Summary of recent advances
Bryostatins are a family of protein kinase C modulators that have potential applications in biomedicine. Found in miniscule quantities in a small marine invertebrate, lack of supply has hampered their development. In recent years, bryostatins have been shown to have potent bioactivity in the central nervous system, an uncultivated marine bacterial symbiont has been shown to be the likely natural source of the bryostatins, the bryostatin biosynthetic genes have been identified and characterized, and bryostatin analogues with promising biological activity have been developed and tested. Challenges in development of bryostatins for biomedical and biotechnological application include the cultivation of the bacterial symbiont and heterologous expression of bryostatin biosynthesis genes. Continued exploration of the biology and the symbiotic origin of the bryostatins presents promising opportunities for discovery of additional bryostatins, and new functions for bryostatins.
Introduction
Microbial symbionts have often been suggested to be the source of natural products isolated from marine invertebrates, usually based on structural similarity of compounds to known microbial secondary metabolites. Structural similarity is a valid argument only for structurally complex molecules assembled through many biosynthetic steps. The bryostatins were originally isolated from the bryozoan Bugula neritina, and the bacterial symbiont of B. neritina appears to be responsible for bryostatin biosynthesis (see “Evidence for symbiotic origin of bryostatins” below). The B. neritina symbiosis represents one of the best documented examples of bioactive metabolite symbiosis. In this model system, the ecological roles and biomedical potential of the compound are well described, and the symbiotic origin of the compound has been explored. This review provides an overview of the origin, biological context, and biotechnological prospects of bryostatins, and covers the period from 2005–2010.
Bryostatins: Discovery and structures
The bryostatins are a family of structurally related cyclic polyketides originally isolated from the marine bryozoan Bugula neritina (reviewed in [1]). All known bryostatins share a common macrolactone core with three tetrahydropyran rings (Figure 1); they differ predominantly in their substituents at C-7 and C-20 positions, and whether a γ-lactone ring is fused to the C-19 to C-23 tetrahydropyran ring. The bryostatins can also be categorized by the presence or absence of a 2,4-octadienoate moiety at their C-20 positions. The first bryostatins were discovered using an antineoplastic bioassay-guided fractionation approach [2]. Bryostatins are present in very low amounts in B. neritina colonies: nearly all of the bryostatins were isolated at a yield of 10−5 and 10−7 % B. neritina adult colony wet weight from various populations worldwide. Bryostatins 10 and 20 were isolated from B. neritina larvae, but at a thousand-fold higher concentration than the adult colonies, which suggests a significant role of these compounds in the larval stage of B. neritina. Additional bryostatins undoubtedly exist in nature. Using a bioassay, bryostatin activity was observed in a related bryozoan, B. simplex, and the presence of structurally similar but novel bryostatins was detected by chromatography and mass spectrometry [3]. Whether the structural diversity of bryostatins is related to biological function, environmental conditions or artifacts of isolation is not clear [4,5].
Figure 1.

Structures of bryostatins “0”-20 (“bryostatin 0” is the hypothetical precursor to all the known bryostatins). Bryostatins which contain octa-2,4-dienoate moieties (in blue) are found only in the Deep B. neritina sibling species, whereas Shallow B. neritina contains bryostatins with other kinds of esterifications (in green). The oxygen groups highlighted in red represent the pharmacophoric elements involved in binding to PKC.
Clinical status of bryostatins
Most of the pharmacological and clinical research on bryostatins has focused on bryostatin 1, which is noteworthy for its hydrophobic alkyl chain at C-20 (Figure 1). Bryostatins bind to the diacylglycerol binding site of the C-1 regulatory domain of protein kinase C (PKC); most, but not all, of bryostatins’ pharmacological effects are attributed to this interaction [6]. Protein kinase C (PKC) signaling pathways are involved in many regulatory processes in eukaryotic cells. PKC exists in ten isoforms that are differentially expressed in tissues, and variously regulated by diacylglycerol, calcium and phospholipid. Consequently, PKC activators, including bryostatins, have complex effects in animals, and these effects can vary greatly with concentration.
Bryostatins were first isolated due to their inhibition of the growth of murine P388 lymphocytic leukemia cells in vitro and in vivo [2]. After more than thirty phase I and II clinical trials in a variety of cancers, alone and in combination with other chemotherapy agents, bryostatin 1 has not been effective enough to progress to phase III clinical trials as a cancer treatment. More recently, bryostatin 1 has been explored in other contexts. It shows promise in central nervous system applications [7]. In rodent models of learning, depression, stroke and Alzheimer’s disease, bryostatin 1 has produced striking positive results [8–13]. A human Alzheimer’s disease Phase II clinical trial has been initiated. Most recently, bryostatin 1 has been proposed as a possible therapy for human immunodeficiency virus (HIV) [14]. Although current antiviral therapy is effective against HIV, the disease cannot be cured because latent viruses persist in resting cells. Based on in vitro studies, bryostatins may be effective in activating latent viruses so they can be purged from cellular reservoirs, exposed to antiviral therapy, and eliminated.
