Abstract
Polyploidy is observed across the tree of life, yet its influence on evolution remains incompletely understood1–4. Polyploidy, usually whole genome duplication (WGD), is proposed to alter the rate of evolutionary adaptation. This could occur through complex effects on the frequency or fitness of beneficial mutations 2,5–7. For example, in diverse cell types and organisms, immediately after a WGD, newly formed polyploids missegregate chromosomes and undergo genetic instability8–13. The instability following WGDs is thought to provide adaptive mutations in microorganisms13,14 and can promote tumorigenesis in mammalian cells11,15. Polyploidy may also affect adaptation independent of beneficial mutations through ploidy-specific changes in cell physiology16. Here, we performed in vitro evolution experiments to directly test whether polyploidy can accelerate evolutionary adaptation. Compared to haploids and diploids, tetraploids underwent significantly faster adaptation. Mathematical modeling suggested that rapid adaptation of tetraploids was driven by higher rates of beneficial mutations with stronger fitness effects, which was supported by whole-genome sequencing and phenotypic analyses of evolved clones. Chromosome aneuploidy, concerted chromosome loss, and point mutations all provided large fitness gains. We identified several mutations whose beneficial effects were manifest specifically in the tetraploid strains. Together, these results provide direct quantitative evidence that in some environments polyploidy can accelerate evolutionary adaptation.
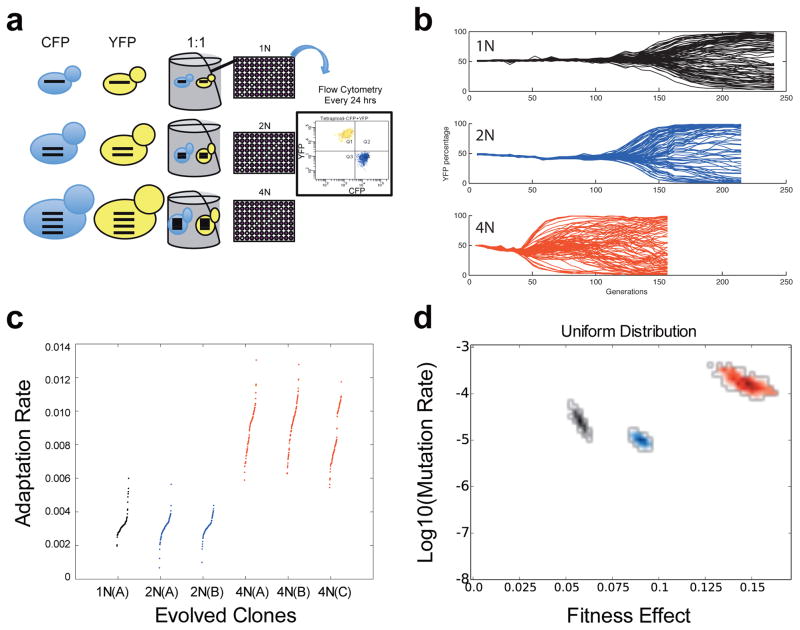
To determine how polyploidy affects the rate of adaptation, we performed hundreds of independent passaging experiments in a poor carbon source medium (raffinose, Fig. 1a), comparing isogenic haploid (1N), diploid (2N), and tetraploid (4N) strains (Extended Data Fig. 1, Extended Data Table 1). The evolution experiments were performed as competitions between equal numbers of CFP and YFP cells of the same ploidy16,17, where the acquisition and spread of beneficial mutations is visualized by divergence from a 50:50 ratio of CFP and YFP-expressing cells (Fig. 1b). The rate of adaptation was determined by measuring the change in fitness relative to the diploid ancestor over time (Methods). Over 250 generations, the tetraploids adapted at a rate that was significantly faster than haploids or diploids (Fig. 1c, t-test, p<1e-10, Methods). This faster rate of adaptation in tetraploids may be due to a higher rate of beneficial mutations, higher fitness effects of the acquired mutations, or both.
Figure 1. Rapid spread of beneficial mutations in tetraploid yeast.
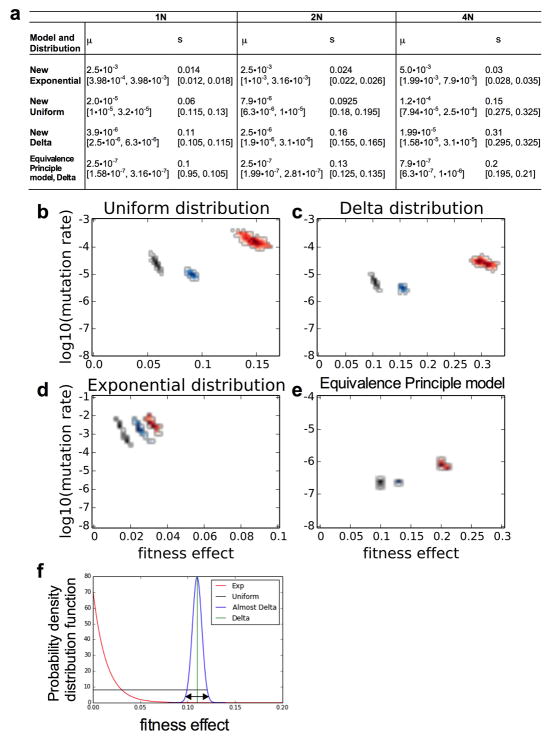
(a) Schematic of the evolution experiment. (b) Flow cytometry analysis of isogenic haploid (black), diploid (blue), and tetraploid (red) populations during adaptation to raffinose medium. Each line is the percentage of YFP cells in an independent population of YFP and CFP cells. Here and below, data from haploids is black, from diploids is blue, and from tetraploids is red. (c) The adaptation rate of the evolved clones relative to the diploid ancestor after 250 generations. Data points are the average rate of adaptation (change in fitness between generation 250 and generation zero, divided by 250 generations) of two replicate fitness measurements for the evolved clones. Clones from replicate evolution experiments (A or B) are indicated. The tetraploids acquired significantly more fitness in the same number of generations as compared to the haploids and diploids (t-test, p<1e-10). (d) Estimates from the branching evolution model of the best-fit value of the selection coefficient and beneficial mutation rate of each ploidy experiment, and their error range, determined using a uniform distribution of acquired mutations (other distributions are analyzed in Extended Data Fig. 2c–d, and the Equivalence Principle model is analyzed in Extended Data Fig. 2e). Error ranges were obtained by parametric bootstrap of 1000 independent realizations (Methods).
To gain insight into the rapid adaptation of tetraploids, we applied two complementary mathematical modeling approaches (see Methods). First, we use a model based on a branching evolutionary process17, designed to closely mimic the divergence experiments. At each time-step, a cell is chosen at random to die or to divide, with a probability corresponding to its fitness. Mutations arise with rate μ. If a mutation occurs, the fitness of the daughter cell may change and the fitness increase is then chosen from a fitness distribution. Second, we use the “Equivalence Principle” model18, which focuses on beneficial mutations that establish in the population, and estimates that these mutations confer a single effective fitness advantage. Proliferation of clonal subpopulations under this model is deterministic. These simplifications are relevant to examples of high clonal interference, and are therefore only appropriate when the population size is large or when the beneficial mutation rate is high19. In both models, we assume no epistasis; the fitness change is independent of whether the cell already had one or more mutations. Furthermore, there is no restriction on the number of cells that acquire beneficial mutations, thus allowing clonal interference to occur20,21.
Both modeling approaches led to the same general conclusion -- the rapid adaptation of tetraploids results from both more frequent beneficial mutations and stronger fitness effects (Extended Data Fig. 2, Methods). For the branching evolution model, these conclusions are independent of the assumed distribution of beneficial mutations, although there are differences in the magnitude of the best-fit values that are expected from the shape of the chosen distribution (Fig. 1d, and Extended Data Fig. 2). Moreover, the conclusions are insensitive to the inclusion of deleterious mutations in the model (Extended Data Fig. 3, Methods).
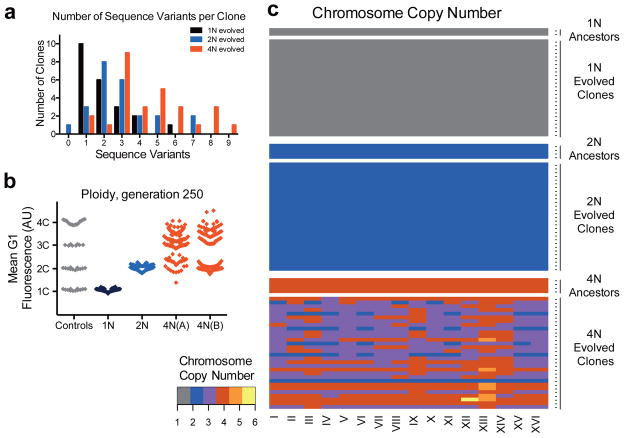
To evaluate these conclusions experimentally, we performed whole genome sequencing (WGS) to compare the frequency of mutations in 1N and 4N ancestors with 74 evolved clones. In total, we identified 240 de novo sequence variants (SNPs and small insertions/deletions): 45 from the 1N, 69 from the 2N, and 126 from the 4N-evolved clones, an average of 2.05, 2.87, and 4.5 variants respectively per cell type (Supplementary Table 1). We observed significantly more variants per 4N clone than per 1N and 2N-evolved clones (Fig. 2a, t-test, p<1e-04 and p=0.0040, respectively). Note that these results are not a direct measurement of the mutation rate or beneficial mutation rate (μ), but rather the total number of mutations acquired during the experiment (see Supplementary discussion).
Figure 2. Tetraploid clones acquire frequent sequence variants, recurrent whole chromosome aneuploidy, and large-scale ploidy shifts during adaptation.
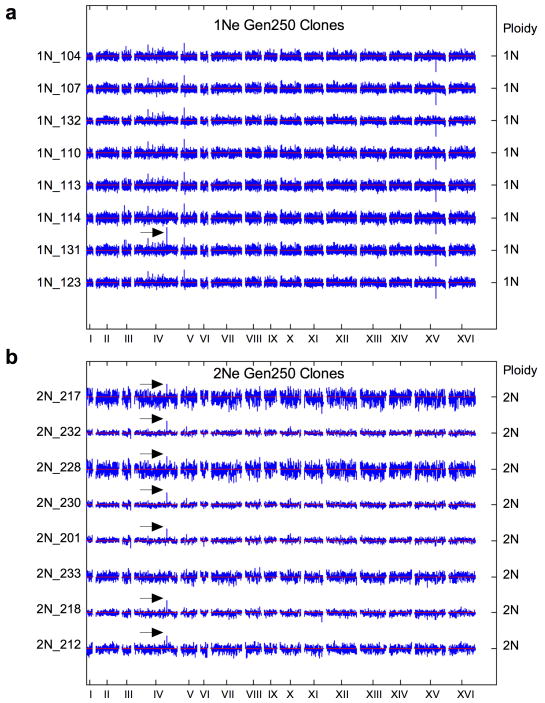
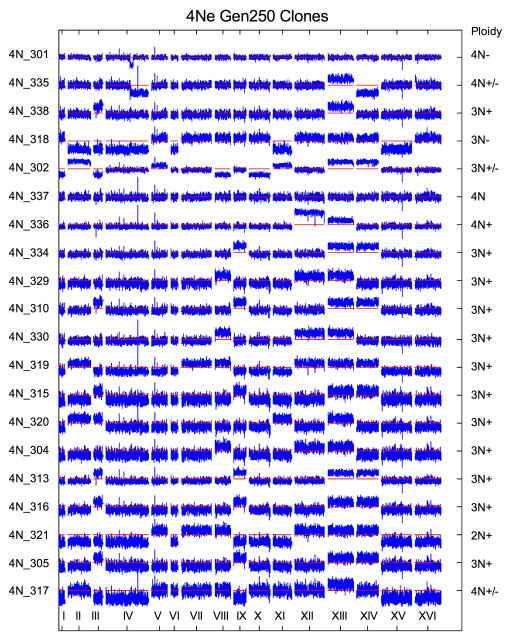
(a) The number of sequence variants per clone was determined with whole genome sequencing of 74 evolved clones (22 haploid, 24 diploid, and 28 tetraploid clones, Supplementary Table 1). The difference between tetraploids and haploids or diploids was significant (t-test, p<1e-04 and p=0.004, respectively). (b) DNA content of evolved clones at generation 250, measured as the mean G1 propidium iodide fluorescence for each evolved clone (n = 192). For reference, the DNA content of ancestral, control strains (1N, 2N, 3N, and 4N) is shown in gray. (c) Heat map of chromosome copy number data obtained from aCGH and WGS for the ancestral and evolved 1N, 2N, and 4N clones at generation 250; color key at left. See Extended Data Figs. 4–6, and Supplementary Table 2 for all individual clones).
Sequence variants frequently occurred in genes encoding proteins in the Snf3/Rgt2 glucose-signaling pathway (SNF3, RGT2, MTH1, RGT1), as expected from previous yeast evolution experiments under carbon-source limitation22–24. Several independent mutations in these genes resulted in either identical base-pair changes or altered the same amino acid (Supplementary Table 1). Nonsynonymous SNF3 mutations were identified in all ploidy types, whereas loss-of-function mutations in MTH1 were observed most frequently in the 1N-evolved clones.
