Summary
The E. coli alternative sigma factor, σE, transcribes genes required to maintain the cell envelope and is activated by conditions that destabilize the envelope. σE is also activated during entry into stationary phase in the absence of envelope stress by the alarmone (p)ppGpp. (p)ppGpp controls a large regulatory network, reducing expression of σ70-dependent genes required for rapid growth and activating σ70-dependent and alternative sigma factor-dependent genes required for stress survival. The DksA protein often potentiates the effects of (p)ppGpp. Here we examine regulation of σE by (p)ppGpp and DksA following starvation for nutrients. We find that (p)ppGpp is required for increased σE activity under all conditions tested, but the requirement for DksA varies. DksA is required during amino acid starvation, but is dispensable during phosphate starvation. In contrast, regulation of σS is (p)ppGpp- and DksA-dependent under all conditions tested, while negative regulation of σ70 is DksA- but not (p)ppGpp-dependent during phosphate starvation, yet requires both factors during amino acid starvation. These findings suggest that the mechanism of transcriptional regulation by (p)ppGpp and/or DksA cannot yet be explained by a unifying model and is specific to individual promoters, individual holoenzymes, and specific starvation conditions.
Introduction
Most bacteria have an intricate array of stress responses that allow them to sense changes in their surroundings and adapt their transcriptional profiles in order to survive. One of the major classes of stress responses relies on the modular nature of the multisubunit RNA polymerase (RNAP). The α, β, β′ and ω subunits of RNAP form the core enzyme (E), which is responsible for transcription elongation, but cannot specifically initiate transcription (Borukhov and Nudler, 2008). The sigma subunit binds to the core enzyme, forming the holoenzyme (Eσ), and confers specific promoter recognition (Burgess et al., 1969). The bulk of bacterial transcription is directed by a housekeeping sigma factor, σ70 in E. coli. However, many bacteria possess an array of alternative sigma factors that are induced by particular stresses or environmental conditions (Gruber and Gross, 2003; Paget and Helmann, 2003). Once activated, they bind to core RNAP and redirect the enzyme to promoters for genes required for adaptation and survival.
The alternative sigma factor system is very efficient, but is essentially reactionary. A stress occurs, a sigma factor is activated, and a transcriptional response is generated. However, when nutrients are in short supply, the bacterium may not have the resources to rapidly mount a response that requires the energy-consuming processes of transcription and translation. In E. coli, a growing body of evidence suggests that as nutrients are depleted, the alarmones pppGpp and ppGpp (collectively referred to as (p)ppGpp herein) co-ordinately activate the stress-responsive alternative sigma factors, σE, σH, σS and σN, essentially preloading the cell with factors to combat stresses should they arise (Gentry et al., 1993; Hirsch and Elliott, 2002; Jishage et al., 2002; Laurie et al., 2003; Costanzo and Ades, 2006; Costanzo et al., 2008; Österberg et al., 2011). At the same time, (p)ppGpp inhibits transcription of genes required for rapid growth (Durfee et al., 2008; Potrykus and Cashel, 2008; Traxler et al., 2008). In effect, (p)ppGpp mediates a switch from a transcriptional program for rapid growth to one optimized for stress survival (Nyström, 2004; Magnusson et al., 2005;Traxler et al.,2008).
The alternative sigma factor, σE, plays an essential role ensuring the integrity of the bacterial cell envelope (De Las Peñas et al., 1997a; Hayden and Ades, 2008). The amount of σE available to form σE holoenzyme (EσE) is regulated by the antisigma factor, RseA. RseA is an inner membrane protein whose cytoplasmic domain binds tightly to σE and prevents it from interacting with core RNAP (De Las Pehas et al., 1997b; Missiakas et al., 1997; Campbell et al., 2003). RseA is proteolytically unstable, and its degradation increases in response to cell envelope stress (Ades et al., 1999; 2003). When RseA is degraded, σE is released, binds to core RNAP, and directs transcription of the genes in its regulon. In addition to the stress-signalling pathway acting through RseA, (p)ppGpp increases transcription by EσE in response to starvation, independently of RseA and cell envelope stress (Costanzo and Ades, 2006; Costanzo et al., 2008). Therefore, the overall amount of σE activity in the cell is determined by a combination of the two regulatory pathways. The stress induction pathway mediated through RseA has been extensively studied (Barchinger and Ades, 2013); however regulation of σE by (p)ppGpp is not well understood.
(p)ppGpp is a general signal of starvation stress. Its levels increase upon nutrient downshifts and are inversely correlated with growth rate (Dennis et al., 2004; Potrykus and Cashel, 2008). In E. coli, (p)ppGpp is synthesized by two enzymes, RelA and SpoT (Xiao et al., 1991). RelA is a strong (p)ppGpp synthase, while SpoT has weak synthase activity (Cashel et al., 1996). SpoT is also a hydrolase and is responsible for hydrolysing (p)ppGpp (Cashel et al., 1996). spoT is essential for viability due to toxicity associated with the high levels of (p)ppGpp that accumulate in its absence (Xiao et al., 1991). Cells lacking both relA and spoT are devoid of (p)ppGpp and referred to as (p)ppGpp0. Changes in the amount of (p)ppGpp in the cell in response to limitation for various nutrients are achieved via regulation of RelA and/or SpoT activity.
The best-studied cellular role of (p)ppGpp is its involvement in balancing the protein synthetic capacity of the cell with nutrient availability by acting on Eσ70 to negatively regulate transcription of stable RNA operons (rRNA and tRNA) and positively regulate promoters of several amino acid biosynthetic operons (Cashel et al., 1996; Dennis et al., 2004; Paul et al., 2004a). (p)ppGpp regulates transcription at these promoters in concert with the protein, DksA, which potentiates the effects of (p)ppGpp (Paul et al., 2004b; 2005). Because DksA levels are thought to be constant, while those of (p)ppGpp change, (p)ppGpp is regarded as the stress signal for the system (Brown et al., 2002; Paul et al., 2004b). Both (p)ppGpp and DksA bind to core RNAP, (p)ppGpp at the interface between the β′ and ω subunits and DksA in the secondary channel (Perederina et al., 2004; Lennon et al., 2012; Mechold et al., 2013; Ross et al., 2013; Zuo et al., 2013). By binding directly to core RNAP, they are able to regulate transcription in a large part by altering the kinetics of transcription initiation (Paul et al., 2004b; 2005). The outcome of this regulation, activation or inhibition, depends on the overall kinetic and thermodynamic properties of individual promoters and whether (p)ppGpp and/or DksA affect a step that is rate-limiting for transcription initiation at that promoter.
(p)ppGpp has been proposed to regulate σE activity both directly and indirectly (Costanzo et al., 2008). ppGpp alone has little effect in vitro on transcription by EσE. However, ppGpp and DksA together directly activate transcription by EσE in multi-round in vitro transcription assays (Costanzo et al., 2008). Direct activation of transcription by EσN, Eσ32 and EσS has not been observed in vitro, so this mode of regulation of alternative sigma factors may be specific to EσE (Jishage et al., 2002; Laurie et al., 2003; Szalewska-Palasz et al., 2007). (p)ppGpp has also been proposed to act indirectly by altering the competition among sigma factors for limiting core RNAP in favour of alternative sigmas through its effects on transcription of stable RNA genes by Eσ70 (Zhou and Jin, 1998; Jishage et al., 2002; Szalewska-Palasz et al., 2007; Potrykus and Cashel, 2008). In rapidly growing cells, ~ 70% of the RNAs transcribed in the cell are stable RNAs encoded in long operons (Dennis et al., 2004). (p)ppGpp together with DksA decreases transcription of these operons, thereby reducing the amount of RNAP actively engaged in their transcription and increasing the size of the pool of free core RNAP available to bind all sigma factors.
To better understand regulation of σE by (p)ppGpp and DksA and the extent to which σE is integrated into the cellular response to starvation, we investigated regulation of σE during entry into stationary phase caused by starvation for specific nutrients. Here, we show that (p)ppGpp is required for activation of σE under all starvation conditions tested. However, the requirement for DksA varies. DksA is dispensable for the increase in σE activity following phosphate starvation, while it is required during entry into stationary phase in rich medium and following amino acid starvation. We further show that the contribution of (p)ppGpp and/or DksA to the regulation of other sigma factors during phosphate starvation is not the same for each sigma factor. DksA, but not (p)ppGpp, is required for decreased transcription of the rrnB P1 rRNA operon promoter by Eσ70, while DksA and (p)ppGpp are both required for increased σS activity. These data suggest that regulation of transcription in response to phosphate starvation is specific to individual sigma factors and promoters, and cannot be explained by a concerted global model for (p)ppGpp-dependent transcriptional regulation that affects all sigma factors equivalently.