Poor availability of bryostatins has hindered clinical development. Mariculture of B. neritina has been accomplished, but has not been commercially implemented [15]. Although total synthesis of naturally occurring bryostatins has been achieved and is steadily being improved, it is not yet practical for industrial production [16]. Synthesis of simplified structural analogues of bryostatins is currently an active area of research [17,18]. Analogues that are more synthetically accessible, yet retain biological activity, have been attained and may ultimately prove clinically useful.
Bugula neritina
Bugula neritina, the source of all fully characterized bryostatins, is a marine bryozoan with a cosmopolitan distribution. Like other bryozoans, it is a colonial organism comprised of individual zooids (Figure 2A). Each feeding zooid in the colony possesses a lophophore, a ring of tentacles used to capture plankton from seawater (Figure 2B). Zooids in a colony are interconnected by a funicular system, which transports nutrients from feeding zooids to non-feeding zooids and to the brood chambers called ovicells (Figure 2B) [19]. Embryos develop from fertilized eggs in the ovicells, and when mature, they are released as non-feeding larvae into the seawater (Figure 2C). B. neritina larvae typically settle on hard substrate, after which they metamorphose into the first feeding zooid, called the ancestrula. The ancestrula then reproduces asexually by budding to form a juvenile colony (Figure 2D).
Figure 2.
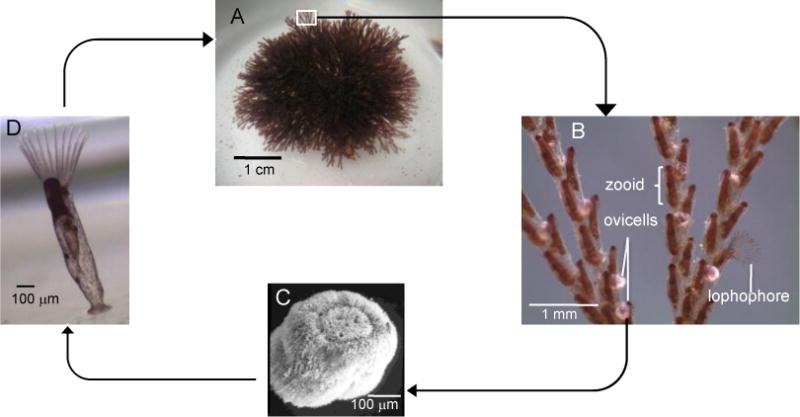
(A) A typical colony of Bugula neritina, which consists of (B) zooids arranged biserially. Ovicells brood (C) ciliated non-feeding larvae, which settled on hard substrate and metamorphose into the first feeding zooid, called the ancestrula, which then reproduces by budding to form a (D) juvenile colony
Ecological role of bryostatins
The first hint that bryostatins may be significant to the ecology of B. neritina larvae came from a study using whole extracts of B. neritina adult and larvae in fish feeding assays [20]. The study demonstrated that adult extracts were palatable to generalist fish, while larval extracts were deterrent. Moreover, it was shown that when predators ingest B. neritina larvae, the larvae are regurgitated and their metamorphosis is not hindered [21]. A follow-up study identified bryostatins 10 and 20 as feeding deterrents when tested at concentrations found in the larvae [22,23]. Ontogenetic localization of bryostatins, using a unique fluorescence technique based on bryostatins’ PKC-binding activity, revealed that the bryostatins are delivered as a coating onto the developing embryo in the ovicell from bacterial aggregates via the funicular system [24]. Upon maturation and release, this “cloak” of bryostatins (Fig. 3E) remains on the larvae until two days after they settle on a substrate and begin metamorphosis. The loss of bryostatins not unexpectedly coincides with the synthesis of a chitin exoskeleton, which would physically protect juvenile B. neritina from predation. The data from the bryostatin localization corroborate previous research demonstrating ontogenetic changes in bioactivity during early B. neritina development [25].
Figure 3.

“Candidatus Endobugula sertula” and bryostatin localization in Bugula neritina larvae. (a) Bacteria in the pallial sinus of a Bugula neritina larva, as shown by simultaneous fluorescence in situ hybridization (FISH) with two probes – the general eubacterial probe CY5-EUB338 (red) and a specific CY3“Candidatus Endobugula sertula” probe (green) – shown as a composite of the two signals (yellow). Bar = 20 μm. The same image is shown in higher magnification in (b)–(d), where (b) is a high magnification image of the pallial sinus with both probes. The symbiont-specific probe (c) and EUB338 (d) hybridize to the same cells, indicating a monoculture of “Candidatus Endobugula sertula” in the larval pallial sinus. Bar = 5 μm. (e) A coating of the bryostatins (blue) is detected around the exterior of a B. neritina larva via the PKC-labeling method [24]. Also visible in the larval pallial sinus is the CY5-EUB338/CY3-“Candidatus Endobugula sertula” FISH signal (yellow). Bar = 50 μm.
Discovery of symbionts
Before the discovery of bryostatins, B. neritina was the subject of numerous studies in developmental biology and natural history. In 1969, during an investigation of the microanatomy of several bryozoans, Lutaud observed the presence of rod-shaped bacteria in the funicular system of adult colonies of a Bugula species, B. turbinata [26]. To our knowledge, this was the first account of bacteria living within a bryozoan. Whether the observed bacteria are a monoculture or are a mixed community could not be determined from microscopic examination. Another microscopic investigation of larvae of different Bugula species revealed that B. neritina, B. pacifica and B. simplex contained rod-shaped bacteria in the larval pallial sinus [25]. Interestingly, the pallial sinuses of B. stolonifera and B. turrita are devoid of bacteria. The morphological similarities of the bacteria in adult B. turbinata observed by Lutaud [26] and in B. neritina, B. pacifica, and B. simplex larvae [27] hint at close evolutionary relationships among the bacteria; however, molecular methods were necessary to uncover the identities and relationships of the bacteria in the Bugula larvae.