In addition to WGS, we used a combination of flow cytometry, microarray comparative genome hybridization (aCGH), and qPCR to measure the frequency of DNA copy number variations (CNV) in the evolved clones. The only CNV that arose in all 3 ploidy types was amplification of two adjacent genes encoding the high-affinity hexose transporters, HXT6 and HXT7, a frequently identified beneficial mutation in low glucose environments20,22,23. The HXT6/7 amplification was significantly more common in 2N and 4N-evolved clones than in 1N clones (t-test, p=0.005 and p=1e-04), respectively, Methods), which may be due to negative epistasis between HXT6/7 amplification and mutations in 1N cells, such as those in MTH124.
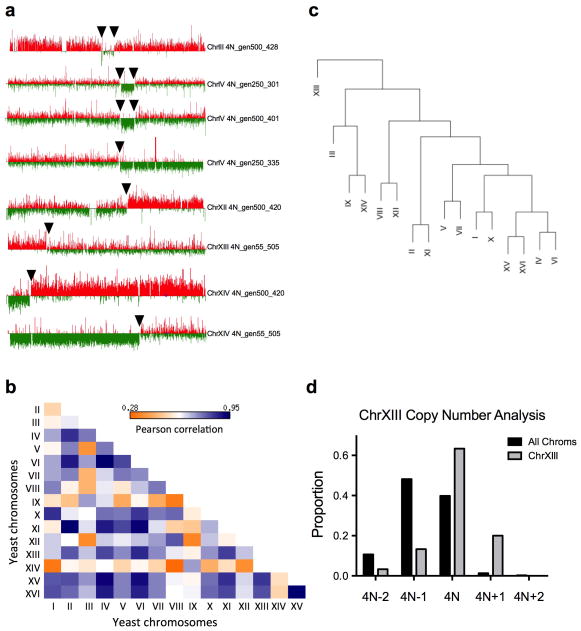
Additional CNVs, including recurrent chromosome aneuploidy, were detected only in the 4N-evolved clones. With the exception of a small segmental amplification in one 2N-evolved clone, there were no CNVs or aneuploidy in the ancestral strains or the 1N- and 2N-evolved clones (Fig. 2b, c, Extended Data Figs. 4, 5Fig. 2b) and all but two of the 4N-evolved clones were aneuploid at generation 250 (n = 30, Figs. 2c, Extended Data Fig. 6 and Supplementary Table 2). These alterations included large segmental aneuploidies with breakpoints at loci of transposable elements (Extended Data Fig. 7a). Pairwise patterns of chromosome copy number alterations were observed, indicating that there is a strong copy number relationship between certain pairs of chromosomes (Extended Data Fig. 7b, c). Notably, increased copy number of ChrXIII was significantly more common than all other aneuploidies (Extended Data Fig. 7d, Cochran Armitage test, p<1e-07). These chromosome-level alterations were present early, at the time of CFP/YFP marker divergence in the 4N populations (~generation 45, Extended Data Fig. 8). Therefore, 4N-evolved clones had a higher frequency and greater diversity of mutations, supporting the inference from our mathematical model that 4N-evolved clones have a relatively higher beneficial mutation rate.
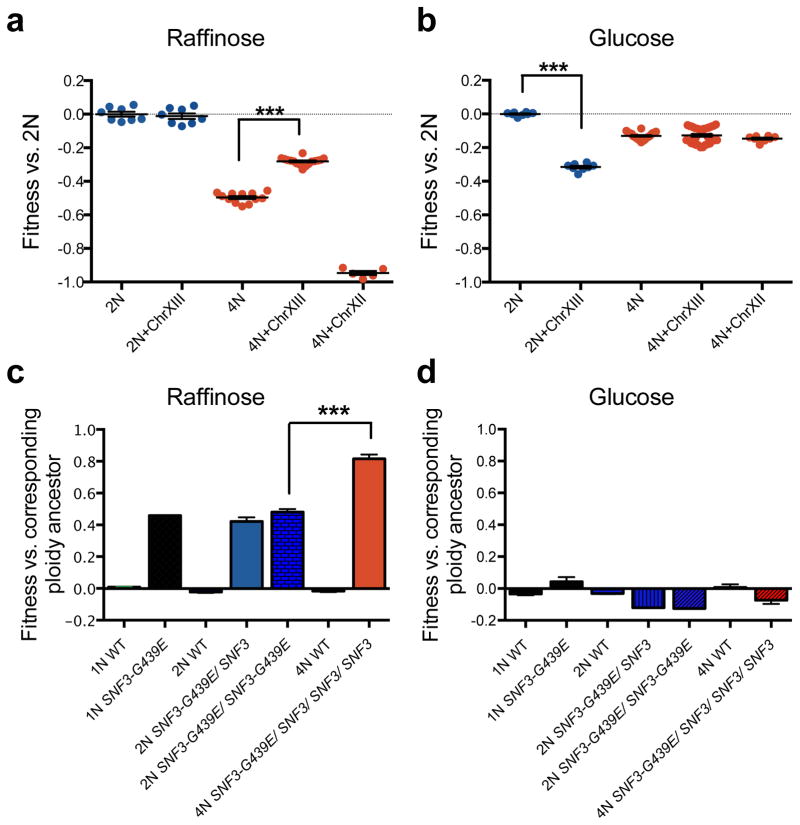
Next, we determined the effects of specific mutations on the fitness of the ancestral cells of differing ploidy. We first determined whether ChrXIII gain contributed directly to the rapid adaptation of 4N cells. Isogenic 2N and 4N strains, with and without an extra copy of ChrXIII, were generated (Methods, Extended Data Fig. 9). The increased copy number of ChrXIII provided a significant fitness increase to 4N strains specifically in raffinose medium relative to the 2N ancestor (Fig. 3a, t-test, p<1e-04), not in glucose (Fig. 3b). This was not a general effect of aneuploidy because the gain of a different chromosome, ChrXII, had the opposite effect on fitness (Fig. 3a). In striking contrast to 4N cells, ChrXIII trisomy was not beneficial to 2N strains in raffinose medium and decreased fitness of 2N cells in glucose. Although increased fitness due to whole and segmental chromosome gain is known to occur during adaptation13,14,23, to our knowledge, this is the first observation of a ploidy-specific fitness advantage for an aneuploid chromosome. Thus, aneuploidy, acquired through high rates of mitotic errors, is one way that 4N cells can acquire more beneficial mutations with higher fitness effects.
Figure 3. Ploidy-specific fitness effects for certain beneficial mutations.
Gain of ChrXIII is beneficial to tetraploid cells grown in raffinose medium but not for diploids. Shown is the fitness of isogenic wild-type 2N and 4N strains, with or without ChrXIII gain, relative to the 2N ancestor in raffinose (a) or glucose (b) medium. Error bars indicate the mean with the S.E.M. of seven individual clones and two technical replicates. (c) Competitive fitness of engineered isogenic strains of the indicated ploidy and genotype, relative to the 2N ancestor, in raffinose and (d) glucose medium. Error bars indicate the mean with the S.E.M. of three independent SNF3-G439E transformants of each ploidy type, t-test ***p<1e-04.
We also characterized how ploidy impacts the fitness effect of recurrently isolated mutations in SNF3, a gene encoding a plasma membrane glucose sensor25. We identified SNPs that changed the codon for the same amino acid in the 9th transmembrane domain of Snf3p (G439E, G439V, G439R, Supplementary Table 1), and increased HXT expression in raffinose (Methods)25. By analyzing the fitness of isogenic SNF3-G439E strains differing only by ploidy, we found that SNF3-G439E had a dominant, raffinose-specific, beneficial effect that was relatively stronger in the 4N strain (Fig. 3c, d, t-test, p<1e-04).
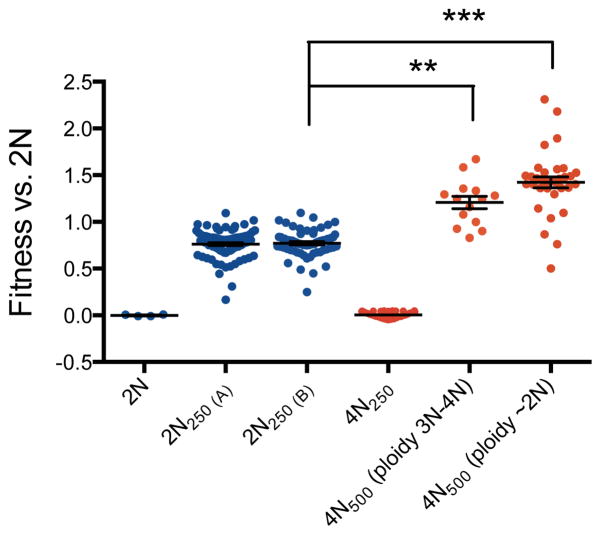
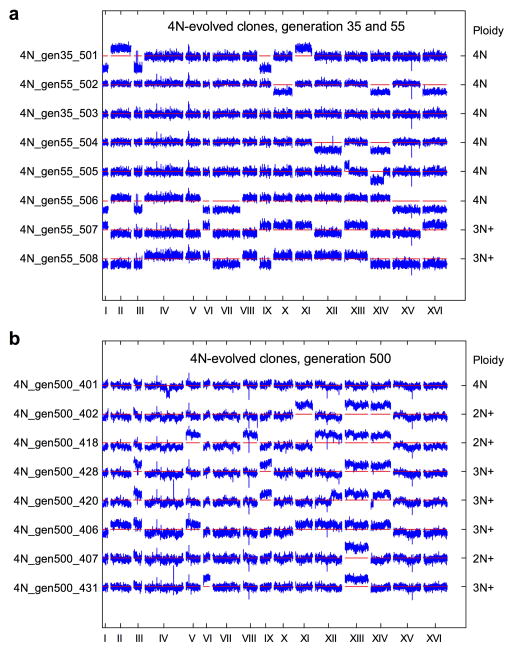
Ploidy-specific effect size of mutations could be an intrinsic property of polyploidy, as was recently suggested in plants16, or it could be related to the fitness of the 4N ancestor relative to the 1N and 2N ancestors26,27. To address the impact of initial fitness generally, we isolated 48 clones from the 4N evolution experiments at generation 250 (4N250) with fitness values equal to the 2N ancestor (competitive fitness difference < 0.05), and determined the speed of their next adaptive step. We compared the fitness acquired by the selected 4N250 clones after an additional 250 generations to that of 2N clones evolved for 250 generations (2N250, n=192). Despite comparable starting fitness, the 4N-derived clones still underwent more rapid adaptation and achieved significantly higher fitness. This occurred irrespective of large-scale shifts in ploidy: 29% of the 4N500 clones maintained a ploidy of 3N-4N and acquired higher fitness than the 2N250 clones (Fig. 4, KS-test, p<1e-06); 71% of the 4N500 clones underwent chromosome loss to become near-diploid and acquired even higher fitness relative to 2N250 clones (Fig. 4, KS-test, p<1e-08). Thus, the rapid adaptation of tetraploid cells was at least partially independent of their initial fitness.
Figure 4. Rapid adaptation of tetraploids normalized for initial fitness.
Fitness of 2N and 4N clones relative to the 2N ancestor. Evolved tetraploids (4N250) with fitness equivalent to the diploid ancestors were identified and passaged for another 250 generations to generate 4N500 clones (n=48). The fitness of these 4N500 clones was then compared to the fitness of evolved diploids after 250 generations (2N250, n= 192, replicate experiments A and B). 4N500 clones reached a higher fitness than 2N250 clones, irrespective of whether the 4N500 clones maintained a 3N-4N DNA content (n=14, KS-test, **p<1e-06) or underwent large-scale chromosome loss to a near diploid chromosome content (n=34, KS-test, ***p<1e-08). Error bars indicate the mean with the S.E.M.
Here, we measured the acquisition and spread of beneficial mutations in isogenic yeast populations that differed only by ploidy. Mathematical modeling enabled us to infer parameters driving the evolutionary dynamics of these strains and indicated that in a poor carbon-source environment, polyploidy increases the rate and fitness effects of the acquired mutations. Polyploidy increased the genetic diversity of the population. We identified examples of mutations that are selectively beneficial in polyploid strains, including whole chromosome aneuploidy. Because aneuploidy itself is mutagenic28, the high rates of aneuploidy induced by whole genome duplication may further increase the rate at which beneficial mutations are acquired. If these mutations are beneficial at lower ploidy states, then the long-term benefit of polyploidy will be preserved, even if polyploidy is transient during adaptation. Indeed, 4N-evolved clones that became near-diploid had higher fitness than the 2N-evolved clones. Moreover, although we only studied one environmental condition, polyploidy buffers the effects of partially recessive deleterious mutations12,29, which in principle can then accumulate2, providing a reservoir of mutations that might be adaptive in a new environment. Interestingly, the evolved tetraploid karyotypes closely resemble the polyploid and aneuploid karyotypes of fermentation, industrial, baking, natural desert isolates30, and antifungal drug-resistant yeasts14, consistent with a role for polyploidization events during adaptation to these stressful environments. Thus, the genetic plasticity of polyploid cells, together with ploidy-specific beneficial effects, can facilitate rapid adaptation.
