Abstract
Sineoculis homeobox homolog 1 (Six1), normally a developmentally restricted transcriptional regulator, is frequently dysregulated in mutiple cancers. Increasing evidences show that overexpression of Six1 plays a key role in tumorigenesis. However, the Six1 expression status and its relationship with the clinicopathological characteristics in prostate cancer were unclear. In this study, the mRNA and protein levels of Six1 in prostate cancer tissues and normal prostate tissues were evaluated. The clinicopathological significance of Six1 was investigated by immunohistochemistry (IHC) on a prostate cancer tissue microarray. The cut-off score for high expression of Six1 was determined by the receiver-operating characteristic (ROC) analysis. The correlation between Six1 protein expression and clinicopathological characteristics of prostate cancer was analyzed by Chi-square test. Increased expression of Six1 protein was observed in the majority of prostate cancer, compared with their paired adjacent normal prostate tissues. When Six1 high expression percentage was determined to be above 55 % (area under ROC curve = 0.881, P = 0.000), high expression of Six1 was observed in 55.6 % (80/144) of prostate cancer tissues and low expression of Six1 was observed in all normal prostate tissues by IHC. Increased expression of Six1 in patients was correlated with high histological grade (χ2 = 58.651, P = 0.00), advanced clinical stage (χ2 = 57.330, P = 0.000), high Gleason score (χ2 = 63.480, P = 0.000), high primary tumor grade (χ2 = 57.330, P = 0.000) and positive regional lymph node metastasis (χ2 = 19.294, P = 0.000). Furthermore, univariate and multivariate survival analysis suggested that Six1 was an independent prognostic indicator for overall survival (P < 0.05). This study suggests that Six1 could be served as an additional biomarker in identifying prostate cancer patients at risk of tumor progression, might potentially be used for predicting survival outcome of patients with prostate cancer.
Keywords: Six1, IHC, Prostate cancer, Prognosis
Introduction
Current diagnosis for prostate cancer includes digital rectal examination (DRE), prostate-specific antigen (PSA) and needle biopsy [1]. The appearance of prostate-specific antigen (PSA) testing has revolutionarily improved early prostate cancer detection. If rising PSA levels are detected, a needle biopsy of the prostate is recommended to inspect for histologic evidence of prostate cancer. If cancer is inspected, the patient can choose either active surveillance or one of several given treatment options, such as surgery, radiotherapy, or brachytherapy. However, current reports have raised concern over the efficacy of PSA testing. The U.S. prostate cancer screening trial report found that the mortality caused by prostate cancer was not reduced by PSA screening [2]. These studies suggested that PSA testing cannot distinguish between aggressive and indolent prostate cancer; therefore, many patients accepted overtreatment. As a result of these kind reports, the U.S. Preventive Services Task Force recommended to stop routine screening by PSA testing on all males [3]. Regardless of how much controversy about the PSA screening, there is an agreement that a definitive test is needed to distinguish patients with aggressive prostate cancer from the patients that have latent or indolent prostate cancer. Therefore, a critical question in the clinical activity of prostate cancer is how to distinguish the patients with indolent prostate cancer from patients with highly aggressive prostate cancer who would benefit from definitive treatment.
Homeobox genes encode transcription factors that are essential for the development of numerous organs and control processes such as proliferation, apoptosis, migration, and invasion [4–7]. The Six1 homeoprotein, a member of the Six family of homeodomain transcription factors, has been found overexpressed in mutiple cancers, including breast [8–10], rhabdomyosarcomas [11–13], hepatocellular carcinomas [14], ovarian [15] and Wilms tumors [16]. Recent evidence demonstrates that Six1 plays a role in cellular migration and invasion during embryogenesis [4, 17–20] and in breast cancer [21, 22] through a mechanism that may involve an EMT, which was associated with increased TGF-β receptor type I (TβRI) expression and Smad-dependent transforming growth factor-beta (TGF-β) signaling. In addition, Six1 overexpression enhances lymph node metastasis by increasing VEGF-C depending on TGF-β signaling in cervical cancer [23–26]. Forementioned studies suggested that Six1 enhances cancer cell proliferation and shortens survival and its overexpression in immortalized mammary epithelial cells induces EMT, leading to highly aggressive and invasive tumors when transplanted into nude mice [7].
Noteworthy, HD Jin et al. reported that Six1 overexpression positively correlated with clinical stage, lymph node metastasis, overall survival (OS) and disease-free survival (DFS) rates of patients with breast cancer. Moreover, patients with high Six1 expression had poorer prognosis than those with low Six1 expression in late stage breast cancer cases [27]. However, the relationship between Six1 and the clinicopathological characteristics of prostate cancer has not previously been examined.