Haygood and Davidson used bacterial small subunit (SSU, 16S) ribosomal RNA gene sequencing to identify bacteria in from B. neritina larvae collected in Southern California [28]. A specific oligonucleotide probe targeting the SSU rRNA sequence hybridizes to rod-shaped bacterial cells in the pallial sinus of B. neritina larvae. Identical SSU rRNA sequence was found in all of the California populations tested in this study, while absent in a co-occurring bryozoan, suggesting that bacterium in the pallial sinus engaged in a specific relationship with B. neritina. Co-hybridization of the specific probe and a universal bacterial probe demonstrates that both probes bind to all the bacterial cells, indicating the pallial sinus bacteria are a monoculture (Figure 3B). This bacterial symbiont was named “Candidatus Endobugula sertula.” The Candidatus designation is used for description of bacteria that have not been cultivated. Two strains of “Candidatus Endobugula sertula” are known (see discussion of sibling species of B. neritina below). The bacterial SSU rRNA sequences from the other symbiotic Bugula species were subsequently obtained and analyzed, and the results demonstrate that they are all closely related and form a unique bacterial group within the gamma proteobacteria, none of which has yet been cultivated [29].
Evidence for a symbiotic origin of bryostatins
The structure of the structurally and biosynthetically complex bryostatins is reminiscent of typical bacterial secondary metabolites, leading to the hypothesis that the symbiotic bacterium “Candidatus Endobugula sertula” carries out bryostatin biosynthesis. However, to move from speculation to confirmation of a microbial origin is challenging and requires rigorous experimental testing (reviewed in [30]). It is particularly challenging when the microbe is uncultivated.
Although definitive proof that the symbiont “Candidatus Endobugula sertula” is the producer of bryostatins is lacking, several lines of evidence support this hypothesis. Perhaps the most suggestive piece of evidence is the reduced bryostatin content in B. neritina colonies that developed from antibiotic-treated larvae [22,31]. The reduction in bryostatin was concomitant with a reduction in “Candidatus Endobugula sertula” levels. In these two studies, the bryozoan host did not appear to be affected by the antibiotic treatment, however, the possibility that the observed decrease in bryostatins was due to the decrease of a cryptic symbiont cannot be eliminated.
Another line of evidence for a symbiotic origin of bryostatins stems from the discovery of B. neritina sibling species. There are three sibling species known to date: the Deep, Shallow [32]and Northern sibling species are readily distinguished by mitochondrial cytochrome oxidase I sequence [33] (Table 1). Because the taxonomy has not been revised, they are all still identified as Bugula neritina. The Deep sibling species is predominantly found in Southern Californian waters below 9 meters in depth, and contains bryostatins 1–3, (in addition to other bryostatins). These bryostatins share the 2,4-octadieonoate moiety at C-20, which is absent in the other bryostatins (Figure 1). The Shallow sibling species, which was further divided into the S1 and S2 genotypes by Mackie et al. [34], in contrast, does not contain bryostatins 1–3, but does contain the other bryostatins. The S1 genotype appears to have a cosmopolitan distribution and is considered an invasive species, while the S2 genotype is restricted to California. Both the Deep and Shallow sibling species contain “Candidatus Endobugula sertula”, and the SSU rRNA sequence of “Candidatus Endobugula sertula” strains from Deep and Shallow sibling species differ by 4 bp. The Northern sibling species was found in coastal waters off Delaware and Connecticut; these animals do not appear to contain “Candidatus Endobugula sertula” or bryostatins [33]. Thus, the presence of bryostatins has always been correlated with the presence of “Candidatus Endobugula sertula”, supporting the hypothesis of a symbiotic origin of bryostatins.
Table 1.
Summary of symbiotic status and bryostatin content in B. neritina
| Sibling species | Location | Symbiotic? | Bryostatins? | Reference |
|---|---|---|---|---|
| Deep (D) | California (> 9 m depth) | Yes | Chemotype O | [32] |
| Shallow (S1) | Cosmopolitan | Yes | Chemotype M | [50,32] |
| Shallow (S2) | California (< 9 m depth) | Yes | Chemotype M | [50,32] |
| Northern | Delaware and Connecticut | None detected | None detected | [33] |
Chemotype O: includes bryostatin 1 and other C-20 2,4-octadienoate bryostatins plus other bryostatins; Chemotype M, lacks bryostatin 1 and other C-20 2,4-octadienoate bryostatins, but contains other bryostatins
Examination of the biosynthesis of bryostatins also provides evidence for the role of “Candidatus Endobugula sertula” in bryostatin production. The putative gene cluster, the bry cluster (described below), is proposed to produce “bryostatin 0”. Expression of mRNA from the cluster was detected by in situ hybridization in “Candidatus Endobugula sertula” cells in B. neritina larvae, and not in host cells [31]. Taken together, these three lines of evidence point to “Candidatus Endobugula sertula” as the producer of “bryostatin 0”.