Supplementary Methods
Batch culture evolution experiment
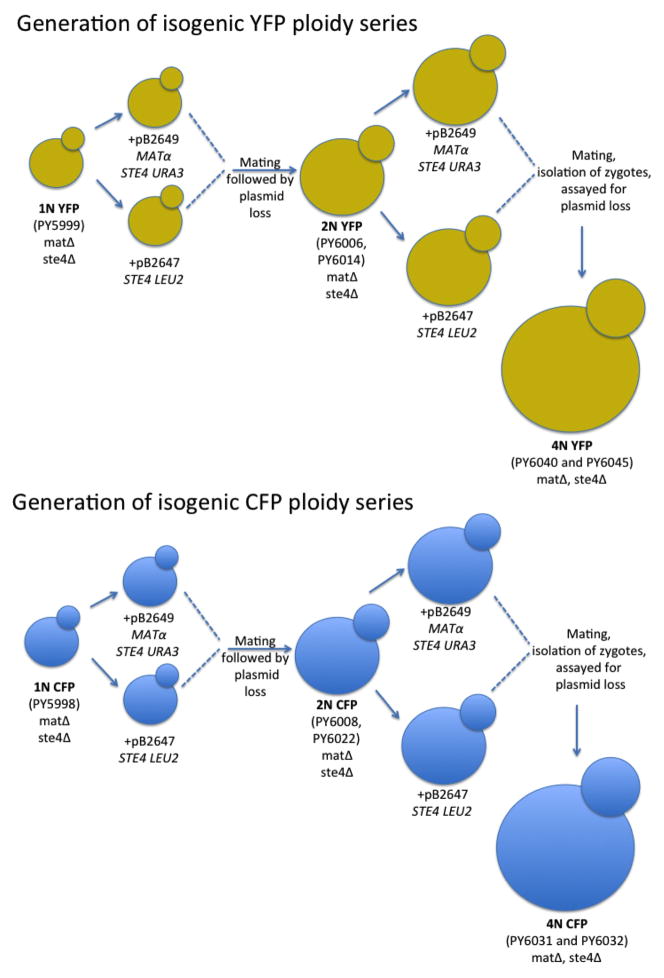
All S. cerevisiae strains used in this study were in the S288c background (detailed information on strain construction is provided below under the heading Yeast Strain Construction). Briefly, the isogenic ploidy series was generated in a matΔ ste4Δ background to eliminate mating and meiosis during the course of the experiment. Either a pGAL-CFP or a pGAL-YFP construct was integrated at the TRP1 locus near the ChrIV centromere in a haploid strain (PY5998 and PY5999, respectively). These haploid strains were used to generate isogenic diploids, from which isogenic tetraploids were then derived (Extended Data Fig. 1). This procedure ensured that all copies of ChrIV had the capacity to express the inducible fluorescent marker even if the strains became aneuploid. Mating-competent haploids were generated from the matΔ ste4Δ ancestor, PY5998, by transformation with either plasmid PB2647 (CEN-LEU2-STE4) or PB2648 (CEN-URA3-STE4-Matα). Zygotes from mating-competent haploids were isolated by micromanipulation to obtain diploid CFP ancestors (PY6008 and PY6022). Similarly, zygotes from mating-competent diploids were isolated by micro-manipulation to obtain tetraploid CFP ancestors (PY6031 and PY6032). The same mating scheme was performed for the YFP lineage starting with PY5999 to generate diploid YFP (PY6006 and PY6014) and tetraploid YFP (PY6040 and PY6045) ancestors.
The ancestor strains were grown to saturation from the −80°C stock, in Synthetic Complete (SC) + 2% glucose. The cell density of each ancestor was determined using a hemocytometer and an automated cell counter (Vi-Cell-XR from Beckman Coulter). An equal number of YFP and CFP cells of the same ploidy were diluted into fresh SC + 2% raffinose medium, and combined into a single tube for an initial concentration of 1×105 cells per ml. The 50:50 YFP:CFP culture was distributed equally into the wells of a 96 deep-well plate (1 ml per well, U-bottom block plate from Qiagen). Seven or eight wells were not inoculated, to detect cross-well contamination during the experiment. The plates were covered with “breathe-EASIER™” tape (Electron Microscopy Science) and incubated at 30°C on a 96-well plate shaker (Union Scientific). Two plates of haploid and three plates of diploid and tetraploid cells were analyzed, representing 173 parallel haploid evolutions, 264 parallel diploid evolutions, and 265 parallel tetraploid evolutions.
At 24 hour intervals, the cells were resuspended (by pipetting) and diluted into fresh SC + 2% raffinose medium. The dilution factor was determined for each ploidy type based on the initial strain fitness in order to maintain an equivalent population size, as reported previously18. The number of cells transferred each day was calculated by counting the number of cells in 10 replicate wells of each ploidy before and after dilution with an automated cell counter (Vi-Cell-XR from Beckman Coulter), and averaged across 3 consecutive days. The dilution factor for the haploid, diploid, and tetraploid experiments was 1/100, 1/50, and 1/33, respectively. This corresponds to 6.64, 5.64, and 5.04 generations per day18. The tetraploid evolution experiment from generation 250 to 500 (Fig. 4) was performed with the same dilution factor as the diploid experiments (1/50).
The number of CFP and YFP cells in each population was measured at the same time each day. First, expression of the fluorescent proteins was induced by transferring 10 μL of the overnight culture into 200 μL SC + 2% galactose medium for 4 hours at 30°C. The number of CFP- or YFP-expressing cells was determined using the BD LSRII flow cytometer high-throughput plate reader (10,000 cells were analyzed from each well). Pacific Blue and FITC filters were used to detect CFP and YFP, respectively. All experiments were passaged for 250 generations, but daily acquisition of CFP:YFP ratios was not always continued to the 250th generation.
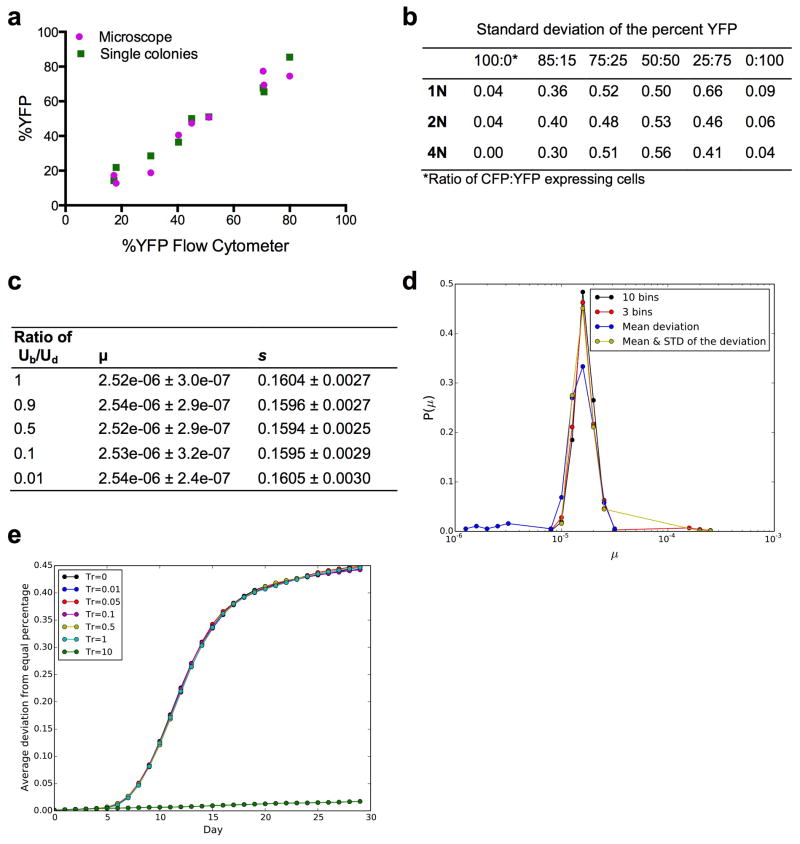
To ensure that the flow cytometer measurement and the galactose induction of CFP and YFP was an accurate reflection of the size of these populations, the ratio of CFP:YFP cells was determined by both flow cytometry and microscopy, and the ratio was determined both before and after galactose induction. To do this, we combined overnight cultures of the 1N, 2N, and 4N ancestor CFP and YFP strains at 3 different ratios (9 populations total) and analyzed the ratios in two ways. First, for an aliquot of the mixture, we induced the expression of the fluorescent proteins with 2% galactose for 4 hours and analyzed 10,000 cells using flow cytometry. In parallel, we also added 2% galactose for 4 hours and then counted ~300 cells by fluorescence microscopy. Finally, to ensure that the induction with 2% galactose did not alter the CFP:YFP ratio, a portion of the population was used to determine the number of CFP and YFP cells in the population before adding galactose to the medium. To do this, cells from each population were struck for single colonies on YPD plates for two days. 96 colonies were chosen randomly from each plate and added to a single well of a 96-well plate containing SC + 2% Galactose. The fluorescence of each colony was determined by flow cytometry, and the %YFP of the initial population was determined. There was a strong correlation between the %YFP-expressing cells obtained from all three measurements (Extended Data Fig. 3a): including the flow cytometer and fluorescence microscopy (Pearson correlation coefficient = 0.979), and both before and after galactose induction (Pearson correlation coefficient = 0.985).
Finally, frozen stocks of the evolution experiments were made at 3–4 day intervals throughout the experiment. At the end of each experiment, single colony clones were isolated and used for competitive fitness assays, flow cytometry analysis of ploidy, and preparation of DNA for aCGH.
We isolated 48 clones from the 4N evolution experiments at generation 250 (4N250) with fitness values equal to the 2N ancestor (competitive fitness difference < 0.05), and determined the rate of adaptation after an additional 250 generations (Fig. 4). Each 4N250 clone was grown to saturation from the −80°C stock in SC + 2% raffinose medium. Cell counts were performed as above, and each population was diluted to an initial concentration of 1×105 cells per ml. At 24 hour intervals, the cells were resuspended (by pipetting) and diluted 1/50 into fresh SC + 2% raffinose medium (the same dilution factor as the diploid experiments). These evolution experiments were not performed as CFP:YFP competitions, so daily flow cytometry was not necessary. After 250 generations, single colony clones (4N500) were isolated on SC + 2% raffinose plates. Each 4N500 clone was cultured overnight in 1ml SC + 2% raffinose medium and aliquots of this culture were immediately used for competitive fitness assays, flow cytometry analysis of ploidy, preparation of DNA for aCGH, and frozen stocks.
Measuring the variation in the flow cytometer measurements
We determined the amount of noise in our flow cytometer measurements by calculating the mean and standard deviation of the %YFP obtained from 48 independent populations at 6 different ratios of CFP:YFP, for each ploidy type. The 1N, 2N, and 4N ancestor strains were cultured separately overnight in 2% raffinose medium and transferred to 2% galactose for 4 hours to induce expression of CFP and YFP. Next, CFP and YFP cells of the same ploidy were combined at ratios of 100:0, 85:15, 75:25, 50:50, 25:75, and 0:100 to reach the same final volume (200 ul). 10,000 cells from each population were analyzed by the LSRII (BD) flow cytometer using the same parameters (ex. gating and flow rate) that we used for the evolution experiments, the total number of CFP and YFP cells were obtained, and the percent YFP was calculated ((#YFP cells)/(#CFP+#YFP cells))*100. The standard deviations are presented in Extended Data Fig. 3b, and indicated that there is little well-to-well variability for the same CFP:YFP ratio (across 48 wells), and that this variability changes only slightly across different CFP:YFP ratios of all three ploidy types. Importantly, it is never greater than 0.66% of the measurement. This small variability has a minimal effect on our analysis because the fitting procedure we used to measure the deviation from an equal percentage of CFP and YFP cells used bins of 5% deviation to combine the number of wells that had deviations between 0%–5%, 5%–10%, and so on, for each experiment.
Statistical analysis of the experimental data
Adaptation rate
The CFP vs. YFP evolution experiments were designed to analyze the dynamics of the adaptation process at the population level within and across wells. The adaptation rate was determined with additional competition experiments that were designed to measure the change in fitness from generation zero to generation 250 for cells of each ploidy. To this end, we isolated single colony clones from each evolved well of different ploidy types at generation 250 and measured their competitive fitness relative to the 2N ancestor (see detailed methods below). Fitness was defined as the slope of the log of the ratio of the evolved clone to the reference strain (2N ancestor) over time; more precisely, we performed a linear least squares fit of log(Nt1/Nt0) over multiple dilution cycles (where Nt1 is the number of cells from the evolved clone, and Nt0 is the number of ancestor cells). The fitness relative to the ancestor is defined as s = d/dt [log2(Nt1/Nt0)], where t is measured in days18. The rate of adaptation was the relative fitness at generation 250 minus the relative fitness of generation zero, divided by 250 generations. The rate of adaptation for each ploidy is shown in Fig. 1c. We found that the tetraploid populations had a significantly larger rate of adaptation (0.009 [0.0062, 0.011]) than haploids (0.0031 [0.0022, 0.0052]) or diploids (0.0031 [0.0018, 0.0041]) during the 250 generations in raffinose medium (values indicate the median rate followed by the 95% confidence interval in brackets, Fig. 1c, t-test, p<1e-10).
