Abstract
Reversible lysine acetylation is a highly regulated post-translational protein modification that is known to regulate several signaling pathways. However, little is known about the radiation-induced changes in the acetylome. In this study, we analyzed the acute post-translational acetylation changes in primary human cardiac microvascular endothelial cells 4 h after a gamma radiation dose of 2 Gy. The acetylated peptides were enriched using anti-acetyl conjugated agarose beads. A total of 54 proteins were found to be altered in their acetylation status, 23 of which were deacetylated and 31 acetylated. Pathway analyses showed three protein categories particularly affected by radiation-induced changes in the acetylation status: the proteins involved in the translation process, the proteins of stress response, and mitochondrial proteins. The activation of the canonical and non-canonical Wnt signaling pathways affecting actin cytoskeleton signaling and cell cycle progression was predicted. The protein expression levels of two nicotinamide adenine dinucleotide (NAD+)-dependent deacetylases, sirtuin 1 and sirtuin 3, were significantly but transiently upregulated 4 but not 24 h after irradiation. The status of the p53 protein, a target of sirtuin 1, was found to be rapidly stabilized by acetylation after radiation exposure. These findings indicate that post-translational modification of proteins by acetylation and deacetylation is essentially affecting the radiation response of the endothelium.
Keywords: acetylation, ionizing radiation, proteomics, endothelial cell, sirtuins, heart
INTRODUCTION
Acetylation of the lysine residue is a reversible post-translational modification (PTM) that plays an essential role in the regulation of protein stability and the activation of several signaling pathways [1]. The regulation of gene expression through the modification of core histone tails by histone acetyltransferases (HATs) or histone deacetylases (HDACs) is well known [2, 3], but lysine acetylation is also essential for p53-associated functions [1]. It has been suggested that the ∼3600 acetylation sites identified so far contribute to the regulation of almost all nuclear functions and to the control of a large array of cytoplasmic functions [1].
The deacetylases responsible for the controlled removal of acetyl groups can be categorized into four subgroups. The sirtuins, representing Class III deacetylases, are nicotinamide adenine dinucleotide (NAD+)-dependent enzymes, and include seven members in humans [4]. The best characterized of these is sirtuin 1 (SIRT1) [5]. SIRT1 functions as a metabolic sensor involved in cell cycle progression, the apoptosis or survival decision, senescence and inflammation [6]. Sirtuin 3 (SIRT3) is located in mitochondria and has been implicated in regulating metabolic processes [7, 8].
The deacetylation targets of SIRT1 have been studied extensively over the recent years. Besides its ability to deacetylate histones H1, H3 and H4 [9–11], SIRT1 was also found to deacetylate the master regulator of DNA damage, the p53 protein, either directly [12, 13] or by inactivating (deacetylating) the histone acetyltransferase TIP60 that is able to acetylate p53 [14]. In the case of DNA damage, such as that triggered by ionizing radiation, p53 is rapidly stabilized by phosphorylation and acetylation [15]. Acetylation competes for lysine residues that are also targets for ubiquitination, sumoylation and methylation [16]. The stabilization of p53 rapidly triggers the transactivation of several genes that are involved in cell cycle arrest, apoptosis and metabolic changes [17–19]. The deacetylation of p53 by SIRT1 has been shown to increase cell survival [20], while SIRT1 deficiency caused accumulation of p53 acetylation, thereby enhancing oxidative stress-induced cellular senescence [21]. In an apparent feedback loop, the transcription of the SIRT1 gene is negatively controlled by active p53 [22]. Thus, SIRT1 plays a major role in its own transcriptional regulation.
In addition to p53 deacetylation, SIRT1 has other essential functions in the repair of DNA double-strand breaks: it contributes to the formation of gamma-H2AX, BRCA1, Rad51 and NBS1 foci after gamma irradiation [23]; it also induces Ku70-dependent DNA repair [24]. Thus, SIRT1 seems to play a dual role in the radiation response by, on the one hand, destabilizing p53 that is crucial for DNA repair but, on the other hand, actively contributing to the formation of DNA repair complexes. Indeed, we have previously shown a rapid alteration in the expression of the Ku protein complex (Ku70/Ku80) in the human endothelial cell line EA.hy926 after exposure to a gamma dose of 2.5 Gy [25].
The endothelial response to ionizing radiation is of particular interest because the epidemiological evidence indicates an enhanced risk of cardiovascular disease after radiotherapy treatment of malignant disease such as breast cancer if the heart is at least partially exposed [26–28]. Recently, in addition to the cardiac macrovascular damage and atherosclerosis, the role of microvascular damage and the subsequent reduction of capillary density has been suggested to be the initial and promoting cause of radiation-induced heart disease [29].
In this study, we have investigated whether the acetylation/deacetylation process plays a role in the activation of known [25] or novel pathways following a single acute radiation dose (2 Gy). For this purpose we used primary human cardiac microvascular endothelial cells (HCMEC) as the cellular model.
MATERIALS AND METHODS
Cell culture, irradiation and harvesting of cells
Primary human microvascular endothelial cells were purchased from Promocell (Berlin, Germany). The cells were originally isolated from multiple donors (at least three); the doubling time of the cells was approximately 31 h. The cells were cultured in T75 culture flasks at 37°C with 5% CO2 in air in Endothelial Cell Growth Medium MV2 (Promocell) containing supplements (5% fetal calf serum, 0.2 µg/ml hydrocortisone, 0.5 ng/ml vascular endothelial growth factor, 10 ng/ml basic fibroblast factor, 20 ng/ml IGF-1, 1 µg/ml ascorbic acid). Ionizing radiation was delivered to exponentially growing cells at the indicated doses using a 137Caesium gamma source (HWM-D 2000, Waelischmüller, Germany) operated at a dose rate of 0.49 Gy/min. At 4 and 24 h after irradiation, the cells were harvested by scraping and rinsing once with 10 mM Tris–250 mM sucrose (pH 7.0). Detached cells were centrifuged 3 min at 220g, the supernatant was removed and the cells were resuspended in 1 ml 10 mM Tris–250 mM sucrose (pH 7.0) and immediately frozen in a pre-cooled (4°C) isopropanol-containing freezing device and stored at −80°C. Cell pellets were lysed with a lysis buffer containing 1% Triton X100, 100 mM Tris HCl pH 7.6, protease and phosphatase inhibitor cocktails (Roche) and lysine deacetylase inhibitors MS275 and suberoylanilide hydroxamic acid (Alexis Biochemicals). The samples were diluted five times with water for the protein concentration measurement in triplicate by the Bradford assay.
