Summary
Multiple studies have consistently established that miR (microRNA)-210 induction is a feature of the hypoxic response in both normal and transformed cells. Here, we discuss the emerging biochemical functions of this miRNA and anticipate potential clinical applications. miR-210 is a robust target of hypoxia-inducible factor, and its overexpression has been detected in a variety of cardiovascular diseases and solid tumors. High levels of miR-210 have been linked to an in vivo hypoxic signature and associated with adverse prognosis in cancer patients. A wide spectrum of miR-210 targets have been identified, with roles in mitochondrial metabolism, angiogenesis, DNA repair, and cell survival. Such targets may broadly affect the evolution of tumors and other pathological settings, such as ischemic disorders. Harnessing the knowledge of miR-210’s actions may lead to novel diagnostic and therapeutic approaches.
Keywords: hypoxia, microRNA, cancer, ischemia, miR-210, mitochondria, apoptosis, metabolism
HYPOXIA-REGULATED NONCODING TRANSCRIPTS
Hypoxia is a hallmark of the neoplastic microenvironment and a well-documented source of therapeutic failure in clinical oncology (1). Low oxygen tension also plays a central role in the pathogenesis of major ischemic disorders, such as myocardial infarction, stroke, and peripheral artery disease (2); therefore, a deeper understanding of cellular adaptation to oxygen deprivation has broad and profound implications for translational medicine. Hypoxia-inducible factors (HIF) represent core components of the hypoxia-sensing machinery and orchestrate a regulatory transcriptional program comprising of hundreds of protein-encoding genes. Although historically gene induction by low oxygen, and in particular by HIF, has dominated hypoxia research, more recently the study of gene repression promoted by low oxygen tension has received increased attention (3, 4). Since 2007 a multitude of reports demonstrated that specific microRNAs (miRNAs) are involved in the hypoxic response and contribute to the repression of biologically important genes by low oxygen tension (5–11).
miRNAs act as regulators of most physiological and pathological cellular processes. In their mature active form, 19- to 24-ribonucleotides long, miRNAs are transferred to the RNA-induced silencing complex (RISC), followed by base pairing to partially complementary sites, primarily within the 3′ untranslated regions (UTRs) of target genes. This generally leads to inhibition of target gene expression, by a combination of translational blockade and/or mRNA degradation. It is currently thought that thousands of classic (protein-encoding) genes are regulated via such mechanisms (12). By virtue of interfering with multiple transcripts, often hundreds, miRNAs have the potential to regulate virtually all cellular mechanisms, such as differentiation, proliferation, death, and metabolism (13, 14).
miR-210’s ROLE IN CANCER
miR-210 is the only miRNA consistently upregulated in all published studies, in both normal and transformed hypoxic cells (5, 8–11, 15). It is also generally recognized as a robust HIF target (10, 16, 17), and its level may be a reflection of HIF activity even under circumstances of normoxic activation of this transcription factor. For example, miR-210 is particularly overexpressed in renal clear cell carcinomas (RCCs) (18), which express abnormally high levels of HIF, because of the genetic inactivation of the von Hippel-Lindau tumor suppressor, the well-recognized E3-ligase for HIF (19). Aside from the particular case of RCC, miR-210 is upregulated in most solid tumors and negatively affects the clinical outcome (8, 9, 20–24). Consistently, at least in breast cancer, miR-210 levels are correlated with a genetic signature of hypoxia, suggesting that tumor overexpression of this miRNA is the direct consequence of decreased oxygen tension in the microenvironment.
On the basis of the newly acquired knowledge of miR-210 targets and biochemical functions, we can begin to assess its involvement in human disease.