Mathematical modeling of population dynamics
Branching Evolution Model
We formulated a mathematical model of the population dynamics of cells that was then used to infer evolutionary parameters using the experimental data. This branching evolution model was designed to closely mimic the divergence experiments containing two equally fit populations (CFP or YFP), each initially consisting of 50,000 cells. The model is based on a stochastic birth and death process called a branching process32. In this process, at each time-step, a cell is chosen to die at random, or to divide with a probability corresponding to its fitness. During each cell division, a mutation arises with mutation rate μ. If no mutation occurs, the fitness of the daughter cell is equal to the fitness of the mother cell. If a mutation does occur, the fitness may change; the additive fitness of the daughter cell is then chosen from a fitness distribution. The fitness change is independent of whether the ancestors of this clone had already obtained one or more mutations. Furthermore, there is no restriction on the number of cells that acquire beneficial mutations, thus allowing clonal interference to occur20,21.
We compared results assuming either a uniform, exponential or delta distribution of fitness values (Extended Data Fig. 2a–d). For the initial formulation of the model, we considered half of the newly arising mutations to be beneficial and half deleterious, and their fitness effects were considered to be additive to the fitness value of the mother cell. Because complete simulations of the branching process would be prohibitively slow, we approximated this branching process with a Wright-Fisher process17 with non-overlapping generations. This process was implemented as a Monte Carlo simulation in C++ and the code is provided as a Supplementary Software file.
In the simulation, each competition experiment was initiated with 1×105 cells (5×104 of each CFP and YFP cell type). At every generation, each cell reproduces and gives birth to a random number of surviving offspring distributed according to a Poisson distribution. The initial population of each ploidy had a different growth rate per day: the haploid population increased ~ 100-fold, the diploids ~ 50-fold, and the tetraploids ~ 30-fold in a 24 hour time interval (described above in the section Batch culture evolution experiment). Assuming a population doubling every generation, the average number of generations per day was 6.64, 5.65, and 5.04 for haploids, diploids and tetraploids, respectively. In the Wright-Fisher model, the number of generations is discrete; thus we rounded these numbers to the closest integers (7,6, and 5, respectively). The initial fitness (f) of each ploidy was chosen to satisfy the growth rate of that ploidy with this number of generations, i.e. (f1N)7 = 100, (f2N)6 = 50, (f4N)5 = 30. The fitness of a cell is the average number of its surviving offspring in the next generation.
During each cell division, a new mutation might arise. In haploid cells, the impact of amutation is given by its additive fitness value s d awn from the fitness distribution. In diploid and tetraploid cells, however, a mutation might have a degree of dominance, be recessive or have a different fitness effect than it would have in a haploid cell. Following Otto and Whitton (2000), we assumed that the effect of a given mutation in a diploid or tetraploid cell as compared to that in a haploid cell is scaled by a certain factor h. Therefore, the fitness effect of a given mutation in one allele will be s̃ = h · s.
In our model, we considered a beneficial mutation to arise independently at rate μ (per whole genome). Because our populations proliferate asexually, we cannot differentiate between the two components of s̃, namely the haploid fitness effect s and the dominance coefficient h, and therefore we can only infer the combined value of s̃. Our simulations were done in exactly the same way for all ploidy types, and the selection coefficient of a new mutation was taken from a given distribution. However, the meaning of the added fitness is different for the haploids, where it is s, than for the diploids and tetraploids, where it is s̃. The assumption of independence of mutations is justified by the low point mutation rate, which is of the order of magnitude of 2×10−10 per base per generation in yeast33. The low per-base mutation rate means that the probability of independently obtaining a second identical mutation is vanishingly small (probability of μ2, or 4×10−20), and therefore we did not consider such events in the model. Similarly, the probability of obtaining a given mutation that is then copied by a recombination-based mechanism such as gene conversion34 is low (2×10−10 × 4×10−5), and was not included. These assumptions are validated by our whole genome sequencing data demonstrating single copies of all point mutations.
Our identification of specific mutations that have a larger fitness effect in the tetraploid strains than in the haploid or diploid strains (Fig. 3) is consistent with the overall larger fitness effect of the tetraploids in our experiment. Note, however, that our parameters describe the entire distribution of mutations, rather than any specific mutation. Thus, it is also possible that some mutations could have a larger fitness effect in the haploids or diploids relative to the tetraploids. Note also that in our model, clonal interference can occur, as there is no limitation on the number of independent mutations that can arise within a population. Thus multiple clones can emerge and compete with each other. As shown previously, clonal interference is an important aspect of microbial population dynamics and thus cannot be ignored18,20,21. Additionally, multiple mutations can exist in the same cell.
As a sensitivity analysis, we also varied the ratio between advantageous and deleterious mutations, and found that even when the fraction of the deleterious mutation is very large, their inclusion has only a negligible effect on the rate and dynamics of adaptation. This is consistent with a large body of prior literature18,35–37. To accomplish this, we fixed the rate of the advantageous mutations and generated datasets with different ratios of beneficial to deleterious mutations: 100:0, 90:10, 50:50, 10:90 and 1:99. For each ratio we generated 100 datasets (each dataset with 264 single deviation experiments) using a beneficial mutation rate of 1.2×10−6 and a fitness effect of s=0.16 (the best-fit values of the delta function for the diploid experiments, as a typical example, we used the same distribution as in Hegreness et al. 2006), and fitted them against the simulations that we used to fit the empirical experiments (see below for the fitting procedure). The mean of the fitted values are presented in Extended Data Fig. 3c. We found no significant difference between the different ratios of beneficial to deleterious mutations (t-test, p-value >0.2 between all ratios tested). Therefore, in our model, we assumed that half of all non-neutral mutations are deleterious and half are beneficial, and that the deleterious and beneficial mutations have the same fitness effect distribution with the same parameters. Note that we also obtained similar results using the Equivalence Principle model18 that does not include deleterious mutations (see below).
The upper bound of cellular fitness
For biological plausibility, it was necessary to set an upper bound for acquired fitness. Based on the well-described growth rates achievable for S. cerevisiae in optimal conditions, we set this boundary at a doubling time of 1 hour (224 per day). Given that g is the number of generations a strain experiences in a day, the upper bound of the fitness f of any ploidy type was therefore set to satisfy the equation fg =224.
The initial growth rate of the tetraploid cells was lower than the growth rate of the haploids and diploids (see section “Batch Culture Evolution Experiment” above). In the simulations we rounded the number of generations to satisfy the assumption of non-overlapping generations in the Wright-Fisher model: the tetraploid cells underwent ~5 generations per day, whereas the haploid cells underwent around ~7 and diploid cells ~6 generations per day. In the simulations, as in the experiments, we diluted the populations every day by choosing at random 1% of the haploid, 2% of diploid cells and 3.3% of the tetraploid cells. This dilution was done by using a hypergeometric random generator for populations smaller than 100 million (http://www.agner.org/random/); for populations larger than 100 million this method is not applicable and we used a direct Bernoulli sampling of cells38 forcing the total sampled cells to be 1%, 2% or 3.3% for haploid, diploid and tetraploid populations, respectively. The concentration of each of the cell types was recorded in the simulation output at the end of every day, after dilution.
In the experiments with the diploid and tetraploid cells, we observed an initial small bias against the YFP-labeled population: in the diploid cells there was a decline in the YFP-labeled population of 0.5% per day, and in the tetraploid cells there was a 1% decline per day (Fig. 1b). We included these biases in the simulations by including this initial small difference in the fitness (i.e. a reduced average number of offspring) of CFP- vs. YFP-labeled cells18,26.
Note that drift cannot feasibly play a role in our experiment, as the time scale (in generations) for fixation of an allele due to drift is approximately equal to the effective population size39. This time is of the order of 106 generations in our experiments18, much larger than the time frame of our experiment (250 generations). Thus the simulations continued until one of the cell types overtook the whole population or until the end of the time that the CFP:YFP data was collected on the 30th day; the first of either event terminated a simulation. In order to increase the efficiency of the simulations, extinction of a certain color (and thus fixation of the other) was defined not by a value of zero frequency, but as a frequency of less than or equal to 1%, as the probability is negligible that a sub-clone present at a frequency of 1% will overtake another sub-clone present at a frequency of 99% in the timeframe of our experiment21. Furthermore, while the variability between flow cytometer measurements was never greater than 1% of the measurement (See above, and Barrick et al. 2010), our ability to detect changes in CFP or YFP populations below a frequency of 1% was a limitation of the flow cytometer.
Fitting procedure
The empirical data for each ploidy type were combined to represent the average deviation from an equal percentage of YFP and CFP cells. We then utilized the combined ploidy data to estimate the best-fit values of mutation rate (μ) and selection coefficient (s), using least squares fitting.
In order to compare data from the experiment and the simulations, we investigated several different summary statistics and used the one that performed best. The summary statistics evaluated were:
Mean deviation from an equal percentage of CFP and YFP cells: For every experiment corresponding to an individual well in a 96-well plate, we calculated the deviation from equal percentage every day. Then, for every dataset generated (experiment and simulation) we calculated the average deviation per day. This procedure created a vector of the mean deviations per day for every dataset.
Mean and standard deviation of the deviation from an equal percentage of CFP and YFP: As in i, except that we also calculated the standard deviation of the deviation from equal percentage for every day. This procedure generated two vectors for every dataset.
A distribution of the deviation from an equal percentage of CFP and YFP with 10 bins: we calculated the absolute value of the deviation from equal percentage for each deviation experiment for each day. Then for every day, we binned the deviation values into 10 bins, each with a size of 5%. i.e. we counted the number of wells that had a deviation between 0%–5%, 5%–10%,..,45%–50%. This procedure generated a matrix of 10 bins*30 days for each dataset.
A distribution of the deviation from equal percentage CFP and YFP with 3 bins: Similar to iii, except we used only three bins, one for the non-deviated wells (defined as 0–0.1), one for fixated wells (defined as 0.4–0.5) and one for those in the middle (0.1–0.4).
The comparison between the experiments and the simulations was done by calculating the sum of squares (SOS) between the summary statistic of the experiment and the summary statistic of the simulations.
Where Ω spans the day and the number of values that each summary statistic has each day. The best-fit pair was the set of μ and s values with the smallest SOS. To determine the performance of the different summary statistic, we generated 1000 datasets with the same parameters (1N, Uniform distribution with s = 0.05, and μ=8*10−5), each containing 264 single deviation experiments, and inferred their values by scanning the parameter space for each of the 1000 artificial datasets. The expected values from the simulations were then generated by 1000 single deviation experiments for each set of parameters.
The distribution of inferred μ values is shown in Extended Data Fig. 3d. Whereas all of the summary statistics had the mutation rate used in the simulations as their mode, the observed means varied modestly. The 10-bin summary statistic had the narrowest range, and therefore this SS was used for further analysis. A similar summary statistic was also used previously40.
The scanned range of mutation rates and fitness effects
The best-fit value was found by scanning a range of mutation rates and fitness effects26; to this end, we scanned the parameter space of the fitness effect in linear steps from 0.005 to 0.35, with increments of 0.005. The mutation rates were scanned in logarithmic steps from log10(μ)=−8 to log10(μ)=−4, with increments of 0.1. Thus, per ploidy per fitness distribution, we scanned 2460 parameter regimes, analyzed 1000 divergence simulations per μ and s combination, and scanned 3 ploidy types and 3 different fitness distributions in total.
For the exponential fitness distribution, larger mutation rates and smaller selection coefficients needed to be investigated in order to find the best-fiting pair: we scanned mutation rates from log10(μ)=−8 to log10(μ)=−3, and selection coefficients from 0.002 to 0.1, with increments of 0.002. Additionally, in the exponential distribution, the majority of mutations have a fitness value which is too small to contribute to the competition between the two cell populations (YFC and CFP). However, increasing the mutation rate will increase the probability that mutations with larger fitness value are obtained. We found that for mutation rates higher than 10−4 per genome per cell division the computational time constraints were substantial, and thus we excluded all mutations that had an s value smaller than 10% of the average fitness effect s. Support for this choice was provided by the finding that for large mutation rates (i.e. N*μ>1), the mutations chosen from an exponential distribution that eventually reach fixation within the population have a larger fitness effect than the distribution average (Barrett et al. 2006, Figure 3)19. The above criteria for large mutation rates are within our experimental regime (N*μ=106*10−4>1). Furthermore, in Extended Data Fig. 3e we present simulation results of the average deviation from equal percentages of CFP and YFP for a given mutation rate and fitness effect drawn from an exponential distribution. We found that the results are robust to including or excluding those mutations with fitness effects smaller than the distribution average. Thus, for high mutation rates (μ*N>1), we can exclude weak mutations, as was shown by Barrett et al. (2006)19.
The fitting of the empirical experiments and the simulations was done as described above. The results of the best-fit values are presented in Extended Data Fig. 2a–d.
The Equivalence Principle model
As a complementary approach we evaluated the Equivalence Principle model developed by Hegreness et al. (2006) to analyze a similar competition experiment in E. coli. This study concluded that the mutations that eventually reach fixation in large microbial populations have a very narrow range of fitness values. Very weak mutations are unlikely to lead to a takeover of one population (CFP or YFP) by the other, whereas very strong mutations are very infrequent. Based on this prediction, the competition experiments were described by assuming that all beneficial mutations have exactly the same fitness value, i.e. assuming that the distribution of fitness effects is a delta function. Based on this assumption, the authors developed a method for inferring the two key parameters of the dynamics of the adaptation process: the mutation rate of a beneficial mutation and the single fitness effect value. Note that the assumption of a single fitness value for beneficial mutations was shown to be true only if the mutation rate is very high19, which cannot be known a priori. This is the reason that the branching evolution model was implemented first and then compared to the Equivalence Principle model. Note also that the Equivalence Principle model analyzes only the initial CFP:YFP divergence phase, rather than using the entire data set as in the branching evolution model. Although this excludes some data, it has the advantage of not necessitating the assumption that the distribution of fitness effects is constant41.
