Abstract
FBXO31 is a member of F-box family which is involved in diverse biological functions and development of disease. Recent reports in breast cancer, hepatocellular carcinoma and ovarian cancer demonstrated inhibitory effect of FBXO31 on proliferation and tumorigenesis. However, the function of FBXO31 is not analyzed in lung cancer so far. In this study, we reported that expression of FBXO31 was higher in lung cancer tissues compared with non-cancerous lung tissues, and that higher expression of FBXO31 was significantly associated with tumor size, tumor infiltration, clinical stages and lymph node metastasis. In addition, exogenous expression of FBXO31 promoted cell growth, metastasis and invasion in A549 cells. Conversely, silencing FBXO31 by specific siRNA caused inhibitory effect on cell growth, metastasis and invasion. Moreover, tumorigenicity assays in nude mice showed FBXO31 promoted tumor growth in vivo. In conclusion, our data suggest FBXO31 promotes cell proliferation, metastasis and invasion in lung cancer.
Keywords: FBXO31, tumor growth, metastasis, invasion, lung cancer
Introduction
Lung cancer is one of the most common malignance in worldwide due to late diagnosis in advanced stages, recurrence, limited efficiency of chemotherapy and radiotherapy. Tumori-genesis of lung cancer is a complicated network involving numerous oncogene [1-5], tumor suppressor [6-9] and their regulators [10,11]. Identifying tumor suppressor may help to establish novel therapeutic strategy for lung cancer.
FBXO31 is a member of F-box family located at chromosome 16q24.3 and comprises SCF complex with SKP1 and CUL1 [12]. FBXO31 serves as the substrate recognition subunit in the SCF(FBXO31) ligase and mediates target protein degradation through ubiquitin proteasome pathway. FBXO31 involves in DNA damage response by mediating cyclin D1 degradation through ubiquitin proteasome pathway and inducing G1 arrest after DNA damage [13-15]. Study in neuronal development proves that FBXO31 involves in neuronal morphogenesis and migration [16]. Recent reports show that FBXO31 is a candidate tumor suppressor in breast cancer [12] and hepatocellular carcinoma [17]. Expression of FBXO31 determines poor prognosis in esophageal squamous cell carcinoma [18]. However, the role of FBXO31 in lung cancer has not been reported yet.
In this study, we investigate expression and effects of FBXO31 on cell growth, metastasis and invasion in A549 cells and in nude mice. This study will provide new insights on the function of FBXO31 in cancer field.
Materials and methods
Cell culture
Human lung cancer cell lines A549 was obtained from Shanghai Cell Institute Country Cell Bank, (Shanghai, China). A549 cells were grown in DMEM medium with 10% fetal bovine serum (FBS) (GIBCo/BRL, MD), supplemented with 100 U/ml penicillin G and 100 lg/ml streptomycin (Sigma-Aldrich Corp., St. Louis, MO). Cells were maintained at 37°C in a humidified 5% CO2 incubator.
Plasmids
EGFP-FBXO31 expressing plasmid used in this manuscript was constructed as previous described [19]. FBXO31 open reading frame (ORF) was PCR amplified from FBXO31 cDNA clone (Open Biosystem) and inserted into the pEGFP-C1 vector (Clontech Laboratories, Inc.) to generate constructs capable of expressing EGFP-FBXO31 protein. The sequences of all constructs were confirmed by DNA sequencing.
Oligonucleotide transfection
siRNA against FBXO31 was purchased from GenePharma Co,Ltd. siRNA sequences are shown in Table 1. Oligonucleotide transfection was done using Lipofectamine 2000 reagents according to the manufacturer’s protocol. Expression of FBXO31 in transfected cells was determined by Real-time quantitative reverse transcription (RT)-PCR (qRT-PCR).
