Abstract
The Duffy Binding protein (DBP) of Plasmodium vivax is vital for host erythrocyte invasion. DBP region II (DBPII) contains critical residues for receptor recognition and anti-DBPII antibodies have been shown to inhibit erythrocyte binding and invasion, thereby making the molecule an attractive vaccine candidate against P. vivax blood stages. Similar to other blood-stage antigens, allelic variation within the DBPII and associated strain-specific immunity is a major challenge for development of a broadly effective vaccine against P. vivax malaria. We hypothesized that immunization with a vaccine composed of multiple DBP alleles or a modified epitope DBP (DEKnull) will be more effective in producing a broadly reactive and inhibitory antibody response to diverse DBPII alleles than a single allele vaccine. In this study, we compared single, naturally occurring DBPII allele immunizations (Sal1, 7.18, P) and DEKnull with a combination of (Sal1, 7.18, P) alleles. Quantitative analysis by ELISA demonstrated that the multiple allele vaccine tend to be more immunogenic than any of the single allele vaccines when tested for reactivity against a panel of DBPII allelic variants whereas DEKnull was less immunogenic than the mixed-allele vaccine but similar in reactivity to the single allele vaccines. Further analysis for functional efficacy by in vitro erythrocyte-binding inhibition assays demonstrated that the multiple allele immunization produced a stronger strain-neutralizing response than the other vaccination strategies even though inhibition remained biased toward some alleles. Overall, there was no correlation between antibody titer and functional inhibition. These data suggest that a multiple allele vaccine may enhance immunogenicity of a DBPII vaccine but further investigation is required to optimize this vaccine strategy to achieve broader coverage against global P. vivax strains.
Keywords: Malaria, Plasmodium vivax, vaccines, multiple alleles
1. Introduction
Plasmodium vivax is the most widely distributed cause of malaria worldwide, with debilitating morbidity and serious economic burden in endemic areas, which for the most part are rural areas of developing countries [1]. Most endemic areas have intermittent or unstable low-level transmission leading to development of a weak immunity commonly resulting in clinical infections in all ages [1]. Despite the complex nature of the malaria parasite’s life cycle, there is significant evidence supporting vaccine development as an integral part of the overall strategy for malaria control [2–4].
The clinical manifestations of malaria are associated with the asexual erythrocytic stages of the parasite and targeting these stages will help reduce clinical symptoms during malaria. Merozoite proteins, which are in direct contact with the host immune system and play a major role in the invasion process, are important candidates for vaccine development to neutralize invasion and limit blood-stage growth. The parasite selectively invades reticulocytes, which accounts for about 1% of total red blood cells in circulation, as a consequence of a family of ligands that controls initial invasion by binding to receptors present on reticulocytes and absent on mature erythrocytes. A secondary receptor for P. vivax is the Duffy Antigen Receptor for Chemokines (DARC) that is recognized by the merozoite microneme ligand Duffy Binding Protein (DBP). It is consensus that DBP plays a critical role in junction formation during the invasion process, since P. vivax infections are absent from most of West Africa where individuals lack DARC on their red blood cells [5, 6]. This dependence of P. vivax on DBP for invasion makes DBP a prime target for vaccine development against vivax malaria. DBP is characterized by a conserved cysteine-rich domain, region II, that contains residues critical for receptor recognition and in DBP is referred to as DBPII [7–10].
Naturally acquired antibodies to DBPII prevalent in residents of P. vivax malaria endemic regions can functionally inhibit its erythrocyte binding and merozoite invasion of human reticulocytes. These individuals develop anti-DBPII antibodies with significant quantitative and qualitative differences in their serological responses [11–15]. Generally, serological responses to DBP and inhibition of DBP-erythrocyte binding activity increases with age as a result of a boosting effect due to recurrent exposure [15–19]. Similarly, vaccine-induced anti-DBP antibodies also inhibit DBP-erythrocyte binding and invasion of human reticulocytes [16, 20–22]. These data support the potential of DBPII as a candidate vaccine for blood stage P. vivax malaria. DBPII also contains a large number of polymorphisms, a pattern consistent with host immune evasion [11, 23, 24] and create a bias towards strain-specific immunity in P. vivax that is typically short-lived [18, 25–27].
An effective DBPII vaccine is expected to overcome strain-specific immunity by inducing the production of broadly neutralizing antibodies capable of inhibiting diverse P. vivax strains. To attain this objective, a vaccine needs to focus immune responses to conserved neutralizing DBPII epitopes similar to strategies for other microbial pathogens [28, 29] and with a P. falciparum AMA1 combination allele vaccine [30, 31]. We hypothesize that immunization with a mixed-allele DBPII vaccine or a synthetic DEKnull vaccine [32] will be more effective in inducing broadly neutralizing antibody responses to diverse DBPII alleles than a single allele vaccine. Our data demonstrates that the mixed-allele vaccine overall produces a stronger binding-inhibitory antibody response, even to alleles not included in the vaccine than the single allele and DEKnull vaccines, but this neutralizing inhibition is stronger to some alleles than others.
2. Materials and Methods
2.1. Recombinant protein expression
The gene encoding the ligand domain of Plasmodium vivax Duffy binding protein region II (DBPII) from three naturally occurring alleles: Sal1, 7.18 and P (accession numbers P22290; AAL79051.1 and AAL7907.3 respectively), a synthetic allele (DEKnull) [32] and PvMSP1-19 were codon-optimized for expression with a C-terminal 6x His-tag (pET21a+, Novagen) in BL21 (DE3) LysE E. Coli (Invitrogen) [22].
2.2. Purification of refolded recombinant DBPII
Expressed proteins were purified from inclusion bodies under denaturing conditions as previously described [21, 33, 34], with some modifications. The final protein concentration in the refolding mixture was adjusted to 50 µg/ml, degassed with nitrogen and the refolding process allowed to proceed for 36 h at 10°C with brief stirring every 12 h. This was followed by dialysis steps ending in PBS and filtered through a 0.2 µm filter. Endotoxins were quantified (ToxinSensor Chromogenic LAL Endotoxin Assay Kit (GenScript)). Final protein concentrations were adjusted to 1 mg/ml and stored in aliquots at −80°C. Denatured forms of the refolded proteins were generated by treating with 10 mM DTT followed by alkylation as previously described [35, 36] and also stored at −80°C.