Results
Regulation of σE during nutrient limitation
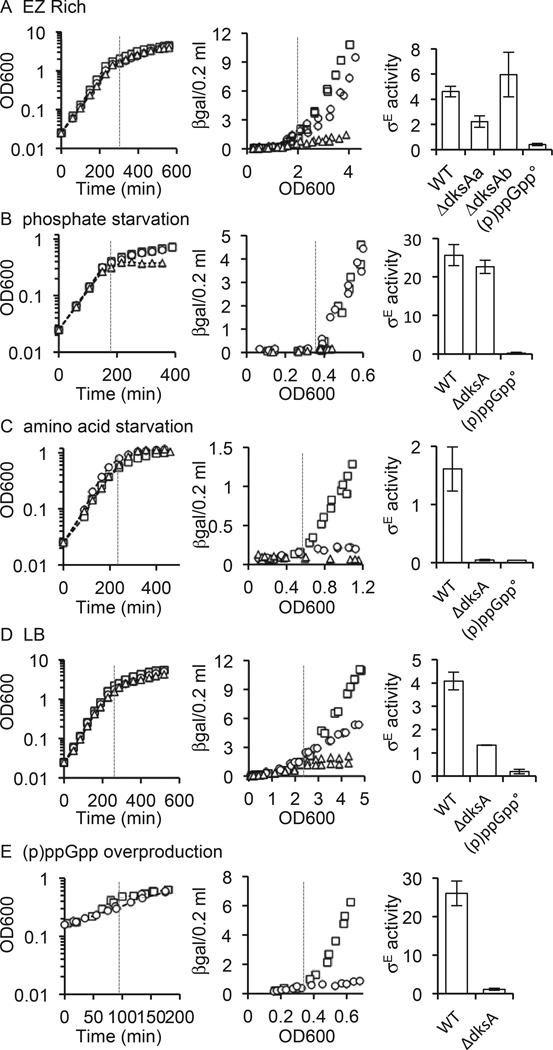
Previous work on the regulation of σE by (p)ppGpp and DksA focused on entry into stationary phase in the rich growth medium, LB (Costanzo and Ades, 2006; Costanzo et al., 2008). To better understand the extent to which (p)ppGpp integrates σE into the cellular response to starvation, we investigated the regulation of σE by (p)ppGpp during entry into stationary phase under several different growth conditions known to be associated with increased (p)ppGpp production. E. coil MG1655 were grown in a MOPS-buffered, rich, defined medium (EZ Rich, Teknova), in EZ Rich with limiting phosphate to induce phosphate starvation or limiting isoleucine to induce amino acid starvation, and in LB. σE activity was measured throughout growth of the cultures using the σE-dependent rpoHP30–lacZ reporter, which has been used extensively as a measure of σE activity. For all culture conditions tested, σE activity increased when growth slowed (Fig. 1A – D). This increase was abrogated in all cases in a strain unable to make (p)ppGpp due to disruption of the relA and spoT genes (Fig. 1A – D), indicating that (p)ppGpp is involved in regulating σE. Because DksA is required for σE to respond to (p)ppGpp in vivo during entry into stationary phase in LB and in vitro in transcription reactions, we next examined σE activity in a strain lacking dksA under the same set of culture conditions (Costanzo et al., 2008). In contrast to (p)ppGpp, the requirement for DksA varied depending on the starvation condition. DksA was required for the increase in σE activity during amino acid starvation and entry into stationary phase in LB (Fig. 1C and D). However, deletion of dksA had little effect on σE activity in response to phosphate starvation (Fig. 1B). In EZ Rich itself, the increase in σE activity in the ΔdksA strain was complex and could be divided into two phases. When the culture first transitioned into stationary phase, σE activity was slightly lower than in the wild-type strain, indicating partial DksA-dependence (Fig. 1A, see ΔdksAa). However, σE activity was comparable to that of the wild-type strain at the end of the transition into stationary phase (Fig. 1A, see ΔdksAb), suggesting that after a brief adjustment period to stationary phase, dksA was no longer required. Similar results were obtained with the σE-dependent PrybB-lacZ reporter (data not shown).
Fig. 1.
The increase in σE activity during entry into stationary phase is dependent on (p)ppGpp under all conditions, but dependent on dksA only under certain conditions. σE activity was measured throughout the growth curve in wild-type (squares), ΔdksA (circles) and (p)ppGpp0 (triangles) cultures grown in (A) EZ Rich, (B) EZ Rich with limiting phosphate (phosphate starvation), (C) EZ Rich with limiting isoleucine (amino acid starvation), (D) LB, and (E) following gratuitous production of (p)ppGpp in the absence of starvation. In E IPTG was added to cultures grown in LB at time 0 (OD600 = 0.15) to induce expression of relA′ and overproduction of (p)ppGpp. Growth curves are shown on the left. σE activity is shown in the right two graphs. The graphs in the middle are differential rate plots in which β-galactosidase activity from the σE-dependent reporter in fixed volume of culture is displayed as a function of culture growth throughout the growth curve. The dashed vertical lines on the left and middle graphs correspond to the OD600 after which σE activity increases. On the right, the increase in σE activity is quantified for each strain, as calculated from the slope on the differential rate plot for the points to the right of the dashed line. In EZ Rich, the increase in σE activity in the ΔdksA strain is biphasic, low during entry into stationary phase (OD600 = 2.0–3.5) then increasing at the end of the transition into stationary phase (OD600 = 3.5–4.5), and both slopes are quantified (ΔdksAa and ΔdksAb respectively). Data from at least two representative experiments are shown in each graph.
σE activity is also known to increase when (p)ppGpp is made in the absence of starvation by overexpression of a fragment of relA, relA’, which constitutively synthesizes (p)ppGpp independently of the ribosome (Svitil et al., 1993; Costanzo et al., 2008). The involvement of DksA in regulating σE activity under these conditions was not examined. Therefore, we measured σE activity following overexpression of relA’ in wild-type and ΔdksA cells. Because high levels of (p)ppGpp are toxic to E. coil, we induced relA with a low concentration of IPTG that does not significantly alter the growth rate. When the relA’gene was overexpressed, σE activity increased in the wild-type strain, but not in the ΔdksA strain, indicating that DksA is required for σE to respond to (p)ppGpp in the absence of a starvation signal (Fig. 1E). These data indicate that σE activity increases under all conditions tested thus far that are known to lead to an increase in (p)ppGpp levels, and that disruption of (p)ppGpp production disrupts the σE response. However, the mechanism of regulation differs with the culture conditions. In some cases DksA is required and in other cases it is not.
Regulation of σS during nutrient limitation
If the alternative sigma factors in E. coil are indeed regulated in concert by (p)ppGpp and DksA via their effects on σ70, then other sigma factors should show the same pattern of regulation as σE. To test this idea, we examined the activity of the general stress factor, σS, in wild-type, (p)ppGpp0 and ΔdksA strains under the same series of culture conditions used for σE. A σS-dependent bolA–lacZ fusion was used to monitor σS activity. Similar to σE, σS activity increased when cells entered stationary phase in the wild-type strain under each of the growth conditions (Fig. 2). However, σS did not exhibit the differential requirement for DksA. σS activity failed to increase in (p)ppGpp0 or ΔdksA strains under all of the tested culture conditions, indicating that both factors are involved in regulating σS and that regulation of σS is therefore distinct from regulation of σE (Fig. 2). In addition, these data show that DksA is active during phosphate starvation.
Fig. 2.
The increase in σS activity during entry into stationary phase is dependent on (p)ppGpp and dksA under all conditions tested. σS activity was measured from the bolA–lacZ fusion throughout the growth curve in wild-type, ΔdksA and (p)ppGpp0 cultures grown in (A) EZ Rich, (B) EZ Rich with limiting phosphate (phosphate starvation), (C) EZ Rich with limiting isoleucine (amino acid starvation), and (D) LB. The increase in σS activity when growth slows is quantified for each strain as described in Fig. 1.
Regulation of σ70 during phosphate starvation
Inhibition of transcription of rRNA promoters by (p)ppGpp and DksA has been proposed to indirectly activate σE and other alternative sigma factors by increasing the amount of free core RNAP (Zhou and Jin, 1998; Costanzo et al., 2008; Österberg et al., 2011). The results with the σS-dependent promoter fusion during phosphate starvation suggest that this indirect mechanism of regulation could still be applicable for alternative sigma factors other than σE, provided that (1) the rRNA operons are subject to regulation by (p)ppGpp and DksA, in which case σE is regulated in a unique manner that does not require DksA, or (2) the rRNA operons are regulated by (p)ppGpp only, in which case σS is regulated in a unique manner that requires DksA.