Table 1.
siRNA sequences
| Cat. No. | Sequences | nt | MW |
|---|---|---|---|
| FBXO31 siRNA 1 | GAGCUCAUCCUGAUGAAGUdTdT | 21 | 204.6 |
| FBXO31 siRNA 1_AS | ACUUCAUCAGGAUGAGCUCdTdT | 21 | 200.8 |
| FBXO31 siRNA 2 | CGAAGCUGCUUCACCGAUAdTdT | 21 | 197.7 |
| FBXO31 siRNA 2_AS | UAUCGGUGAAGCAGCUUCGdTdT | 21 | 202.3 |
| FBXO31 siRNA 3 | CCUGGAUACAGCAGAUGUAdTdT | 21 | 208.4 |
| FBXO31 siRNA 3_AS | UACAUCUGCUGUAUCCAGGdTdT | 21 | 196.6 |
| NC | UUCUCCGAACGUGUCACGUTT | 21 | 196.2 |
| NC_AS | ACGUGACACGUUCGGAGAATT | 21 | 213.6 |
RNA extraction and Real-time quantitative reverse transcription PCR
Total RNA was extracted from clinical samples using the TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. RNA samples were then reverse transcribed into cDNA using a PrimeScriptTM RT reagent kit with gDNA Eraser (TaKaRa) in a total volume of 20 μL according to the manufacturer’s protocol. Equal amounts of cDNA samples were used as a template for real-time PCR to detect the level of FBXO31 expression. Quantitative PCR was performed using a LightCycler480 Real-Time PCR system and a SYBR Premix Ex TaqTM II PCR Kit (Takara); β-actin was used as an endogenous reference, and each sample was normalized to its β-actin content. All experiments were performed in duplicate and repeated twice. Results are represented as fold induction using the ΔCt method. Primers for quantitative PCR are shown in Table 2.
Table 2.
qPCR primer sequences
| Genes | Sequences |
|---|---|
| FBXO31-F | 5’-CCATACGGAGGACTGCTGA-3’ |
| FBXO31-R | 5’-GTACATCCACCCGATGATGA-3’ |
| 18srRNA-F | 5’-CCTGGATACCGCAGCTAGGA-3’ |
| 18srRNA-R | 5’-GCGGCGCAATACGAATGCCCC-3’ |
Wound-healing assays
Cell migration was examined using the wound-healing assay. Briefly, cells were cultured to about 80%-90% confluence in a 6-well plate at 37°C and 5% CO2. A wound about 1 mm width was created by scratching cells with a sterile 100 μL micropipette tip. Cells were washed with PBS (pH 6.8) three times to remove floating cells, then 1 mL serum-free DMEM was added. A computer-based microscopy imaging system was used to determine wound healing at 0 h, 6 h, 24 h and 48 h, with a microscope at 200× magnification. The values of wound-healing were assessed by measuring the pixel of wound area by Photoshop 7.01 software. The experiments were performed in triplicate.
In vitro invasion assays
The assay was done by using chambers with polycarbonate filters (pore size, 8 µm) coated on the upper side with Matrigel (Becton Dickinson Labware). Twenty-four hours after siRNA transfection, A549 cells were harvested and 5×104 transfected cells in 200 µL of 0.1% serum medium were placed in the upper chamber. The lower chamber was filled with 10% fetal bovine serum medium (600 µL). After 24 h incubation and removal of the cells on the upper chamber of the filter with a cotton swab, the cells on the underside were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet in 20% ethanol, and counted in five randomly selected fields under phase contrast microscope. The invasion cells were monitored by photographing at 400× magnification with Olympus Microscope. The assays were performed in triplicate.
Western blot analysis
Western blot analysis was performed according to standard Western blot procedures as previously described. Briefly, proteins were separated by 10% SDS-PAGE and then transferred to nitrocellulose membrane (Bio-Rad). After blocking in 5% nonfat milk, the membranes were incubated with the following primary antibodies: mouse anti-FBXO31 monoclonal antibody (mAb; 1:300; Abcam), mouse anti-β-actin mAb (1:1,000; Abcam). The proteins were visualized with enhanced chemiluminescence reagents (Pierce).
Tumorigenicity assays in nude mice
Six-week-old female athymic nude mice were subcutaneously injected in the right armpit region with 1×107 cells in 0.1 mL of PBS. Three groups of mice (n = 6/group) were tested. Group 1 (vector) was injected with A549 cells transfected with empty vector; and group 2 (FBXO31) was injected with A549 cells stably expressed FBXO31; group 3 (shRNA) was injected with A549 cells transfected with shRNAs targeted FBXO31. The tumor sizes was measured every 7 days with calipers. The tumor volume was calculated with the formula: (L×W2)/2, where L is the length and W is the width of the tumor. After the mice were killed at five weeks, the weights of the tumors were measured. All experimental procedures involving animals were in accordance with the Guide for the Care and Use of Laboratory Animals (NIH publication no. 80-23, revised 1996) and were performed according to the institutional ethical guidelines for animal experiments.