2.3. Purification of rPvMSP1-19
Recombinant PvMSP1-19 was expressed using the same protocol for recombinant DBPII above but purified under native conditions. Enriched protein dialyzed against PBS and endotoxin levels determined as above. Final protein concentrations were adjusted to 1 mg/ml and stored in aliquots at −80°C.
2.3. Western blot analysis
Refolded and denatured antigens were separated on SDS-PAGE, transferred onto nitrocellulose, blocked in PBS/0.05% Tween-20 containing 5% skimmed milk and probed with an anti-DBPII specific monoclonal antibody (mAb-2D10) [22]. Bound antibody was detected with goat anti-mouse HRP-conjugated antibody (KPL) and ECL substrate (GE Healthcare).
2.4. Functional analysis of refolded rPvDBPII antigens
A standard erythrocyte-binding assay [33, 34, 37] tested functionality of refolded antigens. Briefly, 20 µg refolded rDBPII was incubated with 100 µl of DARC+ or DARC-erythrocytes in PBS/1% BSA for 2 h and then separated on 500 µl silicone oil (Dow Corning), 30 sec at 500 × g. Bound protein was eluted by re-suspending cell pellet in 40 µl of 0.3 M NaCl and incubated 10 min at 25°C with agitation every 2–3 min, centrifuged at 500 × g for 5 min and supernatant mixed with SDS-PAGE loading buffer, heated at 65°C for 3 min. Samples were separated on SDS-PAGE, transferred onto nitrocellulose membrane and probed with an anti-DBPII antibody, mAb-3D10 [22].
2.5. Animal handling and Immunizations
BALB/c (Harlan) were handled in compliance with good animal practice and approved IACUC protocol. Endotoxin levels in vaccine preparations were ≤ 5 endotoxin units per 100 µg of protein. Six cohorts of 15 mice each were set up and pre-immune serum was collected from each mouse. Animals in the first five cohorts were immunized twice with 25 µg/dose of recombinant Sal1, 7.18, P, DEKnull or PvMSP1-19, respectively, at three weeks interval, while the sixth cohort received 25 µg/dose of a mixture of Sal1/7.18/P (8.33 µg each). A seventh cohort of 10 mice (control) received adjuvant alone in PBS. A 50 µl antigen-adjuvant mixture (TiterMax Gold) was injected subcutaneously at the base of the tail and serum samples were collected three weeks after second immunization and stored at −20°C until needed.
2.6. Measurement of antibody titers to recombinant proteins
All the individual sera were analyzed by end point titration ELISA to homologous and heterologous recombinant DBPII alleles and PvMSP1-19 [38, 39]. Wells of 96-well plates were coated with 200 ng/well recombinant DBPII, blocked with 5% skimmed milk powder in wash buffer and incubated with 3-fold dilutions of mice sera starting at a 1:4000. Bound antibody was detected with AP-conjugated goat-anti-mouse antibody (KPL). Anti-DBPII monoclonal antibody (3D10), previously demonstrated to have the same binding specificity to variant DBPII alleles [22], was used as standard calibrator on each plate. All OD values were normalized at a point on the standard curve where OD630 ≈ 1.0 and antibody values were expressed as ELISA Units (EU), determined as a ratio of the OD630 generated by the test antibody and OD630 of the standard. Anti-PvMSP1-19 sera was a negative control, while pre-immune sera or sera from mice immunized with adjuvant alone were used as background control. Antibody titers were determined as reciprocal of the serum dilution to achieve EU = 1.5 [40].
2.7. Inhibition of DBPII binding to human erythrocytes
COS7 cells were transfected with the plasmid pEGFP-DBPII designed to express Sal1, 7.18, P, 27.16 and AH alleles of the DBPII (Table 1) prepared as previously reported [8, 41, 42]. Each construct was designed to target the expressed DBPII allele onto the surface of transiently transfected COS7 cells with a C-terminal GFP fusion tag as a transfection marker. Transfected cells were pre-incubated with triple fold dilutions of pooled mouse anti-DBPII sera from each immunization group prior to incubation with human erythrocytes. Rosettes were counted in 30 microscope fields at 200× magnification as positive when adherent erythrocytes covered ≥ 50% of the cell surface [10, 17, 43]. Percent binding-inhibition of each antiserum was determined by assessing the percentage of rosettes in wells of transfected COS7 cells in the presence of test serum (Ts) relative to rosettes in wells of transfected cells in presence of pre-immune serum (PIs). Anti-MSP1-19 serum served as negative control for each experiment.
% Inhibition = [1− (# rosettes in the presence of Ts / # rosettes in the presence of PIs)] × 100
Table 1.
Panel of DBPII alleles used for protein expression and COS 7 assay
| DBPII Variant | Accession # | Residue Position | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 308 | 319 | 333 | 369 | 371 | 375 | 384 | 385 | 386 | 387 | 388 | 389 | 390 | 391 | 392 | 393 | 417 | 424 | 437 | 447 | 454 | 992 | 503 | 505 | ||
| DBPII-Sal 1 | P22290.2 | R | R | L | Y | K | N | D | E | K | A | Q | Q | R | R | K | Q | N | L | W | S | Q | K | I | V |
| DEKnull | n/a | . | . | . | . | . | . | A | A | T | A | A | T | S | R | T | S | . | . | . | . | . | . | . | . |
| DBPII-27.16* | AAL79076 | S | . | . | . | . | . | G | . | . | . | . | . | H | . | . | . | . | I | . | K | . | E | . | . |
| DBPII-7.18 | AAL79051.1 | S | . | . | . | . | . | G | . | Q | . | . | . | . | . | . | . | K | I | R | . | . | . | K | . |
| DBPII-AH* | AAY34130.1 | S | . | . | E | . | G | . | Q | . | . | . | . | . | . | . | K | I | R | . | . | . | K | . | |
| DBPII-P | AAL79073.1 | S | . | F | . | . | D | G | K | N | . | . | . | H | . | . | . | K | I | . | . | . | . | K | . |
Table shows the differences in amino acid residues between the different DBPII alleles. Dot (.) means residue is the same as residue in DBPII-Sal 1.
Alleles used for COS 7 assay only.
2.8. Statistical analyses
Distribution of the antibody titers and inhibition concentrations for each antiserum was compared between all the alleles tested for any statistically significant differences in antibody reactivity and inhibitory responses by 1-way analysis of variance (ANOVA) and multiple comparison analysis by Bonferroni test using SAS software.
