Abstract
Background
Because the future application of cell-free fetal DNA screening is expected to dramatically improve the diagnostic yield and reduce unnecessary invasive procedures, it is time to summarize the indications of invasive prenatal diagnosis. This retrospective study was performed to evaluate the changes and efficacies of indications of invasive procedures for detecting cytogenomic abnormalities from 2000 to 2012.
Material/Methods
From our regional obstetric unit, 7818 invasive procedures were referred by indications of advance maternal age (AMA), abnormal ultrasound findings (aUS), abnormal maternal serum screening (aMSS), and family history (FH). Chromosome, fluorescence in situ hybridization (FISH), and array comparative genomic hybridization (aCGH) analyses were performed on chorionic villus sampling (CVS) and amniotic fluid (AF) specimens at the Yale Cytogenetics Laboratory. The abnormal findings from single or combined indications were compared to evaluate the diagnostic yield.
Results
The annual caseload declined by 57.2% but the diagnostic yield increased from 7.2% to 13.4%. Chromosomal and genomic abnormalities were detected in 752 cases (9.6%, 752/7818) and 12 cases (4%, 12/303), respectively. Significantly decreased AMA referrals and increased aUS and aMSS referrals were noted. The top 3 indications by diagnostic yield were AMA/aUS (51.4% for CVS, 24.2% for AF), aUS (34.7% for CVS, 14.5% for AF), and AMA/aMSS (17.8% for CVS, 9.9% for AF).
Conclusions
Over a period of 13 years, the indication of aMSS and aUS were increasing while AMA was decreasing for prenatal diagnosis of cytogenomic abnormalities, and there was a continuous trend of reduced invasive procedures. Prenatal evaluation using AMA/aUS was the most effective in detecting chromosomal abnormalities, but better indications for genomic abnormalities are needed.
Keywords: Comparative Genomic Hybridization; Genetic Counseling; Prenatal Diagnosis; Surgical Procedures, Minimally Invasive
Background
Invasive procedures – mainly chorionic villus sampling (CVS) and amniocentesis – are very common for prenatal diagnosis. Nowadays, conventional prenatal chromosome analysis, fluorescence in situ hybridization (FISH), and array comparative genomic hybridization (aCGH) analyses depend on samples acquired from invasive procedures [1]. Chromosomal abnormalities that are compatible with life but cause considerable morbidity occur in 0.65% of newborns, and apparently balanced structural chromosomal rearrangements that will eventually affect reproduction occur in 0.2% of newborns [2]. The application of aCGH or the single-nucleotide polymorphism (SNP) chip has allowed the detection of submicroscopic abnormalities and genomic disorders; the most commonly seen recurrent genomic disorders occur in approximately 0.18% of newborns [3]. In 2009, the American College of Obstetricians and Gynecologists (ACOG) recommended that conventional karyotyping should remain the principal cytogenetic tool in prenatal diagnosis, and that aCGH should be an adjunct to prenatal care for women with abnormal ultrasound findings (aUS) and a normal conventional karyotype [4].
However, many factors such as the risk that the fetus will have a chromosomal abnormality from direct fetal ultrasound imaging and indirect maternal serum markers, the risk of procedure-related miscarriage, the consequences of having an affected child, anxiety, ethnic background, and religion can affect a pregnant woman’s decision to accept or reject an invasive procedure [5–7]. We expect that future application of cell-free fetal DNA (cff-DNA) screening for cytogenetic disorders will dramatically improve the diagnostic yield and reduce unnecessary invasive procedures [8,9]; therefore, it is time to summarize the clinical indications of invasive prenatal diagnosis. In addition, cff-DNA screening has many limitations [10] and many cytogenomic abnormalities, including chromosomal structural abnormalities and genomic abnormalities, cannot be screened with it. Therefore, clinical indications accurately predicting the risk of cytogenomic abnormalities play an important role in prenatal genetic counseling and diagnosis. The use of clinical indications constantly changes with time and regional differences [11]. Current prenatal clinical indications include aUS, abnormal maternal serum screening (aMSS), advanced maternal age (AMA), family history (FH) of chromosomal abnormalities, and other events that could affect fetal health [12,13].