Mitochondrial Metabolism and Oxidative Stress
Following exposure to hypoxia, cell metabolism shifts from mitochondrial oxidative phosphorylation to glycolysis (often termed the “Pasteur effect”). This effect is generally thought to be a consequence of HIF activation, which in turn induces most glycolytic enzymes, pyruvate dehydrogenase kinase, and represses mitochondrial biogenesis (25). Very recent data from several groups have demonstrated that miR-210 contributes to this metabolic shift by downregulating several steps of mitochondrial metabolism and, in particular, the electron transport chain (ETC) complexes. Indeed, miR-210 directly represses the expression of iron-sulfur (Fe-S) cluster scaffold proteins, ISCU1 and ISCU2 (26–29). ISCU1/2 facilitates the assembly of Fe-S clusters that are incorporated into enzymes involved in energy production, including aconitase, which catalyses one step of the tricarboxylic acid cycle (TCA), as well as mitochondrial respiratory complexes I, II, and III (30). Other relevant mitochondrial miR-210 targets that have been reported are NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4 (NDUFA4) (9), a subunit of complex I, and succinate dehydrogenase complex, subunit D (SDHD), a subunit of complex II (31). An additional intriguing target is glycerol-3-phosphate dehydrogenase 1-like (GPD1-L) (27), which may be involved in the Pasteur effect by modulating NAD+/NADH ratios (32). Although the biochemical function of GPD1L remains to be elucidated, it is highly homologous to glycerol-3-phosphate dehydrogenase (GPD), the catalyst of the glycerol phosphate shuttle, which transfers electrons from cytoplasmic NADH to the mitochondria electron transport chain. Thus, forced expression of miR-210 in normoxia represses mitochondrial respiration and enhances glycolysis, which leads to decreased ATP; therefore, it is not surprising that miR-210 presence is not beneficial when oxygen supply is optimal. In contrast, upon hypoxia, the repression of the electron transport chain decreases the mismatch with reduced oxygen tension increasing ATP levels (26), minimizing the impact of hypoxia on cell energy production. It is noteworthy that tumors largely rely on glycolysis even when the oxygen supply is normal (Warburg effect). In RCCs, miR-210 is high and the oxygen supply is also high compared with other solid tumors, as these are typically well-vascularized tumors; therefore, miR-210 may be involved in a glycolytic shift in such tumors. Whether miR-210 is more generally involved in the Warburg effect is currently unclear.
The repressive effect of miR-210 on ETC also impacts on mitochondrial ROS production, an expected consequence of electron leakage. Indeed, miR-210 expression increases oxidative stress in normoxic conditions, and this is, at least in part, mediated by ISCU (26, 28). However, conflicting results were reported in hypoxia. Favaro et al. (28), using cancer cell lines, described hypoxic induction of ROS and alleviation with anti-miR-210. On the other hand, Chan et al. (26), using normal endothelial cells, could not measure any significant change in ROS production after exposure to hypoxia, which increased when miR-210 was blocked. The reason for this discrepancy warrants additional investigation but may be a reflection of the underlying differences between normal versus cancer cells employed by the two studies.
Puissegur et al. (31) provided the first evidence that high miR-210 also contributes to maintenance of high HIF during hypoxia; therefore, miR-210 and HIF form a feed-forward loop. This mechanism may be dependent on the suppression of ISCU, SDHD, NDUFA4, and the resulting generation of ROS and TCA metabolites, such as succinate.
Collectively, these data demonstrate that miR-210 coordinates a metabolic shift in tumors toward a high glycolytic phenotype, which has been long thought to benefit tumor growth.
Tumors with high miR-210 may be particularly sensitive to antiglycolytic agents. In vitro, the first evidence was provided by Chen et al. (29). Cells overexpressing miR-210 were significantly more susceptible to killing by 3-bromo-pyruvate, an inhibitor of the glycolytic pathway. Molecules of this class, such as 2-deoxyglucose or dichloroacetate, have been considered as promising therapeutic agents; however, they are yet to fulfill their promise in clinical trials. Identification of molecular signatures of tumors that exhibit high sensitivity to such agents may help identify patients that could benefit from them, and a full understanding of miR-210’s role in this process may be a step in this direction.
Angiogenesis
Given the role of hypoxia in angiogenesis induction, it is not surprising that hypoxia-induced miR-210 seems to play a role in this event. We found that miR-210 overexpression in normoxic endothelial cells stimulated the formation of capillary-like structures as well as VEGF-driven cell migration. The direct involvement of this miRNA in these processes was demonstrated by the inhibition of both capillary-like structures formation and VEGF-induced chemotaxis upon miR-210 blockade. One critical direct target of miR-210 proangiogenic function is Ephrin-A3 (EFNA3) (10, 27). Override of its downmodulation either by miR-210 expression or by hypoxia prevents both tubulogenesis and chemotaxis stimulation. Further studies are ongoing to investigate the role of miR-210 in the stimulation of angiogenesis in vivo.