3. Results
3.1. Production of functional rDBPII immunogen
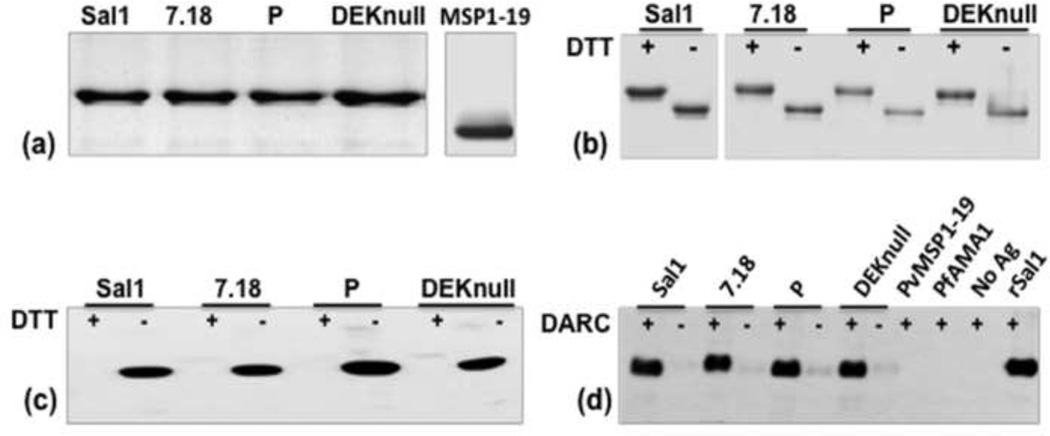
Refolded rDBPII antigens used for vaccination were observed as a single band of 39 kDa on SDS-PAGE gel (Fig. 1a). Mobility shift of refolded and denatured antigens was used as a simple indicator of the presence of disulfide linkages in the refolded antigens (Fig. 1b). The conformational structure of refolded DBPII was confirmed using a conformation-dependent anti- DBPII specific monoclonal antibody 2D10 [22], which demonstrated a strong reactivity with the refolded and little reactivity with denatured antigens (Fig. 1c). To confirm functional integrity, a standard in vitro erythrocyte-binding assay was used to test the ability of refolded rDBPII to bind Duffy positive erythrocytes. Refolded antigens bound to DARC positive erythrocytes with specificity similar to that of the native merozoite antigens, with very little or no binding observed with DARC negative erythrocytes (Fig. 1d). No binding was detected with recombinant PvMSP1-19 and PfAMA1, which were used as control antigens as well as control erythrocytes without bound antigen. This is a good indication that the refolded antigens have conformation similar to the native parasite protein.
Fig. 1. Purification and functional analysis of recombinant proteins.
(a) Coomassie-stained SDS-PAGE gel of recombinant DBPII variants and PvMSP1-19 purified by affinity chromatography on Ni+ column. (b) Differential mobility of refolded rDBPII antigens on SDS-PAGE gel before (−) and after (+) reduction with DTT. (c) Western blot analysis of rDBPII probed with conformation dependent mAb-2D10, shows antibody reacting with refolded (−) but not reduced (+) antigens. (d) Erythrocyte binding assay showing binding of refolded antigens to Duffy positive erythrocytes (+) and reduced or no binding with chymotrypsin-treated Duffy positive erythrocytes (−). Duffy positive erythrocytes incubated with PvMSP1-19 and PfAMA-1 and erythrocytes without bound antigen were used as negative controls.
3.2. Anti-DBPII reactivity profiles
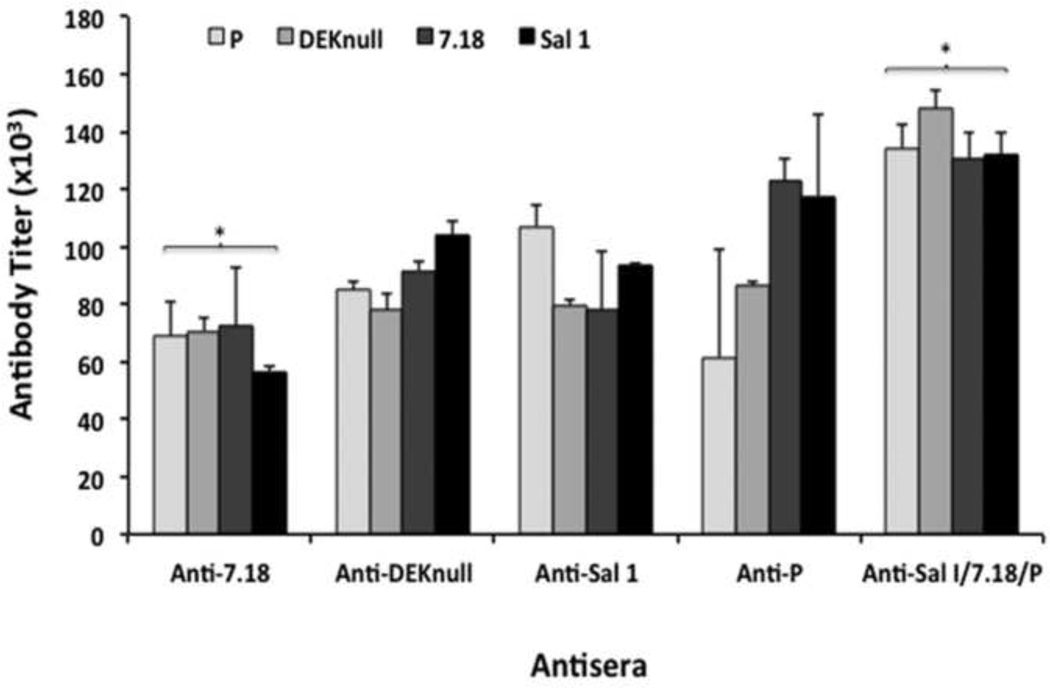
Immunogenicity of rDBPII was evaluated in BALB/c immunized in separate groups with single recombinant rDBPII alleles, Sal1, 7.18, P, DEKnull, or a combination of recombinant Sal1, 7.18 and P (Sal1/7.18/P). PvMSP1-19 or adjuvant alone served as controls. Binding specificity and antibody titers of each individual antiserum were determined by ELISA with the homologous recombinant DBPII alleles as well as cross reactivity with the heterologous antigens used for the immunization and the PvMSP1-19 control. Both single and mixed-allele vaccination strategies elicited high-level anti-DBPII IgG responses in mice against homologous and heterologous rDBPII. No anti-DBPII responses were observed with anti-PvMSP1-19 and pre-immune sera (Fig. S1). Antibody ELISA titers at EU = 1.5 were used as the basis for comparing anti-DBP antibody responses from different immunization groups to different rDBPII alleles. The mixed-allele immunization induced a higher antibody response to all the individual refolded rDBPII immunogens, including the synthetic DEKnull, compared to the single allele immunizations (Fig. 2). However, with exception of the anti-7.18 antibody (p < 0.045), there were no statistically significant differences in antibody titers among the groups. Each immunization group elicited antibody responses of similar magnitude to both homologous and heterologous antigens, with the exception of anti-P, which showed a lower antibody titer to its homologous antigen. Anti-DEKnull response was similar to that of the single naturally occurring alleles.