To the best of our knowledge, regulation of the rRNA promoters during phosphate starvation and the involvement of (p)ppGpp and DksA in any such regulation have not been reported, although (p)ppGpp levels have long been known to increase (Spira et al., 1995). To assess regulation of rRNA transcription following phosphate starvation, transcription of the stringently regulated rrnB P1–lacZ transcriptional reporter fusion was monitored. Decreased transcription is difficult to accurately measure using a β-galactosidase enzyme assay, in contrast to increased transcription, because β-galactosidase is a stable protein. A reduction in its production will be masked by the pre-existing enzyme. Therefore, we used primer extension to assay expression of the short-lived RNA transcript made from the rrnB P1–lacZ reporter (Schneider et al., 2003). rrnB P1 promoter activity decreased in the wild-type strain such that the primer extension product was undetectable after entry into phosphate starvation (Fig. 3). In contrast, promoter activity stayed relatively constant in the ΔdksA strain rather than decreasing, indicating that DksA is important for the observed regulation (Fig. 3A). Surprisingly, promoter activity also decreased in the (p)ppGpp0 strain, suggesting that (p)ppGpp was not required for the response (Fig. 3A). Because strains lacking (p)ppGpp and DksA can accumulate suppressor mutations, the experiments were repeated in independently constructed (p)ppGpp0 and ΔdksA strains and similar results were obtained. To further ensure that the (p)ppGpp-independent, DksA-dependent regulation of rrnB P1 was not an artefact of our experimental system or due to a mutation in our strains, we analysed the response to amino acid starvation caused by the addition of serine hydroxamate. Both (p)ppGpp and DksA were required for negative regulation of rrnB P1 following amino acid starvation, consistent with previous reports (Fig. S1) (Paul et al., 2004b). In addition, we monitored transcription of the fusion in the absence of starvation and it was transcribed equivalently in the wild-type, (p)ppGpp0 and ΔdksA strains (Fig. S1). These data suggest that DksA, but not (p)ppGpp, is necessary for inhibition of transcription of rrnB P1 by Eσ70 during phosphate starvation. The results do not support a model in which negative regulation of stable RNA operons is coupled with regulation of the alternative sigma factors during phosphate starvation, because regulation of transcription directed by σ70 is (p)ppGpp-independent and DksA-dependent, σS is (p)ppGpp- and DksA-dependent, and σE is (p)ppGpp-dependent and DksA-independent.
Fig. 3.
DksA, but not ppGpp, is required for full inhibition of transcription from rrnB P1 by Eσ70 following phosphate depletion.
A. Primer extension was used to measure mRNA production from the rrnB P1–lacZ fusion. The primer extension product is indicated by the closed arrowhead and a recovery marker by the open arrowhead.
B. Growth curves of the reporter strains are shown on the right and the times at which samples were taken for primer extension are indicated.
σE activity, not production, is regulated in response to phosphate limitation and the response is independent of envelope stress
The DksA-independence of the σE response to phosphate starvation indicates that the mechanism of regulation is not the same as the DksA-dependent mechanism that controls σE activity in response to amino acid starvation or entry into stationary phase in LB. To better understand the DksA-independent mode of regulation, we focused on phosphate starvation. Phosphate starvation could cause an envelope stress that is sensed by the RseA-dependent stress signalling pathway such that this pathway is involved in the regulation along with (p)ppGpp. If so, the response of σE to phosphate starvation should be disrupted in a strain lacking rseA. However, σE activity increased when culture growth slowed in the ΔrseA strain, indicating that the envelope stress pathway does not participate in regulation of σE in response to phosphate starvation (Fig. 4).
Fig. 4.
σE activity increases following phosphate depletion independently of rseA. The ΔrseA strain was grown in EZ Rich (open squares) and EZ Rich with limiting phosphate (shaded squares). Growth curves are shown on the left and σE activity is shown on the right in a differential rate plot in which β-galactosidase from the σE-dependent reporter in 0.2 ml culture is displayed as a function of culture growth. The dashed vertical lines correspond to the OD600 after which σE activity increases in EZ Rich with limiting phosphate.
σE activity could increase due to changes in its expression, stability and/or activity. As such, we analysed the steady state level of σE, which reflects its expression and stability, in wild-type and (p)ppGpp0 strains grown in EZ Rich with limiting phosphate. The total amount of σE increased as the cultures transitioned into stationary phase in both strains (Fig. 5A and B). This increase was not specific to phosphate starvation, because a similar accumulation of σE was observed during entry into stationary phase in LB (Costanzo et al., 2008). The regulator responsible for the increase in σE levels has not been identified. The finding that σE accumulates in the wild-type and (p)ppGpp0 strains during phosphate starvation and entry into stationary phase in LB suggests that alterations in the amount of σE are not specific to the (p)ppGpp-dependent, DksA-independent regulation of σE. Therefore regulation is likely to be at the level of activity.
Fig. 5.
σE levels are similar in wild-type and (p)ppGpp0 strains following phosphate depletion. Wild-type and (p)ppGpp0 strains were grown in EZ Rich with limiting phosphate.
A. Samples were taken at the indicated OD600 and cell extracts used for western blotting with an anti-σE polyclonal antibody. The band marked with an arrowhead corresponds to σE, while the top band is a cross-reacting band of unknown identity.
B. Growth curves for the cultures from which samples were taken are shown. Comparable amounts of protein were loaded in each lane. The asterisk marks the point at which growth slows and cultures start to enter stationary phase.
Low levels of phosphate are sensed in E. coli by a signalling complex that includes the PhoR/PhoB two-component system. Although σE has not been implicated as part of the PhoB regulon, transcriptomic and proteomic studies indicate that the expression of 287 genes and up to 400 proteins are affected by phosphate limitation (Hsieh and Wanner, 2010; Yang et al., 2012). A PhoB-regulated protein or sRNA could in turn regulate σE activity. To determine if PhoB contributes to activation of σE, σE activity was analysed in a ΔphoB strain following phosphate starvation. Activity was unchanged compared to the wild-type strain indicating that PhoB does not play a role in the response of σE to phosphate limitation (Fig. S2).
(p)ppGpp synthesis during phosphate starvation
The σE response to starvation is dependent on (p)ppGpp under all conditions tested. It is possible that the lack of requirement for DksA following phosphate starvation could be caused by variations in the amount of (p)ppGpp produced among the strains. Therefore, growth, σE activity, and (p)ppGpp levels were measured in the wild-type, ΔdksA and (p)ppGpp0 strains during phosphate starvation (Fig. 6). σE activity increased when growth slowed in the wild-type and ΔdksA strains, approximately one hour following the shift to medium with limiting phosphate (Fig. 6A). (p)ppGpp levels alone and as a fraction of guanosine nucleotide pools, (p)ppGpp/(GTP + (p)ppGpp), also peaked at the one hour time point and then declined for both wild-type and ΔdksA strains (Fig. 6B – D). The amount of (p)ppGpp was approximately twofold to fourfold higher in the ΔdksA strain than in the wild-type strain, whereas no (p)ppGpp was detected in the (p)ppGpp0 strain (Fig. 6B – D). While measuring (p)ppGpp levels following phosphate starvation, we noticed that the ATP, GTP and CTP levels dropped substantially when growth slowed in all the strains tested, including the (p)ppGpp0 strain (Fig. 6E). In contrast, the levels of these nucleotides stayed constant during amino acid starvation, while (p)ppGpp levels increased, similar to observations by others (Paul et al., 2004a) (Fig. S3).
Fig. 6.
(p)ppGpp levels increase and GTP, ATP and CTP levels decrease following phosphate starvation.
A. Growth (inset graph) and σE activity (differential rate plot) from cultures used to isolate nucleotides are shown. Both graphs start with points after the shift to limiting phosphate (t = 0, OD600~0.15).
B. TLC separation of nucleotides extracted with formic acid is shown for samples taken from cultures of WT, ΔdksA and (p)ppGpp0 strains at the indicated times after a shift to EZ Rich with limiting phosphate.
C and D. (C) (p)ppGpp levels (arbitrary units, a.u.) were quantified from the TLC in part A and normalized to OD600 at the time of sampling or to (D) total (p)ppGpp and GTP pools.
E. GTP, ATP and CTP levels normalized to the level at 15 min. in each of the three strains after a shift to low phosphate. Symbols are the same in all parts of the figure as indicated in the legend.