Acetylome analysis
Protein extract (300 μg) from control and irradiated HCMEC (4 h) in 1% Triton, 100 mM Tris buffer, pH 7.6 was subjected to overnight in-solution tryptic digestion as described previously [30]. Acetylated peptides in the lysate were enriched using agarose-conjugated antibody against acetylated lysine (ImmunChem) and eluted using 0.1% TFA as described previously [31]. Peptides were separated by reversed phase chromatography (PepMap, 15 cm × 75 μm ID, 3 μm/100 Å pore size, liquid chromatography (LC) Packings) operated on a nano-HPLC (Ultimate 3000, Dionex) with a non-linear 170-min gradient with a flow rate of 300 nl/min as described previously [32]. The gradient settings were subsequently: 0 to 140 min: between 2% and 5% to 31% B; 140 to154 min: 31% to 95% B; 145 to150 min: constant at 95% B; 150–155 min 95% to 5% B. The nano-LC was connected to a linear quadrupole ion trap mass spectrometer (LTQ Orbitrap XL, ThermoFisher, Bremen, Germany) equipped with a nano-ESI source. The mass spectrometer was operated in the data-dependent mode to automatically switch between Orbitrap-MS and LTQ-MS/MS acquisition. Survey full-scan mass spectra (from m/z 300 to 1500) were acquired in the Orbitrap with a resolution of R = 60 000 at m/z 400. This method allowed up to 10 of the most intense ions to be isolated sequentially, depending on signal intensity, for fragmentation on the linear ion trap using collision-induced dissociation. High-resolution MS scans in the Orbitrap and MS/MS scans in the linear ion trap were performed in parallel. Target peptides already selected for MS/MS were dynamically excluded for 60 s. The acquired spectra (Thermo raw files) were loaded into the Progenesis LC-MS software (version 3.0, Non-linear) and label-free quantification was performed as described before [32, 33]. Briefly, the profile data of the MS scans as well as MS/MS spectra were transformed to peak lists with Progenesis LC-MS using a proprietary algorithm and then stored in peak lists comprising m/z and abundance. One sample was set as a reference, and the retention times of all other samples within the experiment were aligned (three to five manual landmarks, followed by automatic alignment) to create maximal overlay of the 2D feature maps. At this point, features with only one charge or more than seven charges were masked and excluded from further analyses, and all remaining features were used to calculate a normalization factor for each sample that corrects for experimental variation. Samples were then allocated to their experimental group (control or irradiated).
MS/MS spectra were exported from the Progenesis LC-MS software as a Mascot Generic file (mgf) and used for peptide identification with Mascot (version 2.3) in the Ensembl database for human (release 72, 40 047 703 residues, 105 287 sequences). The following search parameters were used: 10 ppm peptide mass tolerance and 0.6 Da fragment mass tolerance, one missed cleavage was allowed, carbamidomethylation (C) was set as fixed modification and oxidation (M), deamidation (N,Q) as well as acetylation (K and protein N-terminus) were allowed as variable modifications. A MASCOT-integrated decoy database search calculated a false discovery of ≤1% when searching was performed on the concatenated mgf files with an ion score cut-off of 30 and a significance threshold of P ≤ 0.01.
For quantification, all unique peptides (with Mascot score ≥30 and P < 0.01, see above) of an identified protein were included, and the total cumulative abundance was calculated by summing the abundances of all unique acetylated peptides derived from the respective proteins. No minimal thresholds were set for the method of peak picking or selection of data used for quantification.
Bioinformatics analysis
To further analyze the overlapping networks and pathways of radiation-responsive proteins, we imported all identified proteins with their corresponding accession numbers into GeneMANIA (http://www.genemania.org/, 15 July 2014, date last accessed) and Ingenuity Pathway Analysis (IPA) (Ingenuity System, http://www.ingenuity.com, 22 October 2014, date last accessed) bioinformatics software. GeneMANIA was used to identify the affected biological functions. IPA was used to identify the significantly deregulated canonical pathways.
Immunoblotting analysis
For the validation of protein expression changes by immunoblotting, 20 mg of protein extract was separated on 8% and 12% SDS polyacrylamide gels according to Laemmli [34]. Proteins were transferred to nitrocellulose membranes (GE Healthcare) using a semidry blotting system at 100 mA for 90 min. Membranes were saturated for 1 h with 5% advance blocking reagent (GE Healthcare) in TBS (50 mM Tris.HCl, pH 7.6 and 150 mM NaCl) containing 0.1% Tween 20 (TBS/T).
Blots were incubated overnight at +4°C with antibodies against either SIRT1, SIRT3, acetyl-p53, phospho-p53 or p21 protein (Cell Signaling) with alpha-tubulin (Cell Signaling) as the loading control. After washing three times in TBS/T, blots were incubated for 1 h at room temperature with horseradish peroxidase-conjugated anti-mouse or anti-goat secondary antibody (Santa Cruz Biotechnology) in blocking buffer (TBS/T with 5% w/v advance blocking reagent). Immunodetection was performed with ECL advance Western blotting detection kit (GE Healthcare). The protein bands were quantified using ImageQuant 5.2 software (GE Healthcare) by integration of all pixel values in the band area after background correction, and normalized to the alpha-tubulin expression. The level of alpha-tubulin is not affected by irradiation in endothelial cells and was therefore used for normalization [25].