Fig. 2. IgG antibody titers from mice immunized with refolded rDBPII.
A titer for each group (n=15) was determined by end point dilution against homologous and heterologous antigens. All OD values were converted to ELISA Units (EU) by normalizing against the OD of a standard. Antibody titers were determined from a regression curve for each immunization group at EU = 1.5. The bars represent the ELISA titers for each immunization group against the different antigens. Error bars represent the standard deviation and the asterisk (*) indicates that there is a significant difference in antibody titers between the two antibodies. Anti-MSP1-19 antiserum and rMSP1-19 served as negative control antiserum and antigen, respectively.
3.3. Functional assessment of the anti-DBPII antibodies
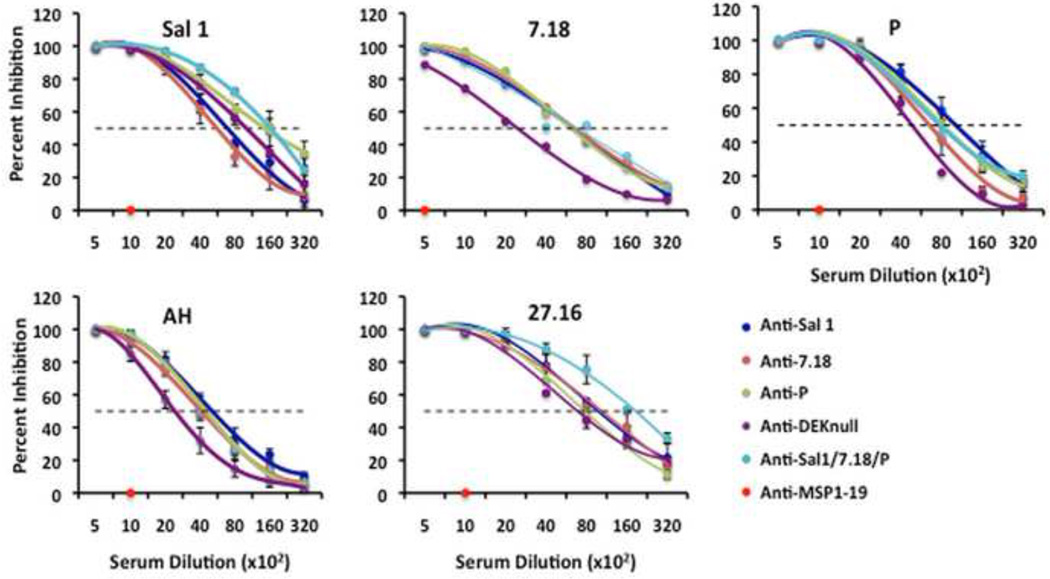
To assess the quality of immune responses generated by the different immunization strategies, we evaluated the potential of anti-DBPII immune sera from each group to inhibit DBPII-erythrocyte binding in a standard in vitro COS7 cell assay. Each antiserum titrated by end point dilution was incubated with COS7 cells expressing one of five naturally occurring DBPII alleles on their surfaces, including two DBPII alleles not used for immunization (Table 1). There was a concentration-dependent inhibition of DBPII-erythrocyte binding with all tested antisera, with 100% inhibition observed at a 1:500 serum dilution while no inhibition was observed with anti-PvMSP1 sera at 1:1000 (Fig. 3). The reciprocal serum dilution to give a 50% inhibition (IC50) of DBPII-erythrocyte binding of the different COS7 expressed alleles was used as a quantitative measure to compare the biological and functional activity of anti-DBPII antibodies from the different immunization groups.
Fig. 3. Inhibition of DBPII binding to DARC on human erythrocytes.
Transfected COS7 cells that express one of the DBPII alleles on their surface were incubated with the different mouse antisera at various dilutions prior to addition of human erythrocytes. Binding was scored after counting rosettes in 30 fields at a magnification of 200×. The percent binding inhibition was determined relative to a 1:1000 dilution of pooled pre-immune sera used as control. Each curve on the charts represents the mean of two independent experiments with each dilution tested in triplicate. Error bars represent ± standard deviation.
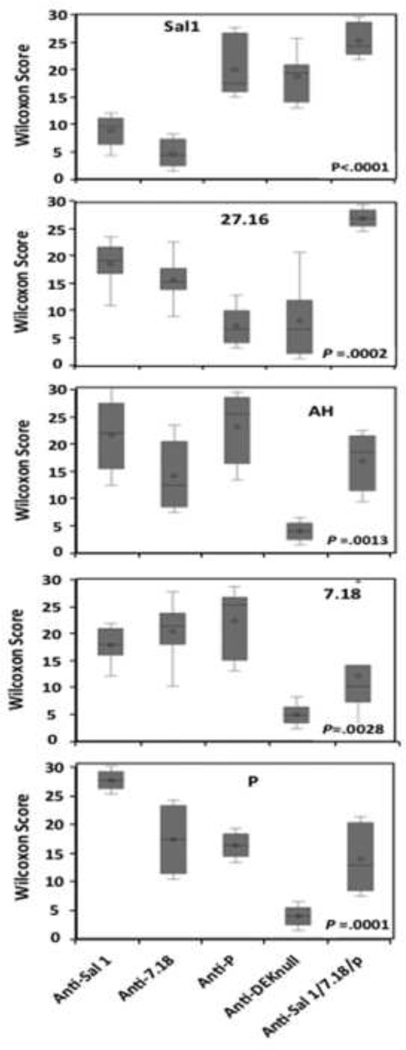
There was considerable variation in the level of inhibitory responses to the different alleles by the various immune sera (Fig. 4). Mixed-allele vaccination induced anti-DBPII highly inhibitory responses skewed towards a Sal1 and 27.16, even though the allele 27.16 was not part of the mixed-allele vaccine. Anti-DEKnull induced a lower inhibitory response to all the different alleles tested with the exception of Sal1. Using the inhibitory patterns to these DBPII, the different alleles could be classified into three antigenic groups, with Sal1 and 27.16 belonging to two separate antigenic groups and 7.18, AH and P to a third group. No difference in inhibitory pattern was observed between sera from the single allele and the mixed-allele vaccination groups with respect to the individual alleles.
