Abstract
Background
Recently, increasing research evidence indicates that miRNA plays important roles in oncogenesis of hepatocellular carcinoma (HCC). The objective of this study was to investigate the potential of plasma miRNAs as biomarkers for HCC determination.
Material/Methods
This trial included 4 phases: (i) miRNAs in tumor tissues were screened with a miRNA array for determining candidate miRNAs. (ii) Candidate miRNAs were measured by RT-qPCR in plasma of 10 HCC patients before and after surgery (7–10 days) for target miRNAs that displayed a pattern of postoperative decrease. (iii) Plasma levels of target miRNAs in 37 HCC patients, 29 cirrhosis patients, and 31 healthy controls were measured by RT-qPCR for determining potential biomarkers. (iv) The powers of biomarkers for differentiating HCC were validated and the correlations with clinicopathological variables of HCC patients were analyzed.
Results
miRNA array demonstrated an abnormal expression of 92 miRNAs in tumor tissues compared to adjacent non-tumor tissues. Of those molecules with an over-expressed level in tumor tissues and preoperative plasmas, a decrease in postoperative plasma was observed in miR-15b-5p, miR-338-5p, and miR-764. Plasma levels of these miRNAs in HCC patients were higher than in the other 2 groups (P<0.05). Receiver-operator characteristic (ROC) curve analyses suggested these plasma miRNAs could be useful biomarkers for determining HCC. miR-338-5p yielded an area under the ROC curve (AUC) of 0.799 (74.5% sensitivity and 82.8% specificity) and 0.909 (72.3% sensitivity and 99.68% specificity) to discriminate HCC patients from cirrhosis patients and healthy controls, respectively. The expression level of miR-338-5p was negatively correlated with the level of AFP (r=−0.306, P=0.036), and the expression level of miR-764 was positively correlated with the tumor size (r=0.371, P=0.01).
Conclusions
Circulating miR-15b-5p, miR-338-5p, and miR-764 may be biomarkers for diagnosis of HCC.
Keywords: Liver Neoplasms; MicroRNAs; Neoplastic Cells, Circulating
Background
Liver cancer is the second most common cause of death from cancer [1]. Hepatocellular carcinoma (HCC) accounts for a large part in all liver cancer cases, and is the fifth most common malignant tumor worldwide [2]. Infections with hepatitis B and C virus are the major risk factors of HCC [3]. Because of the difficulty of clinical diagnosis at the early stage, only 30–40% of cases can undergo curative resection [4]. HCC has an overall 5-year survival rate of 5–9%, and the rate can be raised to 69% in patients receiving curative resection [5]. In the present regimen, a-fetoprotein (AFP) has served as a biomarker to screen HCC. However, AFP only has a sensitivity of 39–56% and a specificity of 76–93%; approximately one-third HCC patients cannot be detected with AFP testing [2].
MicroRNAs (miRNAs) are a class of noncoding, endogenous, single-stranded RNAs with 19–22 nucleotides. They posttranscriptionally regulate gene expression by binding to the untranslated regions (UTRs) of target mRNA and exert their influences on biologic processes, such as the proliferation, differentiation, apoptosis, invasion and metastasis [6]. Over-expression of miR-125b inhibited endometrial cancer cell invasion [7]. Previous researches also indicated many miRNAs were involved in esophageal cancer [8]. Meta-analysis found high expression of miR-21 was associated with poor survival in patients with gastric cancer [9]. The miR-34b/c rs4938723 polymorphism may be associated with the risk of cancers, including nasopharyngeal cancer, osteosarcoma, and renal cell cancer [10]. Dysregulation of miR-15 and miR-16 was observed in chronic lymphocytic leukemia, and investigations have shown that miRNAs were associated with oncogenesis and development of various tumors [11]. miRNAs can exist stably in circulation, and are resistant to RNAase digestion. Moreover, serum miRNAs remained stable even when treated in low (pH=1) or high (pH=13) pH solution for 3 h [12]. Thus, circulating miRNAs have been explored as a biomarker, and some important findings have been reported. Yoshito et al. [13] found miR-21 had an area under the ROC curve (AUC) of 0.953, with a sensitivity of 87.3% and a specificity of 92% for determining HCC from healthy controls; the AUC was 0.773 (61.1% sensitivity and 83.3% specificity) for distinguishing HCC from chronic hepatitis. Liu et al. [5] showed that the combination of miR-15b and miR-130b for HCC detection yielded an AUC of 0.98 (98.2% sensitivity and 91.5% specificity); the sensitivity was 96.7% in a subgroup of HCC with low AFP (<20 ng/ml).
