 2, and one representative result is shown. M = molecular weight marker (Invitrogen catalogue number 15628019).
We screened for mutations in the coding region and exon/intron boundaries of the USH3revised gene of 32 unrelated patients clinically diagnosed with USH3, by PCR amplification of their genomic DNA followed by either heteroduplex analysis or direct sequencing. Heteroduplex analyses were performed using either PAGE or denaturing high performance liquid chromatography (WAVE DNA Fragment Analysis System).
Four novel putative disease-causing mutations were identified (table 3). Seven variants were also detected that were not considered causative for USH3 syndrome, by virtue of their frequency in control individuals, position within the gene, and type of mutation (table 4). Four of these noncausative mutations are SNPs, whereas the fifth is a compound variable dinucleotide repeat.
Table 3.
Summary of Disease-Causing Mutations Identified in the USH3 Gene[Note]
| Mutationa | Exon | Effect on CodingSequence (233aa) | No. of Alleles(of 28 Total)Detected inPresent Study | Primer Usedto Detect | Apparent Ancestry |
| Nonsense: | |||||
| 528T→Gb | 3 | Tyr→stop in codon 176 | 11 | P3F/P3Rc | Finnish/other northern European |
| Deletion or Insertion: | |||||
| *149delCAGG/insTGTCCAAT | 1 | Frameshift in codon 50, stop at codon 61 | 3 | Exl-3′-U3/L3 | Insufficient data |
| *165delC | 1 | Frameshift in codon 55, stop at codon 71 | 1 | Exl-3′-U3/L3 | Insufficient data |
| Missense: | |||||
| *144T→G | 1 | Asn→Lys in codon 48 | 11 | Exl-3′-U3/L3 | Jewish |
| 359T→Ad | Met→Lys in codon 120 | 0 | P2F2/P2R2c | Insufficient data | |
| *449T→C | 3 | Leu→Pro in codon 150 | 1 | P3F/P3R | Insufficient data |
| 459-461delATTe | 3 | Ile-Leu→Met in codons 153–154 | 0 | P3F/P3R | Insufficient data |
Note.— None of these mutations were observed in 200 control alleles from individuals unrelated to probands.
An asterisk (*) indicates a novel mutation found in the present study. Numbering begins with ATG in the longest ORF; assumes (CA)14(TA)5 in 3′ UTR repeat.
Reported by Joensuu et al. (2001) as Y100X (300T→G) in 52 Finnish probands.
Primer designed by Joensuu et al. (2001).
Reported by Joensuu et al. (2001) as M44K (131T→A) in two Finnish probands heterozygous for Y100X.
Reported by Joensuu et al. (2001) as 231-233delATT in an Italian proband.
Table 4.
Summary of Polymorphisms Identified in and near the USH3 Gene
| Name | Location | Results of Heteroduplex Analysis | Primer |
| −617A→G | 326 bp 5′ of 5′ UTR; 617 bp 5′ of codon 1 | Frequency not determined | 5′ UTR-Exl-U1/L1 |
| −71A→G | 5′ UTR | Frequency of −71A is ∼.77 | Exl-3′-U1/L1 |
| 57A→T | Exon 1 | Ala19 is unchanged; frequency of 57A is ∼.89 (57A) | Exl-3′-U1/L1 |
| IVS2-544G→A | 544 bp 5′ of Exon 3 | Frequency not determined | IVS1-Ex2-U1/L1 |
| 965-1008-VNTR1 | 3′ UTR | Nominally (CA)14(TA)5; frequency not determined2 | Ex3-3′ UTR-U1/L1 |
| 1012T→C1 | 3′ UTR | Frequency of 1012T is ∼.77 | Ex3-3′ UTR-U1/L1 |
| 1069T→C1 | 3′ UTR | Frequency of 1069T is ∼.77 | Ex3-3′ UTR-U1/L1 |
1012C and 1069C have been found in cis in 16 chromosomes, when sequenced; in all cases, 1012C+1069C was in trans to 1012T+1069T, and there were no observations of homozygous 1012C or 1069C, nor of either T→C conversion in isolation.