Statistical analysis
Data were shown as mean ± SD unless otherwise noted; the clinical association of FBXO31 with age, gender, histology type, tumor size, clinical stage and lymph node metastasis in lung cancer specimens was analyzed using the two-tailed Student’s t-test. A P value of < 0.05 was considered statistically significant.
Results
Higher expression of FBXO31 correlated with tumor size, clinical stage and lymph node metastasis in lung cancer
We investigated expression of FBXO31 in lung cancer tissues and corresponding non-cancerous tissues. Our data showed significantly higher expression of FBXO31 in lung cancer tissues compared with corresponding non-cancerous lung tissues (Figure 1). We also investigated the association of FBXO31 expression with climatological parameters of lung cancer patients. Our results showed higher expression of FBXO31 is associated with larger tumor size, higher clinical stages and lymph node metastasis, and not with age, gender and histology type (Figure 1, Table 3).
Figure 1.
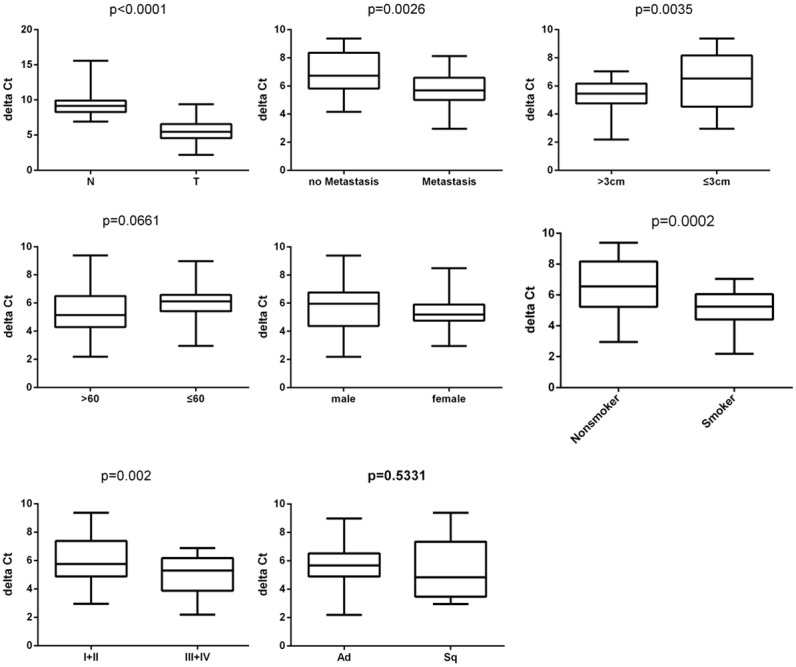
Expression of FBXO31 was assessed by qRT-PCR in lung cancer tumor specimens (T) and normal adjacent lung tissues (N). β-actin served as internal control. Data represent as ΔCT. Clinical association of FBXO31 with age, gender, histology type, tumor size, clinical stages and lymph node metastasis in lung cancer specimens was analyzed using the two-tailed Student’s t-test. A P value of < 0.05 was considered statistically significant.
Table 3.