The aim of this study was to determine if plasma miRNAs can be biomarkers for HCC. The potential relationship between circulating miRNAs levels and existing clinicopathological features of HCC, such as the level of AFP, total bilirubin, transaminase, and tumor size, was analyzed.
Material and Methods
Study design and patient sample
This study included four phases
Phase I: Screening candidate miRNAs in HCC. Tumor and adjacent non-tumor tissues were sampled from 6 HCC patients receiving curative surgery, and submitted to miRNA array analysis. miRNAs with an up-regulated expression level were chosen as candidate miRNAs.
Phase II: Determining the target miRNAs. The plasma levels of candidate miRNAs before and after surgery (7–10 days) in 10 patients were detected by RT-qPCR, and those miRNAs that displayed a postoperative decease pattern were selected as the target miRNAs.
Phase III: Determining biomarkers. The plasma levels of target miRNAs in 37 HCC patients, 29 cirrhosis patients, and 31 healthy controls were measured and those miRNAs with up-regulated expressions in HCC patients were selected as biomarkers.
Phase IV: to validate the abilities of potential biomarkers for screening HCC from cirrhosis and healthy cohort, receiver-operator characteristic curves (ROC curve) were constructed and then the relationship between biomarkers and clinicopathologic parameters was validated.
All cases were from the First Affiliated Hospital of Chongqing Medical University. Cancer and adjacent non-tumor tissues from six HCC patients who received curative resection were sampled in June 2010, and were sent to Western Biotechnology, Inc. (Chongqing, China) for miRNA array analysis. The blood samples of 47 HCC patients, 29 cirrhosis patients, and 31 healthy cases were collected from January 2014 to June 2014. The diagnosis of HCC was confirmed by pathological examinations. The clinicopathological variables are summarized in Table 1. This study was approved ethically and scientifically by the local review board. Informed consent was obtained from all patients.
Table 1.
Demographic and clinicopathologic parameters of patients from different cohorts.
| Parameters | The number of different cohorts | P values | ||||
|---|---|---|---|---|---|---|
| Patients with HCC (n=47) | Patients with cirrhosis (n-29) | Healthy controls (n=31) | miR-15b-5p | miR-338-5p | miR-764 | |
| Sex | ||||||
| Male | 44 | 12 | 18 | |||
| Female | 3 | 17 | 13 | |||
| Age (years) | ||||||
| ≥60 | 7 | 5 | 0 | |||
| <60 | 41 | 24 | 31 | |||
| α-fetoprotein (ng/ml) | 0.595 | 0.039 | 0.102 | |||
| ≥200 | 19 | |||||
| <200 | 28 | |||||
| Alanine aminotransferase (U/l) | 0.312 | 0.822 | 0.875 | |||
| ≥40 | 31 | |||||
| <40 | 16 | |||||
| Aspartate aminotransferase (U/l) | 0.718 | 0.555 | 0.768 | |||
| ≥40 | 29 | |||||
| <40 | 18 | |||||
| Total bilirubin(μmol/L) | 0.644 | 0.426 | 0.54 | |||
| ≥17.1 | 20 | |||||
| <17.1 | 27 | |||||
| Child-Pugh grade | 0.989 | 0.7 | 0.184 | |||
| A | 44 | |||||
| B | 3 | |||||
| C | 0 | |||||
| Tumor size (cm) | 0.346 | 0.336 | 0.026 | |||
| ≥5 | 26 | |||||
| <5 | 21 | |||||
Plasma preparation
We collected 2 ml of peripheral venous blood in a tube containing EDTA, and centrifuged (2000 rpm, 4°C for 10 min) to isolate plasma. The plasma was transferred to a 1.5-ml Eppendorf tube, and centrifuged (3000 rpm, 4°C for 8 min) to precipitate cell debris. The supernatant was then transferred to another Eppendorf tube, and stored at −80°C.
miRNA array
Tumor and matched adjacent non-tumor tissues from HCC patients were analyzed by miRNA array analysis (Western Technology, Inc., Chongqing, China).