Materials and methods
Patient specimens and prostate cancer cohorts
A total of 144 paraffin-embedded tissues diagnosed from 2002 to 2012 were retrieved for tissue microarray (TMA) construction and immunohistochemistry analysis. All the samples were collected from the Department of Urology, Nanfang Hospital (Guangzhou, China). The samples selected were pathologically diagnosed with prostate cancer, having received no prior chemotherapy or radiotherapy before surgery. Ages of the 144 patients with prostate cancer varied from 44 to 77 years (median, 65 years), clinicopathological features of patients including age at diagnosis, histologic grade, clinical stage, gleason score and pTNM stages. Written informed consent was obtained from all patients for use of the tissue samples and clinical records. The study protocol was performed under the approval by the Ethic Committee of the Nanfang Hospital. All cases were evaluated by experienced pathologists for histopathological grading.
Quantitative real-time PCR
qRT-PCR was performed to determine the expression of Six1 messenger RNA (mRNA). Briefly, we isolated total RNAs from frozen tissues using TRIzol Reagent according to the manufacturer’s protocol (Life Technologies) and reverse transcribed to generate cDNA (PrimeScript RT-PCR Kit, Takara Bio). β-Actin was used as an internal control. The levels of mRNA encoding were quantified by real-time PCR with the Applied Biosystems 7500 Fast Real-Time PCR System using SYBR Premix Ex Taq (Applied Takara Bio). The sequences of the primers were as follows: Six1 forward 5’- TTCTCGCCTCACAACCACCCCA-3’ and reverse 5’-TACCACTCCCGCAGGACACCCC-3’ and β-actin forward 5’-TGGCACCCAGCACAATGAA-3’ and reverse 5’-CTAAGTCATAGTCCGCCTAGAAGCA-3’. The PCR conditions were as following procedures: initial step 95 °C for 2 min, followed by 35 cycles of 95 °C for 30 s, 56 °C for 30 s, and 72 °C for 2 min and a final elongation step of 72 °C for 5 min. All qRT-PCRs were repeated three times. Relative quantification of Six1 mRNA expression was calculated using the 2−ΔΔCT method.
Western blotting analysis
Total proteins from 8 pairs of fresh prostate cancer and adjacent normal tissues were extracted by radio-immunoprecipitation assay (RIPA) buffer containing 1 mM phenylmethanesulfonyl fluoride (PMSF) (Beyotime, Haimen, China). After centrifugation, supernatant was collected and treated with BCA Protein Assay Kit (Beyotime, Haimen, China). The protein concentration was then measured at 562 nm -by the TECAN Infinite 200 microplate reader (TECAN, Switzerland). Tissue homogenates (35 μg protein for each sample) were separated by 10 % sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and the resolved proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore, USA) by a Trans-Blot SD Semi-dry transfer cell machine (Bio-Rad, USA). After the blots were washed with 1 × TBST buffer (10 mM Tris-HCl [pH 7.6], 150 mM NaCl, and 0.05 % Tween-20), the membranes were blocked overnight with 5 % skim milk and incubated with the appropriate primary antibody at room temperature for 2 h. Polyclonal rabbit anti-human antibody against Six1 (Atlas antibody, Sweden, 1:500), and monoclonal rabbit anti-human antibody against β-actin (Cell Signaling Technology, Danvers, MA, USA, 1:3000) were used for detecting the protein level of Six1 and β-actin in each sample. The membranes were then washed by 1 × TBST, primary antibodies were detected with goat anti-rabbit IgG conjugated to horseradish peroxidase (HRP) (Santa Cruz Biotechnology, Cal, USA, 1:5000), and the bands were detected by BeyoECL Plus Kit (Beyotime, Haimen, China). Finally, the results of western blotting were visualized by the Image Station 4000R PRO scanner (CareStream Health, Rochester, NY, U.S.A.).
Tissue microarray (TMA) construction and immunohistochemistry (IHC)
Representative sections of prostate cancer or normal prostate tissue in the pre-existing paraffin-embedded tissue blocks were determined according to the overlaid H&E staining slides. The TMA was constructed by using a needle to punch a 1.5 mm diameter cylinder in the representative section of each block, and placing the cylinders into an array on a recipient paraffin block. 5 μm thick multiple sections were cut from the TMA block and mounted on microscope slides for immuohistochemistry analysis. The TMA consisted of a total of 144 cases of prostate cancer and 10 cases of normal control paraffin-embedded tissue. Clinical characteristic about the patients was summarized in Table 1. The TMA slide was dried overnight at 37 °C, deparaffinized in xylene, rehydrated through graded alcohol, and then immersed in 3 % hydrogen peroxide for 20 min to block the endogenous peroxidase activity, and antigen-retrieved by microwave heating with sodium citrate buffer (pH = 6.0) at 100 °C for 30 min. Then the slides were pre-incubated with 10 % normal goat serum at room temperature for 30 min to reduce nonspecific reaction. The primary rabbit anti-Six1 polyclonal antibody (Atlas antibody, Sweden) was diluted (1:1000) with 1 × phosphate buffered saline (PBS) and applied overnight in a humidity chamber at 4 °C. The slide was sequentially incubated with a polymer peroxidaselabeled secondary antibody (ZSGB-Bio, Beijing, China) for 30 min at room temperature, and then stained with DAB Horseradish Peroxidase Color Development Kit (Beyotime, Haimen, China). Finally, the sections were counterstained by hematoxylin. Known IHC positive slide was used as a positive control, and phosphate buffered saline replaced anti-Six1 primary antibody was used as a control.