The association of FBXO31 with clinicopathological features of 50 patients with lung cancer
| Characteristic | No. of patients | FBXO31 ΔCt | P | |
|---|---|---|---|---|
|
| ||||
| No | % | Mean ± SD | ||
| Age, years | ||||
| >60 | 31 | 62% | 5.356 ± 0.2192 | 0.0661 |
| ≤60 | 19 | 38% | 6.015 ± 0.2407 | |
| Sex | ||||
| Male | 36 | 72% | 5.729 ± 0.2423 | 0.2690 |
| Female | 14 | 28% | 5.346 ± 0.2143 | |
| Size | ||||
| >3 cm | 27 | 54% | 5.255 ± 0.1630 | 0.0035 |
| ≤3 cm | 23 | 46% | 6.284 ± 0.3606 | |
| Histology type | ||||
| Adenocarcinoma | 36 | 72% | 5.640 ± 0.1764 | 0.5331 |
| Squamous cancer | 14 | 28% | 5.400 ± 0.4140 | |
| Histological grade | ||||
| I, II | 28 | 56% | 6.055 ± 0.2352 | 0.0020 |
| III, IV | 22 | 44% | 5.027 ± 0.2164 | |
| Lymph node status | ||||
| No Metastasis | 13 | 26% | 6.900 ± 0.2895 | 0.0026 |
| Metastasis | 37 | 74% | 5.701 ± 0.2428 | |
| Smoking history | ||||
| Smoker | 32 | 64% | 5.147 ± 0.1500 | 0.0002 |
| Nonsmoker | 18 | 36% | 6.433 ± 0.3657 | |
Effects of FBXO31 on cell growth and colony formation
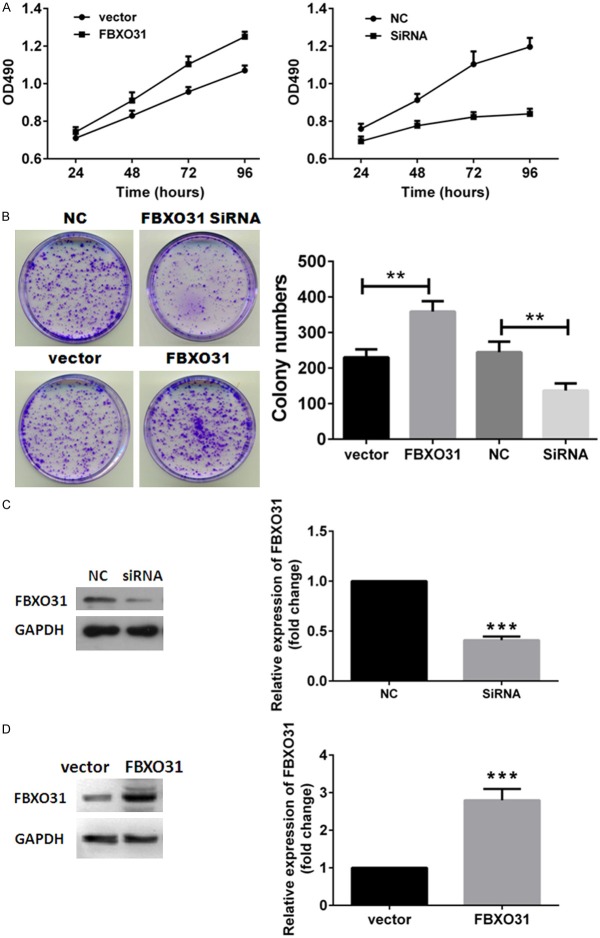
We first investigated effects of FBXO31 on cell growth and colony formation ability of lung cancer cells. Specific siRNA was used to silencing FBXO31 in A549 cells. NC served as control. Enforced expression of FBXO31 was accomplished by transfecting FBXO31-EGFP expressing plasmids into A549 cells with empty vector served as control. Exogenous expression of FBXO31 and silencing efficiency of siRNA were confirmed by western blot (Figure 2C and 2D). MTT assay showed exogenous expression of FBXO31 enhanced and suppression of FBXO31 inhibited cell growth in A549 cells (Figure 2A). Colony formation assay showed enforced expression of FBXO31 increased colony formation and silencing FBXO31 resulted in fewer colonies (Figure 2B). Taken together, these data revealed the oncogenic potential of FBXO31 in lung cancer cells.
Figure 2.
Effects of FBXO31 on cell proliferation and colony formation of lung cancer cells. A. MTT assay of A549 cells. Data represent the means ± SD. B. Colony formation assay of A549 cells. Left, images are representative of three independent experiments. Right, columns, mean of three separate experiments; bars, SD. **P < 0.01. C and D. Expression of FBXO31 in A549 cells. Columns, mean of three separate experiments; bars, SD. ***P < 0.001.