In E. coli, both ppGpp and pppGpp accumulate during starvation, although their relative amounts vary depending on the starvation condition. For example, both pppGpp and ppGpp accumulate during amino acid starvation, while ppGpp primarily accumulates during carbon starvation (Cashel, 1975; Gallant, 1979; Mechold et al., 2013). Our previous work demonstrated that DksA was required for EσE to respond to ppGpp in in vitro transcription reactions. However, the effects of pppGpp on transcription by EσE were not measured, leaving open the possibility that pppGpp may have different effects on EσE than ppGpp and perhaps not require DksA. An accumulation of more pppGpp than ppGpp during phosphate starvation could then provide an explanation for the differential requirement of DksA. We examined the amount of pppGpp that accumulated during phosphate starvation using TLC conditions that separate pppGpp from ppGpp. The levels of pppGpp were very low compared to ppGpp (< 10% of the amount of ppGpp) (Fig. 7) indicating that ppGpp is the main regulator, and pppGpp is unlikely to contribute significantly to regulation of σE during phosphate starvation.
Fig. 7.
ppGpp accumulates to a greater extent than pppGpp following phosphate starvation in wild-type and ΔdksA strains. 1.5 M KH2PO4 was used as the running buffer in the TLC to resolve ppGpp from pppGpp. Extracts from a ppGpp0 strain and a strain overexpressing the relA′ fragment (pALS13) that accumulates both ppGpp and pppGpp are shown for reference.
The source of (p)ppGpp does not affect the increase in σE activity in response to phosphate starvation
(p)ppGpp levels are thought to increase following amino acid starvation and phosphate starvation by different mechanisms. During amino acid starvation RelA is activated by the binding of uncharged tRNA in the A site of the ribosome (Haseltine and Block, 1973; Wendrich et al., 2002). During phosphate starvation, (p)ppGpp levels are thought to increase due to inhibition of the SpoT hydrolase, although the signalling mechanism is not known (Spira et al., 1995). To determine if the mechanism by which (p)ppGpp levels increase influences regulation of σE, σE activity and (p)ppGpp production were measured in a series of strains that differ in their ability to synthesize and hydrolyse (p)ppGpp. It was not possible to use a strain in which (p)ppGpp levels were solely controlled by RelA, because the spoT gene is essential in the presence of a functional relA gene. Instead we used a strain carrying the spoTE319Q (spoTsyn−) mutation that inactivates the SpoT synthase (Harinarayanan et al., 2008). (p)ppGpp levels in this strain are controlled by RelA-dependent synthesis and SpoT-dependent hydrolysis. Growth, σE activity and (p)ppGpp accumulation in the spoTsyn− mutant in response to phosphate starvation were comparable to that measured for the wild-type strain (Fig. 8A – D), indicating that control of (p)ppGpp synthesis by RelA and hydrolysis by SpoT are sufficient for regulation of σE.
Fig. 8.
σE activity increases independently of the source of (p)ppGpp in all strains that make (p)ppGpp following phosphate starvation. The indicated strains were grown in EZ Rich and shifted to EZ Rich with limiting phosphate at time 0.
A and B. (A) σE activity measured when growth slows due to phosphate starvation is shown for each strain and (B) the corresponding growth curves are shown.
C and D. (C) TLC separation of nucleotides and (D) quantification of (p)ppGpp levels (arbitrary units) normalized to OD600 at time of sampling after the shift to low phosphate are shown.
E. (p)ppGpp levels normalized to total (p)ppGpp and GTP pools for wild-type, spoTsyn−, ΔrelA, ΔrelAspoThyd− strains. Symbols are the same in all parts of the figure as indicated in the legend.
σE activity and (p)ppGpp production were also measured in a ΔrelA strain in which (p)ppGpp levels are controlled by SpoT-dependent synthesis and hydrolysis. σE activity increased in the ΔrelA mutant, like it did in the wild-type strain (Fig. 8A). However, growth of the ΔrelA cultures slowed more following phosphate starvation compared to the relA+ strains, suggesting that the ΔrelA mutant adapts less efficiently to lower phosphate levels (Fig. 8B). (p)ppGpp also accumulated somewhat slower than it did in the wild-type strain and the levels were still elevated at the 90 min. time point, whereas they decreased after a peak at 60 min. in the other strains (Fig. 8C–E). Finally, to isolate effects due to regulation of (p)ppGpp levels by control of the SpoT hydrolase, we examined a strain lacking relA and carrying the spoTR39A (spoThyd−) mutation that inactivates the SpoT hydrolase (Harinarayanan et al., 2008). In this strain, (p)ppGpp accumulates from the weak synthase activity of SpoT. Similar to the ΔrelA strain, growth of this strain slowed more during entry into phosphate starvation than strains with relA (Fig. 8B). σE activity increased when culture growth slowed, but about twofold less than in the other strains (Fig. 8A). In the ΔrelA spoThyd− mutant, (p)ppGpp levels stayed relatively constant and were at least 10-fold lower than the peak amount of (p)ppGpp detected in the other strains (Fig. 8C–E). However, the (p)ppGpp/(GTP+(p)ppGpp) ratio increased due to the decrease in GTP levels.
σE activity increased in response to phosphate depletion in all of the strains that were able to synthesize (p)ppGpp. However, activity was not strictly correlated with the amount of (p)ppGpp (compare (p)ppGpp levels and σE activity in the ΔrelA spoThyd− strain to those in the other strains). The (p)ppGpp0 strain was the only mutant examined in which the σE response to phosphate starvation was completely disrupted, and this strain is the only strain lacking spoT. These data suggest that the SpoT protein itself could modulate σE activity, independently of (p)ppGpp production. To test this idea, we combined the spoTsyn− allele with a relA deletion. This strain does not make (p)ppGpp (data not shown), but does express a full-length SpoT protein (Harinarayanan et al., 2008). σE did not respond to phosphate starvation in this strain and (p)ppGpp was not detected, suggesting that synthesis of (p)ppGpp is required for the increase in σE activity (Fig. 8A). Taken together, these data show that σE responds to phosphate starvation independently of the source of (p)ppGpp.
Secondary channel binding proteins and σE activity
DksA exerts its effects on transcription by binding in the secondary channel of core RNAP. E coll possesses several other proteins, GreA, GreB and Rnk, which also bind in the secondary channel (Rutherford et al., 2007; Lamour et al., 2008; Vinella et al., 2012). GreA and GreB are well-characterized for their role in transcriptional pausing (Nickels and Hochschild, 2004). Both factors can also substitute for DksA and regulate transcription initiation by Eσ70 under certain circumstances (Potrykus et al., 2006; Rutherford et al., 2007; Vinella et al., 2012). The Rnk protein is a structural homologue of DksA and the Gre factors. It can compete with DksA for binding to the secondary channel of RNAP, although its role in transcriptional regulation is not clear (Lamour et al., 2008). To determine if any of these factors could substitute for DksA in regulating σE during phosphate starvation, we measured σE activity in strains lacking greA, greB, or rnk alone or in combination with the dksA deletion. σE activity in response to phosphate starvation was similar to that of the wild-type strain for all mutants, except the ΔgreAΔgreB double mutant in which the increase in σE activity was about twofold lower than that of wild type (Figs S4 and S5). These results indicate that no single one of these proteins takes the place of DksA in regulating σE. It is possible that either greA or greB may play a small role in regulating σE, however their effect is significantly less than that observed in the ppGpp0 strain in which σE activity does not increase upon phosphate depletion.
In addition to the known secondary channel binding proteins, we examined two other genes for their effects on σE activity, ybil and crl. Overexpression of ybil alleviates several of the amino acid auxotrophies associated with the ΔdksA strain, suggesting that it is a functional homologue of DksA (Blankschien et al., 2009). Crl is a small protein that facilitates the association of σS and σH with core RNAP In vitro and has global regulatory effects via σS during stationary phase In vivo (Gaal et al., 2006; Typas et al., 2007). We deleted these genes alone and in combination with a deletion of dksA. The σE response to phosphate starvation was similar to that of the wild-type parent for each of these strains (data not shown), indicating that Ybil and Crl are also not necessary for activation of σE and do not substitute for DksA (Fig. S5).
Discussion
In this work we demonstrate that the E. coli extracytoplasmic stress sigma factor, σE, can be activated in the absence of cell envelope stress by the alarmone (p)ppGpp in response to different starvation conditions using at least two mechanisms, one dependent on DksA and the other DksA independent. Our previous model that (p)ppGpp and DksA regulate σE directly by acting on EσE and indirectly via effects on transcription of stable RNA genes by Eσ70 (Costanzo et al., 2008) cannot explain the (p)ppGpp-dependent, DksA-independent regulation of σE during phosphate starvation described here. In addition, the observations that inhibition of σ70-dependent transcription of stable RNA promoters requires only DksA and activation of σS-dependent transcription requires both ppGpp and DksA further invalidate the model that decreased transcription of the stable RNA genes contributes to activation of alternative sigma factors in concert.