Prenatal diagnosis is expected to move forward with more effective non-invasive prenatal indications and genome-wide analysis of cytogenomic abnormalities. However, the efficacy of clinical indications in prenatal diagnosis could vary in different practice settings and different regions. This study aimed to analyze the efficacies of indications of CVS and amniocentesis from a regional obstetric unit. The results provide the diagnostic yields from single or combined clinical indications, and indicate which women need the invasive testing and future direction for better prenatal diagnosis of cytogenomic abnormalities. This information will be useful for the obstetricians, clinical geneticists, and laboratory staff to further improve the quality of prenatal diagnosis.
Material and Methods
Yale Cytogenetics Laboratory is a regional reference laboratory for prenatal diagnosis in New Haven County and surrounding areas. From 2000 to 2012, the laboratory performed karyotype analysis on 3229 CVS and 4589 amniotic fluid (AF) specimens (excluding 17 culture failure samples). FISH using probes for the HIRA gene at 22q11.21 had been performed on 55 patients with aUS of cardiac defects or FH of DiGeorge syndrome. Since 2009, aCGH has been validated and offered to high-risk pregnancies after pre-testing counseling of technical specifications and limitations. A total of 248 aCGH had been performed as an adjunct test. All test results and clinical indications of these 7818 CVS and amniocenteses were compiled from the laboratory’s CytoAccess database [14].
The clinical indications for prenatal diagnosis included: (1) AMA defined by age equal or greater than 35 years for singlet, 33 years for twin, and 31 years for triplet pregnancies; (2) aUS of increased nuchal translucency (≥3 mm), cystic hygroma and other kinds of suspected fetal anomalies; (3) aMSS from first or second trimester maternal serum Quad screen, including human chorionic gonadotropin (hCG), α-fetoprotein (AFP), unconjugated estriol, and inhibin A; (4) FH of a chromosome abnormality in previous pregnancy, parents with abnormal karyotype, or history of abnormal offspring birth; (5) multiple pregnancy (MP), including twins and triplets; and (6) other indications such as intrauterine fetal death or demise, radiation or medication exposure during pregnancy, anxiety, and consanguineous marriage. Of the 7818 cases, 68%, 30%, and 2% were referred by single indication, 2 indications, and 3 indications, respectively. A total of 63 combinations of clinical indications were documented. For practical purpose, single and combined indications occurring in more than 1% of the samples (except for MP) were used to evaluate diagnostic yields.
Conventional chromosome analysis was performed on G-band metaphases prepared from cultured AF and CVS cells. The chromosome abnormalities were classified as numerical and structural abnormalities. FISH tests using Aneuvision probes for rapid screening of aneuploidies of chromosomes X, Y, 21, 13, and 18 and targeted probes for genomic disorders were performed following manufacturer’s protocols (Vysis/Abbott, Abbott Park, IL). Genomic DNA was extracted from CVS and cultured amniocytes using the Gentra Puregene Kit (Qiagen, Valencia, CA). The oligonucleotide aCGH analysis using the Agilent Human Genome CGH microarray 44K kit (containing 44 913 60-mer oligonucleotides, using NCBI36/hg18 human genome assembly), and the 180K kit (177 873 60-mer oligonucleotides, using GRCh37/hg19 assembly) (Agilent Technologies, Inc., Santa Clara, CA) was performed as previously described [12].
Diagnostic yield was defined as percentage of the abnormal cases from cases referred by single, combined, or all indications. Time series regression analyses were performed for variance to evaluate the significance of trends and chi-square tests to compare rates, using SPSS for Windows (SPSS Inc. Chicago, IL). For tests of significance, a P value of less than 0.05 was considered significant.