We followed the procedure outlined by Hegreness et al. (2006), including the following assumptions and modeling steps (see the Supplementary Information of Hegreness et al. for a more complete description18):
Rather than the computationally intensive simulation of growth and dilution we used the effective population size42. The effective population size is: Ne=N0*log(r)*T, where N0 is the initial population size, r is the growth rate of the population, and T is the number of generations between dilutions. In addition to reducing computation time, the use of the effective population size allows for an analytical approximation of certain quantities, such as the probability of escaping drift.
Even large mutations may be eliminated by drift, and only those that escape drift can contribute to the competition between the two labeled cell populations. The probability of emergence of a new mutation that will escape drift can be calculated analytically for a fixed population, making it possible to generate in simulations only those mutations that escaped drift, and contribute to the competitiveness of the population. Under the above simplifying approximations of (1) a fixed population size and (2) a delta distribution of fitness effects, the probability that a mutation will emerge and escape drift (Pescape) was calculated analytically, as a function of the mutation rate, μ, (the rate for a new mutation to appear in a single cell division), the selection coefficient, s, and the effective population size, Ne.
Given that Pescape can be calculated, simulations can be performed rapidly by generating the time in the experiment when such mutations occur, rather than randomly assigning mutations at each cell division.
We applied the Equivalence Principle model as follows. In order to compare the experiments to the simulations, as in Hegreness et al., we used time and initial slope of CFP:YFP ratio divergence as summary statistics for the divergence curves18. Each CFP:YFP divergence experiment (one single well from a 96-well plate) was fit to an exponential growth model, with slope α that starts at a given time τ, using the following expression −log10(1 + 0.5 · exp(α(t − τ))). An exponential model is a good description of the initial divergence phase. The end of the exponential growth phase was defined by the time that has the maximum likelihood to be described by exponential growth (see Hegreness et al. Supplementary Information for more details). The initial CFP:YFP bias that we detected in some 2N and 4N populations was included in cases where it occurred.
Each divergence experiment is thus described by its values τ and α, whether it is a real experiment or a simulated one. Because a single divergence experiment is subject to significant stochasticity, we compared the empirical experiments and the simulations by combining all the wells of each ploidy type. This was done by collecting all of the α values of a given experiment or a given set of simulations (i.e. those simulations that we generated by the same pair of μ and s values) into a distribution of α values. The experimental distribution was compared to the simulations’ distribution by calculating the KS-test between the two. The same was done for the τ values. For each pair of s and μ values we calculated the sum of the KS-test of its α distribution and the KS-test of its τ distribution. The pair of values (μ, s) that had the smallest sum was declared the best fit (Extended Data Fig. 2a). This was done independently for each ploidy type (Extended Data Fig. 2e).
Modeling Results
The fitting of the empirical experiments and the simulations was done as described above. The results of the best-fit values from the branching evolution model and the Equivalence Principle model are presented in Extended Data Fig. 2a. In order to generate the error range for each distribution and ploidy type, we used the parametric bootstrapping method43. For each estimated set of values, we generated 1,000 simulated datasets and compared those to the empirical datasets (dataset sizes are 172, 264, 265 independent deviation experiments for the haploids, diploids and tetraploids, respectively). We then inferred the best-fit values of those datasets. The 95% confidence intervals of μ and s from those 1,000 datasets were defined as the error ranges (Extended Data Fig. 2a–e).
We infer from these results that the tetraploids have a higher mutation rate and these mutations have, on average, a stronger fitness effect as compared to haploids or diploids. The trend for tetraploids to have higher μ and s values occurs independently of the different assumed distributions of fitness effects. However, the different distributions lead to significantly different absolute values. This is expected from the characteristic shape of these distributions. To illustrate why this result is expected, we show a schematic diagram of the three distributions of fitness effects (Extended Data Fig. 2f).
The mutations that mainly govern the adaptation process are hypothesized to come from a relatively narrow range of fitness effect values that are sufficiently strong, but not extremely rare (Extended Data Fig. 2f, double arrow region from the idealized narrow Gaussian curve). The three assumed distributions that we used in our modeling approximate the true distribution but with the following differences. The delta distribution is located in the center of the double arrow region. Thus, by definition, every mutation from the delta distribution will have a fitness effect that is strong enough to have a significant probability of promoting adaptation. If every mutation has the possibility of contributing, then a lower mutation rate suffices for adaptation. By contrast, the exponential distribution is dominated by small-effect or near neutral mutations, with a relatively low fraction of mutations near the value for the delta distribution. Thus, the assumption of an exponential distribution is accompanied by a requirement for a compensatory higher mutation rate to achieve numbers of equivalently beneficial mutations. By the same line of reasoning, the uniform distribution necessitates a rate of beneficial mutations that is intermediate between the exponential and delta distributions. Indeed, our results match all of these expectations (Extended Data Fig. 2a–d). A similar difference in the estimation of μ was also observed in another recent study that modeled this value based on these three assumptions about the distribution of beneficial mutations44.
In terms of the fitness effect values of the different distributions, the uniform distribution is governed by its upper limit, which, by definition, is the strongest mutation allowed. Therefore, it is expected that the uniform distribution mean, which is half of the upper limit, will be half of the fitness effect value of the delta function. Again, this is what we observed (Extended Data Fig. 2a–d). The exponential distribution mean, is much smaller than the strongest mutation that can be generated by the exponential distribution, and the best-fit parameters are expected to be much smaller than the other two distributions, which is also observed (Extended Data Fig. 2a–d). Furthermore, because the exponential distribution has no upper bound on the fitness effects, a larger mutation rate can lead to the emergence of mutations with much larger fitness effects. In this way, under the exponential distribution assumption, the mutation rate affects the range of possible fitness effects, which results in a larger error range for the exponential distribution as compared to the other distributions (Extended Data Fig. 2a, brackets).
In summary, despite expected differences in absolute values, we reach the same overall conclusions with either the branching evolution model with varied assumptions about the distribution of beneficial mutations or Equivalence Principle model18.
Computer code availability
The computer code is available as a Supplementary Software file. The code was complied by g++ (version 4.2.1) and was tested on Unix (CentOS5 operating system) and Mac (OS X Version 10.9.2 operating system) machines.
Plasmid construction
All plasmids used in this study are listed in Extended Data Table 1. To construct plasmids for the inducible expression of either CFP or YFP, the galactose-inducible GAL1 promoter was subcloned into the YFP plasmid PB1500 and the CFP plasmid PB2452. These plasmids were derived from the GFP protein tagging plasmid generated by Longtine et al.45. Both plasmids contain the ADH gene terminator (tADH) after the YFP or CFP gene and the sequence of the SpHIS5 gene of Schizosaccharomyes pombe as a selectable marker. Plasmids PB1500 and PB2452 were digested with BamHI and PacI to introduce the pGAL promoter, 461 bp upstream of the start codon of GAL1 46, which was amplified using the primers pGAL1 BamHI 5′ (5′-ACGGATCCCCGGGTTGAAGTACGGATTAGAAGCCGCCGAG-3′) and pGAL1 PacI 3′ (5′-CGTTAATTAATATAGTTTTTTCTCCTTGACGTTAAAG-3′). Site directed mutagenesis (Quick Change Mutagenesis Kit, Stratagene) was used to introduce an ATG translation start codon to the YFP and CFP genes (using the GAPATGpFA6 primer, 5′-CAATCAATCAATCAATCATCACATAAATTAATTAAATGAGTAAAGGAGAAGAACTTTTCACTGGAGTTGTC-3′). The resulting plasmids PB2694 and PB2697 contain the cassette pGAL1-CFP-tADH-SpHIS5 and pGAL1-YFP-tADH-SpHIS5, respectively.
PB2314 was used to delete the MAT locus as previously described 12. PB1308 was used to perform a URA3 to TRP1 marker swap, as previously described 47. PB1640 (hphMX4) was used for PCR-mediated deletion of STE4. PB2647 (STE4-LEU2) was used to restore mating competency and was constructed by amplifying the STE4 gene with primers STE4 P BamHI 5′ (5′-CCGGATTCTTGTAGCCCTGTTAGGTTTACC-3′) and STE4 T BamHI 3′ (5′-CCGGATTCCAATACATAAGGACGAGCCAGTG-3′), and cloning it into pRS315. PB2649 (STE4 URA3 CEN MATα) was also used to restore mating competency, and was constructed by subcloning the STE4 fragment from PB2647 (digested with SmaI and NotI) into PB2577 (MATα URA3 CEN, digested with SmaI and NotI).
Yeast strain construction
All Saccharomyces cerevisiae strains used in this study are isogenic to PY3295 (BY4741, S288c genetic background MATa his3Δ leu2Δ met15Δ ura3Δ) and listed in Extended Data Table 1. The strategy used to generate the isogenic ploidy series is illustrated in Extended Data Fig. 1 and genotypes of key intermediates are indicated. The CFP and YFP ancestors were derived from the haploid strain PY5997 (matΔ::pSTE5-ura3::TRP1, ste4Δ::HygroR, trp1::NatR, strain construction details available upon request). Isogenic strains with either the CFP or YFP cassettes at the TRP1 locus (ChrIV) were generated (PY5998 and PY5999) as follows: the pGAL1-CFP-tADH-SpHIS5 or pGAL1-YFP-tADH-SpHIS5 cassette was PCR amplified from plasmid pB2694 (CFP) or pB2697 (YFP), respectively, with primers delTRPGFP5′ (5′-TATTGAGCACGTGAGTATACGTGATTAAGCACACAAAGGCAGCTTGGAGTGCAGGTCGACGGATCCCCGGG-3′) and delTRPGFP3′ (5′-GAACGTGCACTGAGTAGTATGTTGCAGTCTTTTGGAAATACGAGTCGAATTCGAGCTCGTTTAAAC-3′) and transformed into PY5997 at the TRP1 locus. The haploid ancestor strains expressing CFP (PY5998) or YFP (PY5999) were confirmed by PCR and fluorescence microscopy. The haploid ancestors were modified to become mating competent by transformation with plasmids PB2647 (LEU2-STE4) and PB2649 (URA3-STE4-MATα). Diploid zygotes were selected on –Ura –Leu plates, and then colony purified on YPD plates to allow plasmid loss. Diploid chromosome content was confirmed by flow cytometry and aCGH, and strains PY6006, PY6008, PY6014, and PY6022 were selected. The diploid ancestors were made mating competent by transformation with plasmids PB2647 and PB2649. Tetraploid zygotes were pulled onto YPD plates using a micromanipulator and after 2 days growth at 30°C, the ploidy of each zygote was determined by flow cytometry and aCGH.
The snf3-G439E mutation was constructed in the haploid YFP strain background (PY5999) using the pCORE counter-selectable reporter system48, a gift from Dr. Michael Resnick (NIEHS). Primers SNF3_pCORE_KAN (5′-TGTTGGGGGTGTTATCATGACTATAGCCAACTTTATTGTGGCCATTGTTGGGAGCTCGTTTTCGACACTGG-3′) and SNF3_pCORE_URA (5′-TATAAATGCTATCATAACTTTTGCGGCCGCTACAGTCTTTAAGGAACACTCCTTACCATTAAGTTGATC-3′) were designed to integrate the CORE sequence at the SNF3 locus; PCR amplification and transformation procedures were followed as detailed previously49. Sanger sequencing was used to identify clones with the desired mutation (chrIV: 112,896 G>A). Diploid snf3-G439E mutants (heterozygous snf3-G439E/SNF3 and homozygous snf3-G439E/snf3-G439E clones) were constructed by mating after introduction of plasmids to confer mating competence (PB2649 or PB2647), as was described above for the construction of the CFP- and YFP- marked strains. An analogous strategy was used to generate tetraploid snf3-G439E strains (heterozygous snf3-G439E/SNF3/SNF3/SNF3).
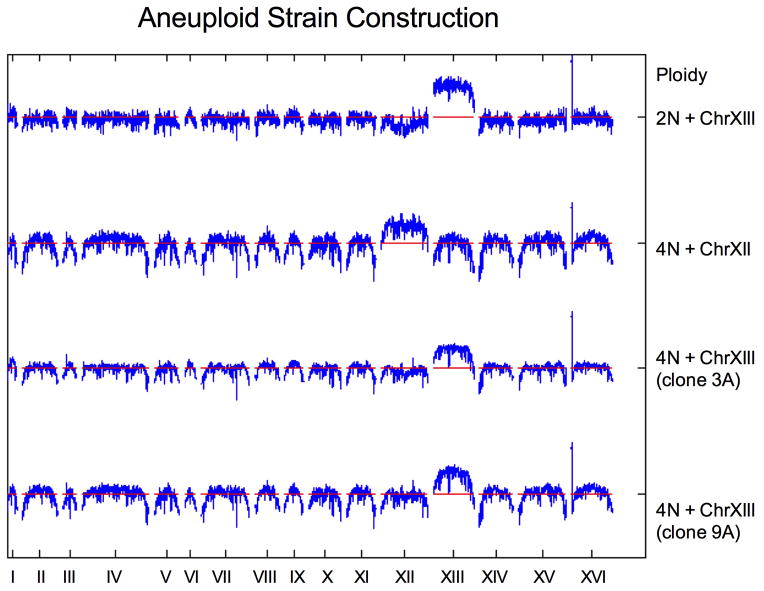
The ChrXIII aneuploid strain series was constructed in the S288c background from the diploid strain PY7245 (RL4737) and the diploid PY7246 (RL4888), which is trisomic for ChrXIII 50. PY7246 was isolated from a triploid meiosis and a minimal number of cell divisions50. We confirmed the ChrXIII trisomy by aCGH (Extended Data Fig. 9). We generated tetraploid clones by mating PY7245 to PY7246, with changes in mating-type accomplished as described previously12. Tetraploid clones were isolated on selective media and analyzed by flow cytometry and aCGH (representative clones Extended Data Fig. 9). Additional details for all yeast strain constructions are available upon request.