RNA extraction
Total RNA in plasma was extracted using TRIzol LS reagent (Invitrogen) according to the manufacturer’s protocols. RNA samples were dissolved in 20 μl of nuclease-free water, and the purity was determined by the ratio of OD260 to OD280 (Nanodrop ND-2000; Thermo Scientific, Worcester, MA, USA).
Determination of miRNA expression by quantitative RT-PCR
RT-qPCR was performed to determine the expression of miRNA using the ALL-in-One™ miRNA qRT-PCR Detection Kit (GeneCopoeia Inc, Ltd, Guangzhou, China) according to the instructions of the manufacturer. Primers of miRNAs were purchased from GeneCopoeia Inc, Ltd (Guangzhou, China). Because of the lack of universally acknowledged internal reference in circulating miRNA research, the expression levels of U6 RNA and miR-16 were detected by quantitative PCR for screening an appropriate internal control. As a result, U6 RNA could not be detected in almost half of plasma samples, while miR-16 was abundant in all plasma samples. Therefore, miR-16 was selected as the internal reference [13,14].
Statistics
Unsupervised hierarchical clustering and correlation analysis were performed to compare miRNA expression profile in tumors and matched adjacent non-tumor tissues from HCC patients. Data were analyzed using the statistics software SPSS 16.0 (SPSS, Inc, Chicago, IL, USA). The Mann-Whitney test was used to compare miRNA levels. ROC was constructed and AUC were calculated to estimate the diagnostic values of biomarkers, and Youden’s index was computed for determining the cutoff value. The clinicopathological variables were divided into subgroups according to the clinical criteria of each variable, and the Mann-Whitney test was used to validate the different expression levels among subgroups. Finally, correlations were analyzed using the Spearman correlation. The relative expression of miRNA was determined by the 2−ΔΔct value [15]. The critical value was set at p<0.05.
Results
miRNA expression profile in tumor and non-tumor tissues
miRNA array showed that the expression level was up-regulated in 40 miRNAs (> 2-fold) and down-regulated in 52 miRNAs (<0.5-fold) in the tumor tissues (partly summarized in Table 2). miR-15b-5p, miR-25-3p, miR-338-5p, and miR-764 were significantly up-regulated and were therefore chosen as candidate miRNAs.
Table 2.
The expression levels of miRNAs in cancer (C) and adjacent non-tumor tissues (N).
| miRNA | C/N | C | N |
|---|---|---|---|
| Up Regulated miRNAs | |||
| miR-15b-5p | 2.541215446 | 451.5 | 188.5 |
| miR-25 | 2.024267353 | 155.5 | 81.5 |
| miR-338-5p | 13.79235537 | 565 | 41 |
| miR-764 | 4.407024793 | 81 | 19.5 |
| hsa-miR-1204 | 2.690657749 | 123 | 48.5 |
| hsa-miR-886-3p | 4.003237315 | 324.5 | 86 |
| hsa-miR-602 | 2.491386045 | 236 | 100.5 |
| hsa-miR-186* | 4.751703446 | 210.5 | 47 |
| hsa-miR-452 | 2.002692915 | 84 | 44.5 |
| hsa-miR-222 | 3.691120507 | 374 | 107.5 |
| Down Regulated miRNAs | |||
| hsa-miR-130a | 0.340420061 | 111.5 | 347.5 |
| hsa-miR-19b | 0.477427686 | 216 | 480 |
| hsa-miR-1201 | 0.393327958 | 38 | 102.5 |
| hsa-miR-185* | 0.225181312 | 26 | 122.5 |
| hsa-miR-1254 | 0.127825351 | 10 | 83 |
| hsa-miR-101 | 0.22825651 | 645 | 2998 |
| hsa-miR-122* | 0.125945081 | 184 | 1550 |
| hsa-miR-1260 | 0.381103061 | 1844 | 5133.5 |
| hsa-miR-33b | 0.368952307 | 108.5 | 312 |
| hsa-miR-26b | 0.436716798 | 708 | 1720 |
Circulating miR-15b-5p in HCC cases has been researched and its expression level was reduced after surgery [5]. miR-15b-5p was chosen for use in this study as an experimental control. Our literature search found no studies on circulating miR-338-5p and miR-764 in HCC; their expression levels in cancer were higher than all other miRNAs compared to non-tumor tissues, and they may be useful for as novel screening biomarkers. miR-25-3p was randomly selected for use in the next stage of the study.