DNA Repair
Recent data indicate that miR-210 targets RAD52 (16, 27), which assists the loading of RAD51 onto DNA to form nucleoprotein filaments involved in homology-dependent repair (HDR) (33, 34). Previous work established that HDR activity is dramatically decreased in hypoxic cells (4, 35), and miR-210-mediated suppression of RAD52 may provide an additional mechanism by which HDR is suppressed in hypoxia. Whether miR-210’s action on RAD52 is enough to measurably affect DNA damage is currently unknown.
Regulation of Apoptosis
Compromised survival of cells devoid of miR-210 in severe hypoxia has been documented (10, 15, 28). However, the normoxic expression of miR-210 appears to be detrimental for cell viability, at least in some cell types. Mechanistically, recent evidence shows that miR-210 directly antagonizes an apoptotic component, CASP8AP2. The relevance of this for cancer is currently unknown, as the investigation was performed in the context of mesenchymal stem cell survival following ischemic preconditioning (36).
Cell Cycle
In contrast to other solid tumors, miR-210 is frequently underexpressed in ovarian cancers, which potentially leads to increased expression of E2F transcription factor 3 (E2F-3) (9). Likewise, miR-210 is also expressed at low levels in esophageal squamous cell carcinomas, derepressing fibroblast growth factor receptor-like 1 that in turn accelerates cell cycle progression (37).
However, one cannot generally state that miR-210 induction in hypoxia negatively regulates cell cycle progression. miR-210 overexpression in cancer cell lines bypassed hypoxia-induced cell cycle arrest and partially reversed the hypoxic gene expression signature (38). In these particular cells, miR-210 activates the myc pathway, via downregulation of the c-Myc antagonist MNT, and loss of MYC abolished miR-210-mediated override of hypoxia-induced cell cycle arrest. Therefore, the net impact of miR-210 on cell proliferation seems to be context dependent.
miR-210’s ROLE IN CEREBROVASCULAR AND CARDIOVASCULAR DISEASES
The significance of HIF regulation of miR-210 is not limited to cancer, as hypoxia is a central component of other clinical conditions, which have major impacts on morbidity and mortality, including cardiac and peripheral ischemia as well as cerebrovascular diseases (2).
Although further work is needed to substantiate a mechanistic role in such disorders, miR-210 was reported to be upregulated in animal models of cardiac hypertrophy, heart failure (39, 40), transient focal ischemia in the brain (41), hindlimb ischemia (11, 41), and in ischemic wounds (42). miR-210 is also induced in the corresponding clinical settings: acute myocardial infarction (43), atherosclerosis obliterans (44), and preeclampsia (45–47). Consistently, select miR-210 targets, and in particular GPD1L and ISCU, exhibit a significant role in cardiovascular biology. GPD1L mutations are associated with a subset of sudden infant cardiac death patients and with certain cases of Brugada syndrome, a disease that induces syncope, ventricular arrhythmias, and sudden death (48, 49). ISCU mutations, on the other hand, are associated with hereditary lactic acidosis, characterized by myopathy and exercise intolerance (50).
In the brain, miR-210 has been linked to the regulation of EFNA3 and neuronal pentraxin 1 (NP1) in the central nervous system (11). As NP1 stimulates ischemic cell death, its experimentally confirmed downregulation by miR-210 during hypoxia and ischemia may be part of a survival mechanism in stroke. Contrary to the expectation of miRNA-mediated repression, EFNA3 was expressed at high levels in postischemic mouse hippocampus, exhibiting a similar trend to miR-210 (11). Although a series of possible explanations may be hypothesized, it is worth noting that EFNA3 transcription is induced by hypoxia (10). Thus, EFNA3 protein levels are determined by the balance of mRNA induction and miRNA repression, and hence, the outcome may change in different pathological contexts.
Preeclampsia is another example of a disorder with a tissue hypoxic component (45). Consistently, miR-210 was found upregulated in these placentas when compared with those of normal pregnant women. (47). However, at this stage, the association is mostly correlative, and a mechanistic involvement of miR-210 in this condition has not been determined (46).
miR-210 experimentally validated targets and their potential regulatory functions are summarized in Fig. 1.
Figure 1.