Results
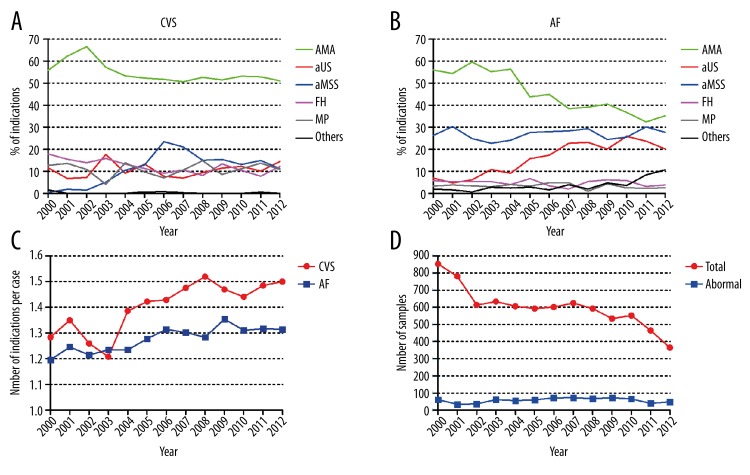
From 2000 to 2012, the most commonly used indications were AMA, aMSS, and aUS. For CVS cases, AMA referrals declined from 55.8% to 51.1% (P<0.05), aMSS referrals increased from 0.5% to 11.2% (P<0.05), and aUS referrals remained at the 10.6±3.3% level (Figure 1A). For AF cases, AMA referrals significantly declined from 55.9% to 35.2% (P<0.001), aMSS referrals remained at 26.8±2.4%, and aUS referrals increased from 5% to 20% (P<0.05) (Figure 1B). The indications of FH, MP, and others remained in less than 12.3±3.1% and showed a relatively stable trend except for a decline in FH referrals for CVS. The usage of multiple indications defined by the average number of indications per case showed an increasing trend during the 13 year period (P<0.01) and CVS referrals required more indications than AF (P<0.01) (Figure 1C).
Figure 1.
The changes of indications of CVS and amniocentesis from 2000 to 2012 at the Yale Cytogenetics Laboratory. (A) The changes of clinical indications of CVS. (B) The changes of clinical indications of amniocentesis. (C) The annual average number of indications per case. (D) The number of total caseload and abnormal samples.
Although the annual caseload of CVS and AF showed a significant 57.2% decrease from 853 cases in 2000 to 365 cases in 2012 (P<0.01), there was no significance change in the number of abnormal findings (Figure 1D). The diagnostic yield for chromosomal abnormalities showed a significant increase from 7.2% in 2000 to 13.4% in 2012 (P<0.01) and the overall diagnostic yield was 9.6%. Clinical indications associated with detected chromosomal abnormalities were also counted and ordered as follows: AMA 57.6%, aUS 40.3%, aMSS 30.5%, FH 9.6%, MP 4.8%, and other indications 2.3%. The efficacy of single or combined indications for detecting chromosomal abnormalities is summarized in Table 1. The 3 highest diagnostic yields were noted in cases with AMA/aUS (51.4% for CVS, 24.2% for AF), aUS (34.7% for CVS, 14.5% for AF), and AMA/aMSS (17.8% for CVS, 9.9% for AF). In this cohort, the most effective single indication was aUS followed by aMSS or FH and then AMA. MP was referred as indication of prenatal diagnosis due to our regional practice. However, there were no chromosomal abnormalities detected from MP referrals.
Table 1.
The efficacy of indications in detecting chromosomal abnormality.
| Clinincal indications | Number of referrals (%) | Abnormality | Diagnostic yield* | ||
|---|---|---|---|---|---|
| All CA (%) | nCA | sCA | |||
| CVS | |||||
| AMA/aUS | 142 (1.8) | 73 (9.7) | 73 | 0 | 51.4% |
| aUS | 262 (3.4) | 91 (12.1) | 85 | 6 | 34.7% |
| AMA/aMSS | 342 (4.4) | 61 (8.1) | 57 | 4 | 17.8% |
| aMSS | 203 (2.6) | 30 (4.0) | 28 | 2 | 14.8% |
| FH | 207 (2.6) | 20 (2.7) | 7 | 13 | 9.7% |
| AMA/FH | 272 (3.5) | 25 (3.3) | 12 | 13 | 9.2% |
| AMA | 1242 (15.9) | 81 (10.8) | 72 | 9 | 6.5% |
| AMA/MP | 388 (5.0) | 12 (1.6) | 8 | 4 | 3.1% |
| MP | 20 (0.3) | 0 | 0 | 0 | 0 |
| Others | 151 (1.9) | 28 (3.7) | 23 | 5 | 18.5% |
| Subtotal** | 3229 (41.3) | 421 (56) | 365 | 56 | 13.0% |
| AF | |||||
| AMA/aUS | 95 (1.2) | 23 (3.1) | 20 | 3 | 24.2% |
| aUS | 571 (7.3) | 83 (11) | 68 | 15 | 14.5% |
| AMA/aMSS | 639 (8.2) | 63 (8.4) | 58 | 5 | 9.9% |
| Other | 92 (1.2) | 6 (0.8) | 3 | 3 | 6.5% |
| FH | 111 (1.4) | 7 (0.9) | 1 | 6 | 6.3% |
| aMSS | 823 (10.5) | 47 (6.3) | 32 | 15 | 5.7% |
| AMA/FH | 131 (1.7) | 6 (0.8) | 4 | 2 | 4.6% |
| AMA | 1782 (22.8) | 61 (8.1) | 43 | 18 | 3.4% |