Fig. 4. Binding-Inhibitory profiles of anti-DBPII immune sera.
A 50% Inhibition dilution (IC50) of each antiserum was compared between COS7-expressed alleles of DBPII by Wilcoxon rank test. The charts show the mean 1C50 dilutions of each antiserum across multiple experiments. The variation in means (represented by diamond) across alleles show significant differences in inhibitory response of each antiserum to the different alleles tested. Box plots indicate the median, lower and upper quartiles of the dilutions.
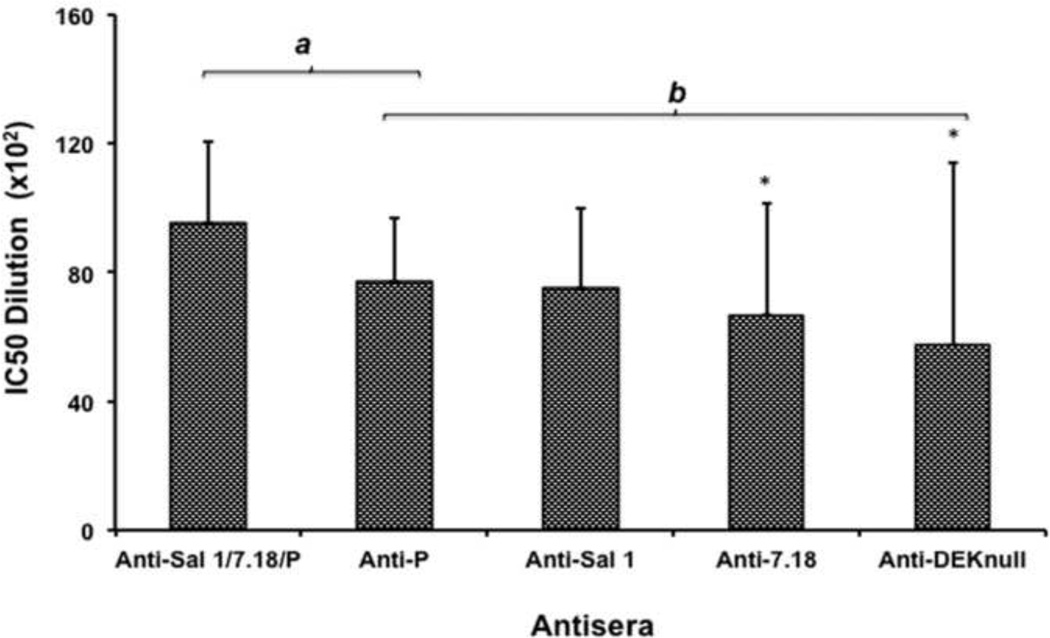
Statistical analysis using Bonferroni multiple comparison adjustment demonstrated that the mixed-allele vaccine overall showed a stronger potential to produce a higher inhibitory response than a single allele vaccine (Fig. 5). The antisera were classified into two statistical groups (a and b), with no significant differences in inhibitory antibody responses observed within each group. However, a significant difference in inhibitory response was observed between the mixed allele vaccine in the ‘a’ group and anti-7.18 and anti-DEKnull antibodies in the ‘b’ group (p<0.05). Quantitative analysis of both ELISA and in vitro binding-inhibition data failed to show any correlation between antibody titer and functional inhibition (P= 0.12, 0.46, 0.91, 0.96 and 1.0 for Anti-Sal1, 7.18, P, DEKnull and the combination Sal1/7.18/P respectively.
Fig. 5. Multiple comparisons of anti-DBPII binding-inhibitory responses.
The overall inhibitory response of each antiserum against all five COS7-expressed DBPII alleles was compared with Bonferroni multiple comparison adjustment. Bars represent the mean IC50 value of each antiserum dilution against all the natural isolates tested in the COS7 assay. Sera were classified into two groups (a and b), indicating that antisera within each group have inhibitory responses that are not statistically different from each other. Asterisk (*) indicates that there is a significant difference in the inhibitory responses between the immune sera from the mixed-allele vaccine and the 7.18 and DEKnull vaccines (P= 0.05).
4. Discussion
The biological role of the P. vivax DBP makes it an important vaccine candidate against asexual stage P. vivax infection, but the polymorphic nature of its ligand domain represents a practical challenge for vaccine to protect against diverse P. vivax strains. In endemic regions, individuals develop naturally acquired immunity, which is weak and biased towards strain specific immunity, with very few individuals developing broadly inhibitory anti-DBP immune antibody responses [15, 18]. Studies with other malaria antigens demonstrated that immunization with a single allele is not sufficient to induce protection against infection by parasites carrying distant alleles [25, 44–47]. Indeed, failure of two leading P. falciparum blood-stage vaccine candidates to protect against clinical malaria can be attributed to strain-specific responses elicited by vaccination with a monovalent vaccine [48, 49]. For a DBP vaccine we are evaluating two strategies, a multi-allele vaccine and a modified vaccine, to focus immune responses to conserved targets of functional anti-DBP protective immunity. Immunization with a multi-allele vaccine may enhance immunogenicity of the individual components by directing a bulk of the immune response against epitopes common to the constituent alleles resulting in a broader specificity, while diluting out the immunodominant effect of variant epitopes in the constituent alleles [30, 50]. The modified synthetic allele vaccine, DEKnull, was designed by ablation of the dominant polymorphic “DEK epitope” most diverse among DBP alleles leaving less immunogenic conserved neutralizing epitopes [32].