Isolation of total RNA and gene expression analysis
The mirVanaTM Isolation Kit (Ambion) was used to isolate and purify total RNA from frozen HCMEC cells according to the manufacturer's protocol. Total RNA was eluted with nuclease-free water. The OD ratio of 260/280 of the lysates estimating the RNA quality was measured using Nanodrop spectrophotometer (PeqLab Biotechnology; Germany). It ranged between 2.0 and 2.1. Lysates were stored at −20°C until quantification of gene expression.
Total RNA isolates (100 ng) from sham-irradiated and 2-Gy–irradiated cells (4 h post-irradiation) were used to quantify the expression of 84 genes related to the p53 signaling pathway (RT2 Profiler PCR array PAHS-027Z – Quiagen). The assays were performed according to the manufacturer's instructions, including genomic DNA elimination, first-strand cDNA synthesis, preamplification of cDNA target templates and real time PCR via RT2 SYBR Green Mastermix. The StepOnePlus device (Applied Biosystems) was used to detect the thermal cycles accompanied with fluorescence emission using the manufacturer's instructions. Normalization of relative expression levels of each mRNA was done against the median of all 84 target genes using the equation 2−ΔΔCt, where:
Statistical analysis
Two biological replicates, each representing a pool of at least three different biological donors, were used for analysis of protein acetylation changes by MS 4 h after irradiation. The mean ratio value of acetylated peptide amounts between controls and irradiated samples were calculated. Proteins showing increased expression levels by ≥1.40 fold or decreased expression levels by ≤0.714 were considered to be deregulated by acetylation/deacetylation.
Western blots were quantified using at least three replicates (two biological and one technical). The differences in the protein amount were considered significant if they reached a P-value of ≤0.05 (unpaired Student's t-test, n = 3).
Gene expression changes were considered significant if they reached a P-value of ≤0.05 (unpaired Student's t-test, n = 3) and had a fold-change of ≥1.2 or ≤−1.2. The threshold of ±1.2 is based on the average experimental technical variance (9.2%) of a set of 14 overlapping target genes from two different RT2 Profiler PCR arrays (unpublished data). Thus, a fold-change of ±1.2 enables confident target gene expression quantification.
Data deposition of proteomics experiments
The MSF files of the obtained MS/MS spectra can be found under http://storedb.org/project_details.php?projectid=50
RESULTS
Changes in the acetylation status of HCMEC after irradiation
Radiation-induced effects on the HCMEC acetylome were investigated using a label-free proteomic quantification approach after enrichment of acetylated peptides. We identified 119 peptides, corresponding to 88 acetylated proteins, of which 54 were changed in their acetylation status significantly (fold change ≥1.30 or ≤0.77). At 4 h after irradiation, 23 proteins were deacetylated, while 31 showed a significant increase in the acetylation status (Supplementary Table S1).
Proteins with a changed acetylation status were imported to GeneMANIA and IPA software tools for network analysis. The imported proteins in GeneMANIA resulted in a creation of a large protein network according to the known interactions between these proteins in the literature. We show here the most significant subnetworks based on biological functions according to the number of affected proteins. The analysis by GeneMANIA showed the initiation of translation to be the most affected pathway involving 21 proteins (6 deregulated proteins from our proteomics results and 15 predicted proteins) with a changed acetylation status as an early response to irradiation (Fig. 1a). The proteins involved in this network were ribosomal proteins and elongation initiation factors. The actin cytoskeleton signaling (5 deregulated and 4 predicted proteins) and nuclear transport (8 deregulated proteins and 1 predicted protein) were also indicated as affected by the GeneMANIA analysis (Supplementary Fig. S1a, S1b).
Fig. 1.
Pathway analysis of radiation-induced acetylated and deacetylated proteins. All differentially regulated acetylated and deacetylated proteins were imported into the GeneMANIA and IPA software tools. (a) GeneMANIA identified translational initiation as the most significant protein class affected by irradiation on the acetylation level. The striped brown and gray circles represent the identified acetylated/deacetylated proteins, whereas the uniquely colored circles represent the predicted interactive proteins. The brown color shows the network involved in protein synthesis; the gray color indicates proteins only indirectly related to protein synthesis. (b) The most affected biological networks from the IPA analysis were cell death and survival, cellular movement, cellular development, cellular morphology and protein trafficking; these are shown as a merged network. The gray molecules represent proteins found to be altered in their acetylation status in this study; the white molecules are interaction partners predicted by the software. The solid arrows and lines represent direct interactions, and the dotted arrows and lines indirect interactions.
The most important biological processes identified by the IPA software were cell death and survival, cellular movement, cellular development, cellular morphology and protein trafficking. The most significantly affected network was the stress-related response (Fig. 1b) with extracellular-signal-regulated kinases (ERK; ERK1/2), p38 mitogen-activated protein kinases (p38 MAPK) and protein kinase B (Akt) as central hubs.
In accordance with the GeneMANIA analysis, IPA identified initiation of translation, Rho and actin cytoskeleton signaling to be among the most affected pathways by radiation-induced changes in the acetylation status (Supplementary Fig. S1c).
Additional proteins showing a change in their acetylation status after radiation exposure included calmodulin 2, beta-catenin, alpha-catenin, actin related 2/3 complex (subunits 1B and 4), actin (gamma 1 and beta), twinfilin, S100 calcium-binding protein, thymosin beta, G protein, protein tyrosine phosphatase-like A domain containing 1 and microtubule-associated protein 4. All these proteins are members or downstream targets of the canonical and non-canonical Wnt pathways. This suggested that the Wnt pathway is highly affected by the acetylation after ionizing radiation.
Radiation-induced alterations in the levels of SIRT1 and SIRT3
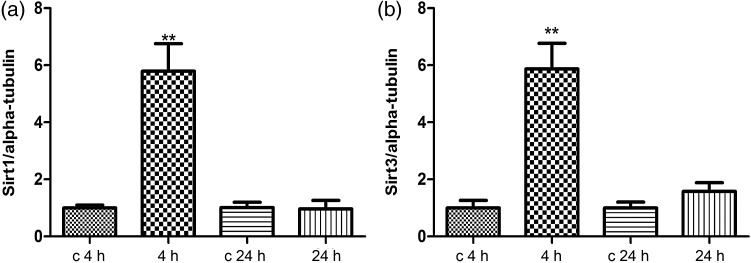
As many ribosomal proteins and elongation factors found to be deacetylated in our study are putative SIRT1 targets [35], we assessed the expression of SIRT1 by immunoblotting. The level of SIRT1 was significantly upregulated several fold 4 h but not 24 h after irradiation (Fig. 2a).