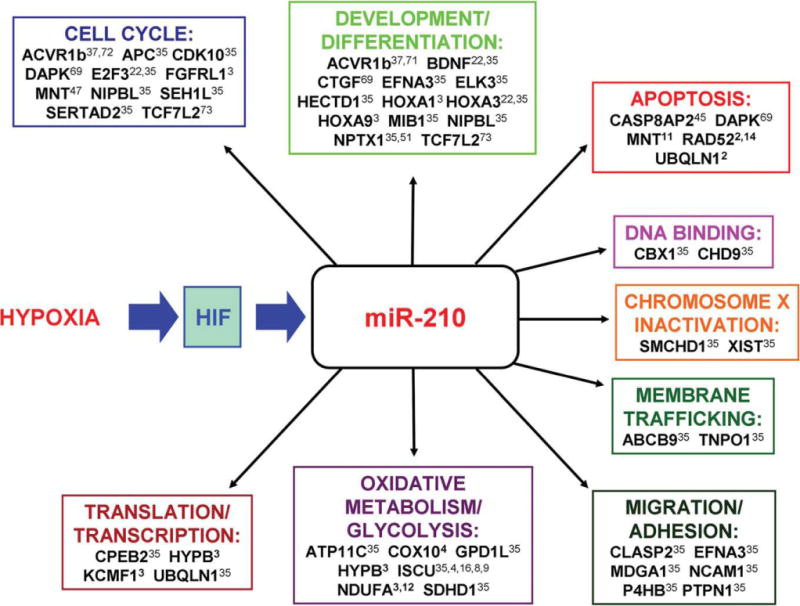
Hypoxia regulates miR-210, which in turn mediates the expression of factors that are implicated in various physiopathological pathways.
IS THE END IN SIGHT FOR miR-210 TARGETS?
A comprehensive understanding of the biological and biochemical roles of miR-210 in hypoxia requires a complete knowledge of its targets; however, the search for targets remains the bottleneck of miRNA research. A continuously increasing number of target identification algorithms have been developed, such as PicTar, TargetScan, DianaLab, micro-RNA.org, and MicroCosm (51).
For miR-210, such an in silico search reveals a dauntingly complex spectrum of candidate targets, including genes involved in proliferation, DNA repair, chromatin remodeling, metabolism, and cell migration (8–11, 16, 26, 29, 52, 53). Most of these programs search for complementarity between the miRNA “seed” sequences and the 3′ UTR of all known genes, and the resulting set of candidates generally contains hundreds of genes. Moreover, when the search is extended to the 5′ UTR and the coding region, the number is even higher. On the other hand, it is becoming increasingly apparent that “seed” binding is not necessarily sufficient, as other features of the surrounding sequences can affect binding efficacy. No specific algorithm is generally acknowledged as the most sensitive or accurate, and to further complicate matters, the lists of candidates generated by these different algorithms usually exhibit very limited overlap. Finally, relying on one particular program may well lead to missing most targets. For example, PicTar and TargetScan predict relatively few targets for human miR-210, and most of the experimentally validated targets are not predicted by any of these programs. Such difficulties prompted the use of integrated bioinformatic and experimental approaches to identify relevant miR-210 targets. One widely used approach takes advantage of the ability of many miRNAs to induce target mRNA destabilization. Gene expression is measured in the whole transcriptome or for a subset of genes in cells where a specific miRNA is either overexpressed or knocked down. Then, only those transcripts modulated in the expected direction are further analyzed with target prediction software programs to distinguish between direct and indirect targets. For instance, both Zhang et al. (38) and Puissegur et al. (31) began their search by identifying transcripts that were downmodulated upon forced miR-210 expression in colorectal and lung adenocarcinoma cancer cell lines, respectively. The downregulated genes that also contained a predicted miR-210-binding site were analyzed further to confirm the identity of MNT (38), NDUFA4, and SDHD (31) as bona fide targets.