We compared immunogenicity of single allele vaccines versus the mixed-allele and DEKnull vaccines in order to assess the antibody specificities and the functional efficacy of vaccine-elicited antibodies. Vaccination strategies elicited high-level antibody responses against homologous and heterologous antigens (Fig. S1). We used antibody titers at EU=1.5 as a quantitative measure to compare reactivity of the different antisera. No significant differences were observed in reactivity of antibodies from the single allele vaccines against heterologous and homologous antigens (Fig. 2), although the P vaccine group showed a trend towards induction of strain-specific immune reactivity with the individual alleles and interestingly, a relatively lower antibody reactivity to its homologous antigen compared to the heterologous antigens. The reactivity of the mixed-allele vaccine did not significantly differ among its component antigens and tended to produce relatively higher antibody responses to all the alleles compared to the single allele vaccines. However, these differences were only significant when compared to the 7.18 immunization group.
In vitro functional assays for DBP enabled us to determine the efficacy of vaccine-elicited antibodies to inhibit DBPII-erythrocyte interaction, using a panel of DBPII allelic variants. The minimum dilution to achieve 50% (IC50) inhibition was used as a quantitative measure for comparing efficacy. The analysis demonstrated specific differences in anti-DBPII inhibitory responses from all vaccination groups against each allele (Fig. 4). This result suggested that the polymorphisms in DBPII play a role in altering the antigenic character of the different alleles and thus confer significant differences in their sensitivity to inhibitory anti-DBPII antibodies as previously reported [22, 25, 27]. The mixed-allele vaccine sera produced similar inhibitory responses to the 7.18, P and AH alleles, but significantly higher inhibitory responses to Sal1 and 27.16, when compare with antisera from single allele vaccines. Multiple comparison analysis demonstrated that the mixed-allele vaccine overall elicited more inhibitory antibodies to the different DBP alleles than the single allele vaccines, with significant differences (p<0.05) observed when compared with anti-7.18 and anti-DEKnull (Fig. 5). However, the level of inhibition varied considerably by the target DBPII allele. We also demonstrate that a synthetic allele can induce anti-DBPII binding antibodies, although anti-DEKnull antibodies showed lower inhibitory titers for most alleles except Sal1 (Fig. 4), suggesting presence of other epitopes. Based on quantitative and qualitative analysis, there was no correlation between antibody titer and inhibition of binding similar to naturally acquired human anti-DBP antibodies [15].
In conclusion, our data established that a vaccine made up of antigenically distinct DBPII maintained specificity to each allele but broadens the number of alleles to which a response was made, suggesting that a mixed-allele vaccine has the potential of generating inhibitory antibodies against a range of DBPII alleles for broader specificity, supporting data from studies of another malaria vaccine candidate, PfAMA-1 [30, 45, 51]. Further investigation is required to optimize such a vaccine especially with respect to the choice and number of alleles required to achieve the broadest specificity.
Supplementary Material
Highlights.
DBP region II is an attractive vaccine candidate against P. vivax blood stages.
A multi-allele DBPII vaccine is more immunogenic than single allele vaccines.
A multi-allele DBPII vaccine induces a strain-limited neutralizing response.
There is no correlation between antibody titer and functional inhibition.
Composition of a DBPII vaccine requires optimization to enhance efficacy.
Acknowledgements
This work is funded by NIAID/DMID contract AI-N01-054210 to Science Application International Cooperation subcontract PO10035958 and grant R01AI064478.
Abbreviations
- AMA1
apical membrane antigen-1
- DARC
Duffy Antigen Receptor for Chemokines
- DBP
Duffy Binding protein (DBP)
- DBPII
DBP region II
- rDBPII
recombinant rDBPII
- MSP1
merozoite surface protein-1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
We have no commercial or other association that poses a conflict of interest.
References
- 1.Mendis K, Sina BJ, Marchesini P, Carter R. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg. 2001;64:97–106. doi: 10.4269/ajtmh.2001.64.97. [DOI] [PubMed] [Google Scholar]
- 2.Jones TR, Hoffman SL. Malaria vaccine development. Clin Microbiol Rev. 1994;7:303–310. doi: 10.1128/cmr.7.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clyde DF, Most H, McCarthy VC, Vanderberg JP. Immunization of man against sporozite-induced falciparum malaria. Am J Med Sci. 1973;266:169–177. doi: 10.1097/00000441-197309000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Rieckmann KH, Carson PE, Beaudoin RL, Cassells JS, Sell KW. Letter: Sporozoite induced immunity in man against an Ethiopian strain of Plasmodium falciparum. Trans R Soc Trop Med Hyg. 1974;68:258–259. doi: 10.1016/0035-9203(74)90129-1. [DOI] [PubMed] [Google Scholar]
- 5.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 6.Barnwell JW, Nichols ME, Rubinstein P. In vitro evaluation of the role of the Duffy blood group in erythrocyte invasion by Plasmodium vivax. J Exp Med. 1989;169:1795–1802. doi: 10.1084/jem.169.5.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams JH, Sim BK, Dolan SA, Fang X, Kaslow DC, Miller LH. A family of erythrocyte binding proteins of malaria parasites. Proc Natl Acad Sci U S A. 1992;89:7085–7089. doi: 10.1073/pnas.89.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chitnis CE, Miller LH. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J Exp Med. 1994;180:497–506. doi: 10.1084/jem.180.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ranjan A, Chitnis CE. Mapping regions containing binding residues within functional domains of Plasmodium vivax and Plasmodium knowlesi erythrocyte-binding proteins. Proc Natl Acad Sci U S A. 1999;96:14067–14072. doi: 10.1073/pnas.96.24.14067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.VanBuskirk KM, Sevova E, Adams JH. Conserved residues in the Plasmodium vivax Duffy-binding protein ligand domain are critical for erythrocyte receptor recognition. Proc Natl Acad Sci U S A. 2004;101:15754–15759. doi: 10.1073/pnas.0405421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xainli J, Baisor M, Kastens W, Bockarie M, Adams JH, King CL. Age-dependent cellular immune responses to Plasmodium vivax Duffy binding protein in humans. J Immunol. 2002;169:3200–3207. doi: 10.4049/jimmunol.169.6.3200. [DOI] [PubMed] [Google Scholar]