Validation of the candidate miRNAs in the preoperative and postoperative plasmas
As illustrated in Table 3, the level of miR-15b-5p, miR-338-5p, and miR-764 decreased post-operatively (P=0.005). The data suggest that those molecules may be primarily derived from tumors. Thus, miR-15b-5p, miR-338-5p, and miR-764 were chosen as the target miRNAs. The expression levels of miR-25-3p were high in some postoperative plasma (P=0.028) and could not be detected in several preoperative plasma samples; therefore, miR-25-3p was excluded.
Table 3.
The expression levels of plasma miRNAs in HCC before and after surgery(n=10).
| miRNA (M(Q1, Q3)) | miR-15b-5p | miR-25-3p | miR-338-5p | miR-764 |
|---|---|---|---|---|
| Preoperation | 3.60 (2.30, 102.71) | 2.57 (0.70, 7.43) | 6.54 (2.46, 17.79) | 2.80 (1.80, 50.18) |
| Postoperation | 1.00 (0.70, 1.00) | 0.84 (0.27, 1.00) | 1.00 (0.45, 1.00) | 1.00 (0.11, 1.00) |
| P value | 0.005 | 0.028 | 0.005 | 0.005 |
The expression levels of target miRNAs in 3 different cohorts
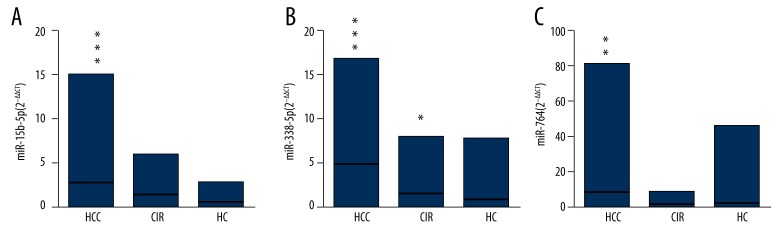
To observe the value of plasma target miRNAs as the biomarkers, plasma levels of target miRNAs in 37 HCC patients, 29 cirrhosis patients, and 31 healthy cases were measured by RT-qPCR. In all, we obtained 107 samples, including 10 preoperative plasma samples. As shown in Table 4 and Figure 1, the plasma expression levels of miR-15b-5p, miR-338-5p, and miR-764 in HCC were higher than that in the other 2 cohorts (P<0.05). The plasma levels of miR-15b-5p and miR-338-5p in CIR were higher than in the HC cohort (P<0.0001, P=0.006), but a similar situation was not found in miR-764 (P=0.667). There were outliers for these 3 miRNAs.
Table 4.
The plasma expression levels of target miRNAs in HCC, cirrhosis (CIR) and healthy controls (HC).
| miRNA P50 (P25, P75) |
HCC (n=47) | CIR (n=29) | HC (n=31) | P value | ||
|---|---|---|---|---|---|---|
| HCC vs. CIR | HCC vs. HC | CIR vs. HC | ||||
| miR-15b-5p | 1.44607 (1.00000, 3.21384) | 1.00000 (1.00000, 1.51870) | 0.41094 (0.18895,0.98531) | 0.0239 | <.0001 | <0.001 |
| miR-338-5p | 4.16015 (1.56735, 8.79197) | 1.00000 (0.77172, 1.51870) | 0.79597 (0.09996,1.00000) | <.0001 | <.0001 | 0.006 |
| miR-764 | 2.49329 (1.03630, 9.01975) | 1.00000 (0.55753, 1.15658) | 1.00000 (0.25045,1.19243) | 0.0002 | 0.0002 | 0.667 |
Figure 1.