Alternative approaches were based on direct analysis of the mRNAs associated with the RISC complex. Our group used a combination of proteomic and transcriptomic techniques to identify miR-210-modulated genes (27). Specifically, proteomic profiling of human primary endothelial cells identified at least 10 downmodulated proteins in miR-210-expressing cells. In additional experiments, 52 transcripts were found to be both induced upon miR-210 knockdown and downmodulated by miR-210 expression. Very few, if any of these genes were found by target identification algorithms. However, a low stringency search revealed that these genes were enriched in miR-210 seed-complementary sequences. Analysis of the mRNAs associated with the RISC loaded with miR-210 and purified by immunoprecipitation revealed that the complex was significantly enriched for 16 candidate targets. Intriguingly, the seed matches for nine of these genes were localized in the 5′ UTR or in the coding region. Furthermore, a noncoding RNA involved in the epigenetic inactivation of the X chromosome (Xist) was identified in the RISC immunoprecipitate, revealing a previously unsuspected layer of interaction between noncoding RNAs. Finally, 15 targets predicted by Pictar and TargetScan were also validated in our study, underlining the usefulness of such programs, especially in conjunction with additional experimental approaches.
Using a similar strategy, Huang et al. (54) compared RISC immunoprecipitates of normoxic and hypoxic breast cancer cells. This resulted in the identification of more than 200 mRNAs that were enriched following cell exposure to hypoxia. In silico search revealed that 50 of these were recognized as direct miR-210 targets by prediction software, and a subset of these genes was further validated by independent techniques.
As expected, because of the clear differences in the above two experimental approaches, there are very few targets in common. However, this may not be surprising because the number of identified targets is still limited, whereas computational predictions estimate that each miRNA can target, on an average, more than 200 genes.
Quo Vadis, miR-210?
The question remains whether miR-210 will ultimately have an exclusive impact on diagnosis or it may also emerge as a therapeutic target. Recent development of anti-miRNA agents such as locked nucleic acid probes (LNAs) represents significant steps for therapeutic targeting of onco-miRNAs (55, 56). It is conceivable that inactivation of a miRNA involved in hypoxic adaptation would be a viable strategy to target a tumor compartment that poses significant therapeutic challenges. From a diagnostic standpoint, it was recently shown that miR-210 is increased in the serum from patients with diffuse large B-cell lymphoma (24) and in the plasma of pancreatic cancer patients (57, 58). Thus, quantification of miR-210 may be further developed into clinical assays to provide valuable information about tumor biology and help guide therapeutic decisions.
Based on its prosurvival and proangiogenic roles, miR-210 may also be therapeutically relevant in cardiovascular diseases. Preclinical work showed that intracardiac injections with a minicircle vector carrying miR-210 in a mouse model of myocardial infarction induced significant improvement of left ventricular fractional shortening, decreased cellular apoptosis, and increased neovascularization (59).
Another major unanswered question is whether HIF is the only regulator of miR-210. If the history of gene regulation has taught us anything, the likely answer will turn out to be “no,” although direct evidence for additional players is currently missing. In our own experience, growth factor deprivation, osmotic stress, acidosis, and oxidative stress did not elicit miR-210 induction (10) (Kulshreshtha and Ivan, unpublished). However, we noticed the presence of multiple conserved transcription factor recognition sites in the vicinity of miR-210 genomic region: Oct-4, AP2, PPARc, and E2F (60). Interestingly, Oct-4 is itself a hypoxia-regulated gene (61) and may add an additional layer of complexity to miR-210 regulation. As a concluding thought, miR-210 has emerged as a nodal molecular component linking microenvironment, metabolism, and clinical course in cancer, the first such case to our knowledge.
Acknowledgments
This work was supported by the American Cancer Society (MI) and by the Italian Ministry of Health (Ministero della Salute).