Relative Fitness assays
Competitive fitness assays were performed using single colony isolates from the evolved populations and a common ancestor. One single colony was isolated from frozen stocks of each well of the evolution experiments (1N(A), 1N(B), 2N(A), 2N(B), 4N(A), 4N(B), 4N(C)) at generation 250. The evolved clones were cultured for 24 hours in 500 μl of SC + 2% raffinose, diluted into fresh medium, and competed with the ancestor expressing the complementary fluorescent protein. Competitions were initially performed using approximately the same number of cells from the ancestor and the evolved clone, but because the evolved clones grew significantly faster than the ancestor strains, the competitions were repeated using approximately 5 times more ancestor cells than evolved clone cells, with an initial population size of 1×105. Serial dilutions were performed each day and the YFP/CFP ratio was determined by flow cytometry, yielding an estimate of the number of evolved (Nt1) cells relative to the ancestors (Nt0) as a function of time. The data were analyzed in Matlab using a custom script that performed a linear least squares fit of log(Nt1/Nt0) over multiple dilution cycles. The fitness relative to the ancestor is defined as s=d/dt [log2(Nt1/Nt0)], where t is measured in days18.
Flow cytometry analysis of DNA content
Cells were prepared for propidium iodide staining as described but with modifications to optimize preparation of samples in 96 well plates. 30,000 cells were analyzed using the BD LSRII HTS. Flow-Jo Cell Cycle analysis was performed using the Dean-Jett-Fox model to estimate the mean G1 and G2 fluorescence peaks of each strain. Control parental 1N, 2N, 3N, and 4N strains were analyzed in triplicate with the evolved strains.
Microarray Comparative Genome Hybridization (aCGH)
Fluorescently labeled DNA was prepared for comparative genome hybridization as described previously51. Genomic DNA from all experimental strains was compared to the same pool of genomic DNA from the ancestral strain background PY3295 (BY4741, Research Genetics). Agilent yeast DNA 4×44K microarrays (ChIP-on-chip Kit) were used for the hybridization according to the manufacturer’s instructions (Agilent Technologies) with several modifications (M. Dunham online protocols, http://dunham.gs.washington.edu/protocols.shtml). Briefly, 2.0 μg of HaeIII-digested (New England Biolabs) genomic DNA was labeled with 2.1 μl of Cy3 or Cy5 (CyDyeTm-Cy3-dUTP or CyDyeTm-Cy5-dUTP, Amersham GE Healthcare). 300 ng of Cy3-labeled DNA (experimental strains) was mixed with 300 ng of Cy5-labeled DNA (control DNA) and the volume was brought to 44 μl with nuclease free water. Blocking Buffer and Hybridization Buffer 2x HiRPM (Agilent Technologies) were added, and 100 μl was applied to each sub-array; the microarray was hybridized at 65°C for 17 hours and then washed, scanned, and analyzed according to the manufacturer’s instructions. Agilent Feature Extraction data were converted from Log10 ratios to Log2 ratios and plotted using Treeview31 and a custom Matlab script. A Log2 ratio of zero (baseline) indicates no difference in DNA copy number between reference and experimental samples14,51.
Quantitative PCR
All quantitative PCR were performed on an Applied Biosystems ViiA-7 real-time PCR machine in 96-well format with Power SYBR Green PCR Master Mix (Applied Biosystems) and 3 technical replicates. HXT6/7 gene copy number was determined relative to the ancestor as previously described52. Genomic DNA was isolated and RNase treated from 30 clones with the highest fitness from each haploid, diploid, and tetraploid experiment. HXT6/7 on ChrIVR was amplified using forward primer 5′-GATTATTGCTGGTCCGATCC-3′ and reverse primer 5′-GAGTAATCGCCAATGGGTCT -3′ and the control loci UBP1 on ChrIVL was amplified using forward primer 5′-GCGCTCTGTCATTGTTCACT-3′ and reverse primer 5′-GACTTTCAGCTTCGTCCACAA-3′. Raw HXT6/7 values were normalized to UBP1 for each clone and then normalized to the ancestor (ΔΔCt). We found the amplification in 3% of the 1N, 30% of the 2N, and 43% of the 4N-evolved clones (n=30). This significant bias for HXT6/7 amplification in the 2N and 4N populations (t-test, p=0.005 and p=1e-04, respectively) may be due to mutations in 1N cells that prevent the acquisition of the HXT6/7 amplification because of negative epistasis24.
SNF3 gain-of-function mutations were previously shown to increase expression of HXT4 25. Therefore we analyzed HXT4 gene expression levels in diploid evolved clone 2N_233 (carrying the snf3-G439E mutation), relative to the diploid ancestor (PY6006). Strains were grown up from −80C stocks overnight in 5 mL SC + 2% raffinose and then diluted into fresh 25 ml SC + 2% raffinose. Cells were cultured at early log phase and RNA was extracted using the RNeasy Mini Kit (Qiagen). cDNA was prepared using SuperScript III First-Strand synthesis system (Life Technologies). HXT4 was amplified using primers 5′-TAAGGTCAGCGCAGACGATCCA-3′ and 5′-TTCACCCCAGGAGGCATTACCA-3′ and ACT1 was amplified using primers 5′-ACGTCGCCTTGGACTTCGAACA-3′ and 5′-TGGAACAAAGCTTCTGGGGCTC-3′. Raw qPCR values were normalized to ACT1 levels and then normalized to the 2N ancestor (PY6006). Relative to the ancestor, the clone bearing SNF3-G439E had 8-fold higher HXT4 expression in raffinose medium.
Whole Genome Sequencing Overview
We performed whole genome sequencing and identified de novo variants for 74 evolved clones and 2 ancestors. Initially 6 evolved clones and one tetraploid ancestor were sequenced on ABI’s SOLiD 4 platform. Subsequent sequencing of 68 evolved clones and the haploid ancestor was performed on an Illumina HiSeq 2500. The specifics of the analysis pipeline for each sequencing platform are provided below. Regardless of the underlying platform, the overall analysis strategy was as follows. Briefly, the raw reads underwent quality analysis and barcode/adapter removal. High quality reads were mapped to the Saccharomyces cerevisiae reference genome (downloaded June 2010). Reads containing PCR-based artifacts were removed and alignments underwent local realignment around insertions and deletions (indels) resulting in the highest quality alignment. Single nucleotide polymorphisms (SNPs) and indels were called and combined across the evolved strains and within the parental strains to identify a set of variants in the strain background relative to the reference. Each evolved strain (all 74) was individually compared to the parental set to identify the set of potential de novo variants. These evolved strain calls were filtered by quality metrics and manually inspected. All variants of moderate or poor quality as well as a few good quality variants were analyzed by Sanger sequencing. Chromosomal aneuploidy was inferred from changes in read depth using a windowing approach.
Per cell, the evolved tetraploids have more mutations that haploids or diploids. However, per haploid genome, the evolved tetraploids on average accumulate a similar number of mutations (1.50 average SNPs per haploid genome, based on final evolved ploidy) as the evolved diploids (1.44 average SNPs per haploid genome) and fewer mutations than the evolved haploids (2.05 average SNPs per haploid genome), however neither comparison is significant. It is likely that the number of mutations in the evolved tetraploids is underestimated because of the high rate of chromosome loss in these strains. Interestingly, there is a higher average for the tetraploid evolved clones that became ~2N in ploidy (2.00 average SNPs per haploid genome) than the diploid evolved clones (1.44 average SNPs per haploid genome), suggesting that near-diploid cells that underwent a tetraploid intermediate may acquire more mutations than cells that remained diploid throughout the experiment. However, despite the trend, this effect did not reach statistical significance (p = 0.22)).
Illumina Sequencing (2×100)
Library Prep
Clones selected for whole genome sequencing were cultured overnight from −80°C stocks in 2 ml SC + 2% raffinose medium. Genomic DNA was isolated using phenol-chloroform- isoamylalcohol (24:25:1) and bead beating. Libraries were prepared as described53. Brielfy, DNA was sheered with Diagenode Bioruptor (UCD-200) to a median size of 300–500 bp, end-repair was performed with NEB Next End repair kit (NEB E6050L) and fragments were A-tailed with Klenow fragment (M0212L). Custom adaptors with in-line barcodes were ligated overnight. Adaptor ligated fragments were size selected on 1% TBE agarose gel stained with Sybr Gold (Invitrogen S-11494) for fragments between 400–600bp and isolated using Qiagen Gel Extraction Kit (28706). Libraries were amplified for 12 cycles with Illumina PE PCR primers 1.0 and 2.0 (Oligonucleotide sequences © 2007–2013 Illumina, Inc. All rights reserved.). Libraries were pooled and underwent additional size selection for fragments of 400–600bp.
Raw data
The genomes were sequenced on an Illumina HiSeq 2500 at the University of Colorado at Denver Next Generation Sequencing Facility. The data, which had an inline barcode, was demultiplexed by the sequencing facility into individual sample R1/R2 files—one file for each read in the pair. The barcodes were removed prior to mapping using Fastx_trimmer (v0.0.13.2, http://hannonlab.cshl.edu/fastx_toolkit/). Read trimming from the 5′ end of the R2 reads was performed on a sample-specific manner trimming anywhere from 0–28 basepairs using in-house script and Fastx_trimmer.
Mapping
Reads were mapped to the Saccharomyces cerevisiae reference sequence for the laboratory yeast strain S288c reference genome (S. cerevisiae genome obtained July 28, 2010 from the Saccharomyces Genome Database, FTP SITE: http://downloads.yeastgenome.org/sequence/S288C_reference/genome_releases/
ACTUAL GENOME: http://downloads.yeastgenome.org/sequence/S288C_reference/genome_releases/S288C_reference_genome_R63-1-1_20100105.tgz). The reads were mapped using the Bowtie2 (v2.0.2)54 local alignment strategy, allowing for multiple mapping, and setting following options: --very-sensitive-local -I 180 -X 1000 --score-min G,70,8. The mapped reads then underwent file format conversion into the binary format for downstream analysis using Samtools view, sort, and index (v0.1.18)55.
Alignment Tailoring
Post-alignment to the genome, duplicate pairs resulting from PCR overamplification were removed using Samtools rmdup, eliminating 1–5% of the paired reads. The reads were realigned over potential indel sites using the Genome Analysis Toolkit (GATK) RealignerTargetCreator and IndelRealigner (v2.4-9)56,57.
Variant Calling and Refinement
Variant calling was performed on the tailored read mappings using GATK UnifiedGenotyper (v2.4-9)56,57. For the haploids and diploids SNPs were called using default parameters, and for the higher ploidy strains the ploidy option was increased to 5N, which allows for identification of mutations at allelic frequencies down to 5% alternate allele representation. Variant lists were combined based on ploidy-type using GATK CombineVariants. SNPs and short indels were compared to the parental set of mutations using in house scripts to generate a set of non-Parental mutations. These mutations were filtered for alternate allele support and allelic frequency (>2 reads supporting alternate allele for coverage 10–20x, and >4 reads supporting alternate allele for coverage >20x). The filtered mutations were manually inspected using the Integrative Genome Viewer (IGV) (v2.1.19)58 to refine the set and further remove mapping artifacts such as strand representation bias, regional mapping quality issues from non-unique mapping, and artifacts of homopolymer and simple repeat alignments. We Sanger sequenced variants with low read support (<5 reads supporting alternate allele), as well as a subset of the other medium and high confidence variants. The final set of evolved variants discovered was annotated versus the gene file specific to the genome using an in house script.
Chromosomal CNV Identification
Identification of chromosomal copy number variations (CNVs) was performed using HTSeq (v0.6.1)59 in conjunction with custom scripts. HTSeq performs coverage estimations on a per-gene basis, and the custom scripts provided normalized Log2FoldChange between each sample and the parental haploid strain. Estimates on chromosomal copy number were inferred using the median value for the Log2FoldChange on a chromosome-by-chromosome basis. We implemented the Cochran Armitage test to determine whether ChrXIII had a trend for higher copy number, relative to the copy number observed for all chromosomes in the tetraploid evolved clones (Fig. 2d and Supplementary Table 2). This trend analysis is similar to a Chi square test, but tests whether there is a significant trend or direction to the observed data set (ChrXIII copy number).