Effects of FBXO31 on metastasis and invasion
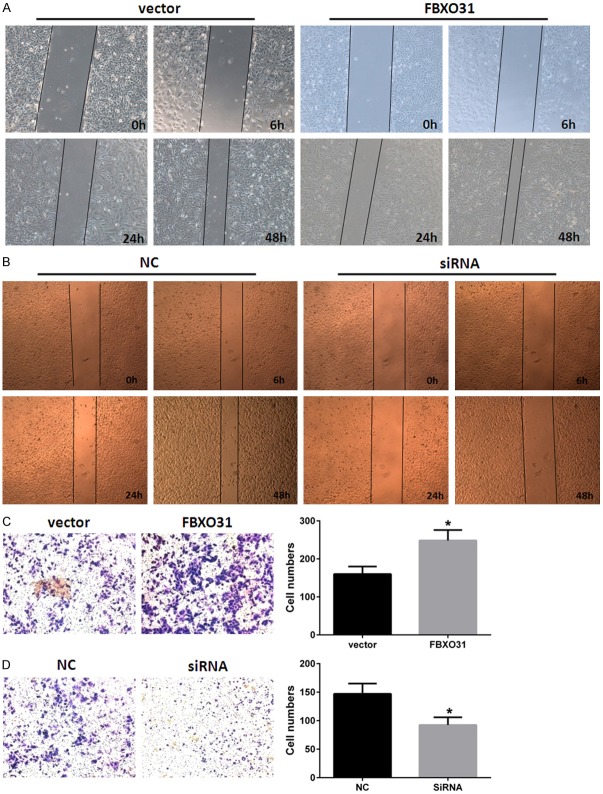
We also examined the effects of FBXO31 on metastasis and invasion capacity of lung cancer cells. The wound-healing assay and the transwell assay were applied in A549 cells. As Figure 3A shown, enforced expression of FBXO31 notably promoted migration of A549 cells on the surface of the tissue culture plate, significantly increased area of wound-healing compared with controls in wound-healing assay. Conversely, silencing FBXO31 led to decreased migration of A549 cells (Figure 3A). Data of transwell assays showed enforced expression of FBXO31 enhanced and suppression of FBXO31 inhibited invasion capacity of A549 cells (Figure 3B). These data suggest FBXO31 plays a promoting role in metastasis and invasion.
Figure 3.
Effects of FBXO31 on metastasis and invasion of lung cancer cells. (A and B) Enforced expression of FBXO31 promotes (A) and silencing FBXO31 inhibits (B) metastasis of A549 cells. Cells were plated in 6-well plates, transfected as indicated. Monolayer was then disrupted with a cell scraper (1 mm width), and photographs were taken at 0, 6, 24 and 48 h in a phase-contrast microscope. Experiments were carried out in triplicate, and four fields of each plate were recorded. (C and D) Enforced expression of FBXO31 promotes (C) and silencing FBXO31 inhibits (D) invasion of A549 cells. The cells that invasion through the pores to the lower surface of the filter were stained with crystal violet and counted under a microscope. The values obtained were calculated by averaging the total number of cells from three separate experiments. Columns, mean of three separate experiments; bars, SD. *P < 0.05.
FBXO31 promotes tumor growth in vivo
To further verify the results of in vitro cell growth assays, tumorigenic assays in nude mice were performed using A549 cells stably expressing or silencing FBXO31. Empty vector served as control. Then we evaluated the tumorigenic effects of each of these cells in BALB/C nude mice. The tumors in mice injected with A549 cells stably expressing FBXO31 showed a proliferative tendency, with significantly bigger tumor volume compared with the control group (Figure 4). Moreover, suppression of FBXO31 in A549 cells led to consequential suppression of tumor growth in nude mice (Figure 4). These results indicate that FBXO31 promotes tumor growth in vivo.
Figure 4.
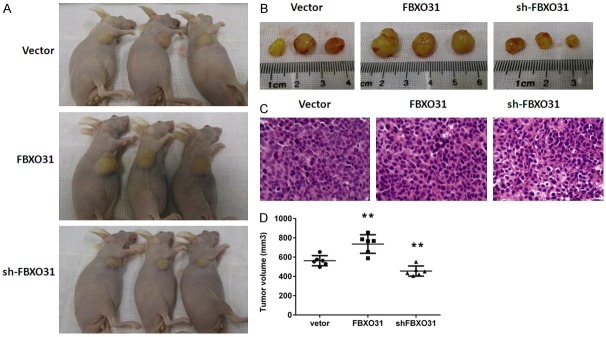
FBXO31 promotes tumor growth in nude mice. A. Images of nude mice that were injected with the three different groups of A549 cells (vector, FBXO31 and shFBXO31. B. Representative images of tumors isolated from nude mice. The images shown are representative of the results from 6 mice. C. HE staining of tumors tissues. Images are representative of three independent experiments. Original magnification 200×. D. The tumor volumes were estimated using calipers. Data represent the means ± SD of 6 mice per group. **p < 0.001.