Fig. 2.
Immunoblot analysis of the SIRT1 and SIRT3 expression. The extracted proteins from control and irradiated cells at 4 or 24 h post-irradiation were separated by 12% SDS-PAGE and analyzed by immunoblotting with antibodies against (a) SIRT1; (b) SIRT3. The columns represent the average ratios with standard errors of the mean (SEMs) of relative protein expression in sham-irradiated and irradiated cells. The protein bands (E) were quantified using ImageQuant 5.2 software (GE Healthcare) by integration of all the pixel values in the band area after background correction and normalized to alpha-tubulin expression. Three biological replicates were used for each experiment. *P ≤ 0.05; **P ≤ 0.01 (Student's t-test).
Some mitochondrial proteins (cyclophilin A, fission 1 and NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9) were changed in their acetylation status after radiation exposure (Supplementary Table S1). Therefore, the level of the mitochondrial deacetylase SIRT3 was also studied 4 and 24 h after irradiation. Figure 2b shows that, similar to SIRT1, SIRT3 was significantly upregulated 4 but not 24 h after radiation exposure.
The acetylation and phosphorylation status of p53 and the expression of p21
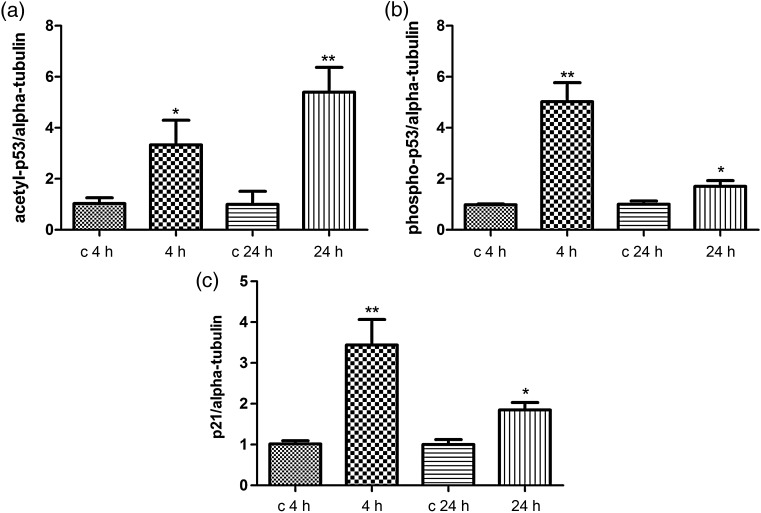
We also investigated the acetylation status of p53 by performing immunoblotting against its acetylated form (lysine-382) (Fig. 3a). We found an upregulation of the acetylated form at both time-points (4 and 24 h) after irradiation. The phosphorylated p53 (serine-15) was also found to be upregulated at both time-points (Fig. 3b). The p21 protein, the expression of which is tightly controlled by p53, was also upregulated 4 and 24 h after irradiation (Fig. 3c). The immunoblots are shown in Supplementary Fig. S1.
Fig. 3.
Immunoblot analysis of the radiation-induced changes in the p53 acetylation and phosphorylation status and in the p21 expression level. The extracted proteins from control and irradiated cells at 4 or 24 h after exposure were separated by 12% SDS-PAGE and analyzed by immunoblotting with antibodies against (a) anti-acetyl-p53; (b) anti-phospho-p53; (c) p21. The columns represent the average ratios with standard errors of the mean (SEMs) of relative protein expression/modification in sham- and irradiated cells. The protein bands (E) were quantified using ImageQuant 5.2 software (GE Healthcare) by integration of all the pixel values in the band area after background correction and normalized to the alpha-tubulin (αTub) expression. Three biological replicates were used for each experiment. *P ≤ 0.05; **P ≤ 0.01 (Student's t-test).
Cell cycle/p53 gene expression profiles
Acetylation and phosphorylation of p53 leads to activation of p53-related pathways. We tested 84 genes that are involved in cell cycle/p53 signaling as regulators or downstream targets. A total of 13 genes were found to be significantly upregulated and 4 downregulated (P < 0.05) 4 h after irradiation (Supplementary Table S2). However, at 24 h there were no significant radiation-induced changes in the 84 investigated genes (data not shown).
The gene expression levels of three proteins (SIRT1, HDAC1, KAT2B) involved in the acetylation/deacetylation process were found to be upregulated 4 h after irradiation. The upregulation of SIRT1 gene expression was in agreement with the immunoblotting results at this time-point. However, it is conceivable that p53 is not a target of SIRT1 in this study because both SIRT1 and the level of acetylation of p53 are simultaneously upregulated.
To further analyze the interaction of the p53-dependent network, we imported the deregulated genes from gene expression data in the IPA software tool (Fig. 4). In accordance with the immunoblotting results, the gene expression data predicted a significant activation of the p53 gene (2.156 activation z-score and P-value of 5,39E10).
Fig. 4.