Table 1.
Association of Six1 expression with patients’ clinicopathologic features in prostate cancer
| Six1 protein | |||||
|---|---|---|---|---|---|
| Variable | All Cases | Low | High | χ2 | P valuea |
| Ageb | |||||
| ≤65 | 68 | 31 | 37 | 0.794 | 0.068 |
| >65 | 76 | 33 | 43 | ||
| Histological grade | |||||
| G1 + G2 | 77 | 57 | 20 | 58.651 | 0.000 |
| G3 | 67 | 7 | 60 | ||
| Clinical stage | |||||
| I + II | 40 | 38 | 2 | 57.330 | 0.000 |
| III + IV | 104 | 26 | 78 | ||
| Gleason score | |||||
| 1–7 | 54 | 47 | 7 | 63.480 | 0.000 |
| 8–10 | 90 | 17 | 73 | ||
| pT status | |||||
| T1 + T2 | 40 | 38 | 2 | 57.330 | 0.000 |
| T3 + T4 | 104 | 26 | 78 | ||
| pN status | |||||
| N0 | 107 | 59 | 48 | 19.294 | 0.000 |
| N1 + N2 | 37 | 5 | 32 | ||
| pM status | |||||
| M0 | 105 | 48 | 57 | 0.253 | 0.615 |
| M1 | 39 | 16 | 23 |
aChi-squared test
bMean age
Evaluation of IHC
Immunoreactivity for the Six1 protein was scored by a semi-quantitative method by recording the proportion of positive tumor cells over the total number of tumor cells. Scores were assigned by using 5 % increments (0, 5…100 %). The reproducibility of the scoring method between pathologists has been described previously for TMAs [28–31]. The scores were accepted if two of the three investigators (M.W, R.S and J.Z) agree with the values. Otherwise, the values were re-estimated until a consensus was reached. Our conclusions were in complete agreement in 80 % of the cases, which indicated that the scoring method was highly reproducible.
Selection of Cut-off scores using receiver- operating characteristic (ROC) curves
Receiver operating characteristic curve analysis was utilized to determine the cut-off score by using the 0, 1-criterion [32]. At the Six1 score, the sensitivity and specificity for each outcome under study was plotted, thus generating various ROC curves. The score was selected as the cut-off score, which was closest to the point with both maximum sensitivity and specificity. Tumors designated as “low” for Six1 were those with scores below and equal to the cut-off value, whereas “high” tumors were those with scores above the value [29]. In order to perform ROC curve analysis, the clinicopathological features were dichotomized: age (≤65 or >65), histological grade (low [G1 + G2] or high G3), clinical stage ([I + II] or above [III + IV]), Gleason stage (1–7 or 8–10), T stage (early [T1 + T2] or late [T3 + T4]), N stage (N0 [no lymph node involvement] or N1 + N2 [any lymph node involvement]), M stage (M0 [no distant metastasis] or M1 [distant metastasis]).
Statistical analysis
Statistical analysis was performed by using the SPSS statistical software program (standard version 18.0; SPSS, Chicago, USA). The relationship between Six1 protein expression and clinicopathological data of Prostate cancer patients was estimated using the χ2 test. ROC curve analysis was applied to determine the cut-off score for Six1 positivity, and the areas under curves (AUCs) were then calculated. The association between survival and each variable was determined with the Kaplan-Meier method (the log-rank test). Multiple Cox proportional hazards regression was carried out to identify the independent prognostic factors. P < 0.05 (two-tailed) denotes the presence of a statistically significant difference.
Results
The expression level of Six1 in prostate cancer and adjacent normal prostate tissues detected by western blotting and IHC
In this study, the protein and/or mRNA expression of Six1 was first examined by Western blotting and/or qRT-PCR in 8 pairs of primary prostate cancer and adjacent normal prostate tissues. A significantly increase in both protein and mRNA expression of Six1 was detected in prostate cancer tissues compared to adjacent nontumorous tissues (Fig. 1a and b). For Six1 IHC staining in prostate cancer tissues and normal prostate tissues, immunoreactivity was seen primarily in the cytoplasm and perinucleus of prostate cancer cells (Fig. 1). Six1 expression could be evaluated informatively in 144 prostate cancer cases by the TMA constructed previously and in 10 normal prostates. Immunoreactivity ranged from 0 to 100 %. According to ROC analysis, expression percentage for Six1 above the critical value 55 % was defined as high expression. The high expression of Six1 was detected in 80 ⁄ 144 (55.6 %) of prostate cancer cases. The decreasing frequency of Six1 high expression was detected in normal prostate tissues (Fig. 1).