Biosynthesis of bryostatins
Because “Candidatus Endobugula sertula” is as yet uncultivated, conventional biosynthetic studies using isotope labeling, mutants and in vitro studies of purified enzymes are not feasible. Kerr and coworkers purified radiochemically pure bryostatin 1 by incubating a B. neritina cell-free enzyme preparation with radiolabeled precursors. The authors showed the incorporation of acetate, glycerol, and SAM-adenosylmethionine, indicating these moieties as probable precursors of bryostatin biosynthesis [34].
Other inferences about bryostatin biosynthesis have been made from bioinformatic analyses. Modular polyketides synthases (PKS) consist of modules containing a set of catalytic domains for each extension cycle. Using primers based on conserved sequence signatures of modular PKS ketosynthase (KS) domains, a ketosynthase gene fragment that was consistently present in Shallow and Deep B. neritina metagenomic DNA was obtained [31]. The fragment was used to screen libraries prepared with metagenomic DNA isolated from Shallow and Deep B. neritina and enriched in “Candidatus Endobugula sertula” DNA. Several PKS-encoding clones were obtained which, when sequenced, converged to a single genomic region with features of a bacterial gene cluster, named the bry cluster [31,35,36]. The bry cluster spans ~80Kb in size and contains two parts, both entirely conserved in both Shallow and Deep “Candidatus Endobugula sertula” strains; the bry cluster does not reveal the origin of the different bryostatin profiles among B. neritina sibling species. The two parts are a 71kb bryBCXDA PKS operon, formed by five giant type I PKS genes, and the 6kb bryPQRS operon, with four smaller accessory genes (Figure 4) [36]. Genes for the oxidation of C-20 and modification of C-7 and C-20 have not been identified in the bry region. In the Deep “Candidatus Endobugula sertula” strain the two parts are not contiguous, while in the Shallow strain they form a unified and possibly ancestral locus (Figure 4). The three large identical sequence repeats in the bryBCXDA PKS operon must represent duplication events that would normally be resolved by recombination, suggesting that, like other obligate symbionts, “Candidatus Endobugula sertula” is recombination deficient.
Figure 4.

The organization of the bry cluster. Light gray box and dotted line emphasize differences found in Deep and Shallow strains of “Candidatus Endobugula sertula”. Modular polyketide synthase genes are shown as open arrows while accessory functions, primary metabolism and transposase genes are shown as orange, dark gray and black arrows respectively. The three large DNA repeats in the bryBCXDA PKS operon are represented as light blue, gold and pink boxes. PKSs modules proposed to be involved in “bryostatin 0” assembling are shown as black bars and numbered according to their order of action. Adapted from [37].
Analysis of the order and sequence features of catalytic domains shows that bry cluster enzymes lack acyltransferase (AT) domains within modules, and thus belong to the class of trans-AT type I PKSs. Except for the enigmatic bryX, the predicted enzymatic reactions are strikingly correlated with the biosynthetic steps required for the synthesis of “bryostatin 0” (Figure 5) [35,36]. Particularly, the hypothesis of D-lactate starter unit formation by bryA_LM [35], the match between the introduction of gem-dimethyl groups with the location of methyltransferase (MT) domains in modules 4 and 9, as well as pyran ring formation with the pyran-synthase (PS) domain in module 8 [36] gave credence to the biosynthetic proposal (Figure 5). Moreover, recent in vitro characterization of BryP, the tandem-AT encoded by bryP, conclusively showed this enzyme’s ability to selectively acylate malonyl-CoA on native and heterologous acyl-carrier-protein (ACP) domains, as well as in entire trans-AT PKS modules, corroborating the proposed role of BryP in the trans-loading of all extending bry cluster modules [37].
Figure 5.

Proposed biosynthesis of bryostatins. A – BryKS domains’ substrate specificity. Except for the D-lactate starter unit, all hypothetically recognized substrates are highlighted in colors. B –precursor chain elongation. Added substrates are shown after modifications, highlighted following the color code presented in A. Catalytic domains are shown in gray or, when hypothesized to be inactive, in white bubbles. Acyl-carrier-protein domains are shown as small filled circles with growing chain attached by the thiol group. Two proteins with split modules are shown in pink. The box details the HMG-CoA synthase-driven acetyl-CoA condensation onto C13/C20 beta-keto groups. Abbreviations not included in review’s text: KR, ketoreductase; OMT, O – methyltransferase; ER, enoyl reductase; DH, dehydratase; C, condensation. DH* and KR* are respectively DH and KR-like domains involved in the biosynthesis of D-lactate starter unit from the3 carbon precursor loaded by the FkbH-like domain, as proposed [36].