| AMA/MP | 103 (1.3) | 0 | 0 | 0 | 0 |
| MP | 11 (0.1) | 0 | 0 | 0 | 0 |
| Others | 231 (3.0) | 35 (4.7) | 26 | 9 | 15.2% |
| Subtotalb | 4589 (58.7) | 331 (44) | 255 | 76 | 7.2% |
| Total | 7818 (100.0) | 752 (100.0) | 620 | 132 | 9.6% |
Diagnostic Yield=all CA/number of referrals;
Statistically significant intergroup comparison (p<0.001) is seen with CVS; CA – chromosomal abnormality; nCA – numerical CA; sCA – structural CA; CVS – chorionic villus sampling; AF – amniotic fluid; AMA – advanced maternal age; aUS – abnormal ultrasound findings; aMSS – abnormal maternal serum screening; FH – family history; MP – multiple pregnancy; others – all other combinations except the indications in the table.
FISH tests were used to detect 22q11.2 deletion on prenatal cases with aUS of cardiac defects and FH of DiGeorge syndrome. Of the 55 cases tested, 6 were diagnosed with DiGeorge syndrome. Previous validation of aCGH analysis on 11 cases with prenatally detected structural abnormalities successfully defined the genomic imbalances and gene content [12]. The aCGH analysis performed on 237 prenatal cases with normal chromosome findings detected 6 pathogenic and likely pathogenic genomic abnormalities. Recurrent genomic disorders of 16p13.11 deletions inherited from a paternal carrier were detected in 2 unrelated families and a 22q11.2 duplication was seen in 1 case. The aUS findings of IUGR, cystic hygroma, echogenic bowel, and tetralogy of Fallot (TOF) were noted in cases with recurrent genomic disorders. The clinical indications and the genomic abnormalities from FISH and aCGH analyses are shown in Table 2. Variants of unknown significance (VOUS) were detected in 5 cases; 2 of them pursued parental study and were confirmed as familial variants of maternal origin (data not shown). The estimated diagnostic yield from the FISH and aCGH analyses for genomic abnormalities on tested prenatal cases was 4% (12/303).
Table 2.
Genomic abnormalities detected by aCGH and their indications.
| Case# | Indications | Sampling | Karyotype | FISH or aCGH findings |
|---|---|---|---|---|
| Pathogenic | ||||
| 1 | aUS (Tetralogy of Fallot) | AF | 46,XX | ish del(22)(q11.2q11.2)(TUPLE1-) |
| 2 | aUS (Tetralogy of Fallot) | AF | 46,XY | ish del(22)(q11.2q11.2)(TUPLE1-) |
| 3 | aUS (congenital heart disease) | AF | 46,XX | ish del(22)(q11.2q11.2)(TUPLE1-) |
| 4 | aUS (congenital heart disease) | AF | 46,XX | ish del(22)(q11.2q11.2)(TUPLE1-) |
| 5 | aUS (fetal anomalies) | AF | 46,XX | ish del(22)(q11.2q11.2)(TUPLE1-) |
| 6 | FH of DiGeorge syndrome | AF | 46,XY | ish del(22)(q11.2q11.2)(TUPLE1-)mat |
| 7 | aUS (cystic hygroma, fetal demise) | AF | 46,XY | [hg18]16p13.11(14,876,156-16,174,951)x1pat |
| 8 | aUS (IUGR, echogenic bowel) | AF | 46,XY | [hg19]16p13.11(14,910,205-16,586,915)x1pat |
| 9 | aUS (Tetralogy of Fallot) | AF | 46,XX | [hg18]22q11.21(17,274,635-18,589,433)x3 |
| 10 | aUS (IUGR) | AF | 46,XY | [hg18]Xq25q26.3(129,092,340-133,914,595)x2dn |
| Likely pathogenic | ||||
| 11 | aMSS (increased DS risk) | CVS | 46,XY | [hg19]12q24.13(112,713,491-112,942,507)x3mat |
| 12 | aMSS (increased DS risk) | AF | 46,XX | [hg19]Xq28(154,118,643-154,560,375)x3mat |
FISH – fluorescence in situ hybridization; aCGH – array comparative genomic hybridization; aUS – abnormal ultrasound findings; AF – amniotic fluid; CVS – chorionic villus sampling; FH – family history; aMSS – abnormal maternal serum screening; IUGR – intrauterine growth retardation; DS – Down syndrome.