Sequencing Quality Assessment
Because our sequencing was highly multiplexed, quality assessment on the sequencing data was necessary to eliminate strains without adequate genome coverage. For the haploids and diploids, we determined the adequate depth of coverage to recover mutations in two ways. First, we took the set of “strain-background” mutations, which were identified by filtering the parental variant calls for a conservative, high quality (qual > 100), homozygous set of locations. Each strain was then queried for the ability to recapitulate these variants, reporting a percentage overlap between each strain’s variant calls and the set of background variants. Any strain with less than 97% of the background mutations was dropped from consideration. Additionally, we examined the impact of subsampling down to various depths to investigate the impact of lower coverage on recovering variants. This was done using Picard’s DownsampleSam.jar (v1.72, http://broadinstitute.github.io/picard/) on two higher coverage diploid strains to randomly down-sample the coverage to 100x, 50x, 25x, and 10x coverage. We examined the SNP call overlap and found that for the strain-unique SNPs they could be captured even at a level of 10x coverage. Using this information, we set minimum coverage requirements for each strain on a genome wide scale to eliminate strains without adequate genomic representation. Depth of coverage analysis was performed on all of the mapped data using Bedtools genomeCoverageBed (v2.16.2)60. The per-base coverage was then analyzed using an in house script to produce statistics on minimum coverage per allele, average coverage, etc.
SOLiD Sequencing (2×50)
Library Prep
A pilot experiment was performed on 7 strains using SOLiD paired-end sequencing (Supplementary Table 1). Clones selected for SOLiD sequencing were cultured overnight from −80°C stocks in 4 ml SC + 2% raffinose medium. Genomic DNA was isolated using QIAGEN Genomic-Tip 100 according to the manufacturer’s instructions. SOLiD library preparation and sequencing was performed by the Molecular Biology Core Facility at Dana-Farber Cancer Institute according to the manufacturer’s instructions (Applied Biosystems, Life Technologies).
Mapping
The sequencing reads were mapped to the Saccharomyces cerevisiae reference genome (See Illumina Mapping) using multiple different mapping software including BWA (v.0.5.9)61, NovoAlignCS (v1.01.05)62, Bfast(v0.6.5a)63, and BowtieCS (v 0.12.7)64. BowtieCS and BWA were used in the downstream variant calling and copy number changes, while NovoAlignCS and Bfast served as added support in manual inspection of variants.
Alignment Tailoring
After mapping, the reads were post-processed for local realignment using SRMA (v0.1.15)65 and Samtools BAQ (v0.1.18)55.
Variant Calling and Refinement
Single nucleotide polymorphisms (SNPs), small insertions and deletions (indels) were called from the post-processed reads using Samtools Mpileup(v0.1.18)55, VARiD (v1.0.7f)66, and Freebayes (v0.8.9, http://bioinformatics.bc.edu/marthlab/FreeBayes). Samtools and VARiD variant calls were used to identify the strain background (parental variants relative to the reference). These variants were filtered on the basis of reads supporting the allele in both directions, quality score of the call, and adequate read coverage over the call. Once filtered, all of the variant calls for the evolved strains were merged and compared to the parent. Variations were verified by manual inspection followed by Sanger validation for both a set of randomly sampled loci and regions of disagreement between different combinations of the mapping software and the variant callers (i.e. dinucleotide SNPs and multiple indels within a single read). The resulting set is later used for identification of strain-unique variants in the evolved strains.
To identify strain-unique variants, Freebayes, a variant caller capable of higher-ploidy (ploidy > 2N), was used. Freebayes has the ability to set the assumed ploidy over a genomic region to adjust the expected distribution for allelic frequency. The assumed ploidy was determined using aCGH as well as the copy number changes. Freebayes’ called variants on each evolved progeny were then cross-referenced with the parental variants to produce strain-unique variants (Supplementary Table 1). These variants were then manually examined in IGV58 and validated by Sanger sequencing. PCR amplification and Sanger sequencing of ~200 bp on either side of the sequence variants was performed using DNA from the evolved clone and the ancestor.
Chromosomal Copy Number Variations
Copy number changes, first identified in the aCGH data, were confirmed in the gDNA sequencing using BedTools genomeCoverageBed60 in combination with custom in-house scripts and DESeq (v1.10.1)67. Briefly, the normalized genomic copy number of all annotated genes in each strain was compared back to the parent. These comparisons then were plotted using an in house script (See Illumina Chromosomal CNV Identification).
Extended Data
Extended Data Figure 1. A schematic representation of the construction of isogenic haploid, diploid, and tetraploid strains used in this study.
Relevant strain numbers are indicated for the CFP-containing and YFP-containing ancestors.
Extended Data Figure 2. Estimates from our mathematical modeling of the best-fit value of the beneficial mutation rate (μ) and the selection coefficient (s) of each ploidy evolution experiment.
(a) Table of μ and s values that had the best-fit between the simulations and the experimental data, brackets indicate 95% confidence intervals. Values were determined based on different assumptions about the underlying distribution of beneficial mutations, which included: (b) uniform, (c) exponential, and (d) delta distributions. Estimates of μ and s were also obtained with (e) the Equivalence Principle model18 that assumes a delta distribution of beneficial mutations. Each two-dimensional plot includes the error range obtained by parametric bootstrap of 1000 independent simulated datasets (Methods). The 95% confidence intervals of μ and s from those 1,000 datasets were defined as the error ranges. (f) A schematic diagram of the three distributions of fitness effects that we used in our mathematical modeling: exponential (red), uniform (black), and delta (green) distributions. Just for illustration, we also provide a narrow Gaussian distribution (blue) that is close to a delta function. The real distribution of fitness effects probably has a more complex structure than any of the examples shown. The diagram illustrates the fact that the shape of the assumed distribution mandates differences in mutation rates. For example, if the mutations that mainly drive adaptation fall within the region of the double arrow, only a small proportion of the mutations from the exponential distribution will fall within this range, necessitating a much higher mutation rate to generate mutations in this region. By contrast, the delta distribution lies in the middle of the double arrow range; therefore, all of the mutations that arise from this distribution are strong enough to contribute to adaptation, resulting in a relatively lower mutation rate. The uniform distribution is intermediate between these two extremes. Only a small portion of the mutations of the uniform distribution is within the double arrow region, but the probability of these mutations is orders of magnitude larger than the exponential. Therefore, the mutation rate of the uniform is closer to the delta than to the exponential distribution. The values used to generate this figure are the best-fit values of μ and s of the haploid populations in the different three distributions. See Methods for more details.
Extended Data Figure 3. Experimental and computational analyses of the noise in our experimental measurements and of the methods used in our mathematical modeling (see Supplementary Information).
(a) Three different methods were used to determine the percent of YFP-expressing cells in mixtures of the 1N, 2N, and 4N CFP and YFP ancestor strains. Cells were analyzed by flow cytometry (10,000 cells) and fluorescence microscopy (300 cells), and by single colony analysis (96 colonies) of the mixture before galactose induction. The percent YFP determined by all three methods was highly correlated (Pearson correlation coefficient = 0.98). (b) Table showing variation in flow cytometry replicate measurements. The standard deviation of the percent YFP obtained from 48 replicate populations of 6 different CFP:YFP ratios, for each ploidy type. (c) Table showing the average and standard deviation of the best-fit values for different ratios between beneficial (Ub) and deleterious (Ud) mutations, obtained from 100 independent datasets. (d) Evaluation of different summary statistics by calculating the distribution of best-fit values from 1000 replicate simulations. Four different summary statistics were used to analyze 1000 replicates of a parameter pair, s and μ (see Methods). The summary statistic using 10 bins has the highest mode and no outliers and was used to generate our best-fit values. (e) Criteria for exclusion of near-neutral mutations for implementation of the branching evolutionary model with an exponential distribution of mutations. Shown is the average deviation from equal percentages of YFP and CFP-expressing cells with different thresholds for neutral mutations. The threshold (Tr) represents the fraction of the average fitness effect (s), meaning every mutation whose absolute value is smaller than Tr*s was excluded. For this scenario (with parameters μ=2*10−5 and s=0.08), we can exclude every mutation with a fitness effect smaller than s (i.e. Tr=1, light blue) without changing the outcome relative to excluding no mutations (Tr=0). However, when excluding all mutations with fitness effects smaller than ten times s (Tr=10, dark green), the result changes substantially. Thus, for high mutation rates (μ*N>1), we can exclude weak mutations19.
Extended Data Figure 4. aCGH karyotype of the ancestor strains used in this study.
Aneuploidy was not detected in the parental 1N, 2N, or 4N strains. Genomic DNA from each strain was compared to that of an isogenic ancestor PY3295 (BY4741 MATa ura3 his3 trp1 leu2 LYS2) and log2 DNA copy number ratios were plotted using a custom Matlab script. To account for regions of complete deletion, the data were cropped at log2 ratios of ± 2.0 and averaged across each chromosome using a sliding window of nine oligos. A log2 ratio of zero is indicated by the red line. Loci altered during strain construction are indicated (TRP1, pSTE5, URA3, STE4). Strain ploidy, determined by flow cytometry, is indicated on the right.
Extended Data Figure 5. aCGH karyotype of haploid and diploid evolved clones at generation 250.
(a) aCGH of eight haploid evolved clones. Data are displayed as in Extended Data Fig. 4. No aneuploidy was detected. Clone 1N_131 acquired the HXT6/7 amplification (arrow). (b) aCGH of eight diploid evolved clones. No aneuploidy was detected, but all clones except 2N_233 acquired the HXT6/7 amplification. Log2 ratios were averaged across each chromosome using a sliding window of twenty-nine oligos. The ploidy of the evolved clone, determined by flow cytometry, is indicated on the right.
Extended Data Figure 6. aCGH karyotype for twenty tetraploid evolved clones at generation 250.
aCGH data are displayed as in Extended Data Fig. 4. Note that whole chromosome or large segmental chromosome gain and loss events are observed in all clones except clone 4N_337. Ploidy of the evolved clone, determined by flow cytometry, is indicated on the right, with +/− indicating chromosome aneuploidy. Some highly aneuploid clones had widely different chromosome copy numbers for different chromosomes (e.g. some chromosomes were disomic, others trisomic and tetrasomic).
Extended Data Figure 7. Analysis of recurrent and concerted chromosome loss events in the tetraploid evolved clones.
(a) Evolved tetraploids acquired large segmental aneuploidies (regions greater than the ~7kb HXT6/7 amplification). aCGH data for individual chromosomes with large segmental aneuploidies in 4N-evolved clones (plotted using Treeview31). All breakpoints occurred at or near Ty sequences (arrowheads). (b) The pairwise patterns (Pearson correlation) of all chromosome copy number alterations in the 4N-evolved clones at generation 250 (n = 30, Supplementary Table 2). The copy number of some chromosomes were correlated (e.g. ChrXV and chrXVI), whereas others were anti-correlated (e.g. ChrVIII and ChrIX), possibly reflecting the need for gene expression balance. (c) Hierarchical clustering showing the copy number relationship among the chromosomes. (d) Proportion of all chromosomes in the evolved tetraploid clones with the indicated copy number (black). The copy number of ChrXIII (grey) in the 4N-evolved clones at generation 250 was significantly different from that of all other aneuploid chromosomes (Cochran Armitage test, p<1e-07).
Extended Data Figure 8. aCGH karyotype for tetraploid evolved clones at generations 35, 55, and 500.
All 4N-evolved clones at (a) generations 35 and 55 and (b) generation 500 are aneuploid for multiple chromosomes or carry large segmental chromosome aneuploidies, except for clone 4N_503, which remained tetraploid. Data are displayed as in Extended Data Fig. 4. Ploidy of the evolved clone, determined by flow cytometry, is indicated on the right, with +/− indicating chromosome aneuploidy.
Extended Data Figure 9. aCGH from isogenic 2N and 4N strains with an extra copy of ChrXIII or ChrXII.
Data are displayed as in Extended Data Fig. 5b.
Extended Data Table 1.