The plasma levels of miR-15b-5p (A), miR-338-5p (B), and miR-764 (C) in HCC, CIR, and HC. (A) The different plasma expression levels of miR-15b-5p in 3 independent cohorts; (B) The different plasma expression levels of miR-338-5p in 3 independent cohorts; (C) The different plasma expression levels of miR-764 in 3 independent cohorts.“*” for outliers.
Diagnostic value analysis
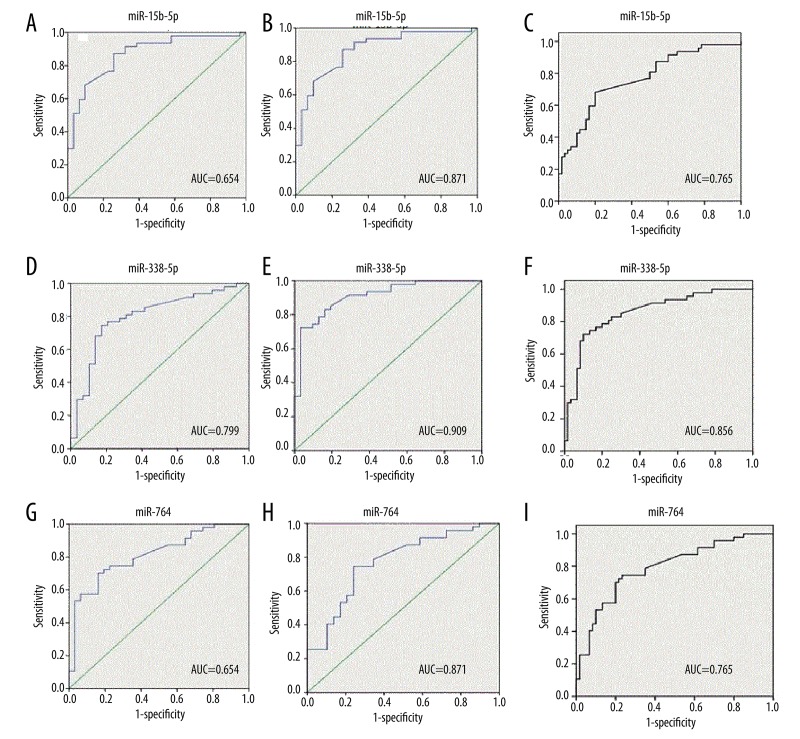
The differentiating power of plasma miR-15b-5p, miR-338-5p, and miR-764 expressed in patients with HCC, cirrhosis, and healthy controls was validated. ROC was constructed using 3 models: HCC vs. CIR, HCC vs. HC, and HCC vs. CIR and HC. AUC and the cutoff values (using the highest Youden’s index) were computed. Consequently, we found these 3 miRNAs could differentiate HCC from CIR and HC. Especially, the plasma level of miR-338-5p was helpful for differentiating HCC from CIR and HC, with AUC of 0.799 (74.5% sensitivity, 82.8% specificity), and 0.909 (72.3% sensitivity, 99.68%), respectively. Furthermore, at the cutoff value of 1.784, miR-338-5p yielded an AUC of 0.856 (72.3% sensitivity, 90% specificity) for differentiating HCC samples from a total of 107 samples. Similarly, we found miR-764 and miR-15b-5p yielded an AUC of 0.791(74.5% sensitivity, 77% specificity), 0.765 (68.1% sensitivity, 80% specificity), respectively, for differentiating HCC samples from all samples. Moreover, differentiating power of the 3 plasma miRNAs was better in the HCC vs. HC model than in the HCC vs. CIR model. Details of ROC analysis are summarized in Table 5 and Figure 2.
Table 5.