Discussion
Our cytogenetics laboratory has provided prenatal chromosome, FISH, and microarray analyses for Yale affiliated hospitals with stable referrals from New Haven County and surrounding areas. Consistent with previous studies [13,15], our data demonstrated a continuous trend of reduced invasive procedures. The caseload reduction and procedure shifting were largely attributed to better knowledge and more informative counseling from clinical indications. The revised 2007 ACOG screening and invasive testing guidelines recommended that genetic screening and invasive diagnostic testing for aneuploidy should be available to all pregnant women, regardless of maternal age [2]. AMA, a known factor for increased risk of Down syndrome, is still the leading single indication in many reports [13,15,16]. In our practice, AMA is the most common indication, followed by aMSS in second place and aUS in third place. However, due to the development of sensitive ultrasonic technology and maternal serum markers, the indication of aMSS for CVS and aUS for AF increased from 2000 to 2012, with a significant decrease in AMA.
Analysis of diagnostic yields from single and combined clinical indications can provide useful information for prenatal genetic counseling and diagnosis. For single indications for CVS and AF, AMA, aMSS, aUS, and FH accounted for 39%, 13%, 11%, and 4% of referrals, but 19%, 10%, 23%, and 4% of total abnormal findings, respectively. For combined indications, AMA/aUS, AMA/aMSS, and AMA/FH accounted for 3%, 13%, and 5% of referrals, but 13%, 17%, and 4% of total abnormal findings, respectively (Table 1). It was obvious that AMA/aUS, aUS, and AMA/aMSS are the most effective indications and together they accounted for 26% of total referrals and 52% of total abnormal findings. However, the remaining 48% of chromosomal abnormalities from other single and combined indications are indispensable in prenatal practice. In the present cohort, only 40% of cases detected with chromosome abnormalities had aUS findings, suggesting the absence of ultrasound visible fetal structural anomalies in about 60% of prenatal chromosomal abnormalities. Other groups reported aUS findings in 9–30% of fetuses with chromosomal abnormalities [13,15,16]. Further advancement of prenatal ultrasonography with better imaging and the inclusion of soft markers will improve the diagnostic yield. However, some fetuses with chromosomal abnormalities could lack ultrasound-detectable fetal anomalies.
With a population of 0.86 million in New Haven County and 2.17 million in the surrounding counties and a 1.3% birth rate by the 2012 United States Census, it is estimated that each year there are 11 180 newborns in New Haven County and 27 430 newborns in the surrounding counties. In this prenatal cohort during a 13-year period, chromosome analysis detected 292 fetuses (average 22 cases per year) with trisomy 21 for the diagnosis of Down syndrome; FISH and aCGH detected 7 fetuses with deletions and duplication in the DiGeorge syndrome region (0.5 case per year). In a general population, the occurrence of Down syndrome (OMIM#190685) is about 1/650 to 1/1000 live births and occurrence of DiGeorge syndrome (OMIM#188400) is 1 in 4000, indicating a 6:1 to 4:1 ratio of Down vs. DiGeorge. Using a 1/800 occurrence for Down syndrome, prenatal diagnosis of 22 fetuses inferred an annual basis of 17 600 pregnant women, which could be translated to a population of 1.35 million, consisting of the 0.86 million in the New Haven county and 23% of the 2.17 million from the surrounding counties. From the same population basis, postnatal cytogenomic analysis was performed on 1354 pediatric patients from 2006 to 2009 and detected 7 cases with trisomy 21 (1.75 cases per year), 13 cases with the 22q11.21 deletion, and 3 cases with the reciprocal duplication (4 cases per year) [17]. Combined the prenatal and postnatal data, the annual detection of 24 Down syndrome and 4~5 DiGeorge region deletions and duplications matches the occurrence and Down vs. DiGeorge ratio from the estimated 17 600 pregnancies. This observation indicated that about 92% (22/24) of Down syndrome cases but only 10% (0.5/4~5) of DiGeorge deletions and duplications were detected prenatally in the current setting. The lower diagnostic yield of genomic disorders in prenatal practice is likely due to the lack of sensitive and specific clinical indications.