- 12.Xainli J, Cole-Tobian JL, Baisor M, Kastens W, Bockarie M, Yazdani SS, et al. Epitope-specific humoral immunity to Plasmodium vivax Duffy binding protein. Infect Immun. 2003;71:2508–2515. doi: 10.1128/IAI.71.5.2508-2515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole-Tobian JL, Cortes A, Baisor M, Kastens W, Xainli J, Bockarie M, et al. Age-acquired immunity to a Plasmodium vivax invasion ligand, the duffy binding protein. J Infect Dis. 2002;186:531–539. doi: 10.1086/341776. [DOI] [PubMed] [Google Scholar]
- 14.Fraser T, Michon P, Barnwell JW, Noe AR, Al-Yaman F, Kaslow DC, et al. Expression and serologic activity of a soluble recombinant Plasmodium vivax Duffy binding protein. Infect Immun. 1997;65:2772–2777. doi: 10.1128/iai.65.7.2772-2777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chootong P, Ntumngia FB, VanBuskirk KM, Xainli J, Cole-Tobian JL, Campbell CO, et al. Mapping epitopes of the Plasmodium vivax Duffy binding protein with naturally acquired inhibitory antibodies. Infect Immun. 2010;78:1089–1095. doi: 10.1128/IAI.01036-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grimberg BT, Udomsangpetch R, Xainli J, McHenry A, Panichakul T, Sattabongkot J, et al. Plasmodium vivax invasion of human erythrocytes inhibited by antibodies directed against the Duffy binding protein. PLoS Med. 2007;4:e337. doi: 10.1371/journal.pmed.0040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michon P, Fraser T, Adams JH. Naturally acquired and vaccine-elicited antibodies block erythrocyte cytoadherence of the Plasmodium vivax Duffy binding protein. Infect Immun. 2000;68:3164–3171. doi: 10.1128/iai.68.6.3164-3171.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King CL, Michon P, Shakri AR, Marcotty A, Stanisic D, Zimmerman PA, et al. Naturally acquired Duffy-binding protein-specific binding inhibitory antibodies confer protection from blood-stage Plasmodium vivax infection. Proc Natl Acad Sci U S A. 2008;105:8363–8368. doi: 10.1073/pnas.0800371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dutta S, Daugherty JR, Ware LA, Lanar DE, Ockenhouse CF. Expression, purification and characterization of a functional region of the Plasmodium vivax Duffy binding protein. Mol Biochem Parasitol. 2000;109:179–184. doi: 10.1016/s0166-6851(00)00244-9. [DOI] [PubMed] [Google Scholar]
- 20.Singh AP, Puri SK, Chitnis CE. Antibodies raised against receptor-binding domain of Plasmodium knowlesi Duffy binding protein inhibit erythrocyte invasion. Mol Biochem Parasitol. 2002;121:21–31. doi: 10.1016/s0166-6851(02)00017-8. [DOI] [PubMed] [Google Scholar]
- 21.Yazdani SS, Shakri AR, Chitnis CE. A high cell density fermentation strategy to produce recombinant malarial antigen in E. coli. Biotechnol Lett. 2004;26:1891–1895. doi: 10.1007/s10529-004-6040-4. [DOI] [PubMed] [Google Scholar]
- 22.Ntumngia FB, Schloegel J, Barnes SJ, McHenry AM, Singh S, King CL, et al. Conserved and variant epitopes of Plasmodium vivax Duffy binding protein as targets of inhibitory monoclonal antibodies. Infect Immun. 2012;80:1203–1208. doi: 10.1128/IAI.05924-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuboi T, Kappe SH, al-Yaman F, Prickett MD, Alpers M, Adams JH. Natural variation within the principal adhesion domain of the Plasmodium vivax duffy binding protein. Infect Immun. 1994;62:5581–5586. doi: 10.1128/iai.62.12.5581-5586.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cole-Tobian J, King CL. Diversity and natural selection in Plasmodium vivax Duffy binding protein gene. Mol Biochem Parasitol. 2003;127:121–132. doi: 10.1016/s0166-6851(02)00327-4. [DOI] [PubMed] [Google Scholar]
- 25.VanBuskirk KM, Cole-Tobian JL, Baisor M, Sevova ES, Bockarie M, King CL, et al. Antigenic drift in the ligand domain of Plasmodium vivax duffy binding protein confers resistance to inhibitory antibodies. J Infect Dis. 2004;190:1556–1562. doi: 10.1086/424852. [DOI] [PubMed] [Google Scholar]
- 26.Ceravolo IP, Sanchez BA, Sousa TN, Guerra BM, Soares IS, Braga EM, et al. Naturally acquired inhibitory antibodies to Plasmodium vivax Duffy binding protein are short-lived and allele-specific following a single malaria infection. Clin Exp Immunol. 2009;156:502–510. doi: 10.1111/j.1365-2249.2009.03931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell B, Suwanarusk R, Borlon C, Costa FT, Chu CS, Rijken MJ, et al. A reliable ex vivo invasion assay of human reticulocytes by Plasmodium vivax. Blood. 2011;118:e74–e81. doi: 10.1182/blood-2011-04-348748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tobin GJ, Trujillo JD, Bushnell RV, Lin G, Chaudhuri AR, Long J, et al. Deceptive imprinting and immune refocusing in vaccine design. Vaccine. 2008;26:6189–6199. doi: 10.1016/j.vaccine.2008.09.080. [DOI] [PubMed] [Google Scholar]