The differentiating powers of miR-15b-5p, miR-338-5p and miR-764.
| miRNA | HCC vs. CIR | HCC vs. HC | HCC vs. CIR and CH |
|---|---|---|---|
| Cutoff value, AUC (sensitivity, specificity) | |||
| miR-15b-5p | 1.007, 0.654 (68.1%, 79%) | 0.807, 0.871 (87.2%, 74.2%) | 1.007, 0.765 (68.1%, 80%) |
| miR-338-5p | 1.743, 0.799 (74.5%, 82.8%) | 1.784, 0.909 (72.3%, 99.68%) | 1.784, 0.856 (72.3%, 90%) |
| miR-764 | 1.181, 0.770 (74.5%, 75.9%) | 1.465, 0.811 (70.2%, 83.9%) | 1.199, 0.791 (74.5%, 77%) |
Figure 2.
The performance of miR-15b-5p, miR-338-5p, and miR-764 for detecting HCC from different cohorts. (A–C): ROC curve analysis using plasma miR-15b-5p for discriminating HCC from CIR, HC, CIR, and HC; (D–F): ROC curve analysis using plasma miR-338-5p for discriminating HCC from CIR, HC, CIR, and HC; (G–I): ROC curve analysis using plasma miR-764 for discriminating HCC from CIR, HC, CIR, and HC.
Correlation between miRNAs and clinicopathological variables
Further analysis (Table 6) revealed that miR-15b-5p, miR-338-5p and miR-764 were useful biomarkers for determining HCC cases in patients with AFP<200 ng/ml. At the cutoff value of 1.007 (for miR-15b-5p), 1.784 (for miR-338-5p), and 1.199 (for miR-764) the sensitivities were 71.4%, 78.6% and 82.1%, respectively. When defining the cutoff value of AFP at 200 ng/ml, the sensitivity was 40.4% for determining HCC cases from all patients with HCC.
Table 6.
The differentiating power of plasma miR-15b-5p, miR-338-5p and miR-764 in HCC patients with AFP<200 ng/ml.
| miRNA | Cutoff value | Sensitivity |
|---|---|---|
| miR-15b-5p | 1.007 | 71.4% |
| miR-338-5p | 1.784 | 78.6% |
| miR-764 | 1.199 | 82.1% |
Next, we verified the correlation between the 3 target miRNAs and clinicopathological variables (Table 1). The level of miR-764 was significantly upregulated in HCC patients with a tumor size of >5 cm (P=0.026). The expression level of miR-338-5p was higher in plasma of HCC patients with AFP<200 ng/ml than ones with high AFP levels (P=0.039). However, the expression levels of these miRNAs were uncorrelated with the levels of total bilirubin, transaminase, and HBsAg positivity. The correlation coefficient between plasma miR-338-5p level and the level of AFP, plasma miR-764 level, and the tumor size was −0.306 (r=−0.306) (P=0.036), and 0.371(r=0.371) (P=0.01), respectively. Because of the number of cases in HCC subgroups divided according to agent, age, and Child-Pugh classification was obviously different, correlation analysis was not taken into account.
Discussion
Our study was divided into 4 phases according to international classical cases of screening potential biomarkers. Interestingly, we found: (i) the plasma levels of miR-764 in the HCC cohort were higher than that in the other 2 cohorts, while no difference occurred between cirrhosis and healthy cohorts. This phenomenon indicated that miR-764 did not affect the transformation from healthy liver to cirrhosis; instead, it played a role in the transformation from cirrhosis to HCC. Plasma levels of miR-15b-5p and miR-338-5p increased gradually from healthy controls to HCC patients, which indicated that they played roles in the malignant transformation of liver cells. (ii): miR-764 yielded the highest sensitivity for determining HCC cases with AFP<200 ng/ml. This might be useful for determining HCC in early stages when the level of AFP is lower than the diagnostic standard.