Since the publication of the bry cluster, Piel’s group discovered that the KS domains of trans-AT PKSs can be grouped into types based on sequence similarity [38]. The types reflect the structure of the incoming precursor at the beta carbon, and thus the specific reaction catalyzed by the preceding module. Piel’s analysis [39] of the KS domains of the bry cluster supports the hypothesis proposed by Sudek et al. [36] that the bry cluster codes for “bryostatin 0” biosynthesis. In fact, only two biosynthetic steps were modified in the revised scheme, and these differences reflect the steps in which branching occurs in the acyl units loaded by the virtually identical modules 3 and 7 of the bryostatin route (respectively Bry_M3, and Bry_M7) [39]. In this analysis, the condensation of an acetyl-CoA moiety proposed to be catalyzed by the hydroxy-methyl-glutaryl-CoA synthase (HMG-CS), BryR, occurs on the β-keto group of loaded malonyl-CoA substrates, during chain elongation instead of after macrocyclization [39] (Figure 5B, substrates in red). This is in agreement with the grouping of BryKS4 and BryKS8 into the clade Ib (Figure 5A) for KS domains with specificity for β-branched substrates. Continuing advances in catalytic domain annotation are improving the tools for chemical structure predictions of trans-AT systems. In the case of the bry cluster, these tools might be helpful in pinpointing the role of BryX, the PKS with an intriguingly aberrant catalytic domain constellation and with no obvious role in bryostatin precursor biosynthesis (Figures 4 and 5). However, bryX is completely conserved in bry clusters from both Deep and Shallow “Candidatus Endobugula sertula” strains, and is expressed in the symbiotic state, suggesting it is probably functional [36].
Challenges and opportunities in future research on bryostatins
While gaps remain in our understanding of the B. neritina-“Candidatus Endobugula sertula” system, it is perhaps the most well-rounded example of a bioactive metabolite symbiosis. The uncultivated symbiont has been identified by molecular methods, the bioactive metabolites are characterized, a probable biosynthetic pathway has been postulated, and the ecological underpinnings of the symbiosis have been established. However, there are still many questions that need to be addressed.
Cultivation of symbionts
Cultivation of “Candidatus Endobugula sertula” would facilitate bryostatin research, but may prove difficult or impossible. Over evolutionary time, obligate symbionts tend to undergo genomic degradation resulting in impaired recombination and repair, reduced genome size, increased AT content of the genome and loss of non-essential genes [40]. “Candidatus Endobugula sertula” appears to fall in the middle of the degradation spectrum. The AT content of the bry cluster is high, exceeding 70% in some regions. The large perfect repeats in the cluster suggest impaired recombination and repair. The genome size is approximately 2 Mb based on flow cytometry (C.M Anderson and M.G. Haygood, unpublished data), compared to 5 Mb for its close relative Teredinibacter turnerae [41]. Metabolic genes upstream of the bry cluster appear to be nonfunctional (C.M Anderson and M.G. Haygood, unpublished data). A genome size of 2 Mb, while small, does not necessarily preclude growth in free-living conditions. However, on the whole, the genome of “Candidatus Endobugula sertula” appears to be that of an obligate symbiont, which should be difficult to grow outside the host, since it would have poor adaptability to environmental change.
Expression of bry genes
Expressing the bry cluster in another host and producing an indisputable bryostatin precursor would be conclusive proof of the symbiotic origin of bryostatins. Such proof has been achieved in a different system via heterologous production of the patellamides, which are synthesized by the cyanobacterial symbiont Prochloron didemni in didemnid ascidians [42]. There are major obstacles to the heterologous expression of the bry cluster: i) the cluster size, which challenges any strategy for manipulation, ii) the bryA loading-module’s impressively high AT content (~70%), which renders this DNA segment highly unstable and toxic for commonly used expression vectors/hosts systems (eg, E. coli/Bacillus sp. expression vectors), iii) the large DNA repeats in the PKS operon, which favor recombination and collapse, finally iv) the mechanisms of expression regulation which are still obscure for bry cluster. However, major advances have been made in all steps of heterologous expression in the last decade, and similar issues have been overcome in other systems. Manipulation of large DNA fragments for example, were achieved in the reconstitution of the myxothiazol gene cluster [43] and in the transfer of a 100Kb DNA segment in B. subtilis 168 [44]. Problems of DNA toxicity and codon usage bias, on the other hand, can be circumvented by de novo design and artificial DNA synthesis [45,46].
How is bryostatin biosynthesis regulated?
Several studies have documented the changes in bryostatin content across life stages in B. neritina [22,24,25], and regulatory regions have been identified upstream of the bryBCXDA operon (Christine M Anderson, PhD thesis, University of California, San Diego, 2006), but it is still unknown what factors regulate bryostatin biosynthesis in nature. It is also unclear if the increase in bryostatin content in the ovicells is due to a stoichiometric increase in “Candidatus Endobugula sertula” levels, or to density-dependent gene expression (ie, quorum sensing) of bryostatin biosynthetic genes. One tantalizing piece of evidence in support of the quorum sensing hypothesis is the lack of bryostatin signal in B. neritina juveniles which contain a small inoculum of “Candidatus Endobugula sertula”, and the subsequent colocalization of bryostatins with “Candidatus Endobugula sertula” when the population density of “Candidatus Endobugula sertula” increases [24]. It is also unclear if an intermediary host factor made by B. neritina is used to transduce the signal from the environment to the symbionts.