References
- 1.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Semenza GL. Vascular responses to hypoxia and ischemia. Arterioscler Thromb Vasc Biol. 2010;30:648–652. doi: 10.1161/ATVBAHA.108.181644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakamura H, Tanimoto K, Hiyama K, Yunokawa M, Kawamoto T. Human mismatch repair gene, MLH1, is transcriptionally repressed by the hypoxia-inducible transcription factors, DEC1 and DEC2. Oncogene. 2008;27:4200–4209. doi: 10.1038/onc.2008.58. [DOI] [PubMed] [Google Scholar]
- 4.Bindra RS, Schaffer PJ, Meng A, Woo J, Maseide K. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol Cell Biol. 2004;24:8504–8518. doi: 10.1128/MCB.24.19.8504-8518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hua Z, Lv Q, Ye W, Wong CK, Cai G. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donker RB, Mouillet JF, Nelson DM, Sadovsky Y. The expression of Argonaute2 and related microRNA biogenesis proteins in normal and hypoxic trophoblasts. Mol Hum Reprod. 2007;13:273–279. doi: 10.1093/molehr/gam006. [DOI] [PubMed] [Google Scholar]
- 7.Guimbellot JS, Erickson SW, Mehta T, Wen H, Page GP. Correlation of microRNA levels during hypoxia with predicted target mRNAs through genome-wide microarray analysis. BMC Med Genomics. 2009;2:15. doi: 10.1186/1755-8794-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camps C, Buffa FM, Colella S, Moore J, Sotiriou C. hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 9.Giannakakis A, Sandaltzopoulos R, Greshock J, Liang S, Huang J. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther. 2008;7:255–264. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fasanaro P, D’Alessandra Y, Di Stefano V, Melchionna R, Romani S. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pulkkinen K, Malm T, Turunen M, Koistinaho J, Yla-Herttuala S. Hypoxia induces microRNA miR-210 in vitro and in vivo ephrin-A3 and neuronal pentraxin 1 are potentially regulated by miR-210. FEBS Lett. 2008;582:2397–2401. doi: 10.1016/j.febslet.2008.05.048. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leung AK, Sharp PA. microRNAs: a safeguard against turmoil? Cell. 2007;130:581–585. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 15.Kulshreshtha R, Ferracin M, Negrini M, Calin GA, Davuluri RV, Ivan M. Regulation of micro-RNA expression: the hypoxic component. Cell Cycle. 2007;6:1426–1431. [PubMed] [Google Scholar]
- 16.Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69:1221–1229. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juan D, Alexe G, Antes T, Liu H, Madabhushi A, Delisi C, Ganesan S, Bhanot G, Liou LS. Identification of a microRNA panel for clear-cell kidney cancer. Urology. 2010;75:835–841. doi: 10.1016/j.urology.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 19.Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 20.Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci USA. 2008;105:13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gee HE, Camps C, Buffa FM, Patiar S, Winter SC. hsa-mir-210 is a marker of tumor hypoxia and a prognostic factor in head and neck cancer. Cancer. 2010;116:2148–2158. doi: 10.1002/cncr.25009. [DOI] [PubMed] [Google Scholar]
- 22.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 23.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 24.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 2009;10:273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fasanaro P, Greco S, Lorenzi M, Pescatori M, Brioschi M. An integrated approach for experimental target identification of hypoxia-induced miR-210. J Biol Chem. 2009;284:35134–35143. doi: 10.1074/jbc.M109.052779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Favaro E, Ramachandran A, McCormick R, Gee H, Blancher C. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and Krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS One. 2010;5:e10345. doi: 10.1371/journal.pone.0010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Z, Li Y, Zhang H, Huang P, Luthra R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene. 2010;29:4362–4368. doi: 10.1038/onc.2010.193. [DOI] [PubMed] [Google Scholar]
- 30.Tong WH, Rouault TA. Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab. 2006;3:199–210. doi: 10.1016/j.cmet.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Puissegur MP, Mazure NM, Bertero T, Pradelli L, Grosso S. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2010 Oct 1; doi: 10.1038/cdd.2010.119. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu M, Liu H, Dudley SC., Jr Reactive oxygen species originating from mitochondria regulate the cardiac sodium channel. Circ Res. 2010;107:967–974. doi: 10.1161/CIRCRESAHA.110.220673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 34.Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 35.Bindra RS, Gibson SL, Meng A, Westermark U, Jasin M. Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res. 2005;65:11597–11604. doi: 10.1158/0008-5472.CAN-05-2119. [DOI] [PubMed] [Google Scholar]