Numerous studies have revealed that circulating miRNAs can be potential markers for HCC determination. Liu et al. [5] indicated that the combination of miR-15b and miR-130 was a useful biomarker for screening HCC. Li et al. [16] found circulating miR-18b could be a potential HBV-related HCC marker. For searching more convenient HCC markers, Moemen et al. [17] detected the expression levels of miRNAs in urine samples. MiR-618 and miR-650 were selected out, but further analysis revealed the predictive values were not improved compared to the traditional AFP level-based detection method. In addition to the miRNAs mentioned above, miR-221, miR-885-5p, and miR-122a have been identified to be potential biomarkers for HCC determination [18–20]. We found no published research about circulating miR-338-5p and miR-764 in HCC. Moreover, studies on miR-15b in HCC are limited and the role of miR-15b remains largely unknown. Dai et al. [21] reported that miR-15b promoted HBV replication by targeting hepatocyte nuclear factor 1α. Wu et al. [22] indicated the expression of miR-15b can be repressed by HBX, and then enhanced hepatocellular carcinoma proliferation through fucosyltransferase2-induced Globo H expression.
A growing body of evidence shows that circulating miRNAs are probable markers of various other tumors, including thyroid cancer [23], breast cancer [24], testicular germ cell cancer [25], pancreatic cancer [26], lung cancer [27], and gastric cancer [28].
The main limitation of this study was that the sample size was small. The present findings therefore should be validated in trials with more cases. Because of only 3 female cases were included into our study, the difference between males and females should also be explored in subsequent trials.
Conclusions
Our study found that miR-338-5p and miR-764 hold promise as valuable biomarkers for HCC detection, even in HCC patients with low AFP levels. We also validated the differentiating power of miR-15b-5p. In summary, the present study results suggest that miR-15b-5p, miR-338-5p, and miR-764 might be used as biomarkers for HCC.
Footnotes
Source of support: Departmental sources
References
- 1.Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qi J, Wang J, Katayama H, et al. Circulating microRNAs (cmiRNAs) as novel potential biomarkers for hepatocellular carcinoma. Neoplasma. 2013;60(2):135–42. doi: 10.4149/neo_2013_018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Martel C, Ferlay J, Franceschi S, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13(6):607–15. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 4.Zhou J, Yu L, Gao X, et al. Plasma microRNA panel to diagnose hepatitis B virus – related hepatocellular carcinoma. Clinical Oncology. 2011;29(36):4781–88. doi: 10.1200/JCO.2011.38.2697. [DOI] [PubMed] [Google Scholar]
- 5.Liu AM, Yao TJ, Wang W, et al. Circulating miR-15b and miR-130b in serum as potential markers for detecting. hepatocellular carcinoma: a retrospective cohort study. BMJ Open. 2012;2(2):e000825. doi: 10.1136/bmjopen-2012-000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Z, Zhang X, Wang G, et al. Role of MicroRNAs in Hepatocellular Carcinoma. Hepat Mon. 2014;14(8):e18672. doi: 10.5812/hepatmon.18672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shang C, Lu YM, Meng LR. MicroRNA-125b down-regulation mediates endometrial cancer invasion by targeting ERBB2. Med Sci Monit. 2012;18(4):BR149–55. doi: 10.12659/MSM.882617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang Y, Fang D, Hu J. MicroRNA and its roles in esophageal cancer. Med Sci Monit. 2012;18(3):RA22–30. doi: 10.12659/MSM.882509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Cai Q, Jiang Z, et al. Prognostic Role of MicroRNA-21 in Gastric Cancer: A Meta-Analysis. Med Sci Monit. 2014;20:1668–74. doi: 10.12659/MSM.892096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Lu X, Fang Y, et al. Association between miR34b/c polymorphism rs4938723 and cancer risk: A meta-analysis of 11 studies including 6169 cases and 6337 controls. Med Sci Monit. 