- 29.Welsh RM, Fujinami RS. Pathogenic epitopes, heterologous immunity and vaccine design. Nat Rev Microbiol. 2007;5:555–663. doi: 10.1038/nrmicro1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kusi KA, Faber BW, Thomas AW, Remarque EJ. Humoral immune response to mixed PfAMA1 alleles; multivalent PfAMA1 vaccines induce broad specificity. PLoS One. 2009;4:e8110. doi: 10.1371/journal.pone.0008110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kusi KA, Faber BW, Riasat V, Thomas AW, Kocken CH, Remarque EJ. Generation of humoral immune responses to multi-allele PfAMA1 vaccines; effect of adjuvant and number of component alleles on the breadth of response. PloS one. 2010;5:e15391. doi: 10.1371/journal.pone.0015391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ntumngia FB, Adams JH. Design and immunogenicity of a novel synthetic antigen based on the ligand domain of the Plasmodium vivax duffy binding protein. Clin Vaccine Immunol. 2012;19:30–36. doi: 10.1128/CVI.05466-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran TM, Moreno A, Yazdani SS, Chitnis CE, Barnwell JW, Galinski MR. Detection of a Plasmodium vivax erythrocyte binding protein by flow cytometry. Cytometry A. 2005;63:59–66. doi: 10.1002/cyto.a.20098. [DOI] [PubMed] [Google Scholar]
- 34.Singh S, Pandey K, Chattopadhayay R, Yazdani SS, Lynn A, Bharadwaj A, et al. Biochemical, biophysical, and functional characterization of bacterially expressed and refolded receptor binding domain of Plasmodium vivax duffy-binding protein. J Biol Chem. 2001;276:17111–17116. doi: 10.1074/jbc.M101531200. [DOI] [PubMed] [Google Scholar]
- 35.Anders RF, Crewther PE, Edwards S, Margetts M, Matthew ML, Pollock B, et al. Immunisation with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine. 1998;16:240–247. doi: 10.1016/s0264-410x(97)88331-4. [DOI] [PubMed] [Google Scholar]
- 36.Crewther PE, Matthew ML, Flegg RH, Anders RF. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infect Immun. 1996;64:3310–3317. doi: 10.1128/iai.64.8.3310-3317.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wickramarachchi T, Devi YS, Mohmmed A, Chauhan VS. Identification and characterization of a novel Plasmodium falciparum merozoite apical protein involved in erythrocyte binding and invasion. PLoS One. 2008;3:e1732. doi: 10.1371/journal.pone.0001732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Devi YS, Mukherjee P, Yazdani SS, Shakri AR, Mazumdar S, Pandey S, et al. Immunogenicity of Plasmodium vivax combination subunit vaccine formulated with human compatible adjuvants in mice. Vaccine. 2007;25:5166–5174. doi: 10.1016/j.vaccine.2007.04.080. [DOI] [PubMed] [Google Scholar]
- 39.Harris KS, Casey JL, Coley AM, Masciantonio R, Sabo JK, Keizer DW, et al. Binding hot spot for invasion inhibitory molecules on Plasmodium falciparum apical membrane antigen 1. Infect Immun. 2005;73:6981–6989. doi: 10.1128/IAI.73.10.6981-6989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mullen GE, Giersing BK, Ajose-Popoola O, Davis HL, Kothe C, Zhou H, et al. Enhancement of functional antibody responses to AMA1-C1/Alhydrogel, a Plasmodium falciparum malaria vaccine, with CpG oligodeoxynucleotide. Vaccine. 2006;24:2497–2505. doi: 10.1016/j.vaccine.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 41.Fraser TS, Kappe SH, Narum DL, VanBuskirk KM, Adams JH. Erythrocyte-binding activity of Plasmodium yoelii apical membrane antigen-1 expressed on the surface of transfected COS-7 cells. Mol Biochem Parasitol. 2001;117:49–59. doi: 10.1016/s0166-6851(01)00326-7. [DOI] [PubMed] [Google Scholar]
- 42.Michon P, Woolley I, Wood EM, Kastens W, Zimmerman PA, Adams JH. Duffy-null promoter heterozygosity reduces DARC expression and abrogates adhesion of the P. vivax ligand required for blood-stage infection. FEBS Lett. 2001;495:111–114. doi: 10.1016/s0014-5793(01)02370-5. [DOI] [PubMed] [Google Scholar]
- 43.Mayer DC, Mu JB, Kaneko O, Duan J, Su XZ, Miller LH. Polymorphism in the Plasmodium falciparum erythrocyte-binding ligand JESEBL/EBA-181 alters its receptor specificity. Proc Natl Acad Sci U S A. 2004;101:2518–2523. doi: 10.1073/pnas.0307318101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hodder AN, Crewther PE, Anders RF. Specificity of the protective antibody response to apical membrane antigen 1. Infect Immun. 2001;69:3286–3294. doi: 10.1128/IAI.69.5.3286-3294.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Remarque EJ, Faber BW, Kocken CH, Thomas AW. A diversity-covering approach to immunization with Plasmodium falciparum apical membrane antigen 1 induces broader allelic recognition and growth inhibition responses in rabbits. Infection and immunity. 2008;76:2660–2670. doi: 10.1128/IAI.00170-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kennedy MC, Wang J, Zhang Y, Miles AP, Chitsaz F, Saul A, et al. In vitro studies with recombinant Plasmodium falciparum apical membrane antigen 1 (AMA1): production and activity of an AMA1 vaccine and generation of a multiallelic response. Infect Immun. 2002;70:6948–6960. doi: 10.1128/IAI.70.12.6948-6960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Renia L, Ling IT, Marussig M, Miltgen F, Holder AA, Mazier D. Immunization with a recombinant C-terminal fragment of Plasmodium yoelii merozoite surface protein 1 protects mice against homologous but not heterologous P. yoelii sporozoite challenge. Infect Immun. 1997;65:4419–4423. doi: 10.1128/iai.65.11.4419-4423.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Genton B, Betuela I, Felger I, Al-Yaman F, Anders RF, Saul A, et al. A recombinant blood-stage malaria vaccine reduces Plasmodium falciparum density and exerts selective pressure on parasite populations in a phase 1–2b trial in Papua New Guinea. J Infect Dis. 2002;185:820–827. doi: 10.1086/339342. [DOI] [PubMed] [Google Scholar]
- 49.Thera MA, Doumbo OK, Coulibaly D, Laurens MB, Ouattara A, Kone AK, et al. A field trial to assess a blood-stage malaria vaccine. N Engl J Med. 2011;365:1004–1013. doi: 10.1056/NEJMoa1008115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dutta S, Lee SY, Batchelor AH, Lanar DE. Structural basis of antigenic escape of a malaria vaccine candidate. Proc Natl Acad Sci U S A. 2007;104:12488–12493. doi: 10.1073/pnas.0701464104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duan J, Mu J, Thera MA, Joy D, Kosakovsky Pond SL, Diemert D, et al. Population structure of the genes encoding the polymorphic Plasmodium falciparum apical membrane antigen 1: implications for vaccine design. Proc Natl Acad Sci U S A. 2008;105:7857–7862. doi: 10.1073/pnas.0802328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.