Fig. 1.
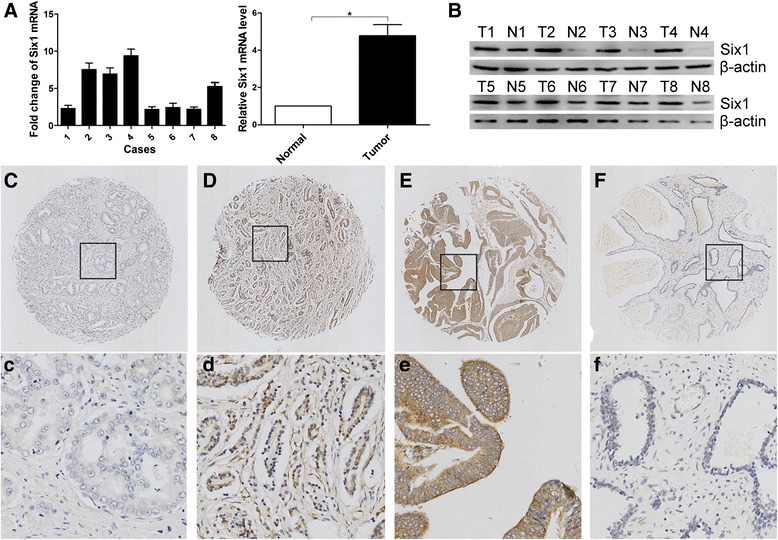
Expression of Six1 in prostate cancer tissues and normal prostate tissues detected by western blotting and immunochemistry. a Relative mRNA expression of Six1 normalized to β-actin was calculated (n = 8), ★ indicates P < 0.05. b Expressions of Six1 protein in 8 paired tissues were examined by western blotting. Micrographs showed weak (c, c), moderate (d, d), and strong (e, e) staining of Six1 in prostate cancer tissues, as well as low (f, f) expression of Six1 in normal prostate tissues (upper magnification × 100, lower panel: magnification × 400.)
Selection of Six1 cut-off scores
The ROC for each clinicopathological parameter (Fig. 2) clearly show the point on the curve closest to (0.0, 1.0) which maximizes both sensitivity and specificity for the outcome. The analysis of ROC for each clinicopathological feature and Six1 expression (AUC = 0.674, P = 0.001) is carried out to evaluate the patients’ survival status (Fig. 3). The cut-off score was determined to be above 55 % for Six1 high expression. Tumor tissues with scores above the determined cut-off values were considered high expression of Six1 protein leading to the greatest number of tumor cases correctly classified as having or not having the clinical outcome. The corresponding AUCs (95 % confidence interval [CI]) are showed in Table 2.
Fig. 2.
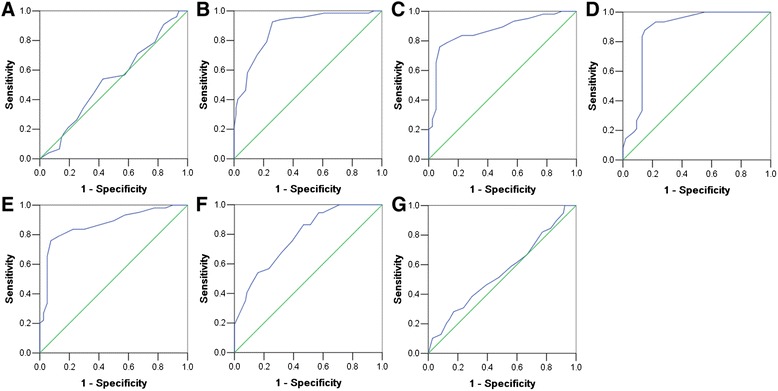
Determination of the cutoff value of high Six1 expression in prostate cancer tissues by ROC curves. The sensitivity and 1-specificity were plotted for each clinical characteristic, including age (a), pathological grade (b), clinical stage (c), Gleason grade (d), pT status (e), pN status (f), pM status (g)
Fig. 3.
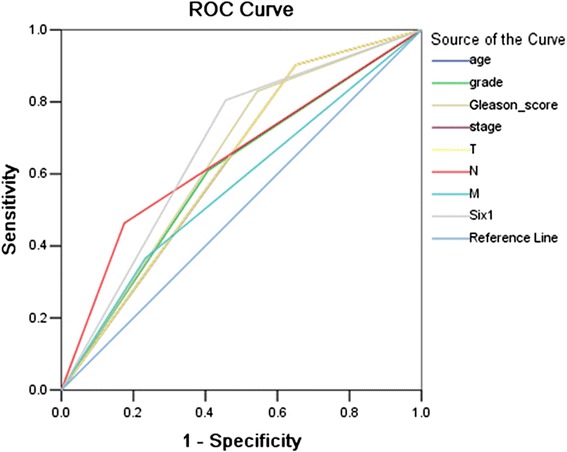
Receiver–operator curve (ROC) analysis for different clinicopathological parameters and Six1 expression was used to evaluate the survival status. clinical stage (AUC = 0.626, P = 0.019), Gleason_score (AUC = 0.643, P = 0.008), T stage (AUC = 0.626, P = 0.019), N stage (AUC = 0.644, P = 0.007), Six1 expression (AUC = 0.674, P = 0.001) implied significant statistical associations with the survival
Table 2.