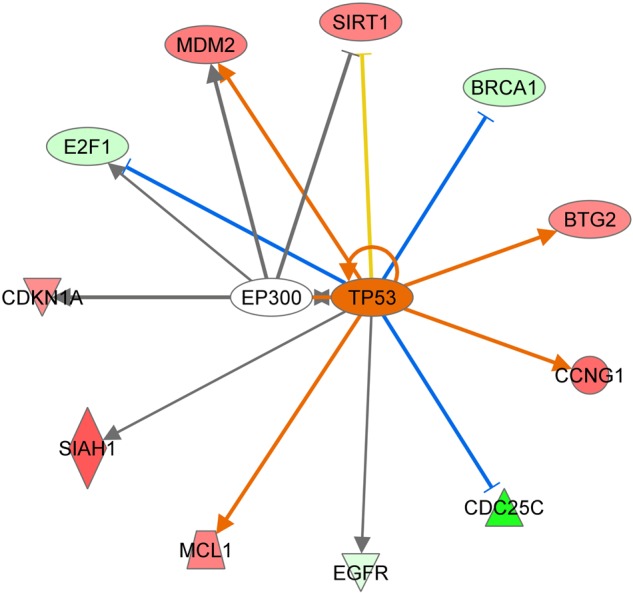
p53-related deregulated target genes using IPA software tool. The predicted interactions between TP53 (p53) and its target genes are shown by arrow (activation) or blocked arrow (inactivation). SIRT1 = sirtuin 1, CDC25C = cell division cycle 25C, EGFR = epidermal growth factor receptor, MCL1 = myeloid cell leukemia 1, SIAH1 = E3 ubiquitin-protein ligase 1; CDKNA1 = cyclin-dependent kinase inhibitor 1A, E2F1 = transcription factor E2F1, MDM2 = E3 ubiquitin-protein ligase, BRCA1 = breast cancer 1, BTG2 = BTG family member 2, anti-proliferative protein, CCNG1 = cyclin G1, EP300 = E1A-binding protein p300. Ingenuity upstream analysis predicted activation of the p53 gene by 2.156 activation score and overlap P-value of 5,39E10. Given the observed differential regulation of a gene (‘up’ or ‘down’) in the dataset, the activation state of an upstream regulator is determined by the regulation direction associated with the relationship from the regulator to the gene. The overlap P-value measures whether there is a statistically significant overlap between the dataset genes and the genes that are regulated by a transcriptional regulator. It is calculated using Fisher's Exact Test, and significance is generally attributed to P-values < 0.01.
DISCUSSION
Lysine acetylation is a major post-translational modification involved in a broad array of physiological responses and it coregulates key cellular functions [31], including DNA damage [36–38]. In this study we aimed to study the acetylation changes of human cardiac microvascular cells after an acute gamma radiation exposure of 2 Gy. A total of 54 proteins were found to be altered in their acetylation status, 31 of which were acetylated and 23 deacetylated. The deacetylases SIRT1 and SIRT3 were transiently upregulated 4 but not 24 h after the exposure.
Acetylation in protein synthesis and stress signaling
The acetylation status of a number of proteins involved in translational initiation was altered after irradiation. As protein synthesis is the most energy-consuming cellular process [39], translational regulation has to be strictly controlled under cellular growth and stress [40–42]. Consequently, an inhibition of global protein synthesis by blocking cap-dependent initiation will reduce energy demand during cellular stress [43], such as that following irradiation. It has been reported, however, that ionizing radiation stimulates protein synthesis immediately and transiently in non-transformed cells through activation of the mTOR pathway [40]. This initial activation of de novo translation is rapidly lost through assembly of the DNA damage-response apparatus and activation of ATM and p53 [40]. We have shown previously that the majority (81%) of proteins showing immediate (4 h) significant changes in expression after ionizing radiation (2.5 Gy) are downregulated [25]. However, while global translation is blocked by inhibition of cap-dependent initiation, translation of cap-independent transcripts involved in survival still takes place, making the latter the predominant mode of translation under stress [43]. We observed a rapid stabilization of the p53 protein by acetylation, its subsequent phosphorylation and activation of p53-regulated gene expression.
EIF1, EIF4E and EIF5A, all translational initiation factors, were found to be changed in their acetylation status in our study (EIF1 deacetylated; EIF4E and EIF5A acetylated). Whereas the effect of EIF1 and EIF4E acetylation in protein synthesis is not known, acetylation of EIF5A regulates its subcellular localization, increased acetylation leading to nuclear accumulation [44].
In total, four ribosomal proteins were found increasingly acetylated and one deacetylated after irradiation. It is suggested that ribosomal protein N-terminal acetylation is necessary for maintaining the ribosome's protein synthesis function, whereas deacetylation of ribosomal proteins may have an inhibitory effect on protein synthesis, because the acetylation is a prerequisite for both the rate and fidelity of translation [45].
The stress response in general is known to lead to the formation of cytoplasmic RNA–protein complexes referred to as stress granules [46]. The EIF5A and karyopherin (importin) that were found to be acetylated in this study are required for the formation of stress granules [47, 48]. The RhoA signaling pathway that was also shown to be affected by acetylation changes is involved in promoting stress granule formation or initiating apoptosis during stress [49]. In addition, KAT2B, a histone acetyl transferase that was found upregulated on the gene level, is required to stabilize stress granules [50].
Acetylation of RNA splicing proteins
Removal of introns by splicing from the primary RNA transcript is an essential process in the biosynthesis of mature mRNAs in eukaryotic cells. This process is performed by a large complex, the spliceosome, consisting of five small nuclear RNAs and more than 150 proteins. We found two spliceosome proteins to be changed in their acetylation status after irradiation, U2 small nuclear RNA auxiliary factor 1 by deacetylation and DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B by increased acetylation. Previous proteomic analyses have shown that numerous spliceosomal proteins are acetylated [31] and that small-molecule inhibitors of acetylation block spliceosome assembly in vitro at distinct stages before activation [51]. This, together with our data, suggests that acetylation plays a role in splicing. However, due to the small number of proteins being altered in our study it is not possible to say how the spliceosome is affected by irradiation.
Acetylation and mitochondrial proteins
The NDUFB9 subunit of the mitochondrial respiratory Complex I showed a significantly increased level of deacetylation (fold change 0.095) 4 h after irradiation. Acetylation of respiratory chain subunits, including the NDUFB9 subunit, is known to regulate the production of ATP and efficiency of the respiratory chain [52]. In fact, a proteomic analysis of intracellular proteins that had internal acetylation residues demonstrated that a disproportionate fraction of identified proteins were in the mitochondria and/or associated with energy metabolism [53]. The mitochondrial SIRT3 has been shown to be a stress-responsive deacetylase, and its increased expression protects cardiomyocytes from genotoxic and oxidative stress-mediated cell death [54]. Mice lacking SIRT3 showed hyperacetylation of mitochondrial proteins and decreased levels of ATP by 50% [55]. Our data suggest that increased SIRT3 levels and deacetylation of NDUFB9 result in increased activity of Complex I immediately after irradiation. Our previous results showed increased amounts of some respiratory chain subunits of Complex I in the heart muscle shortly after irradiation [56]. It is, therefore, tempting to suggest a direct link between SIRT3 and deacetylation of key proteins in mitochondrial respiration.