2014;20:1977–82. doi: 10.12659/MSM.892350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014 doi: 10.1016/j.molmed.2014.06.005. pii: S1471-4914(14)00101-4. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 13.Tomimaru Y, Eguchi H, Nagano H, et al. Circulating microRNA-21 as a novel biomarker for: hepatocellular carcinoma. J Hepatol. 2012;56(1):167–75. doi: 10.1016/j.jhep.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 14.Qi P, Cheng SQ, Wang H, et al. Serum MicroRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS One. 2011;6(12):e28486. doi: 10.1371/journal.pone.0028486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karakatsanis A, Papaconstantinou I, Gazouli M, et al. Expression of microRNAs, miR-21, miR-31,miR-122, miR-145, miR-146a, miR-200c,miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog. 2013;52(4):297–303. doi: 10.1002/mc.21864. [DOI] [PubMed] [Google Scholar]
- 16.Li L, Guo Z, Wang J, et al. Serum miR-18a: a potential marker for hepatitis B virus-related hepatocellular carcinoma screening. Dig Dis Sci. 2012;57(11):2910–16. doi: 10.1007/s10620-012-2317-y. [DOI] [PubMed] [Google Scholar]
- 17.Abdalla MA, Haj-Ahmad Y. Promising Candidate Urinary MicroRNA Biomarkers for the Early Detection of Hepatocellular Carcinoma amongHigh-Risk Hepatitis C Virus Egyptian Patients. J Cancer. 2012;3:19–31. doi: 10.7150/jca.3.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Wang Y, Yu W, et al. Expression of serum miR-221 in human hepatocellular carcinoma and its prognostic significance. Biochem Biophys Res Commun. 2011;406(1):70–73. doi: 10.1016/j.bbrc.2011.01.111. [DOI] [PubMed] [Google Scholar]
- 19.Gui J, Tian Y, Wen X, et al. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin Sci (Lond) 2011;120(5):183–93. doi: 10.1042/CS20100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo J, Chen M, Huang H, et al. Circulating microRNA-122a as a diagnostic marker for hepatocellular carcinoma. Onco Targets Ther. 2013;6:577–83. doi: 10.2147/OTT.S44215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai X, Zhang W, Zhang H, et al. Modulation of HBV replication by microRNA-15b through targeting hepatocyte nuclear factor 1α. Nucleic Acids Res. 2014;42(10):6578–90. doi: 10.1093/nar/gku260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu CS, Yen CJ, Chou RH, et al. Downregulation of microRNA-15b by hepatitis B virus X enhances hepatocellular carcinoma proliferation via fucosyltransferase 2-induced Globo H expression. Int J Cancer. 2014;134(7):1638–47. doi: 10.1002/ijc.28501. [DOI] [PubMed] [Google Scholar]
- 23.Cantara S, Pilli T, Sebastiani G, et al. Circulating miRNA95 and miRNA190 are sensitive markers for the differential diagnosis of thyroid nodules in a Caucasian population. J Clin Endocrinol Metab. 2014;24:jc20141923. doi: 10.1210/jc.2014-1923. [DOI] [PubMed] [Google Scholar]
- 24.Shen J, Hu Q, Schrauder M, et al. Circulating miR-148b and miR-133a as biomarkers for breast cancer detection. Oncotarget. 2014;5(14):5284–94. doi: 10.18632/oncotarget.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Syring I, Bartels J, Holdenrieder S, et al. Circulating serum microRNA (miR-367-3p, miR-371a-3p, miR-372-3p, miR-373-3p) as biomarkers for patients with testicular germ cell cancers. J Urol. 2014 doi: 10.1016/j.juro.2014.07.010. pii: S0022-5347(14)03957-3. [DOI] [PubMed] [Google Scholar]
- 26.Slater EP, Strauch K, Rospleszcz S, et al. MicroRNA-196a and -196b as potential biomarkers for the early detection of familial pancreatic cancer. Transl Oncol. 2014 doi: 10.1016/j.tranon.2014.05.007. pii: S1936-5233(14)00062-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ulivi P, Zoli W. miRNAs as non-invasive biomarkers for lung cancer diagnosis. Molecules. 2014;19(6):8220–37. doi: 10.3390/molecules19068220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsujiura M, Komatsu S, Ichikawa D, et al. Circulating miR-18a in plasma contributes to cancer detection and monitoring in patients with gastric cancer. Gastric Cancer. 2014 doi: 10.1007/s10120-014-0363-1. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]