- 36.Kim HW, Haider HK, Jiang S, Ashraf M. Ischemic preconditioning augments survival of stem cells via miR-210 expression by targeting caspase-8-associated protein 2. J Biol Chem. 2009;284:33161–33168. doi: 10.1074/jbc.M109.020925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsuchiya S, Fujiwara T, Sato F, Shimada Y, Tanaka E. MicroRNA-210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor-like 1 (FGFRL1) J Biol Chem. 2011;286:420–428. doi: 10.1074/jbc.M110.170852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z, Sun H, Dai H, Walsh RM, Imakura M. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle. 2009;8:2756–2768. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]
- 39.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 41.Jeyaseelan K, Lim KY, Armugam A. Micro-RNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39:959–966. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- 42.Biswas S, Roy S, Banerjee J, Hussain SR, Khanna S. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc Natl Acad Sci USA. 2010;107:6976–6981. doi: 10.1073/pnas.1001653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bostjancic E, Zidar N, Glavac D. MicroRNA microarray expression profiling in human myocardial infarction. Dis Markers. 2009;27:255–268. doi: 10.3233/DMA-2009-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li T, Cao H, Zhuang J, Wan J, Guan M. Identification of miR-130a, miR-27b and miR-210 as serum biomarkers for atherosclerosis obliterans. Clin Chim Acta. 2010;412:66–70. doi: 10.1016/j.cca.2010.09.029. [DOI] [PubMed] [Google Scholar]
- 45.Pineles BL, Romero R, Montenegro D, Tarca AL, Han YM. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am J Obstet Gynecol. 2007;196:261.e1–261.e6. doi: 10.1016/j.ajog.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 46.Enquobahrie DA, Abetew DF, Sorensen TK, Willoughby D, Chidambaram K, Williams MA. Placental microRNA expression in pregnancies complicated by preeclampsia. Am J Obstet Gynecol. 2011;204:178, e-12–21. doi: 10.1016/j.ajog.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu XM, Han T, Sargent IL, Yin GW, Yao YQ. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am J Obstet Gynecol. 2009;200:661.e1–661.e7. doi: 10.1016/j.ajog.2008.12.045. [DOI] [PubMed] [Google Scholar]
- 48.London B, Michalec M, Mehdi H, Zhu X, Kerchner L. Mutation in glycerol-3-phosphate dehydrogenase 1 like gene (GPD1-L) decreases cardiac Na+ current and causes inherited arrhythmias. Circulation. 2007;116:2260–2268. doi: 10.1161/CIRCULATIONAHA.107.703330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Norstrand DW, Valdivia CR, Tester DJ, Ueda K, London B. Molecular and functional characterization of novel glycerol-3-phosphate dehydrogenase 1 like gene (GPD1-L) mutations in sudden infant death syndrome. Circulation. 2007;116:2253–2259. doi: 10.1161/CIRCULATIONAHA.107.704627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mochel F, Knight MA, Tong WH, Hernandez D, Ayyad K. Splice mutation in the iron-sulfur cluster scaffold protein ISCU causes myopathy with exercise intolerance. Am J Hum Genet. 2008;82:652–660. doi: 10.1016/j.ajhg.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saito T, Saetrom P. MicroRNAs—targeting and target prediction. New Biotechnol. 2010;27:243–249. doi: 10.1016/j.nbt.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 52.Mizuno Y, Tokuzawa Y, Ninomiya Y, Yagi K, Yatsuka-Kanesaki Y. miR-210 promotes osteoblastic differentiation through inhibition of AcvR1b. FEBS Lett. 2009;583:2263–2268. doi: 10.1016/j.febslet.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 53.Qin L, Chen Y, Niu Y, Chen W, Wang Q. A deep investigation into the adipogenesis mechanism: profile of microRNAs regulating adipogenesis by modulating the canonical Wnt/beta-catenin signaling pathway. BMC Genomics. 2010;11:320. doi: 10.1186/1471-2164-11-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang X, Ding L, Bennewith KL, Tong RT, Welford SM. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009;35:856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M. Therapeutic silencing of micro-RNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ho AS, Huang X, Cao H, Christman-Skieller C, Bennewith K. Circulating miR-210 as a novel hypoxia marker in pancreatic cancer. Transl Oncol. 2010;3:109–113. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, Chen J, Chang P, LeBlanc A, Li D. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res. 2009;2:807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu S, Huang M, Li Z, Jia F, Ghosh Z. MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation. 2010;122:S124–S131. doi: 10.1161/CIRCULATIONAHA.109.928424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ivan M, Harris AL, Martelli F, Kulshreshtha R. Hypoxia response and microRNAs: no longer two separate worlds. J Cell Mol Med. 2008;12:1426–1431. doi: 10.1111/j.1582-4934.2008.00398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Covello KL, Kehler J, Yu H, Gordan JD, Arsham AM. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 2006;20:557–570. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]