In addition, two other mitochondrial proteins had increased levels of acetylation: cyclophilin A and fission protein 1. Cyclophilin A is a pro-inflammatory mediator secreted as a response to reactive oxygen species in a highly regulated manner. Extracellular cyclophilin A activates vascular smooth muscle cells (VSMCs) and endothelial cells (ECs) promoting inflammation, cell growth and cell death. Recently, it was shown that ROS-dependent acetylation of cyclophilin A is required for its import to the extracellular space, where it regulates VSMC and EC activation [57].
Acetylation in Rho and Wnt signaling
We found several members of the Rho signaling pathway to have different acetylation levels after irradiation (calmodulin 2, alpha-catenin, beta-catenin, gamma 1 and beta forms of actin, actin-related 2/3 complex, twinfilin, S100 calcium-binding protein, thymosin beta 10, protein tyrosine phosphatase-like A domain containing 1 and G protein). Several members of RhoGDI/actin cytoskeleton signaling have been previously shown to be regulated on the level of acetylation [58]. Deacetylation means in this case activation of the signaling pathway [59]. Previous studies suggest that activation of this pathway by inflammatory cytokines and agonists of G-protein-coupled receptors effectively triggers vascular disease [60]. We have previously shown that the Rho signaling pathway is affected by irradiation in endothelial cells [25, 61], even with doses as low as 200 mGy [62]. In addition, with regard to the response of Rho/cytoskeleton signaling, we found highly deacetylated cystatin B protein. Cystatin B is thought to play a role in protecting cytosolic and cytoskeletal proteins against the cysteine proteases accidentally released from lysosomes.
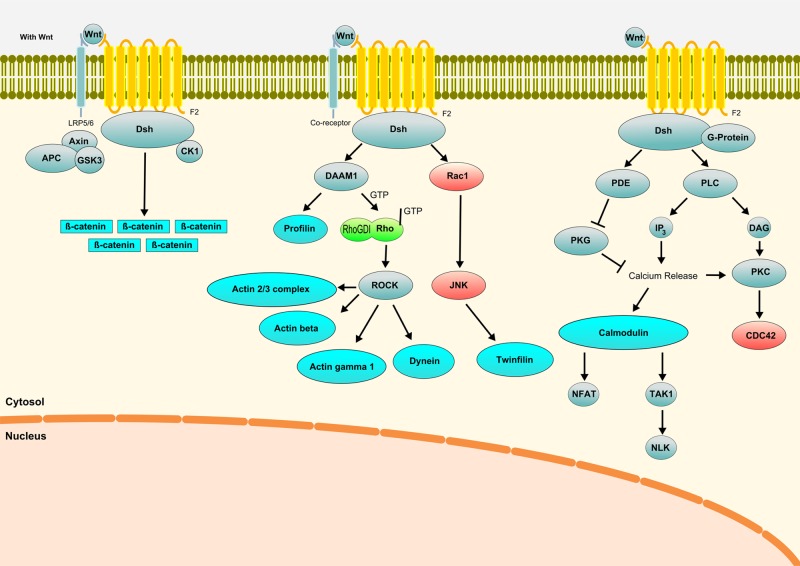
Rho GTPases are key mediators of Wnt pathway signals that promote morphological and transcriptional changes affecting cell behavior [63, 64]. In this study, several proteins belonging either to canonical and non-canonical Wnt signaling pathways were found to be changed in their acetylation status: G protein, profilin, beta-catenin, calmodulin 2, S100 calcium-binding protein, nucleophosmin, and transforming growth factor beta-1-induced transcript 1 protein (Fig. 5). Beta-catenin, a key protein in the Wnt pathway, showed an increased level of acetylation after irradiation. The acetyltransferase p300/CBP-associated factor (PCAF) directly binds to and acetylates beta-catenin, leading to its stabilization and activation of transcriptional activity [65]. SIRT1, which was shown to be upregulated after irradiation, promotes transient and constituent Wnt signaling through regulation of Dishevelled proteins [66]. KRAS, HDAC1 and BTG2 (upregulated in the gene expression analysis) and p21 (upregulated using immunoblotting) are known to stimulate Wnt signaling [67–69]. SIAH-1 (upregulated in the gene expression analysis) is also activating Wnt signaling through activation of ß-catenin by ubiquitination [70]. In addition, negative regulators of Wnt signaling, E2F1 and CDC25C, were downregulated at the gene expression level [71, 72]. These data suggest radiation-induced activation of Wnt signaling via acetylation/deacetylation processes.
Fig. 5.
Wnt canonical and non-canonical pathways affected 4 h after irradiation in HCMEC. Blue color represents proteins regulated by acetylation; green color represents proteins regulated at the level of total expression in endothelial cells [25, 73]; and red color represents proteins predicted activated using bioinformatics tools. Gray color represents proteins not found to be deregulated after irradiation.
CONCLUSION
This study highlights the role of post-translational modification of proteins by changing their acetylation status as an immediate endothelial response to ionizing radiation. Several pathways previously shown to be radiation responsive may be regulated by this mechanism. Such pathways include initiation of translation, stress signaling, energy metabolism, and Rho signaling. The involvement of Wnt signaling in the radiation response suggested by these data needs to be confirmed in future studies. Increased knowledge concerning the regulation of the radiation-induced acetylation process may help in preventing cellular and tissue injury after exposure situations.
FUNDING
This work was supported by a grant from the European Community's Seventh Framework Program (EURATOM), Contract No. 295823 (PROCARDIO). Funding to pay the Open Access publication charges was provided by a grant from the European Community's Seventh Framework Program (EURATOM), Contract No. 295823 (PROCARDIO).