Bryostatin diversity
Though the genetic architecture of the bry cluster is consistent with that of the hypothetical compound, “bryostatin 0”, we still do not understand how this putative precursor is elaborated to form the 20 known bryostatins. While “bryostatin 0” appears to be symbiotically produced, it is possible that the oxidation of C-20, esterifications at C-7 and C-20, and the γ-lactone at C-19 and C-23, are the result of enzymatic modifications by the host bryozoan, other microbes or due to abiotic processes. One possible abiotic process was documented by Abadi et al. who detected esterification products, similar in structure to some bryostatins, by mass spectrometry when bryostatin 1 was incubated with a carboxylic acid [5]. Furthermore, investigation of the mechanism by which bryostatins are transported and attached to the larvae may reveal new and ecologically relevant bryostatin variants. Exploration of the relationships between B. neritina and its various predators and competitors in different habitats should also reveal new information about bryostatins’ structures, activities and regulation. For example a nudibranch predator of B. neritina on the U.S. West coast, Polycera atra, contains bryostatins in its body and egg masses (Seana K Davidson, PhD thesis, University of California, San Diego, 1999; Grace E Lim, PhD thesis, University of California, San Diego, 2004). Are these known or novel bryostatins, and do they function in chemical defense?
Summary
The Bugula-Endobugula-bryostatin system is a complex tapestry of biology, ecology, microbiology and chemistry that will continue to captivate researchers. Given the increasing interest in bryostatins for clinical use, both the synthetic and heterologous expression strategies for generating supplies sufficient of active compounds will likely be pursued. Further insights gained from investigation of bryostatins in their natural biological context will advance marine biology as well as inform biotechnological studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Amaro E. Trindade-Silva, Email: amarots@iqsc.usp.br.
Grace E. Lim-Fong, Email: glim@rmc.edu.
Koty H. Sharp, Email: kotysharp@gmail.com.
Margo G. Haygood, Email: haygoodm@ebs.ogi.edu.
References
- 1.Hale K, Hummersone M, Manaviazar S, Frigerio M. The chemistry and biology of the bryostatin antitumour macrolides. Nat Prod Rep. 2002;19:413–453. doi: 10.1039/b009211h. [DOI] [PubMed] [Google Scholar]
- 2.Pettit G, Herald C, Doubek D, Herald D, Arnold E, Clardy J. Isolation and structure of bryostatin 1. J Am Chem Soc. 1982;104:6846–6848. [Google Scholar]
- 3.Lim GE, Haygood MG. “Candidatus Endobugula glebosa,” a specific bacterial symbiont of the marine bryozoan Bugula simplex. Appl Environ Microbiol. 2004;70:4921–9. doi: 10.1128/AEM.70.8.4921-4929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manning TJ, Rhodes E, Land M, Parkman R, Sumner B, Lam TT, Marshall AG, Phillips D. Impact of environmental conditions on the marine natural product bryostatin 1. Nat Prod Res. 2006;20:611–628. doi: 10.1080/14786410500462645. [DOI] [PubMed] [Google Scholar]
- 5.Abadi G, Manning TJ, McLeod K, Phillips D, Groundwater P, Noble L, Potter T. Naturally occurring esterification reactions with bryostatin. Nat Prod Res. 2008;22:865–878. doi: 10.1080/14786410701642466. [DOI] [PubMed] [Google Scholar]
- 6.Nelson TJ, Alkon DL. Neuroprotective versus tumorigenic protein kinase C activators. Trends Biochem Sci. 2009;34:136–145. doi: 10.1016/j.tibs.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Sun M, Alkon D. Protein kinase C activators as synaptogenic and memory therapeutics. Archive der Pharmazie. 2009;342:689–698. doi: 10.1002/ardp.200900050. [DOI] [PubMed] [Google Scholar]
- 8.Wang D, Darwish DS, Schreurs BG, Alkon DL. Analysis of long-term cognitive-enhancing effects of bryostatin-1 on the rabbit (Oryctolagus cuniculus) nictitating membrane response. Behav Pharmacol. 2008;19:245–256. doi: 10.1097/FBP.0b013e3282feb0d2. [DOI] [PubMed] [Google Scholar]
- 9.Sun MK, Alkon DL. Dual effects of bryostatin-1 on spatial memory and depression. Eur J Pharmacol. 2005;512:43–51. doi: 10.1016/j.ejphar.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 10*.Sun M, Hongpaisan J, Alkon D. Postischemic PKC activation rescues retrograde and anterograde long-term memory. Proc Nat Acad Sci USA. 2009;106:14676–14680. doi: 10.1073/pnas.0907842106. Rats were subjected to experimental ischemia. Bryostatin was administered for 4 months starting 24 hours post-ischemia. When tested in a water maze the bryostatin-treated animals were indistinguishable from uninjured controls; ischemic rats without bryostatin were severely impaired. This is a convincing demonstration of long-term protection of both learning and memory in an animal model of stroke. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun M, Hongpaisan J, Nelson T, Alkon D. Poststroke neuronal rescue and synaptogenesis mediated in vivo by protein kinase C in adult brains. Proc Nat Acad Sci USA. 2008;105:13620–13625. doi: 10.1073/pnas.0805952105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun M, Alkon DL. Synergistic effects of chronic bryostatin-1 and alpha-tocopherol on spatial learning and memory in rats. Eur J Pharmacol. 2008;584:328–337. doi: 10.1016/j.ejphar.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Etcheberrigaray R, Tan M, Dewachter I, Kuiperi C, Van der Auwera I, Wera S, Qiao LX, Bank B, Nelson TJ, Kozikowski AP, et al. Therapeutic effects of PKC activators in Alzheimer’s disease transgenic mice. Proc Nat Acad Sci USA. 2004;101:11141–11146. doi: 10.1073/pnas.0403921101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Mehla R, Bivalkar-Mehla S, Zhang R, Handy I, Albrecht H, Giri S, Nagarkatti P, Nagarkatti M, Chauhan A. Bryostatin modulates latent HIV-1 infection via PKC and AMPK signaling but inhibits acute infection in a receptor independent manner. PLoS ONE. 2010;5:e11160. doi: 10.1371/journal.pone.0011160. The effect of bryostatin on HIV-infected cultured immune cells was tested in vitro. Latent virus was reactivated, suggesting that bryostatin may have promise in treatment of HIV infection by activating latent virus to allow antivirals to take effect. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendola D. Aquaculture of three phyla of marine invertebrates to yield bioactive metabolites: process developments and economics. Biomol Eng. 2003;20:441–458. doi: 10.1016/s1389-0344(03)00075-3. [DOI] [PubMed] [Google Scholar]
- 16.Hale KJ, Manaviazar S. New approaches to the total synthesis of the bryostatin antitumor macrolides. Chem Asian J. 2010;5:704–754. doi: 10.1002/asia.200900634. [DOI] [PubMed] [Google Scholar]
- 17.Wender PA, Verma VA, Paxton TJ, Pillow TH. Function-oriented synthesis, step economy, and drug design. Acc Chem Res. 2008;41:40–49. doi: 10.1021/ar700155p. [DOI] [PubMed] [Google Scholar]
- 18*.Keck GE, Poudel YB, Welch DS, Kraft MB, Truong AP, Stephens JC, Kedei N, Lewin NE, Blumberg PM. Substitution on the A-ring confers to bryopyran analogues the unique biological activity characteristic of bryostatins and distinct from that of the phorbol esters. Org Lett. 2009;11:593–596. doi: 10.1021/ol8027253. In this study, complex analogues of bryostatin 1 were tested using cell-based assays that distinguish between bryostatin-type and undesirable phorbol-type activities, unlike protein kinase C binding assays which do not. The A ring of bryostatins is implicated in conferring bryostatin’s beneficial effects. A nuanced exploration of the subleties of structure-activity relationships. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woollacott R, Zimmer R. Simplified placenta-like system for transport of extraembryonic nutrients during the embryogenesis of Bugula neritina (bryozoa) J Morphol. 1975;147:355–377. doi: 10.1002/jmor.1051470308. [DOI] [PubMed] [Google Scholar]
- 20.Lindquist N, Hay M. Palatability and chemical defense of marine invertebrate larvae. Ecological Monographs. 1996;66:431–450. [Google Scholar]
- 21.Lindquist N. Palatability of invertebrate larvae to corals and sea anemones. Mar Biol. 1996;126:745–755. [Google Scholar]
- 22.Lopanik N, Lindquist N, Targett N. Potent cytotoxins produced by a microbial symbiont protect host larvae from predation. Oecologia. 2004;139:131–139. doi: 10.1007/s00442-004-1487-5. [DOI] [PubMed] [Google Scholar]
- 23.Lopanik N, Gustafson K, Lindquist N. Structure of bryostatin 20: a symbiont-produced chemical defense for larvae of the host bryozoan, Bugula neritina. J Nat Prod. 2004;67:1412–1414. doi: 10.1021/np040007k. [DOI] [PubMed] [Google Scholar]
- 24.Sharp K, Davidson S, Haygood M. Localization of Candidatus Endobugula sertula and the bryostatins throughout the life cycle of the bryozoan Bugula neritina. ISME Journal. 2007;1:693–702. doi: 10.1038/ismej.2007.78. [DOI] [PubMed] [Google Scholar]
- 25.Lopanik N, Targett N, Lindquist N. Ontogeny of a symbiont-produced chemical defense in Bugula neritina (bryozoa) Marine Ecology Progress Series. 2006;327:183–191. [Google Scholar]
- 26.Lutaud G. La nature des corps funiculaires des cellularines, bryozoaires chilostomes. Archives de zoologie expérimentale et générale. 1969;110:5. [Google Scholar]
- 27.Woollacott R. Association of bacteria with bryozoan larvae. Mar Biol. 1981;65:155–158. [Google Scholar]
- 28.Haygood MG, Davidson SK. Small-subunit rRNA genes and in situ hybridization with oligonucleotides specific for the bacterial symbionts in the larvae of the bryozoan Bugula neritina and proposal of “Candidatus Endobugula sertula”. Appl Environ Microbiol. 1997;63:4612–4616. doi: 10.1128/aem.63.11.4612-4616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim-Fong GE, Regali LA, Haygood MG. Evolutionary relationships of “Candidatus Endobugula” bacterial symbionts and their Bugula bryozoan hosts. Appl Environ Microbiol. 2008;74:3605–3609. doi: 10.1128/AEM.02798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hildebrand M, Waggoner L, Lim G, Sharp K, Ridley C, Haygood M. Approaches to identify, clone and express symbiont bioactive metabolite genes. Nat Prod Rep. 2004;21:122–142. doi: 10.1039/b302336m. [DOI] [PubMed] [Google Scholar]