Because DksA and (p)ppGpp often work together and DksA was thought to be required for σE to respond to (p)ppGpp, the differential requirement for DksA during phosphate starvation was surprising. However, a number of reports indicate that the effects of DksA and (p)ppGpp on both cellular physiology and transcription can differ. The phenotypes of ppGpp0 and ΔdksA mutants are similar, but not completely overlapping (Magnusson et al., 2007; Aberg et al., 2009). Similarly, the changes in gene expression in ppGpp0 and ΔdksA mutants are also largely, but not completely overlapping, suggesting that (p)ppGpp and DksA can have independent and even opposing effects on transcription (Aberg et al., 2009). Opposing effects of ppGpp and DksA have also been demonstrated for the phage λ pR promoter (Magnusson et al., 2007). The extent of differential regulation by (p)ppGpp and DksA is not known and adds another level of complexity, as demonstrated here, to their ability to sculpt the transcriptome.
Regulation of EσE by (p)ppGpp and DksA during phosphate starvation
Our data with the ΔrseA strain indicate that phosphate starvation does not create a cell envelope stress to activate σE, nor does it increase the expression of rpoE. Therefore, the observed increase in σE activity is most likely due to (p)ppGpp-dependent increased transcription by EσE. How does (p)ppGpp control σE activity without DksA in response to phosphate starvation? A potential explanation is that the twofold to fourfold increased amount of (p)ppGpp in the ΔdksA strain compensates for the absence of DksA, and DksA is still required in wild-type cells with lower amounts of (p)ppGpp. We think this model unlikely because it predicts that once a threshold amount of (p)ppGpp accumulates, DksA is no longer required for increased σE activity. However, DksA is required for the σE response to amino acid starvation, and our work, together with other studies, indicates that (p)ppGpp levels are actually somewhat higher during amino acid starvation than during phosphate starvation (unpublished observations and Rao et al., 1998; Spira et al., 1995). Additionally, in vitro multi-round transcription assays with high amounts of (p)ppGpp in the absence of DksA show no increase in EσE-dependent transcription (Costanzo et al., 2008). Another explanation for DksA-independence of the response is that the decrease in NTP levels caused by phosphate starvation is important for regulation. Yet, it is difficult to envision a mechanism by which low NTP availability would increase transcription.
We propose that an as yet undiscovered factor participates in the control of σE during phosphate starvation along with (p)ppGpp. Results from experiments with the ΔrelA spoThyd− mutant indicate that the levels of (p)ppGpp in this mutant increased very little, if at all, suggesting that the signal for phosphate starvation is not simply increased levels of (p)ppGpp, but a function of this missing factor. Taking a candidate approach, we tested whether the missing factor was a transcriptional regulator known to respond to phosphate starvation (phoB), one of a series of structural and/or functional homologues of DksA (greA, greB, ybil and rnk), or an additional modulator of polymerase activity (crl). None of these regulators appear to be involved, so the identity of the factor remains a mystery.
Regulation of Eσ70 in response to phosphate starvation
Although transcription of the rrnB P1 promoter by (p)ppGpp and DksA has been extensively studied during amino acid starvation, at different growth rates, throughout the growth curve, and in response to nutrient upshifts and downshifts, phosphate starvation had not been investigated to our knowledge (Dennis et al., 2004; Paul et al., 2004a; Potrykus and Cashel, 2008). Because (p)ppGpp levels increase, we assumed that (p)ppGpp would be required for downregulation of transcription by E σ 70 at the rrnB P1 promoter. Instead, (p)ppGpp was found to be dispensable, while DksA was required. The explanation for why (p)ppGpp is not needed may lie in the observation that nucleotide levels drop significantly when phosphate is depleted from the growth medium. At the rrnB P1 promoter and other stringently regulated rRNA promoters, Eσ70 forms unstable open complexes, which are stabilized by binding of initiating nucleotides (iNTPs) and destabilized by DksA and (p)ppGpp (Gaal et al., 1997; Barker et al., 2001; Murray et al., 2003; Murray and Gourse, 2004; Paul et al., 2004b; Kolmsee et al., 2011). A higher concentration of iNTP is required for transcription initiation at rrnB P1 in the presence of DksA than in its absence (Paul et al., 2004b). Because nucleotide levels drop during phosphate starvation, the decreased availability of nucleotides in combination with destabilization of open complexes by DksA may reduce transcription initiation, eliminating the requirement for (p)ppGpp. Because most other rrn promoters form similarly unstable open complexes limited by iNTP concentrations, this mechanism of regulation is likely to be shared and not particular to rrnB P1 (Gaal et al., 1997; Barker et al., 2001; Murray et al., 2003; Murray and Gourse, 2004; Paul et al., 2004a; Kolmsee et al., 2011).
Regulation of EσS in response to phosphate starvation
σS is regulated by a complex network of signalling pathways that are activated under different conditions and affect σS at the transcriptional, translational and post-translational levels (Battesti et al., 2011). Translation of rpoS increases and proteolysis of σS decreases in response to phosphate limitation, presumably leading to increased levels of σS and increased expression of the σS regulon (Gentry et al., 1993; Bougdour and Gottesman, 2007). The stability of σS is modulated by the expression of the adaptor protein RssB, which delivers σS to the ClpXP protease for degradation (Muffler et al., 1996; Pratt and Silhavy, 1996; Zhou and Gottesman, 1998). A series of anti-adaptor proteins, expressed under different physiological conditions, prevent RssB from binding σS, thereby stabilizing σS (Bougdour et al., 2008). The IraP anti-adaptor is responsible for stabilization of σS during phosphate starvation, and its transcription is reduced in strains lacking (p)ppGpp and DksA (Bougdour et al., 2008). Therefore, the requirement for DksA and (p)ppGpp for activation of σS is likely to be due, at least in part, to positive regulation of Eσ70–dependent transcription of iraP by DksA and (p)ppGpp. DksA and (p)ppGpp may also contribute to translational regulation of rpoS (Battesti et al., 2011). Both factors have been shown to be important for upregulation of rpoS translation during entry into stationary phase (Brown et al., 2002). However, their role in translational regulation of rpoS during phosphate starvation has not been examined.
Production of (p)ppGpp in response to phosphate starvation
The prevailing model for the increased amounts of (p)ppGpp during phosphate starvation is that phosphate levels are sensed by the SpoT hydrolase through an unknown mechanism that results in decreased hydrolysis of (p)ppGpp. This model is based in a large part on observations that during phosphate starvation (p)ppGpp levels increased in a ΔrelA strain and (p)ppGpp-dependent regulation of several factors including σS activity and iraP expression was not disrupted (Spira et al., 1995; Rao et al., 1998; Bougdour and Gottesman, 2007). However, the observation that strains lacking relA stop growing abruptly at the onset of phosphate starvation and that the rise in (p)ppGpp follows different kinetics in the ΔrelA strain than in the wild-type strain (also seen by Spira et al., 1995) suggest a role for RelA in phosphate starvation. Because RelA shares a similar domain structure with SpoT, although the hydrolase of RelA is inactive, it is possible that the production of (p)ppGpp by RelA is subject to regulation during phosphate starvation along with SpoT and regulation of both factors yields the optimal response (Gentry and Cashel, 1996; Gropp et al., 2001; Mechold et al., 2002).
Global regulation of alternative sigma factors by (p)ppGpp and DksA
In E. coli, the activity of the alternative sigma factors, σE, σH, σN and σS, have all been shown to increase under a variety of nutrient limitation conditions or entry into stationary phase in a (p)ppGpp-dependent manner (Gentry et al., 1993; Jishage et al., 2002; Laurie et al., 2003; Costanzo and Ades, 2006; Costanzo et al., 2008; Österberg et al., 2011). The current model explaining how this happens is that (p)ppGpp activates alternative sigma factor-dependent transcription by altering the competition among sigma factors for limiting core RNAP in favour of the alternative sigma factors. As described above, this model does not explain the results presented in this work. A model for the mechanism of action of (p)ppGpp that explains its ability to regulate all alternative sigma factors in concert is certainly attractive. Nevertheless, our results suggest that there may not be a global mechanism by which (p)ppGpp orchestrates changes in the transcriptome that encompasses all sigma factors. Instead, (p)ppGpp may directly regulate transcription of individual promoters by different holoenzymes, with or without DksA and other transcription factors depending on the physiological conditions. (p)ppGpp and/or DksA may also control the expression of specific regulators of a particular sigma factor associated with a particular starvation condition (i.e. regulation of RpoS via IraP). As such, (p)ppGpp may achieve global changes in transcription by acting locally on individual promoters, sigma factors and holoenzymes.