SUPPLEMENTARY DATA
Supplementary data are available at the Journal of Radiation Research online.
ACKNOWLEDGEMENTS
We thank Stefanie Winkler and Anna Schwarz for excellent technical assistance.
REFERENCES
- 1.Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009;325:834–40. [DOI] [PubMed] [Google Scholar]
- 2.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol 2007;8:284–95. [DOI] [PubMed] [Google Scholar]
- 3.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem 2007;76:75–100. [DOI] [PubMed] [Google Scholar]
- 4.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun 2000;273:793–8. [DOI] [PubMed] [Google Scholar]
- 5.Morris BJ. Seven sirtuins for seven deadly diseases of aging. Free Radic Biol Med 2013;56:133–71. [DOI] [PubMed] [Google Scholar]
- 6.Vijg J, Maslov AY, Suh Y. Aging: a sirtuin shake-up? Cell 2008;135:797–8. [DOI] [PubMed] [Google Scholar]
- 7.Onyango P, Celic I, McCaffery JM, et al. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci U S A 2002;99:13653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He W, Newman JC, Wang MZ, et al. Mitochondrial sirtuins: regulators of protein acylation and metabolism. Trends Endocrinol Metab 2012;23:467–76. [DOI] [PubMed] [Google Scholar]
- 9.Das C, Lucia MS, Hansen KC, et al. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 2009;459:113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaquero A, Scher M, Lee D, et al. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell 2004;16:93–105. [DOI] [PubMed] [Google Scholar]
- 11.Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene 2007;26:5505–20. [DOI] [PubMed] [Google Scholar]
- 12.Vaziri H, Dessain SK, Ng Eaton E, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 2001;107:149–59. [DOI] [PubMed] [Google Scholar]
- 13.Luo J, Nikolaev AY, Imai S, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 2001;107:137–48. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Chen J. SIRT1 regulates autoacetylation and histone acetyltransferase activity of TIP60. J Biol Chem 2010;285:11458–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakaguchi K, Herrera JE, Saito S, et al. DNA damage activates p53 through a phosphorylation–acetylation cascade. Genes Dev 1998;12:2831–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 1997;90:595–606. [DOI] [PubMed] [Google Scholar]
- 17.Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol 2010;20:427–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell 1997;88:323–31. [DOI] [PubMed] [Google Scholar]
- 19.Levine AJ, Hu W, Feng Z. The p53 pathway: what questions remain to be explored? Cell Death Differ 2006;13:1027–36. [DOI] [PubMed] [Google Scholar]
- 20.Lee JT, Gu W. SIRT1: Regulator of p53 deacetylation. Genes Cancer 2013;4:112–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furukawa A, Tada-Oikawa S, Kawanishi S, et al. H2O2 accelerates cellular senescence by accumulation of acetylated p53 via decrease in the function of SIRT1 by NAD+ depletion. Cell Physiol Biochem 2007;20:45–54. [DOI] [PubMed] [Google Scholar]
- 22.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science 2004;306:2105–8. [DOI] [PubMed] [Google Scholar]
- 23.Bosch-Presegue L, Vaquero A. The dual role of sirtuins in cancer. Genes Cancer 2011;2:648–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sawada M, Sun W, Hayes P, et al. Ku70 suppresses the apoptotic translocation of Bax to mitochondria. Nat Cell Biol 2003;5:320–9. [DOI] [PubMed] [Google Scholar]
- 25.Sriharshan A, Boldt K, Sarioglu H, et al. Proteomic analysis by SILAC and 2D-DIGE reveals radiation-induced endothelial response: four key pathways. J Proteomics 2012;75:2319–30. [DOI] [PubMed] [Google Scholar]
- 26.Adams MJ, Hardenbergh PH, Constine LS, et al. Radiation-associated cardiovascular disease. Crit Rev Oncol Hematol 2003;45:55–75. [DOI] [PubMed] [Google Scholar]
- 27.Darby S, McGale P, Peto R, et al. Mortality from cardiovascular disease more than 10 years after radiotherapy for breast cancer: nationwide cohort study of 90 000 Swedish women. BMJ 2003;326:256–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darby SC, Ewertz M, McGale P, et al. Risk of ischemic heart disease in women after radiotherapy for breast cancer. N Engl J Med 2013;368:987–98. [DOI] [PubMed] [Google Scholar]
- 29.Darby SC, Cutter DJ, Boerma M, et al. Radiation-related heart disease: current knowledge and future prospects. Int J Radiat Oncol Biol Phys 2010;76:656–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azimzadeh O, Barjaktarovic Z, Aubele M, et al. Formalin-fixed paraffin-embedded (FFPE) proteome analysis using gel-free and gel-based proteomics. J Proteome Res 2010;9:4710–20. [DOI] [PubMed] [Google Scholar]
- 31.Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009;325:834–40. [DOI] [PubMed] [Google Scholar]
- 32.Hauck SM, Dietter J, Kramer RL, et al. Deciphering membrane-associated molecular processes in target tissue of autoimmune uveitis by label-free quantitative mass spectrometry. Mol Cell Proteomics 2010;9:2292–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azimzadeh O, Scherthan H, Yentrapalli R, et al. Label-free protein profiling of formalin-fixed paraffin-embedded (FFPE) heart tissue reveals immediate mitochondrial impairment after ionising radiation. J Proteomics 2012;75:2384–95. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970;227:680–5. [DOI] [PubMed] [Google Scholar]
- 35.Peng L, Ling H, Yuan Z, et al. SIRT1 negatively regulates the activities, functions, and protein levels of hMOF and TIP60. Mol Cell Biol 2012;32:2823–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Y, Xu Y, Roy K, et al. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol Cell Biol 2007;27:8502–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solomon JM, Pasupuleti R, Xu L, et al. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol 2006;26:28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beli P, Lukashchuk N, Wagner SA, et al. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol Cell 2012;46:212–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buttgereit F, Brand MD. A hierarchy of ATP-consuming processes in mammalian cells. Biochem J 1995;312:163–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braunstein S, Badura ML, Xi Q, et al. Regulation of protein synthesis by ionizing radiation. Mol Cell Biol 2009;29:5645–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trivigno D, Bornes L, Huber SM, et al. Regulation of protein translation initiation in response to ionizing radiation. Radiat Oncol 2013;8:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 2005;6:318–27. [DOI] [PubMed] [Google Scholar]