Since 2008–2010 we found that the number of invasive procedures began to change significantly, perhaps because non-invasive prenatal testing became popular from that time [8]. Recently, a non-invasive prenatal screening method using massive parallel sequencing of maternal plasma cff-DNA has been validated and rapidly integrated into prenatal genetic evaluation [8,9,18]. A rapid ACMG statement for this newly introduced cff-DNA-based screening has been issued [19]. Currently, this newly integrated procedure has demonstrated high sensitivity and specificity in detecting common aneuploidies [20,21]. This progress has also re-energized efforts to develop non-invasive prenatal diagnosis using maternal circulating fetal cells [8,19–22]. Further improvement of cff-DNA based screening for genomic imbalances is technically feasible and is expected to become a direct indication of genomic abnormalities in prenatal counseling for recommending further invasive procedures.
Conclusions
From 2000 to 2012, with the development of sensitive ultrasonic technology and maternal serum markers, the indication of aMSS and aUS were increasing while AMA were significantly decreasing, although AMA is the most common indication. Largely due to this change, it demonstrated a continuous trend of reduced invasive procedures. Over a period of13 years, prenatal evaluation using AMA/aUS was the most effective in detecting chromosomal abnormalities, but better indications for genomic abnormalities are needed.
The occurrence of chromosomal morbidity, reproductive defects, and common genomic disorders in 0.65%, 0.2%, and 0.18%, respectively, of newborns indicated that the baseline of prenatal cytogenomic diagnosis should be set on at least 1% but most likely up to 3% of pregnant women if including other genomic abnormalities and gene mutations [2,3]. Diagnostic prenatal cytogenomic tests should be performed on 176 to 528 fetuses from 17 600 pregnancies; the 2012 caseload of 365 in this laboratory is within the baseline level.
The goal of invasive procedures for prenatal diagnosis is to achieve much higher diagnostic yield for cytogenomic abnormalities. Toward that end, prenatal genetic evaluation using multiple screening methods of AMA, aUS, aMSS, and cff-DNA and combined diagnostic chromosome and aCGH tests will become highly effective in detecting cytogenomic abnormalities.
Footnotes
Source of support: This work was partially supported by Shandong Population and Family Planning Science and Technology Project, Shandong, China (2012-10)
Conflict of interest
The authors confirm that they have no conflicts of interest.
References
- 1.Simpson JL. Invasive procedures for prenatal diagnosis: any future left? Best Pract Res Clin Obstet Gynaecol. 2012;26(5):625–38. doi: 10.1016/j.bpobgyn.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 2.ACOG Committee on Practice Bulletins: ACOG Practice Bulletin No. 77: screening for fetal chromosomal abnormalities. Obstet Gynecol. 2007;109(1):217–27. doi: 10.1097/00006250-200701000-00054. [DOI] [PubMed] [Google Scholar]
- 3.Wei Y, Xu F, Li P. Technology-driven and evidence-based genomic analysis for integrated pediatric and prenatal genetics evaluation. J Genet Genomics. 2013;40(1):1–14. doi: 10.1016/j.jgg.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 4.ACOG Committee Opinion No. 446: array comparative genomic hybridization in prenatal diagnosis. Obstet Gynecol. 2009;114(5):1161–63. doi: 10.1097/AOG.0b013e3181c33cad. [DOI] [PubMed] [Google Scholar]
- 5.Dar H, Zuck C, Friedman S, et al. Chorionic villous sampling: differences in patients’ perspectives according to indication, ethnic group and religion. Isr Med Assoc J. 2006;8(8):536–38. [PubMed] [Google Scholar]
- 6.Bot-Robin V, Sendon S, Bourzoufi K, et al. Maternal anxiety and pain during prenatal diagnostic techniques: a prospective study. Prenat Diagn. 2012;32(6):562–68. doi: 10.1002/pd.3857. [DOI] [PubMed] [Google Scholar]