The CA and TA repeats are both polymorphic, making characterization difficult; however, this variable number of tandem repeats (VNTR) does not appear to be grossly expanded.
Patients from 11 families had homozygous pathological mutations, and patients from 2 families were confirmed to be compound heterozygotes. For one family, the second disease allele remained unidentified. As mentioned earlier in this report, the 528T→G mutation (300T→G or Finnmajor, in USH3Joensuu) occurred in six of the families in our study. Three of the new mutations occurred in the first exon of the USH3revised gene, which is not part of USH3Joensuu. None of these mutations—missense, deletion, or nonsense—occurred in 200 control chromosomes. The predicted effects of these mutations on the structure of the USH3revised protein, the primers used to detect them, and the apparent ancestry of the mutations are summarized in table 3.
Two of the new mutations were deletions. The Scottish-Irish patient who carried the 528T→G mutation also carried, in the alternative allele, a deletion (CAGG) at nucleotide position 149–152, with a concomitant insertion of eight nucleotides (TGTCCAAT). This same deletion/insertion event occurred as a homozygous mutation in a patient with ancestry in the United Kingdom. The second deletion, 165delC, was observed in one allele of affected members of a family with Dutch ancestry living in the United States; the other disease allele remains unidentified.
Two new missense mutations were observed in patients from six families with Ashkenazi Jewish ancestry. The substitution 144T→G (Asn48Lys) occurs in all six families, and affected members of five of these are homozygous for the mutation. The alternative disease allele of the remaining Jewish family contains the other missense mutation, 449T→C (Leu150Pro), which was unique to one patient in the present study. Both of these missense mutations occur at evolutionarily conserved residues (fig. 4).
The family with the 165delC mutation and an unidentified mutation in the other allele has been closely followed clinically and has a large pedigree with five affected members across two sibships (double first cousins), showing linkage to the USH3 region (Z>4.5). Therefore, we designed expanded primer sets to screen for their second disease-causing mutation in the intervening genomic sequence. This additional testing did not allow us to find the other diseased allele.
We identified putative disease-causing mutations in 14 (44%) of 32 patients. We cannot exclude the possibility of false diagnoses in some individuals with the USH3 phenotype, because uncommon mutations in other Usher genes can mimic the USH3 phenotype (Astuto et al. 2000; Liu et al. 1998). In addition, other USH3 genes may exist, although we have no direct evidence to support this speculation. Our data suggest that USH3Joensuu and its isoform bJoensuu are rare splice variants that occur at extremely low levels of expression, relative to USH3revised, in retina. An understanding of the relationship between these alternative forms of the USH3 message may help in clarifying the role of the USH3 gene in normal and abnormal development of the ear and eye.
Acknowledgments
We thank the patients and their families, for their cooperation; Lisa Astuto, for genotyping and clinical coordination; Michael D. Weston, for mentorship; and the physicians and staff, for providing support in obtaining samples and diagnostic records. This study was supported by National Institutes of Health grants NIH/NIDCD P01 DC01813, NIH/NIDCD 1R01 DCO3162-01A2, NIH EY-05627, EY-13385, and EY-13729; by the Foundation Fighting Blindness; and by a grant from the Morris J. and Betty Kaplun Foundation.