- 43.Stickel S, Gomes N, Su T. The role of translational regulation in survival after radiation damage; an opportunity for proteomics analysis. Proteomes 2014;2:272–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishfaq M, Maeta K, Maeda S, et al. Acetylation regulates subcellular localization of eukaryotic translation initiation factor 5A (eIF5A). FEBS Lett 2012;586:3236–41. [DOI] [PubMed] [Google Scholar]
- 45.Lee SB, Park JH, Folk JE, et al. Inactivation of eukaryotic initiation factor 5A (eIF5A) by specific acetylation of its hypusine residue by spermidine/spermine acetyltransferase 1 (SSAT1). Biochem J 2011;433:205–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell 2009;36:932–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kedersha NL, Gupta M, Li W, et al. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol 1999;147:1431–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujimura K, Suzuki T, Yasuda Y, et al. Identification of importin alpha1 as a novel constituent of RNA stress granules. Biochim Biophys Acta 2010;1803:865–71. [DOI] [PubMed] [Google Scholar]
- 49.Tsai NP, Wei LN. RhoA/ROCK1 signaling regulates stress granule formation and apoptosis. Cell Signal 2010;22:668–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohn T, Anderson P. The role of posttranslational modifications in the assembly of stress granules. Wiley Interdiscip Rev RNA 2010;1:486–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuhn AN, van Santen MA, Schwienhorst A, et al. Stalling of spliceosome assembly at distinct stages by small-molecule inhibitors of protein acetylation and deacetylation. RNA 2009;15:153–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahn B-H, Kim H-S, Song S, et al. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A 2008;105:14447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim SC, Sprung R, Chen Y, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell 2006;23:607–18. [DOI] [PubMed] [Google Scholar]
- 54.Sundaresan NR, Samant SA, Pillai VB, et al. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol Cell Biol 2008;28:6384–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahn BH, Kim HS, Song S, et al. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A 2008;105:14447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Azimzadeh O, Scherthan H, Sarioglu H, et al. Rapid proteomic remodeling of cardiac tissue caused by total body ionizing radiation. Proteomics 2011;11:3299–311. [DOI] [PubMed] [Google Scholar]
- 57.Soe NN, Sowden M, Baskaran P, et al. Acetylation of cyclophilin A is required for its secretion and vascular cell activation. Cardiovasc Res 2014;101:444–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sadoul K, Wang J, Diagouraga B, et al. The tale of protein lysine acetylation in the cytoplasm. J Biomed Biotechnol 2011;2011:970382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sadoul K, Wang J, Diagouraga B, et al. The tale of protein lysine acetylation in the cytoplasm. J Biomed Biotechnol 2011;2011:970382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seasholtz TM, Brown JH. Rho signaling in vascular diseases. Mol Interv 2004;4:348–57. [DOI] [PubMed] [Google Scholar]
- 61.Sriharshan A, Kraemer A, Atkinson MJ, et al. Radiation-induced crosstalk between microRNAs and proteins of the endothelium: in silico analysis. J Proteomics Bioinform 2014;7:327–31. [Google Scholar]
- 62.Pluder F, Barjaktarovic Z, Azimzadeh O, et al. Low-dose irradiation causes rapid alterations to the proteome of the human endothelial cell line EA.hy926. Radiat Environ Biophys 2011;50:155–66. [DOI] [PubMed] [Google Scholar]
- 63.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell 2012;149:1192–205. [DOI] [PubMed] [Google Scholar]
- 64.Schlessinger K, Hall A, Tolwinski N. Wnt signaling pathways meet Rho GTPases. Genes Dev 2009;23:265–77. [DOI] [PubMed] [Google Scholar]
- 65.Ge X, Jin Q, Zhang F, et al. PCAF acetylates {beta}-catenin and improves its stability. Mol Biol Cell 2009;20:419–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holloway KR, Calhoun TN, Saxena M, et al. SIRT1 regulates Dishevelled proteins and promotes transient and constitutive Wnt signaling. Proc Natl Acad Sci U S A 2010;107:9216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li J, Mizukami Y, Zhang X, et al. Oncogenic K-ras stimulates Wnt signaling in colon cancer through inhibition of GSK-3beta. Gastroenterology 2005;128:1907–18. [DOI] [PubMed] [Google Scholar]
- 68.Goh KY, Ng NW, Hagen T, et al. p21-activated kinase interacts with Wnt signaling to regulate tissue polarity and gene expression. Proc Natl Acad Sci U S A 2012;109:15853–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kamaid A, Giraldez F. Btg1 and Btg2 gene expression during early chick development. Dev Dyn 2008;237:2158–69. [DOI] [PubMed] [Google Scholar]
- 70.Nambiar RM, Ignatius MS, Henion PD. Zebrafish colgate/hdac1 functions in the non-canonical Wnt pathway during axial extension and in Wnt-independent branchiomotor neuron migration. Mech Dev 2007;124:682–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dimitrova YN, Li J, Lee YT, et al. Direct ubiquitination of beta-catenin by Siah-1 and regulation by the exchange factor TBL1. J Biol Chem 2010;285:13507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu Z, Zheng S, Li Z, et al. E2F1 suppresses Wnt/beta-catenin activity through transactivation of beta-catenin interacting protein ICAT. Oncogene 2011;30:3979–84. [DOI] [PubMed] [Google Scholar]
- 73.Barjaktarovic Z, Anastasov N, Azimzadeh O, et al. Integrative proteomic and microRNA analysis of primary human coronary artery endothelial cells exposed to low-dose gamma radiation. Radiat Environ Biophys 2013;52:87–98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.