Area under the ROC curve values for each clinicopathological feature
| Feature | AUC (95 % CI) | p value |
|---|---|---|
| Age | 0.531 | 0.518 |
| Histological grade | 0.881 | 0.000 |
| Clinical stage | 0.869 | 0.000 |
| Gleason score | 0.874 | 0.000 |
| pT | 0.869 | 0.000 |
| pN | 0.784 | 0.000 |
| pM | 0.505 | 0.373 |
AUC, Area under the curve; CI, Confidence interval
Correlations between Six1 protein expression and clinicopathological parameters of prostate cancer
Six1 protein overexpression was significantly correlated with histological grade, clinical stage, Gleason score, primary tumor statues and regional lymph node metastasis of prostate cancer (P < 0.001). However, Six1 protein expression was not related with patient age, distant metastasis in prostate cancer (P > 0.05) (Table 1). For histological grades, the high expression rate of Six1 protein was significantly higher in late grade (G3) prostate cancers (89.6 %, 60/67) than in early histological grade cases (G1 + G2) (26.0 %, 20/77) (P < 0.001). Similarly, the high expression rate of Six1 protein was higher in breast cancers with high Clinical stage (75.0 %, 78/104) compared with those with low Clinical stage (0.5 %, 2/40) (P < 0.001). The high expression rate of Six1 protein was significantly higher in high Gleason score (8–10) prostate cancers (81.1 %, 73/90) than in low Gleason score cases (1–7) (13.0 %, 7/54) (P < 0.001). The high expression rate of Six1 protein was 75.0 % (78/104) in high pT grade, which was significantly higher than in low pT grade (0.5 %, 2/40) (P < 0.001). Meanwhile, the high expression rate of Six1 protein was significantly higher in high pN grade (N1 + N2) prostate cancers (81.1 %, 73/90) than in low pN grade cases (N0) (13.0 %, 7/54) (P < 0.001).
Relationship between clinicopathologic variables, Six1 protein expression and prostate patients’ survival
In univariate analysis, Kaplan–Meier survival curves and the P-values for these curves were determined by log-rank method. Above all, to confirm the representativeness of the prostate cancer in the study, we analyzed established prognostic factors of patient survival. The univariate analysis demonstrated a significant impact of well-known clinicopathological prognostic parameters, such as histolotical grade, clinical stage, Gleason score, pT status, pN status and Six1 expression on patient overall survival (Table 3). Assessment of survival in all specimens demonstrated that increased expression of Six1 protein was associated with worse overall survival (P < 0.001, Fig. 4), and the mean survival time for patients with tumors having low Six1 expression was 87.4 months compared to 42.8 months for patients with tumors having high Six1 expression (Table 3).
Table 3.
Univariate and Multivariate Analysis of Different Prognostic Features in 144 Patients with prostate cancer
| Univariate analysisa | Multivariate analysisb | |||||
|---|---|---|---|---|---|---|
| Variable | All case | Mean (months) | χ 2 | P value | HR (95 % CI) | P value |
| Agec | 6.487 | 0.011 | 2.220 (1.146–4.300) | 0.018 | ||
| ≤65 | 68 | 111.567 | ||||
| >65 | 76 | 96.475 | ||||
| Histological grade | 4.312 | 0.038 | 0.696 (0.335–1.446) | 0.331 | ||
| G1 + G2 | 77 | 106.956 | ||||
| G3 | 67 | 99.493 | ||||
| Clinical stage | 9.381 | 0.002 | 1.369 (0.312–6.004) | 0.677 | ||
| I + II | 40 | 114.868 | ||||
| III + IV | 104 | 99.086 | ||||
| Gleason score | 9.273 | 0.002 | 1.246 (0.391–3.969) | 0.710 | ||
| 1–7 | 54 | 111.779 | ||||
| 8–10 | 90 | 98.610 | ||||
| pT status | 9.381 | 0.002 | 1.369 (0.312–6.004) | 0.677 | ||
| T1 + T2 | 40 | 114.868 | ||||
| T3 + T4 | 104 | 99.086 | ||||
| pN status | 13.772 | 0.000 | 1.710 (0.803–3.642) | 0.164 | ||
| N0 | 107 | 107.933 | ||||
| N1 + N2 | 37 | 90.843 | ||||
| pM status | 1.529 | 0.216 | 1.162 (0.554–2.437) | 0.692 | ||
| M0 | 107 | 104.893 | ||||
| M1 | 37 | 100.275 | ||||
| Six1 expression | 16.051 | 0.000 | 3.434 (1.168–10.100) | 0.025 | ||
| Low | 64 | 113.943 | ||||
| High | 80 | 94.992 | ||||
HR indicates hazards ratio; CI indicates confidence interval
aLog-rank test; bCox regression model; cMean age
Fig. 4.