Yeast strains and plasmids used in this study
| Strain (Ploidy) or Plasmid | Parental strain | Relevant genotype | Source |
|---|---|---|---|
| BY3295 (1N) | BY4741 | MATa his3Δ leu2Δ met15Δ ura3Δ | Pellman collections |
| PY5997 (1N) | BY3295 | matΔ::pSTE5-ura3::TRP1 ste4Δ::HygroR trp1::NatR | This study |
| PY5998 (1N) | PY5997 | matΔ::pSTE5-ura3::TRP1 ste4Δ::HygroR trp1::NatR::pGAL-ceCFP-tADH-SpHIS5 | This study |
| PY5999 (1N) | PY5997 | matΔ::pSTE5-ura3::TRP1 ste4Δ::HygroR trp1::NatR::pGAL-eYFP-tADH-SpHIS5 | This study |
| PY6006 (2N) | PY5999 | (2x) matΔ::pSTE5-ura3::TRP1 ste4Δ::HygroR trp1::NatR::pGAL-eYFP-tADH-SpHIS5 | This study |
| PY6008 (2N) | PY5998 | (2x) matΔ::pSTE5-ura3::TRP1 ste4Δ::HygroR trp1::NatR::pGAL-ceCFP-tADH-SpHIS5 | This study |
| PY6014 (2N) | PY5999 | (2x) matΔ::pSTE5-ura3::TRP1 ste4Δ::HygroR trp1::NatR::pGAL-eYFP-tADH-SpHIS5 | This study |
| PY6022 (2N) | PY5998 | (2x) matΔ::pSTE5-ura3::TRP1 ste4Δ::HygroR trp1::NatR::pGAL-ceCFP-tADH-SpHIS5 | This study |
| PY6031 (4N) | PY6008 | (4x) matΔ::pSTE5-ura3::TRP1 ste4Δ::HygroR trp1::NatR::pGAL-ceCFP-tADH-SpHIS5 | This study |
| PY6032 (4N) | PY6022 | (4x) matΔ::pSTE5-ura3::TRP1 ste4Δ::HygroR trp1::NatR::pGAL-ceCFP-tADH-SpHIS5 | This study |
| PY6040 (4N) | PY6006 | (4x) matΔ::pSTE5-ura3::TRP1 ste4Δ::HygroR trp1::NatR::pGAL-eYFP-tADH-SpHIS5 | This study |
| PY6045 (4N) | PY6014 | (4x) matΔ::pSTE5-ura3::TRP1 ste4Δ::HygroR trp1::NatR::pGAL-eYFP-tADH-SpHIS5 | This study |
| PY7232 (4N) | PY5999 | SNF3-G439E | This study |
| PY7237–PY7238 (2N) | PY5999 | SNF3-G439E/SNF3 | This study |
| PY7233–PY7236 (2N) | PY5999 | SNF3-G439E/SNF3-G439E | This study |
| PY7241–PY7244 (4N) | PY5999 | SNF3-G439E/SNF3/SNF3/SNF3 | This study |
| PY7245 (2N) | S288c | RLY4737 MATa/α ura3Δ his3Δ trp1Δ leu2Δ | 50 |
| PY7246 (2N) | PY7245 | RLY4888 MATa/α + ChrXIII trisomy | 50 |
| PY7247–PY7249 (4N) | PY7245 | MAT a/a/α/α | This study |
| PY7250–PY7252 (4N) | PY7245 | MATa/a/α/+ ChrXIII pentasomy | This study |
| PY7253–PY7255 (4N) | PY7245 | MATa/a/α/+ ChrXII pentasomy | This study |
| PB1500 | YFP-tADH-SpHIS5, AmpR | Yeast Resource Center | |
| PB1499 | CFP-tADH KanR AmpR | Yeast Resource Center | |
| PB2452 | CFP-tADH SpHIS5, AmpR | Pellman collection | |
| PB2694 | pGAL1-ceCFP-tADH-SpHIS5, AmpR | This study | |
| PB2697 | pGAL1-eYFP-tADH-SpHIS5, AmpR | This study | |
| PB2314 | MATa::pSTE5-URA3, AmpR | 12 | |
| PB1308 | ura3::TRP1 AmpR | 47 | |
| PB2577 | MATα URA3 CEN AmpR | This study | |
| B1819 | LEU2 CEN AmpR | Pellman collection | |
| PB2647 | STE4 LEU2 CEN AmpR | This study | |
| PB2649 | STE4 URA3 CEN MATα, AmpR | This study | |
| PB1640 | hphMX4 AmpR | 68 | |
| PB1942 | pGAL-HO HIS3 AmpR | Gift of the Fink lab | |
| PB1650 | pGAL-HO URA3 LEU2 | Gift of the Elion lab | |
| pCORE | kanMX4 KIURA3 | 48 |
Supplementary Material
Supplementary Table 1. Variants identified by whole genome sequencing
Supplementary Table 2. Summary of the ploidy and chromosome copy number data from aCGH and whole genome sequencing of the evolved haploid, diploid, and tetraploid clones.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute, the National Institutes of Health (R37 GM61345), the G. Harold & Leila Y. Mathers Charitable Foundation, the Dana-Farber Cancer Institute Physical Sciences-Oncology Center (U54CA143798), the Boettcher Foundation’s Webb-Waring Biomedical Research Program, the National Science Foundation (NSF 1350915), the National Institutes of Health (R01 GM081617), and an American Cancer Society Postdoctoral Fellowship.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author Contributions:
A.M.S., M.G., N.S., R.K., and D.P. contributed to the overall study design. A.M.S. and M.G. performed the experiments. Y.E.M. implemented the mathematical modeling with contributions from N.S., R.K., and F.M. A.M.S., P.A.R., and A.L.S. generated WGS libraries, P.A.R. developed the sequencing pipeline and analyzed the WGS data with help from A.M.S. and A.L.S, under the supervision of R.D.D. Data analysis was carried out by A.M.S., Y.E.M., P.A.R., M.G., N.S., A.L.S., S.D., F.M. and D.P. The manuscript was written primarily by A.M.S., Y.E.M., and D.P. with contributions from the other authors.
All aCGH data are available at NCBI’s GEO database under accession number GSE51017 and all whole genome sequence data are available at NCBI’s SRA database under accession number SRP047435.
The authors have no competing financial interests.
References
- 1.Ohno S, Wolf U, Atkin NB. Evolution from fish to mammals by gene duplication. Hereditas. 1968;59:169–187. doi: 10.1111/j.1601-5223.1968.tb02169.x. [DOI] [PubMed] [Google Scholar]
- 2.Otto SP, Whitton J. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- 3.Semon M, Wolfe KH. Consequences of genome duplication. Curr Opin Genet Dev. 2007;17:505–512. doi: 10.1016/j.gde.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Hufton AL, Panopoulou G. Polyploidy and genome restructuring: a variety of outcomes. Curr Opin Genet Dev. 2009;19:600–606. doi: 10.1016/j.gde.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Paquin C, Adams J. Frequency of fixation of adaptive mutations is higher in evolving diploid than haploid yeast populations. Nature. 1983;302:495–500. doi: 10.1038/302495a0. [DOI] [PubMed] [Google Scholar]
- 6.Anderson JB, Sirjusingh C, Ricker N. Haploidy, diploidy and evolution of antifungal drug resistance in Saccharomyces cerevisiae. Genetics. 2004;168:1915–1923. doi: 10.1534/genetics.104.033266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zorgo E, et al. Ancient Evolutionary Trade-Offs between Yeast Ploidy States. PLoS genetics. 2013;9:e1003388. doi: 10.1371/journal.pgen.1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayer VW, Aguilera A. High levels of chromosome instability in polyploids of Saccharomyces cerevisiae. Mutation research. 1990;231:177–186. doi: 10.1016/0027-5107(90)90024-x. [DOI] [PubMed] [Google Scholar]
- 9.Bennett RJ, Johnson AD. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 2003;22:2505–2515. doi: 10.1093/emboj/cdg235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerstein AC, Chun HJ, Grant A, Otto SP. Genomic convergence toward diploidy in Saccharomyces cerevisiae. PLoS genetics. 2006;2:e145. doi: 10.1371/journal.pgen.0020145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujiwara T, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 12.Storchova Z, et al. Genome-wide genetic analysis of polyploidy in yeast. Nature. 2006;443:541–547. doi: 10.1038/nature05178. [DOI] [PubMed] [Google Scholar]
- 13.Rancati G, et al. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135:879–893. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nature genetics. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chao DY, et al. Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis. Science. 2013;341:658–659. doi: 10.1126/science.1240561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haccou P, Jagers P, Vatutin VA International Institute for Applied Systems Analysis. . Branching processes: variation, growth, and extinction of populations. Cambridge University Press; 2005. [Google Scholar]
- 18.Hegreness M, Shoresh N, Hartl D, Kishony R. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science. 2006;311:1615–1617. doi: 10.1126/science.1122469. [DOI] [PubMed] [Google Scholar]
- 19.Barrett RD, M’Gonigle LK, Otto SP. The distribution of beneficial mutant effects under strong selection. Genetics. 2006;174:2071–2079. doi: 10.1534/genetics.106.062406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kao KC, Sherlock G. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nature genetics. 2008;40:1499–1504. doi: 10.1038/ng.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang GI, et al. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature. 2013;500:571–574. doi: 10.1038/nature12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown CJ, Todd KM, Rosenzweig RF. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Molecular biology and evolution. 1998;15:931–942. doi: 10.1093/oxfordjournals.molbev.a026009. [DOI] [PubMed] [Google Scholar]
- 23.Gresham D, et al. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS genetics. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kvitek DJ, Sherlock G. Reciprocal sign epistasis between frequently experimentally evolved adaptive mutations causes a rugged fitness landscape. PLoS genetics. 2011;7:e1002056. doi: 10.1371/journal.pgen.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozcan S, Dover J, Rosenwald AG, Wolfl S, Johnston M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci USA. 1996;93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrick JE, Kauth MR, Strelioff CC, Lenski RE. Escherichia coli rpoB mutants have increased evolvability in proportion to their fitness defects. Molecular biology and evolution. 2010;27:1338–1347. doi: 10.1093/molbev/msq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kryazhimskiy S, Rice DP, Jerison ER, Desai MM. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science. 2014;344:1519–1522. doi: 10.1126/science.1250939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheltzer JM, et al. Aneuploidy drives genomic instability in yeast. Science. 2011;333:1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dewhurst SM, et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer discovery. 2014;4:175–185. doi: 10.1158/2159-8290.CD-13-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ezov TK, et al. Molecular-genetic biodiversity in a natural population of the yeast Saccharomyces cerevisiae from “Evolution Canyon”: microsatellite polymorphism, ploidy and controversial sexual status. Genetics. 2006;174:1455–1468. doi: 10.1534/genetics.106.062745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 32.Durrett R, Foo J, Leder K, Mayberry J, Michor F. Intratumor heterogeneity in evolutionary models of tumor progression. Genetics. 2011;188:461–477. doi: 10.1534/genetics.110.125724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu YO, Siegal ML, Hall DW, Petrov DA. Precise estimates of mutation rate and spectrum in yeast. Proc Natl Acad Sci USA. 2014;111:E2310–2318. doi: 10.1073/pnas.1323011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barbera MA, Petes TD. Selection and analysis of spontaneous reciprocal mitotic cross-overs in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2006;103:12819–12824. doi: 10.1073/pnas.0605778103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rouzine IM, Wakeley J, Coffin JM. The solitary wave of asexual evolution. Proc Natl Acad Sci USA. 2003;100:587–592. doi: 10.1073/pnas.242719299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desai MM, Fisher DS, Murray AW. The speed of evolution and maintenance of variation in asexual populations. Curr Biol. 2007;17:385–394. doi: 10.1016/j.cub.2007.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fogle CA, Nagle JL, Desai MM. Clonal interference, multiple mutations and adaptation in large asexual populations. Genetics. 2008;180:2163–2173. doi: 10.1534/genetics.108.090019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vetterling WT. Numerical recipes example book (C) 2. Cambridge University Press; 1992. [Google Scholar]
- 39.Wakeley J. Coalescent theory: an introduction. Roberts & Co. Publishers; 2009. [Google Scholar]
- 40.Moura de Sousa JA, Campos PR, Gordo I. An ABC method for estimating the rate and distribution of effects of beneficial mutations. Genome biology and evolution. 2013;5:794–806. doi: 10.1093/gbe/evt045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goyal S, et al. Dynamic mutation-selection balance as an evolutionary attractor. Genetics. 2012;191:1309–1319. doi: 10.1534/genetics.112.141291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ewens WJ. Mathematical population genetics. 2. Springer; 2004. [Google Scholar]
- 43.Efron B, Tibshirani R. An introduction to the bootstrap. Chapman & Hall; 1993. [Google Scholar]
- 44.Frenkel EM, Good BH, Desai MM. The fates of mutant lineages and the distribution of fitness effects of beneficial mutations in laboratory budding yeast populations. Genetics. 2014;196:1217–1226. doi: 10.1534/genetics.113.160069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 46.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 47.Cross FR. ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast. 1997;13:647–653. doi: 10.1002/(SICI)1097-0061(19970615)13:7<647::AID-YEA115>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 48.Storici F, Lewis LK, Resnick MA. In vivo site-directed mutagenesis using oligonucleotides. Nature biotechnology. 2001;19:773–776. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
- 49.Storici F, Resnick MA. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods in enzymology. 2006;409:329–345. doi: 10.1016/S0076-6879(05)09019-1. [DOI] [PubMed] [Google Scholar]
- 50.Pavelka N, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Selmecki A, Bergmann S, Berman J. Comparative genome hybridization reveals widespread aneuploidy in Candida albicans laboratory strains. Molecular microbiology. 2005;55:1553–1565. doi: 10.1111/j.1365-2958.2005.04492.x. [DOI] [PubMed] [Google Scholar]
- 52.Wenger JW, et al. Hunger artists: yeast adapted to carbon limitation show tradeoffs under carbon sufficiency. PLoS genetics. 2011;7:e1002202. doi: 10.1371/journal.pgen.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hittinger CT, et al. Remarkably ancient balanced polymorphisms in a multi-locus gene network. Nature. 2010;464:54–58. doi: 10.1038/nature08791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in bioinformatics. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Novocraft. Novocraft short read alignment package. 2009 http://www.novocraft.com.
- 63.Homer N, Merriman B, Nelson SF. BFAST: an alignment tool for large scale genome resequencing. PloS one. 2009;4:e7767. doi: 10.1371/journal.pone.0007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Homer N, Nelson SF. Improved variant discovery through local re-alignment of short-read next-generation sequencing data using SRMA. Genome biology. 2010;11:R99. doi: 10.1186/gb-2010-11-10-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dalca AV, Rumble SM, Levy S, Brudno M. VARiD: a variation detection framework for color-space and letter-space platforms. Bioinformatics. 2010;26:i343–349. doi: 10.1093/bioinformatics/btq184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Variants identified by whole genome sequencing
Supplementary Table 2. Summary of the ploidy and chromosome copy number data from aCGH and whole genome sequencing of the evolved haploid, diploid, and tetraploid clones.