Experimental procedures
Media, strains and plasmids
Strains used in this paper are derivatives of the MG1655 E. coli K12 strain and are listed in Table 1. Cells were grown at 30°C or 37°C, as indicated, with aeration in LB (0.5% yeast extract, 1% tryptone, 0.5% NaCI, pH 7.4) or EZ Rich [a MOPS-based medium (Neidhardt et al., 1974) purchased from Teknova]. Antibiotics were used at the following concentrations: ampicillin 100 µg ml−1, kanamycin 15 µg ml−1, tetracycline 10 µg mL−1 and chloramphenicol 20 µg ml−1. For phosphate limitation experiments, EZ Rich was prepared with 0.132 mM K2HPO4, instead of the standard 1.32 mM K2HPO4. For amino acid limitation experiments, isoleucine was included at 0.06 mM, instead of the standard 0.4 mM. The limiting isoleucine medium was prepared using Teknova components with the exception of the 5× supplement, which was prepared according to the recipe from the E. coli Genome Project (http://www.genome.wisc.edu/resources/protocols/ezmedium.htm). The amino acid starvation system utilizing limiting isoleucine is based on that described in Traxler et al. (2008). For E. coli K12, excess valine in the presence of low concentrations of isoleucine leads to feedback inhibition of enzymes responsible for synthesizing isoleucine. As a result, E. coli K12 strains grown in low concentrations of isoleucine will slowly enter amino acid starvation when isoleucine is depleted and de novo synthesis is inhibited. An advantage to this system is that protein synthesis can continue, enabling use of β-galactosidase reporters, and the multi-auxotrophic ppGpp0 strain can grow because all amino acids are present.
Table 1.
Strains and plasmids used in this study.
| Strain/plasmid | Genotype | Source, Reference, P1 donor strain |
|---|---|---|
| Strains | ||
| SEA001 | MG1655 ΔlacX74 λ[rpoH P3::lacZ] | Costanzo and Ades (2006) |
| SEA2010 | SEA001 ΔrelA251::kan ΔspoT207::chlor | Costanzo and Ades (2006) |
| SEA4041 | ΔrpoE::kan Suppressor+ | This work, P1 donor CAG43113 |
| SEA6020 | SEA001 ΔdksA::tet | Costanzo and Ades (2006) |
| SEA6328 | MG1655 ΔlacX74 λ[bolA::lacZ] | provided by S. Finkel |
| SEA6329 | SEA6328 ΔdksA::tet | This work, P1 donor SEA6020 |
| SEA6401 | SEA6328 ΔrelA251::kan, ΔspoT207::chlor | This work, P1 donor SEA2010 |
| SEA6462 | SEA001 ΔrseA yfic::kan | This work |
| SEA6483 | MG1655 λ[rybB::lacZ] pALS13 | This work |
| SEA6487 | SEA6483 ΔdksA::tet | This work |
| SEA6513 | SEA001 ΔrelA251::kan | Costanzo and Ades (2006) |
| SEA6565 | SEA6513 spoTE319 zib563::Tn10 | This work, P1 donor CF11608 provided by M. Cashel |
| SEA6575 | SEA001 spotE319 zib563::Tn10 | This work, P1 donor CF11608 provided by M. Cashel |
| SEA6583 | VH1000 λ[rrnB P1–lacZ] | Provided by R. Gourse, RLG6583 |
| SEA7019 | SEA6513 spoTR39A zib563::Tn10 | This work, P1 donor CF11605 provided by M. Cashel |
| SEA7025 | SEA6583 ΔdksA::tet | This work, P1 donor SEA6020 |
| SEA7027 | SEA6583 ΔrelA251::kan ΔspoT207::chlor | This work, P1 donor SEA2010 |
| SEA7029 | SEA001 ΔgreA::chlor | This work, P1 donor RLG7239 |
| SEA7030 | SEA001 ΔgreB::kan | This work, P1 donor RLG7240 |
| SEA7031 | SEA7029 ΔgreB::kan | This work, P1 donor RLG7240 |
| SEA7032 | SEA7029 ΔdksA::tet | This work, P1 donor SEA6020 |
| SEA7033 | SEA7030 ΔdksA::tet | This work, P1 donor SEA6020 |
| SEA7040 | SEA7031 ΔdksA::tet | This work, P1 donor SEA6020 |
| SEA7060 | SEA001 Δrnk::kan | This work, P1 donor JW0602 |
| SEA7061 | SEA7060 ΔdksA::tet | This work, P1 donor SEA6020 |
| SEA7062 | SEA001 Δybil::chlor | This work |
| SEA7063 | SEA7062 Δ dksA::tet | This work, P1 donor SEA6020 |
| SEA7070 | VH1000 λ[lacUV5–lacZ] | Provided by R. Gourse RLG4993 |
| SEA7149 | SEA001 Δcrl::kan | This work, P1 donor JW0230 |
| SEA7152 | SEA7149 ΔdksA::tet | This work, P1 donor SEA6020 |
| SEA7166 | SEA001 ΔphoB::kan | This work, P1 donor JW0389, Baba et al. (2006) |
| Plasmids | ||
| pALS13 | ptac truncated relA, active protein, ApR | Svitil et al. (1993) |
Mutant alleles were moved into appropriate strains by transduction with P1vir according to standard techniques (Miller, 1992). Transductants were isolated by selection on medium containing the appropriate antibiotic. Experiments with ppGpp0 and spoThyd− strains were performed with at least three independent transductants to ensure that the results were not affected by spontaneous suppressor mutations. In addition, ppGpp0 strains were tested for their inability to grow on minimal media lacking amino acids, because many spontaneous suppressor mutations restore amino acid prototrophy. SEA6462 (ΔrseA yfic::kan) was made by transduction of the yfiC::kan allele from the Keio collection (Baba et al., 2006) into SEA2000 (SEA001 nadB::Tn10 ΔrseA). Transductants were screened for loss of tetracycline sensitivity and increased σE activity due to loss of rseA. SEA7062 was made by targeted disruption of the ybil gene in SEA001 according to the procedure of Datsenko and Wanner (2000).
β-Galactosidase assays
β-galactosidase assays were performed as described (Costanzo and Ades, 2006). Data are presented as differential rate plots in which β-galactosidase activity in a fixed volume of culture is plotted versus the optical density (OD600) of the sample. The slope of the linear portions of the curve represents the accumulation of σ-dependent β-galactosidase activity as a function of increased cell density. To calculate how σE or σS activity changes during entry into stationary phase in rich media or in response to nutrient limitation, the slope of the linear portion of the differential rate plot for the relevant points corresponding to entry into stationary phase was determined. All experiments were performed at least three times with independent cultures and activity varied by < 10%.
Western blotting
Western blotting was performed as described (Costanzo et al., 2008). Briefly, whole cell extracts were precipitated with acetone and resuspended in 2% SDS. Protein concentrations were determined using the BCA protein assay (Pierce) and 10 µg of total protein from each sample were loaded. σE-containing bands were detected using a polyclonal antibody (gift from CA Gross) and alkaline phosphatase conjugated secondary antibody in conjunction with the ECF reagent (GE Healthcare) followed by scanning with the Typhoon 8600 Imager in fluorescence mode.
Primer extension
Primer extension was performed as described (Ross and Gourse, 2009). SEA6583 was grown with shaking to an OD600 of 0.25–0.3 in EZ Rich. For phosphate limitation experiments, cells were then pelleted by centrifugation, resuspended to the same OD600 in EZ Rich with limiting phosphate, and growth resumed. For amino acid starvation, serine hydroxamate was added to 1 mg ml−1 final concentration. Samples were taken at the indicated times after the change in growth medium or addition of serine hydroxamate, and total RNA prepared by the boiling lysis method. To control for sample degradation and variations in gel loading, 5 µg of recovery marker RNA was added to each sample during the phenol-chloroform extraction step of the boiling lysis RNA preparation protocol. Recovery marker RNA was prepared from strain SEA7070, which carries a lacUV5–lacZ promoter fusion. The 5414 primer used for primer extension anneals to this fusion as well as the rm6P1–lacZ fusion and produces primer extension products of different sizes from each reporter. For the primer extension reactions, 15 µg of total RNA was annealed to [32]P end-labelled primer 5414 (5′-TGGTGTTCGTCCCGGCTGTAATGTTCTGGC-3′) and the extension reaction was carried out with Moloney Murine Leukemia Virus reverse transcriptase (M-MLV RT from Promega) for 30 min. at 42°C. Reactions were stopped with formamide loading buffer. Extension products were separated by electrophoresis on denaturing 6% polyacrylamide gels and visualized by phosphorimaging.