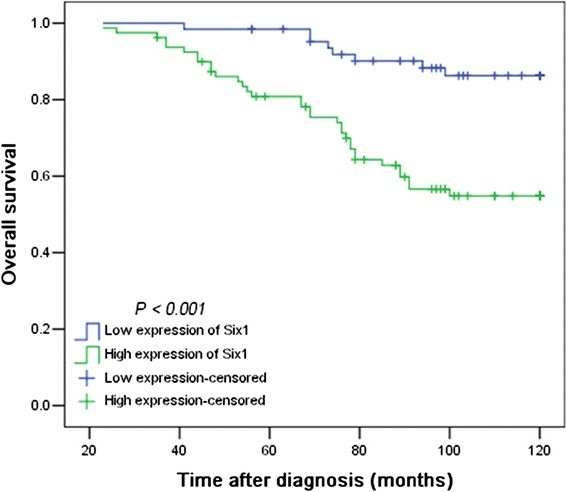
Kaplan–Meier survival analysis of Six1 expression in 144 patients of prostate cancer (log-rank test). Relationship of Six1 expression and patient overall survival: low expression, n = 64 and high expression, n = 80 (P < 0.001)
Independent prognostic factors of prostate cancer: multivariate survival analysis
A multivariate analysis based on the Cox proportional hazard model was applied to test the independent value of each parameter predicting overall survival (Table 3). Expressions of Six1 as well as other clinicopathological features that were significant by univariate analysis (histological grade, clinical stage, Gleason score, pT stage and pN stage) were included in multivariate analysis. The expression of Six1 was found to be an independent prognostic factor for adverse overall survival (relative risk, 2.322; 95 % CI, 1.163-4.636; P = 0.017). Of the other parameters, Clinical stage (P = 0.044), pT stage (P = 0.044) were demonstrated as well an independent prognostic factor for overall survival.
Discussion
Prostate cancer is one of the most frequently diagnosed malignancies and second only to lung and bronchus cancer in men worldwide. Approximately 258,400 men dead from this disease in 2008 [33]. At current rates of diagnosis, one-sixth of men will be diagnosed with prostate cancer during his lifetime [34]. From the clinical perspective, the tumors in different patients must be varying in the molecular level and the aim of personalized medicine is to generate individual risk profiles from the primary prostate cancer that could distinguish high-risk individuals who should accept aggressive therapeutic treatment and clinical follow-up from those who have indolent prostate cancer to avoid these individuals from undergoing overtreatment. Recently, several studies have investigated molecular and genetic characteristics of prostate cancer to develop both predictive and prognostic biomarkers [35–37].
It was reported that in multiple types of human cancer, such as breast, rhabdomyosarcomas, hepatocellular carcinomas, ovarian, and Wilms tumors, overexpression of Six1 was frequently identified [8–16]. In the study, we investigated the expression patterns of Six1, by Western blotting using fresh prostate tissues, and by IHC using TMA containing a large cohort of prostate tumor samples (144 cases) with complete clinicopathological and follow-up data. Western blotting revealed that up-regulation expression of Six1 was detected in prostate cancer, when compared with adjacent normal prostate tissues. Moreover, the IHC results demonstrated that an increasing expression of Six1 was observed from low malignant tumors to high malignant tumors. Six1 immunoreactivity was assessed using a scoring method described in previous study [29]. The reliability of this scoring method for Six1 was assessed by three investigators and was again found to be highly reproducible. To select the IHC cut-off scores for Six1 positivity, ROC analysis was carried out for each of the clinicopathological parameters, including age, histological grade, clinical stage, Gleason score, pTNM stage. ROC analysis for different clinicopathological features and Six1 expression was also used to evaluate the survival status. Six1 expression demonstrated significant statistical associations with the survival status of patients with prostate cancer. These data provided evidence that the unregulated expression of Six1 played an important role in tumorigenic process of multiple human cancers, including prostate cancer.
In summary, the study suggests that increased expression of Six1 may represent a more aggressive status of prostate cancer. In addition, these results show that increased expression of Six1 predicts poorer outcome of the cancer for the patients compared to those patients with decreased expression of Six1. Further studies are needed to clarify the precise mechanism of Six1 in prostate cancer and to develop potential therapies targeting Six1 in prostate cancer.
Footnotes
Competing interest
The authors declare that they have no competing interest.
Authors’ contributions
JZ, WLM and MW carried out the experimental design, participated in the data analysis and drafted the manuscript; RS and CXC participated in the experiments and joined into drafting the manuscript; XRL and YBS participated in the experiments and data analysis. All authors have read and approved the final manuscript.