Footnotes
Nucleotide sequence data reported herein are available in the DDBJ/EMBL/GenBank databases; for details, see the Electronic-Database Information section of this article.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Blast the Mouse Genome, http://www.ncbi.nlm.nih.gov/genome/seq/MmBlast.html (for mouse genome BLAST versus Arachne [Whole Genome Shotgun assembly by the Whitehead Institute])
- DNASTAR, http://www.dnastar.com/ (for Lasergene 5.0 software)
- Ensembl-Arachne Mouse Contigs, http://www.ensembl.org/Mus_musculus/contigview/ (for contigs 176095 and 334287; query format is contig_######)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for 207-kb genomic contig sequence [accession number AF388363], BAC clone RP11-251C9 [accession number AC020636], USH3Joensuu cDNA [accession numbers AF388366 and AF388368], putative USH3 paralogue peptide sequences [accession numbers XP_058398 and XP_068256], USH3revised cDNA [accession number AF482697], USH3 (rat) cDNA [accession number AF482698], and assembly of putative USH3 (Mouse) cDNA [accession numbers BB630393, BB638319 and BB639483])
- Genome Survey Sequences Database, http://www.ncbi.nlm.nih.gov/dbGSS/
- High-Throughput Genomic Sequences, http://www.ncbi.nlm.nih.gov/HTGS/
- MouseBLAST, http://mouseblast.informatics.jax.org/prototype/
- Multalin, http://prodes.toulouse.inra.fr/multalin/multalin.html (for Multalin version 5.4.1)
- Multiple Align Show, Baylor College of Medicine Search Launcher, http://searchlauncher.bcm.tmc.edu/ (for MatInspector/TRANSFAC and Neural Network Promoter Input programs)
- NCBI BLAST Home Page, http://www.ncbi.nlm.nih.gov/BLAST/
- NCBI Expressed Sequence Tags Database, http://www.ncbi.nlm.nih.gov/dbEST/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for USH3 [MIM #276902])
- SignalP Server, http://www.cbs.dtu.dk/services/SignalP-2.0/ (for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites) [DOI] [PubMed]
- TMPred: Prediction of Transmembrane Regions and Orientation, http://www.ch.embnet.org/software/TMPRED_form.html
- UCSC Human Genome Project Working Draft, http://genome.cse.ucsc.edu/
- Virtual Comparative Mapping, Rat Genome Database http://rgd.mcw.edu/tools/banner_ads/ad_redirect.pl?/VCMAP/
References
- Astuto LM, Weston MD, Carney CA, Hoover DM, Cremers CW, Wagenaar M, Moller C, Smith RJ, Pieke-Dahl S, Greenberg J, Ramesar R, Jacobson SG, Ayuso C, Heckenlively JR, Tamayo M, Gorin MB, Reardon W, Kimberling WJ (2000) Genetic heterogeneity of Usher syndrome: analysis of 151 families with Usher type I. Am J Hum Genet 67:1569–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16:10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joensuu T, Blanco G, Pakarinen L, Sistonen P, Kaariainen H, Brown S, Chapelle A, Sankila EM (1996) Refined mapping of the Usher syndrome type III locus on chromosome 3, exclusion of candidate genes, and identification of the putative mouse homologous region. Genomics 38:255–263 [DOI] [PubMed] [Google Scholar]
- Joensuu T, Hamalainen R, Lehesjoki AE, de la Chapelle A, Sankila EM (2000) A sequence-ready map of the Usher syndrome type III critical region on chromosome 3q. Genomics 63:409–416 [DOI] [PubMed] [Google Scholar]
- Joensuu T, Hamalainen R, Yuan B, Johnson C, Tegelberg S, Gasparini P, Zelante L, Pirvola U, Pakarinen L, Lehesjoki AE, de la Chapelle A, Sankila EM (2001) Mutations in a novel gene with transmembrane domains underlie Usher syndrome type 3. Am J Hum Genet 69:673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberling WJ, Orten D, Pieke-Dahl S (2000) Genetic heterogeneity of Usher syndrome. Adv Otorhinolaryngol 56:11–18 [DOI] [PubMed] [Google Scholar]
- Liu XZ, Hope C, Walsh J, Newton V, Ke XM, Liang CY, Xu LR, Zhou JM, Trump D, Steel KP, Bundey S, Brown SD (1998) Mutations in the myosin VIIA gene cause a wide phenotypic spectrum, including atypical Usher syndrome. Am J Hum Genet 63:909–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Engineering 10:1–6 [DOI] [PubMed] [Google Scholar]