Measurement of (p)ppGpp and NTP levels
To measure NTP and (p)ppGpp levels following starvation for phosphate, cultures were grown in EZ Rich at 37°C with shaking to an OD600 of approximately 0.2. Cells were collected by centrifugation, and pellets resuspended in EZ Rich with limiting phosphate to the same optical density and divided between two flasks. [32]P-orthophosphoric acid (specific activity 8500 Ci mmol−1, Perkin-Elmer) was added to one culture to a concentration of 20 µCi ml−1. The other culture was used to monitor OD600 and activity of the rpoHP3–lacZ reporter. At the indicated times, samples were taken from the [32]P labelled culture and nucleotides were isolated by formic acid extraction (Schneider et al., 2003). Nucleotides were separated by PEI cellulose thin-layer chromatography using 0.85 M KH2PO4 as the running buffer. Because pppGpp and ppGpp are not resolved using 0.85 M KH2PO4, a running buffer with 1.5 M KH2PO4 was used to separate pppGpp from ppGpp for the TLC presented in Fig. 7. Nucleotides were detected by phospho-imaging and quantified using Image-Quant 5.2 (Molecular Dynamics). Because equal volumes of samples were spotted onto the TLC plates, the intensities of the nucleotide spots were normalized to the OD of the cultures at the time of sampling during the data analysis. These values are presented in Figs 6A and 8B. To compare samples from different cultures, the amounts of (p)ppGpp as a fraction of (p)ppGpp and GTP were calculated (Figs 6D and 8E).
Supplementary Material
Acknowledgements
We thank Richard Gourse for providing the rrnB P1–lacZ fusion strain RLG6583 and lacUV5–lacZ fusion strain RLG4993, Michael Cashel for the spoTsyn− and spoThyd− mutant strains CF11605 and CF11608, and Steve Finkel for the bolA–lacZ reporter. We would also like to thank Richard Gourse, Tomas Gaal, Karen Wassarman and Jade Wang for advice on nucleotide extraction experiments, and the Gourse lab for advice on the primer extension experiments. This project was supported by Award number NSF0347302 from the National Science Foundation and Award number GM097365 from the National Institute of General Medicine.
Footnotes
Supporting information
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Aberg A, Fernández-Vázquez J, Cabrer-Panes JD, Sánchez A, Balsalobre C. Similar and divergent effects of ppGpp and DksA deficiencies on transcription in Escherichia coli. J Bacteriol. 2009;191:3226–3236. doi: 10.1128/JB.01410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ades SE, Connolly LE, Alba BM, Gross CA. The Escherichia coli sigma(E)-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 1999;13:2449–2461. doi: 10.1101/gad.13.18.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ades SE, Grigorova IL, Gross CA. Regulation of the alternative sigma factor sigma(E) during initiation, adaptation, and shutoff of the extracytoplasmic heat shock response in Escherichia coli. J Bacteriol. 2003;185:2512–2519. doi: 10.1128/JB.185.8.2512-2519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006–2008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchinger SE, Ades SE. Regulated proteolysis: control of the Escherichia coli σ(E)-dependent cell envelope stress response. Subcell Biochem. 2013;66:129–160. doi: 10.1007/978-94-007-5940-4_6. [DOI] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankschien MD, Potrykus K, Grace E, Choudhary A, Vinella D, Cashel M, Herman C. TraR, a homolog of a RNAP secondary channel interactor, modulates transcription. PLoS Genet. 2009;5:e1000345. doi: 10.1371/journal.pgen.1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S, Nudler E. RNA polymerase: the vehicle of transcription. Trends Microbiol. 2008;16:126–134. doi: 10.1016/j.tim.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Bougdour A, Gottesman S. ppGpp regulation of RpoS degradation via anti-adaptor protein IraP. Proc Natl Acad Sci USA. 2007;104:12896–12901. doi: 10.1073/pnas.0705561104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougdour A, Cunning C, Baptiste PJ, Elliott T, Gottesman S. Multiple pathways for regulation of sigmaS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol Microbiol. 2008;68:298–313. doi: 10.1111/j.1365-2958.2008.06146.x. [DOI] [PubMed] [Google Scholar]
- Brown LL, Gentry DD, Elliott TT, Cashel MM. DksA affects ppGpp induction of RpoS at a translational level. J Bacteriol. 2002;184:4455–4465. doi: 10.1128/JB.184.16.4455-4465.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess R, Travers AA, Dunn JJ, Bautz EK. Factor stimulating transcription by RNA polymerase. Nature. 1969;221:43–46. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Tupy JL, Gruber TM, Wang S, Sharp MM, Gross CA, Darst SA. Crystal structure of Escherichia coli sigmaE with the cytoplasmic domain of its anti-sigma RseA. Mol Cell. 2003;11:1067–1078. doi: 10.1016/s1097-2765(03)00148-5. [DOI] [PubMed] [Google Scholar]
- Cashel M. Regulation of bacterial ppGpp and pppGpp. Annu Rev Microbiol. 1975;29:301–318. doi: 10.1146/annurev.mi.29.100175.001505. [DOI] [PubMed] [Google Scholar]
- Cashel M, Gentry DR, Hernandez VJ, Vinella D. The stringent response. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin E, Low KB, Magasanik B, et al., editors. Escherichia coli and Salmonella. Washington, DC: American Society for Microbiology (ASM) Press; 1996. pp. 1458–1496. [Google Scholar]
- Costanzo A, Ades SE. Growth phase-dependent regulation of the extracytoplasmic stress factor, sigmaE, by guanosine 3′,5′-bispyrophosphate (ppGpp) J Bacterial. 2006;188:4627–4634. doi: 10.1128/JB.01981-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo A, Nicoloff H, Barchinger SE, Banta AB, Gourse RL, Ades SE. ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor sigmaE in Escherichia coliby both direct and indirect mechanisms. Mol Microbiol. 2008;67:619–632. doi: 10.1111/j.1365-2958.2007.06072.x. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Las Peñas A, Connolly L, Gross CA. SigmaE is an essential sigma factor in Escherichia coli. J Bacteriol. 1997a;179:6862–6864. doi: 10.1128/jb.179.21.6862-6864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Las Peñas A, Connolly L, Gross CA. The sigmaE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of sigmaE. Mol Microbiol. 1997b;24:373–385. doi: 10.1046/j.1365-2958.1997.3611718.x. [DOI] [PubMed] [Google Scholar]
- Dennis PP, Ehrenberg M, Bremer H. Control of rRNA synthesis in Escherichia coli: a systems biology approach. Microbiol Mol Biol Rev. 2004;68:639–668. doi: 10.1128/MMBR.68.4.639-668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Hansen A-M, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaal T, Bartlett MS, Ross W, Turnbough CL, Gourse RL. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- Gaal T, Mandel MJ, Silhavy TJ, Gourse RL. Crl facilitates RNA polymerase holoenzyme formation. J Bacteriol. 2006;188:7966–7970. doi: 10.1128/JB.01266-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant JA. Stringent control in E. coli. Annu Rev Genet. 1979;13:393–415. doi: 10.1146/annurev.ge.13.120179.002141. [DOI] [PubMed] [Google Scholar]
- Gentry DR, Cashel M. Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol Microbiol. 1996;19:1373–1384. doi: 10.1111/j.1365-2958.1996.tb02480.x. [DOI] [PubMed] [Google Scholar]
- Gentry DR, Hernandez VJ, Nguyen LH, Jensen DB, Cashel M. Synthesis of the stationary-phase sigma factor sigmaS is positively regulated by ppGpp. J Bacteriol. 1993;175:7982–7989. doi: 10.1128/jb.175.24.7982-7989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gropp M, Strausz Y, Gross M, Glaser G. Regulation of Escherichia coli RelA requires oligomerization of the C-terminal domain. J Bacteriol. 2001;183:570–579. doi: 10.1128/JB.183.2.570-579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- Harinarayanan R, Murphy H, Cashel M. Synthetic growth phenotypes of Escherichia coli lacking ppGpp and transketolase A (tktA) are due to ppGpp-mediated transcriptional regulation of tktB. Mol Microbiol. 2008;69:882–894. doi: 10.1111/j.1365-2958.2008.06317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseltine WA, Block R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci USA. 1973;70:1564–1568. doi: 10.1073/pnas.70.5.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden JD, Ades SE. The extracytoplasmic stress factor, sigmaE, is required to maintain cell envelope integrity in Escherichia coli. PLoS ONE. 2008;3:e1573. doi: 10.1371/journal.pone.0001573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch M, Elliott T. Role of ppGpp in rpoS stationary-phase regulation in Escherichia coli. J Bacteriol. 2002;184:5077–5087. doi: 10.1128/JB.184.18.5077-5087.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh Y-J, Wanner BL. Global regulation by the seven-component Pi signaling system. Curr Opin Microbiol. 2010;13:198–203. doi: 10.1016/j.mib.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M, Kvint K, Shingler V, Nystrom T. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 2002;16:1260–1270. doi: 10.1101/gad.227902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmsee T, Delic D, Agyenim T, Calles C, Wagner R. Differential stringent control of Escherichia coli rRNA promoters: effects of ppGpp, DksA, and the initiating nucleotides. Microbiology. 2011;157:2871–2879. doi: 10.1099/mic.0.052357-0. [DOI] [PubMed] [Google Scholar]