- 31.Davidson SK, Allen SW, Lim GE, Anderson CM, Haygood MG. Evidence for the biosynthesis of bryostatins by the bacterial symbiont “Candidatus Endobugula sertula” of the bryozoan Bugula neritina. Appl Environ Microbiol. 2001;67:4531–4537. doi: 10.1128/AEM.67.10.4531-4537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davidson SK, Haygood MG. Identification of sibling species of the bryozoan Bugula neritina that produce different anticancer bryostatins and harbor distinct strains of the bacterial symbiont “Candidatus Endobugula sertula”. The Biological Bulletin. 1999;196:273. doi: 10.2307/1542952. [DOI] [PubMed] [Google Scholar]
- 33.McGovern TM, Hellberg ME. Cryptic species, cryptic endosymbionts, and geographical variation in chemical defences in the bryozoan Bugula neritina. Molecular Ecology. 2003;12:1207–15. doi: 10.1046/j.1365-294x.2003.01758.x. [DOI] [PubMed] [Google Scholar]
- 34.Mackie JA, Keough MJ, Christidis L. Invasion patterns inferred from cytochrome oxidase I sequences in three bryozoans, Bugula neritina, Watersipora subtorquata, and Watersipora arcuata. Mar Biol. 2006;149:285–295. [Google Scholar]
- 35.Kerr R, Lawry J, Gush K. In vitro biosynthetic studies of the bryostatins, anti-cancer agents from the marine bryozoan Bugula neritina. Tetrahedron Lett. 1996;37:8305–8308. [Google Scholar]
- 36.Hildebrand M, Waggoner LE, Liu H, Sudek S, Allen S, Anderson C, Sherman DH, Haygood M. BryA, an unusual modular polyketide synthase gene from the uncultivated bacterial symbiont of the marine bryozoan Bugula neritina. Chemistry & Biology. 2004;11:1543–1552. doi: 10.1016/j.chembiol.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 37.Sudek S, Lopanik NB, Waggoner LE, Hildebrand M, Anderson C, Liu H, Patel A, Sherman DH, Haygood MG. Identification of the putative bryostatin polyketide synthase gene cluster from “Candidatus Endobugula sertula”, the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J Nat Prod. 2007;70:67–74. doi: 10.1021/np060361d. [DOI] [PubMed] [Google Scholar]
- 38**.Lopanik N, Shields J, Buchholz T, Rath C, Hothersall J, Haygood M, Hakansson K, Thomas C, Sherman D. In vivo and in vitro trans-acylation by bryP, the putative bryostatin pathway acyltransferase derived from an uncultured marine symbiont. Chemistry & Biology. 2008;15:1175–1186. doi: 10.1016/j.chembiol.2008.09.013. The authors expressed BryP, the proposed acyltransferase of the bry cluster and demonstrated its activity in vitro. This is the first functional expression of any enzyme from the bry cluster. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39**.Nguyen T, Ishida K, Jenke-Kodama H, Dittmann E, Gurgui C, Hochmuth T, Taudien S, Platzer M, Hertweck C, Piel J. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat Biotechnol. 2008;26:225–233. doi: 10.1038/nbt1379. Using phylogenetic analysis of ketosynthase domains of trans-AT modular polyketides synthases, the authors discovered that ketosynthase clades reflect the structure of the incoming precursor. This provides a powerful new approach for predicting compound structure from gene sequences of this enzyme class. [DOI] [PubMed] [Google Scholar]
- 40.Piel J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat Prod Rep. 2010;27:996–1047. doi: 10.1039/b816430b. [DOI] [PubMed] [Google Scholar]
- 41.Moran N. Tracing the evolution of gene loss in obligate bacterial symbionts. Curr Opin Microbiol. 2003;6:512–518. doi: 10.1016/j.mib.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Yang JC, Madupu R, Durkin AS, Ekborg NA, Pedamallu CS, Hostetler JB, Radune D, Toms BS, Henrissat B, Coutinho PM, et al. The complete genome of Teredinibacter turnerae T7901: an intracellular endosymbiont of marine wood-boring bivalves (shipworms) PLoS ONE. 2009;4:e6085. doi: 10.1371/journal.pone.0006085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Nat Acad Sci USA. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perlova O, Fu J, Kuhlmann S, Krug D, Stewart A, Zhang Y, Müller R. Reconstitution of the myxothiazol biosynthetic gene cluster by red/ET recombination and heterologous expression in Myxococcus xanthus. Appl Environ Microbiol. 2006;72:7485–7494. doi: 10.1128/AEM.01503-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsuge K, Itaya M. Recombinational transfer of 100-kilobase genomic DNA to plasmid in Bacillus subtilis 168. J Bacteriol. 2001;183:5453–5458. doi: 10.1128/JB.183.18.5453-5458.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Welch M, Govindarajan S, Ness JE, Villalobos A, Gurney A, Minshull J, Gustafsson C. Design parameters to control synthetic gene expression in Escherichia coli. PLoS ONE. 2009;4:e7002. doi: 10.1371/journal.pone.0007002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kudla G, Murray A, Tollervey D, Plotkin J. Coding-sequence determinants of expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]