Contributor Information
Min Wei, Email: weimin78@126.com.
Wen-li Ma, Email: Wenlimat@126.com.
References
- 1.Velonas VM, Woo HH, Remedios CG, Assinder SJ. Current status of biomarkers for prostate cancer. Int J Mol Sci. 2013;14(6):11034–11060. doi: 10.3390/ijms140611034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andriole GL, Crawford ED, Grubb RL, III, Buys SS, Chia D, Church TR, et al. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360(13):1310–1319. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moyer VA. Screening for prostate cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2012;157(2):120–134. doi: 10.7326/0003-4819-157-2-201207170-00459. [DOI] [PubMed] [Google Scholar]
- 4.Zheng W, Huang L, Wei ZB, Silvius D, Tang B, Xu PX. The role of Six1 in mammalian auditory system development. Development. 2003;130(17):3989–4000. doi: 10.1242/dev.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ikeda K, Kageyama R, Suzuki Y, Kawakami K. Six1 is indispensable for production of functional progenitor cells during olfactory epithelial development. Int J Dev Biol. 2010;54(10):1453–1464. doi: 10.1387/ijdb.093041ki. [DOI] [PubMed] [Google Scholar]
- 6.Xu PX, Zheng W, Huang L, Maire P, Laclef C, Silvius D. Six1 is required for the early organogenesis of mammalian kidney. Development. 2003;130(14):3085–3094. doi: 10.1242/dev.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coletta RD, Christensen KL, Micalizzi DS, Jedlicka P, Varella-Garcia M, Ford HL. Six1 overexpression in mammary cells induces genomic instability and is sufficient for malignant transformation. Cancer Res. 2008;68(7):2204–2213. doi: 10.1158/0008-5472.CAN-07-3141. [DOI] [PubMed] [Google Scholar]
- 8.Ford HL, Kabingu EN, Bump EA, Mutter GL, Pardee AB. Abrogation of the G2 cell cycle checkpoint associated with overexpression of HSIX1: a possible mechanism of breast carcinogenesis. Proc Natl Acad Sci U S A. 1998;95(21):12608–12613. doi: 10.1073/pnas.95.21.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coletta RD, Christensen K, Reichenberger KJ, Lamb J, Micomonaco D, Huang L, et al. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc Natl Acad Sci U S A. 2004;101(17):6478–6483. doi: 10.1073/pnas.0401139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reichenberger KJ, Coletta RD, Schulte AP, Varella-Garcia M, Ford HL. Gene amplification is a mechanism of Six1 overexpression in breast cancer. Cancer Res. 2005;65(7):2668–2675. doi: 10.1158/0008-5472.CAN-04-4286. [DOI] [PubMed] [Google Scholar]
- 11.Khan J, Bittner ML, Saal LH, Teichmann U, Azorsa DO, Gooden GC, et al. cDNA microarrays detect activation of a myogenic transcription program by the PAX3-FKHR fusion oncogene. Proc Natl Acad Sci U S A. 1999;96(23):13264–13269. doi: 10.1073/pnas.96.23.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu Y, Khan J, Khanna C, Helman L, Meltzer PS, Merlino G. Expression profiling identifies the cytoskeletal organizer ezrin and the developmental homeoprotein Six-1 as key metastatic regulators. Nat Med. 2004;10(2):175–181. doi: 10.1038/nm966. [DOI] [PubMed] [Google Scholar]
- 13.Yu Y, Davicioni E, Triche TJ, Merlino G. The homeoprotein six1 transcriptionally activates multiple protumorigenic genes but requires ezrin to promote metastasis. Cancer Res. 2006;66(4):1982–1989. doi: 10.1158/0008-5472.CAN-05-2360. [DOI] [PubMed] [Google Scholar]
- 14.Ng KT, Man K, Sun CK, Lee TK, Poon RT, Lo CM, et al. Clinicopathological significance of homeoprotein Six1 in hepatocellular carcinoma. Br J Cancer. 2006;95(8):1050–1055. doi: 10.1038/sj.bjc.6603399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Behbakht K, Qamar L, Aldridge CS, Coletta RD, Davidson SA, Thorburn A, et al. Six1 overexpression in ovarian carcinoma causes resistance to TRAIL-mediated apoptosis and is associated with poor survival. Cancer Res. 2007;67(7):3036–3042. doi: 10.1158/0008-5472.CAN-06-3755. [DOI] [PubMed] [Google Scholar]
- 16.Li CM, Guo M, Borczuk A, Powell CA, Wei M, Thaker HM, et al. Gene expression in Wilms’ tumor mimics the earliest committed stage in the metanephric mesenchymal-epithelial transition. Am J Pathol. 2002;160(6):2181–2190. doi: 10.1016/S0002-9440(10)61166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozaki H, Nakamura K, Funahashi J, Ikeda K, Yamada G, Tokano H, et al. Six1 controls patterning of the mouse otic vesicle. Development. 2004;131(3):551–562. doi: 10.1242/dev.00943. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426(6964):247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- 19.Grifone R, Demignon J, Houbron C, Souil E, Niro C, Seller MJ, et al. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development. 2005;132(9):2235–2249. doi: 10.1242/dev.01773. [DOI] [PubMed] [Google Scholar]