- Lamour V, Rutherford ST, Kuznedelov K, Ramagopal UA, Gourse RL, Severinov K, Darst SA. Crystal structure of Escherichia coli Rnk, a new RNA polymerase-interacting protein. J Mol Biol. 2008;383:367–379. doi: 10.1016/j.jmb.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie AD, Bernardo LMD, Sze CC, Skärfstad E, Szalewska-Palasz A, Nyström T, Shingler V. The role of the alarmone (p)ppGpp in sigma N competition for core RNA polymerase. J Biol Chem. 2003;278:1494–1503. doi: 10.1074/jbc.M209268200. [DOI] [PubMed] [Google Scholar]
- Lennon CW, Ross W, Martin-Tumasz S, Toulokhonov I, Vrentas CE, Rutherford ST, et al. Direct interactions between the coiled-coil tip of DksA and the trigger loop of RNA polymerase mediate transcriptional regulation. Genes Dev. 2012;26:2634–2646. doi: 10.1101/gad.204693.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson LU, Farewell A, Nyström T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Magnusson LU, Gummesson B, Joksimović P, Farewell A, Nyström T. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J Bacteriol. 2007;189:5193–5202. doi: 10.1128/JB.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechold U, Murphy H, Brown L, Cashel M. Intramolecular regulation of the opposing (p)ppGpp catalytic activities of Rel(Seq), the Rel/Spo enzyme from Streptococcus equisimilis. J Bacteriol. 2002;184:2878–2888. doi: 10.1128/JB.184.11.2878-2888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechold U, Potrykus K, Murphy H, Murakami KS, Cashel M. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 2013;41:6175–6189. doi: 10.1093/nar/gkt302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. A Short Course in Bacterial Genetics. Cold Spring Harbor, NY: CSHL Press; 1992. [Google Scholar]
- Missiakas D, Mayer MP, Lemaire M, Georgopoulos C, Raina S. Modulation of the Escherichia coli sigmaE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol Microbiol. 1997;24:355–371. doi: 10.1046/j.1365-2958.1997.3601713.x. [DOI] [PubMed] [Google Scholar]
- Muffler A, Fischer D, Altuvia S, Storz G, Hengge-Aronis R. The response regulator RssB controls stability of the sigma(S) subunit of RNA polymerase in Escherichia coli. EMBOJ. 1996;15:1333–1339. [PMC free article] [PubMed] [Google Scholar]
- Murray HD, Gourse RL. Unique roles of the rrn P2 promoters in Escherichia coli. Mol Microbiol. 2004;52:1375–1387. doi: 10.1111/j.1365-2958.2004.04060.x. [DOI] [PubMed] [Google Scholar]
- Murray HD, Schneider DA, Gourse RL. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol Cell. 2003;12:125–134. doi: 10.1016/s1097-2765(03)00266-1. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickels BE, Hochschild A. Regulation of RNA polymerase through the secondary channel. Cell. 2004;118:281–284. doi: 10.1016/j.cell.2004.07.021. [DOI] [PubMed] [Google Scholar]
- Nyström T. Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol Microbiol. 2004;54:855–862. doi: 10.1111/j.1365-2958.2004.04342.x. [DOI] [PubMed] [Google Scholar]
- Österberg S, del Peso-Santos T, Shingler V. Regulation of alternative sigma factor use. Annu Rev Microbiol. 2011;65:37–55. doi: 10.1146/annurev.micro.112408.134219. [DOI] [PubMed] [Google Scholar]
- Paget MSB, Helmann JD. The sigma70 family of sigma factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu Rev Genet. 2004a;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004b;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Berkmen MB, Gourse RL. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci USA. 2005;102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the secondary channel-structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Vinella D, Murphy H, Szalewska-Palasz A, D’ari R, Cashel M. Antagonistic regulation of Escherichia coli ribosomal RNA rrnB P1 promoter activity by GreA and DksA. J Biol Chem. 2006;281:15238–15248. doi: 10.1074/jbc.M601531200. [DOI] [PubMed] [Google Scholar]
- Pratt LA, Silhavy TJ. The response regulator SprE controls the stability of RpoS. Proc Natl Acad Sci USA. 1996;93:2488–2492. doi: 10.1073/pnas.93.6.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao NN, Liu S, Kornberg A. Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J Bacteriol. 1998;180:2186–2193. doi: 10.1128/jb.180.8.2186-2193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Gourse RL. Analysis of RNA polymerase-promoter complex formation. Methods. 2009;47:13–24. doi: 10.1016/j.ymeth.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. The magic spot: a ppGpp binding site on E. coil RNA polymerase responsible for regulation of transcription initiation. Mol Cell. 2013;50:420–429. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Lemke JJ, Vrentas CE, Gaal T, Ross W, Gourse RL. Effects of DksA, GreA, and GreB on transcription initiation: insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J Mol Biol. 2007;366:1243–1257. doi: 10.1016/j.jmb.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DA, Murray HD, Gourse RL. Measuring control of transcription initiation by changing concentrations of nucleotides and their derivatives. Methods Enzymol. 2003;370:606–617. doi: 10.1016/S0076-6879(03)70051-2. [DOI] [PubMed] [Google Scholar]
- Spira B, Silberstein N, Yagil E. Guanosine 3’,5-’bispyrophosphate (ppGpp) synthesis in cells of Escherichia coli starved for Pi. J Bacteriol. 1995;177:4053–4058. doi: 10.1128/jb.177.14.4053-4058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitil AL, Cashel M, Zyskind JW. Guanosine tetraphosphate inhibits protein synthesis in vivo. A possible protective mechanism for starvation stress in Escherichia coli. J Biol Chem. 1993;268:2307–2311. [PubMed] [Google Scholar]
- Szalewska-Palasz A, Johansson LUM, Bernardo LMD, Skärfstad E, Stec E, Brännström K, Shingler V. Properties of RNA polymerase bypass mutants: implications for the role of ppGpp and its co-factor DksA in controlling transcription dependent on sigma54. J Biol Chem. 2007;282:18046–18056. doi: 10.1074/jbc.M610181200. [DOI] [PubMed] [Google Scholar]
- Traxler MF, Summers SM, Nguyen H-T, Zacharia VM, Hightower GA, Smith JT, Conway T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Barembruch C, Possling A, Hengge R. Stationary phase reorganisation of the Escherichia coli transcription machinery by Crl protein, a fine-tuner of sigmas activity and levels. EMBO J. 2007;26:1569–1578. doi: 10.1038/sj.emboj.7601629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinella D, Potrykus K, Murphy H, Cashel M. Effects on growth by changes of the balance between GreA, GreB, and DksA suggest mutual competition and functional redundancy in Escherichia coli. J Bacteriol. 2012;194:261–273. doi: 10.1128/JB.06238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH. Dissection of the mechanism for the stringent factor RelA. Mol Cell. 2002;10:779–788. doi: 10.1016/s1097-2765(02)00656-1. [DOI] [PubMed] [Google Scholar]
- Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- Yang C, Huang T-W, Wen S-Y, Chang C-Y, Tsai S-E, Wu W-E, Chang C-H. Genome-wide PhoB binding and gene expression profiles reveal the hierarchical gene regulatory network of phosphate starvation in Escherichia coli. PLoS ONE. 2012;7:e47314. doi: 10.1371/journal.pone.0047314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Gottesman S. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J Bacteriol. 1998;180:1154–1158. doi: 10.1128/jb.180.5.1154-1158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YN, Jin DJ. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like ‘stringent’ RNA polymerases in Escherichia coli . Proc Natl Acad Sci USA. 1998;95:2908–2913. doi: 10.1073/pnas.95.6.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Wang Y, Steitz TA. The mechanism of E. coil RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol Cell. 2013;50:430–436. doi: 10.1016/j.molcel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.