Discussion
FBXO31 is a member of F-box protein family, which contains a F-box motif and functions as a substrate recognized unit in ubiquitin proteasome system [12]. Recent studies in breast cancer [12], hepatocellular carcinoma [19] and gastric cancer [20] indicated FBXO31 functions as a tumor suppressor. The expression of FBXO31 is relatively lower in tumor tissues compared to their corresponding non-cancerous tissues. However, a study in esophageal squamous cell carcinoma (ESCC) showed a different story about the function and expression profile of FBXO31. They showed the expression of FBXO31 was associated with tumor invasion depth and clinical stage [18]. Moreover, higher expression of FBXO31 correlated with poor prognosis in ESCC patients [18]. Therefore, the expression and function of FBXO31 is controversial in different type of cancers. Until recently, the expression profile, clinical significance and biological function of FBXO31 in lung cancer remains unclear. In the present study, we investigated the expression of FBXO31 in lung cancer tissues and corresponding non-cancerous lung tissues. The association between the expression of FBXO31 and clinical parameters was analyzed. We further evaluated the effects of FBXO31 on cell growth, colony formation, metastasis and invasion in lung cancer cells and nude mice xenograft model.
Our results showed significantly higher expression of FBXO31 in lung cancer tissues compared with corresponding non-cancerous tissues. Further analysis showed expression of FBXO31 correlated with tumor size, clinical stage and lymph node metastasis in lung cancer patients, suggesting that FBXO31 might play an important role in tumor development and progression. Cell proliferation assays and colony formation assays showed enforced expression of FBXO31 promoted and silencing of FBXO31 suppressed cell growth and colony formation in lung cancer cells. Wound healing assays and matrigel-based in vitro invasion assays showed overexpression of FBXO31 increased and suppression of FBXO31 decreased metastasis and invasion abilities of lung cancer cells. Moreover, tumorigenicity assays in nude mice verified the finding in vitro proliferation assays and demonstrated FBXO31 promoted tumor growth in vivo. Taken together, our data indicated FBXO31 might play an oncogenic role in lung cancer development and progression.
In conclusion, FBXO31 promotes tumor growth, invasion and metastasis in lung cancer. Our results demonstrated the oncogenic potential and clinical significance of FBXO31 in lung cancer. This study provides new insights on the function of FBXO31 in cancer field and help establishing novel strategy for lung cancer therapy.
Acknowledgements
This project is supported by Science andTechnology Innovation Fund of Guangdong Medical College (STIF201109), the Natural Science Foundation of China project (NSFC81172615), the natural science foundation of Guangdong province (S2012040006537), medical science research foundation of Guangdong province (B2014309) and medical science foundation of Guangdong Medical College (XB1224).
Disclosure of conflict of interest
None.
References
- 1.Velu TJ, Lowy DR. Proto-oncogene activation in lung cancer. N Engl J Med. 1988;318:992. [PubMed] [Google Scholar]
- 2.Little CD, Nau MM, Carney DN, Gazdar AF, Minna JD. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature. 1983;306:194–196. doi: 10.1038/306194a0. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Z, Hao Y, Liu N, Raptis L, Tsao MS, Yang X. TAZ is a novel oncogene in non-small cell lung cancer. Oncogene. 2011;30:2181–2186. doi: 10.1038/onc.2010.606. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki C, Takahashi K, Hayama S, Ishikawa N, Kato T, Ito T, Tsuchiya E, Nakamura Y, Daigo Y. Identification of Myc-associated protein with JmjC domain as a novel therapeutic target oncogene for lung cancer. Mol Cancer Ther. 2007;6:542–551. doi: 10.1158/1535-7163.MCT-06-0659. [DOI] [PubMed] [Google Scholar]