- 20.Ikeda K, Ookawara S, Sato S, Ando Z, Kageyama R, Kawakami K. Six1 is essential for early neurogenesis in the development of olfactory epithelium. Dev Biol. 2007;311(1):53–68. doi: 10.1016/j.ydbio.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 21.Micalizzi DS, Wang CA, Farabaugh SM, Schiemann WP, Ford HL. Homeoprotein Six1 increases TGF-beta type I receptor and converts TGF-beta signaling from suppressive to supportive for tumor growth. Cancer Res. 2010;70(24):10371–10380. doi: 10.1158/0008-5472.CAN-10-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Micalizzi DS, Christensen KL, Jedlicka P, Coletta RD, Baron AE, Harrell JC, et al. The Six1 homeoprotein induces human mammary carcinoma cells to undergo epithelial-mesenchymal transition and metastasis in mice through increasing TGF-beta signaling. J Clin Invest. 2009;119(9):2678–2690. doi: 10.1172/JCI37815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng XH, Liang PH, Guo JX, Zheng YR, Han J, Yu LL, et al. Expression and clinical implications of homeobox gene Six1 in cervical cancer cell lines and cervical epithelial tissues. Int J Gynecol Cancer. 2010;20(9):1587–1592. [PubMed] [Google Scholar]
- 24.Tan J, Zhang C, Qian J. Expression and significance of Six1 and Ezrin in cervical cancer tissue. Tumour Biol. 2011;32(6):1241–1247. doi: 10.1007/s13277-011-0228-8. [DOI] [PubMed] [Google Scholar]
- 25.Liu D, Li L, Zhang XX, Wan DY, Xi BX, Hu Z, et al. SIX1 Promotes Tumor Lymphangiogenesis by Coordinating TGFbeta Signals That Increase Expression of VEGF-C. Cancer Res. 2014;74(19):5597–5607. doi: 10.1158/0008-5472.CAN-13-3598. [DOI] [PubMed] [Google Scholar]
- 26.Wang CA, Jedlicka P, Patrick AN, Micalizzi DS, Lemmer KC, Deitsch E, et al. SIX1 induces lymphangiogenesis and metastasis via upregulation of VEGF-C in mouse models of breast cancer. J Clin Invest. 2012;122(5):1895–1906. doi: 10.1172/JCI59858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin H, Cui M, Kong J, Cui X, Lin Z, Wu Q, et al. Sineoculis homeobox homolog 1 protein is associated with breast cancer progression and survival outcome. Exp Mol Pathol. 2014;97(2):247–252. doi: 10.1016/j.yexmp.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 28.Zlobec I, Minoo P, Baker K, Haegert D, Khetani K, Tornillo L, et al. Loss of APAF-1 expression is associated with tumour progression and adverse prognosis in colorectal cancer. Eur J Cancer. 2007;43(6):1101–1107. doi: 10.1016/j.ejca.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 29.Zlobec I, Steele R, Terracciano L, Jass JR, Lugli A. Selecting immunohistochemical cut-off scores for novel biomarkers of progression and survival in colorectal cancer. J Clin Pathol. 2007;60(10):1112–1116. doi: 10.1136/jcp.2006.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zlobec I, Vuong T, Hayashi S, Haegert D, Tornillo L, Terracciano L, et al. A simple and reproducible scoring system for EGFR in colorectal cancer: application to prognosis and prediction of response to preoperative brachytherapy. Br J Cancer. 2007;96(5):793–800. doi: 10.1038/sj.bjc.6603619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zlobec I, Steele R, Michel RP, Compton CC, Lugli A, Jass JR. Scoring of p53, VEGF, Bcl-2 and APAF-1 immunohistochemistry and interobserver reliability in colorectal cancer. Mod Pathol. 2006;19(9):1236–1242. doi: 10.1038/modpathol.3800642. [DOI] [PubMed] [Google Scholar]
- 32.Cai MY, Zhang B, He WP, Yang GF, Rao HL, Rao ZY, et al. Decreased expression of PinX1 protein is correlated with tumor development and is a new independent poor prognostic factor in ovarian carcinoma. Cancer Sci. 2010;101(6):1543–1549. doi: 10.1111/j.1349-7006.2010.01560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 34.Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ. Cancer statistics, 2003. CA Cancer J Clin. 2003;53(1):5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- 35.Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest. 2004;113(6):913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Markert EK, Mizuno H, Vazquez A, Levine AJ. Molecular classification of prostate cancer using curated expression signatures. Proc Natl Acad Sci U S A. 2011;108(52):21276–21281. doi: 10.1073/pnas.1117029108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470(7333):269–273. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]