- 7.Hill M, Fisher J, Chitty LS, et al. Women’s and health professionals’ preferences for prenatal tests for Down syndrome: a discrete choice experiment to contrast noninvasive prenatal diagnosis with current invasive tests. Genet Med. 2012;14(11):905–13. doi: 10.1038/gim.2012.68. [DOI] [PubMed] [Google Scholar]
- 8.Chiu RW, Chan KC, Gao Y, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci USA. 2008;105(51):20458–63. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao C, Zhengfeng X, Zhang K. DNA sequencing versus standard prenatal aneuploidy screening. N Engl J Med. 2014;371(6):577–78. doi: 10.1056/NEJMc1405486. [DOI] [PubMed] [Google Scholar]
- 10.Hardisty EE, Vora NL. Advances in genetic prenatal diagnosis and screening. Curr Opin Pediatr. 2014;26(6):634–38. doi: 10.1097/MOP.0000000000000145. [DOI] [PubMed] [Google Scholar]
- 11.Ciach K, Swiatkowska-Freund M, Preis K. Influence of place of residence on indications for genetic amniocentesis in the Pomeranian region of Poland before and after introduction of the Prenatal Screening Program in 2008. Med Sci Monit. 2014;20:720–24. doi: 10.12659/MSM.890159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li P, Pomianowski P, DiMaio MS, et al. Genomic characterization of prenatally detected chromosomal structural abnormalities using oligonucleotide array comparative genomic hybridization. Am J Med Genet A. 2011;155A(7):1605–15. doi: 10.1002/ajmg.a.34043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benn PA, Egan JF, Fang M, et al. Changes in the utilization of prenatal diagnosis. Obstet Gynecol. 2004;103(6):1255–60. doi: 10.1097/01.AOG.0000127008.14792.14. [DOI] [PubMed] [Google Scholar]
- 14.Xiang B, Hemingway S, Qumsiyeh M, et al. CytoAccess: A relational laboratory information management system for a clinical cytogenetics laboratory. J Assoc Genet Technol. 2006;32(4):168–70. [PubMed] [Google Scholar]
- 15.Chang YW, Chang CM, Sung PL, et al. An overview of a 30-year experience with amniocentesis in a single tertiary medical center in Taiwan. Taiwan J Obstet Gynecol. 2012;51(2):206–11. doi: 10.1016/j.tjog.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Siljee JE, Knegt AC, Knapen MF, et al. Positive predictive values for detection of trisomies 21, 18 and 13 and termination of pregnancy rates after referral for advanced maternal age, first trimester combined test or ultrasound abnormalities in a national screening programme (2007–2009) Prenat Diagn. 2014;34(3):259–64. doi: 10.1002/pd.4302. [DOI] [PubMed] [Google Scholar]
- 17.Xu F, Li L, Schulz VP, et al. Cytogenomic mapping and bioinformatic mining reveal interacting brain expressed genes for intellectual disability. Mol Cytogenet. 2014;7(1):4. doi: 10.1186/1755-8166-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vlková B, Szemes T, Minárik G, et al. Advances in the research of fetal DNA in maternal plasma for noninvasive prenatal diagnostics. Med Sci Monit. 2010;16(4):RA85–91. [PubMed] [Google Scholar]
- 19.Gregg AR, Gross SJ, Best RG, et al. ACMG statement on noninvasive prenatal screening for fetal aneuploidy. Genet Med. 2013;15(5):395–98. doi: 10.1038/gim.2013.29. [DOI] [PubMed] [Google Scholar]
- 20.Jiang F, Ren J, Chen F, et al. Noninvasive Fetal Trisomy (NIFTY) test: an advanced noninvasive prenatal diagnosis methodology for fetal autosomal and sex chromosomal aneuploidies. BMC Med Genomics. 2012;5:57. doi: 10.1186/1755-8794-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bianchi DW, Platt LD, Goldberg JD, et al. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012;119(5):890–901. doi: 10.1097/AOG.0b013e31824fb482. [DOI] [PubMed] [Google Scholar]
- 22.Xu ZY, Xie JS, Meng JL, et al. Non-invasive prenatal diagnosis: A comparison of cell free fetal DNA (cffDNA) based screening and fetal nucleated red blood cell (fnRBC) initiated testing. NAJ Med Sci. 2013;6:194–99. [Google Scholar]