- 5.Regala RP, Weems C, Jamieson L, Khoor A, Edell ES, Lohse CM, Fields AP. Atypical protein kinase C iota is an oncogene in human non-small cell lung cancer. Cancer Res. 2005;65:8905–8911. doi: 10.1158/0008-5472.CAN-05-2372. [DOI] [PubMed] [Google Scholar]
- 6.Kabbout M, Garcia MM, Fujimoto J, Liu DD, Woods D, Chow CW, Mendoza G, Momin AA, James BP, Solis L, Behrens C, Lee JJ, Wistuba II, Kadara H. ETS2 mediated tumor suppressive function and MET oncogene inhibition in human non-small cell lung cancer. Clin Cancer Res. 2013;19:3383–3395. doi: 10.1158/1078-0432.CCR-13-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang R, Wang ZX, Yang JS, Pan X, De W, Chen LB. MicroRNA-451 functions as a tumor suppressor in human non-small cell lung cancer by targeting ras-related protein 14 (RAB14) Oncogene. 2011;30:2644–2658. doi: 10.1038/onc.2010.642. [DOI] [PubMed] [Google Scholar]
- 8.Shimizu E, Sone S. Tumor suppressor genes in human lung cancer. J Med Invest. 1997;44:15–24. [PubMed] [Google Scholar]
- 9.Kuramochi M, Fukuhara H, Nobukuni T, Kanbe T, Maruyama T, Ghosh HP, Pletcher M, Isomura M, Onizuka M, Kitamura T, Sekiya T, Reeves RH, Murakami Y. TSLC1 is a tumor-suppressor gene in human non-small-cell lung cancer. Nat Genet. 2001;27:427–430. doi: 10.1038/86934. [DOI] [PubMed] [Google Scholar]
- 10.Chen S, Xu Y, Chen Y, Li X, Mou W, Wang L, Liu Y, Reisfeld RA, Xiang R, Lv D, Li N. SOX2 gene regulates the transcriptional network of oncogenes and affects tumorigenesis of human lung cancer cells. PLoS One. 2012;7:e36326. doi: 10.1371/journal.pone.0036326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar MS, Hancock DC, Molina-Arcas M, Steckel M, East P, Diefenbacher M, Armenteros-Monterroso E, Lassailly F, Matthews N, Nye E, Stamp G, Behrens A, Downward J. The GATA2 transcriptional network is requisite forRAS oncogene-driven non-small cell lung cancer. Cell. 2012;149:642–655. doi: 10.1016/j.cell.2012.02.059. [DOI] [PubMed] [Google Scholar]
- 12.Kumar R, Neilsen PM, Crawford J, McKirdy R, Lee J, Powell JA, Saif Z, Martin JM, Lombaerts M, Cornelisse CJ, Cleton-Jansen AM, Callen DF. FBXO31 is the chromosome 16q24.3 senescence gene, a candidate breast tumor suppressor, and a component of an SCF complex. Cancer Res. 2005;65:11304–11313. doi: 10.1158/0008-5472.CAN-05-0936. [DOI] [PubMed] [Google Scholar]
- 13.Jia L, Sun Y. F-box proteins FBXO31 and FBX4 in regulation of cyclin D1 degradation upon DNA damage. Pigment Cell Melanoma Res. 2009;22:518–519. doi: 10.1111/j.1755-148X.2009.00611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santra MK, Wajapeyee N, Green MR. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459:722–725. doi: 10.1038/nature08011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiloh Y. FBXO31: a new player in the ever-expanding DNA damage response orchestra. Sci Signal. 2009;2:pe73. doi: 10.1126/scisignal.296pe73. [DOI] [PubMed] [Google Scholar]
- 16.Vadhvani M, Schwedhelm-Domeyer N, Mukherjee C, Stegmuller J. The centrosomal E3 ubiquitin ligase FBXO31-SCF regulates neuronal morphogenesis and migration. PLoS One. 2013;8:e57530. doi: 10.1371/journal.pone.0057530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silvers CR, Williams K, Salamone L, Huang J, Jordan CT, Zhou H, Palapattu GS. A novel in vitro assay of tumor-initiating cells in xenograft prostate tumors. Prostate. 2010;70:1379–1387. doi: 10.1002/pros.21171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kogo R, Mimori K, Tanaka F, Komune S, Mori M. FBXO31 determines poor prognosis in esophageal squamous cell carcinoma. Int J Oncol. 2011;39:155–159. doi: 10.3892/ijo.2011.1018. [DOI] [PubMed] [Google Scholar]
- 19.Huang HL, Zheng WL, Zhao R, Zhang B, Ma WL. FBXO31 is down-regulated and may function as a tumor suppressor in hepatocellular carcinoma. Oncol Rep. 2010;24:715–720. doi: 10.3892/or_00000912. [DOI] [PubMed] [Google Scholar]
- 20.Tomoya Sudo RK, Nishida N, Takahahi K, Sawada G, Ishibashi M, Kurashige J, Uchi R, Matsumura T, Ueo H, Mima K. The Significance of the Expression of FBXO31 in Gastric Cancer. Journal of Cancer Therapy. 2013;4:75–79. [Google Scholar]