Abstract
The remodeling of the promoter chromatin structure is a key event for the induction of the PHO5 gene. Two DNA-binding proteins Pho2 and Pho4 are critical for this step. We found that the NuA4 histone acetyltransferase complex is essential for PHO5 transcriptional induction without affecting Pho4 translocation upon phosphate starvation. Our data also indicate that NuA4 is critical for the chromatin remodeling event that occurs over the PHO5 promoter prior to activation. Using Chromatin IP analysis, we found that Esa1-dependent histone H4 acetylation at the PHO5 promoter correlates with specific recruitment of the NuA4 complex to this locus under repressing conditions. We demonstrate that the homeodomain transcriptional activator Pho2 is responsible for this recruitment in vivo and interacts directly with the NuA4 complex. Finally, we show that Pho4 is unable to bind the PHO5 promoter without prior action of NuA4. These results indicate that, before induction, NuA4 complex recruitment by Pho2 is an essential event that presets the PHO5 promoter for subsequent binding by Pho4, chromatin remodeling and transcription.
Keywords: chromatin remodeling, histone acetylation, NuA4, Pho2, PHO5
Introduction
In Saccharomyces cerevisiae, phosphate metabolism is under tight control of the PHO system, which helps to adapt the cell to the consequence of external changes in inorganic phosphate availability. Phosphate deprivation from the culture medium results in an important induction of secreted acid phosphatase enzymes. The most abundant enzyme is the tightly regulated PHO5 gene product whose expression is repressed at the transcriptional level by a particular promoter chromatin structure, making the regulation of this gene one of the most popular models to study the relation between chromatin structure and transcription (Svaren and Horz, 1997). Four stably positioned nucleosomes are present on the PHO5 promoter and the induction of this gene correlates with the alteration of the structure of these nucleosomes. Two positive regulators, homeodomain protein Pho2 (Bas2) and basic helix–loop–helix factor Pho4, are essential for induction and for remodeling of the promoter chromatin structure. Pho4 binding to both of the UAS(s) is essential for chromatin remodeling and is observed only after phosphate starvation (Svaren and Horz, 1997). The critical binding of Pho4 to UAS2 is prevented by nucleosome −2, whose remodeling is essential to allow this interaction and subsequent transcriptional induction. The Pho4 protein is subjected to post-translational regulation by the Pho80–85 Cyclin–CDK complex that phosphorylates it in phosphate-rich media and prevents its nuclear localization (Svaren and Horz, 1997). The exact role of Pho2 in the PHO5 transition is less clear. Pho2 was shown to interact and cooperate with Pho4 for binding at UAS1 and for an efficient transactivation at UAS2 (Barbaric et al, 1998).
In addition to Pho2 and 4, other activities play an important role in the chromatin remodeling at the PHO5 promoter, which is the key event in the transition from repressed to activated state. The INO80 ATP-dependent chromatin remodeling complex is required for full activation, and the SWI/SNF complex has also been implicated by itself or in association with the histone variant Htz1 (Santisteban et al, 2000; Steger et al, 2003). Recently, two studies demonstrated that specific nucleosomes are not only remodeled during activation but in fact also displaced from the promoter DNA (Boeger et al, 2003; Reinke and Horz, 2003).
Studies have also pointed out the importance of histone acetylation in PHO5 regulation; however, none demonstrated an absolute requirement for a specific histone acetyltransferase (HAT) in the transition from repressed to derepressed state. It was shown that the histone H3-specific Gcn5 HAT is not essential for derepression of the PHO5 gene, but could affect the chromatin structure in the constitutively derepressed Pho80 mutant (Gregory et al, 1998). Although Gcn5 does not affect the final PHO5-activated steady-state level, it seems to increase the rate of gene induction by accelerating PHO5 chromatin remodeling (Barbaric et al, 2001). Deletion of the Rpd3 histone deacetylase loosens the repression by increasing PHO5 expression in phosphate-rich media, and delays the inactivation after shifting from inducing to non-inducing medium (Svaren and Horz, 1997; Vogelauer et al, 2000). Importantly, direct evidence of histone tail involvement in the control of PHO5 expression was obtained by genetic analysis. The activation of the PHO5 promoter is significantly and specifically reduced after deletion of the histone H4 tail or mutation of the acetylatable lysines (Durrin et al, 1991). It was also shown that deletion of histone H4 tail, like deletion of Gcn5, delays the transition from repressed to activated PHO5 (Barbaric et al, 2001). In contrast to PHO5, the coregulated PHO8 gene is fully dependent on both Gcn5 and SWI/SNF for chromatin remodeling over its promoter upon induction (Gregory et al, 1999). Thus, chromatin acetylation by Gcn5 plays different roles in the induction of the coregulated PHO5 and PHO8 genes.
In the present work, we demonstrate that the NuA4 HAT complex is essential for PHO5 transition from transcriptionally repressed to activated state and for the chromatin remodeling step over the promoter region. NuA4 becomes dispensable once PHO5 is induced, arguing for an early role of presetting the promoter for activation. We demonstrate that the NuA4 complex is present at the PHO5 promoter under repressive conditions, which parallels the reported presence of Esa1-dependent acetylated histone H4 isoforms under the same conditions (Vogelauer et al, 2000). We also found that the Pho2 homeodomain protein interacts specifically with the NuA4 complex and is responsible for specific recruitment of NuA4 to the PHO5 promoter under uninduced conditions. Moreover, we show that, in the absence of NuA4, Pho4 is unable to bind the PHO5 promoter in vivo. Altogether, these results suggest a mechanism in which NuA4 recruitment by Pho2 leads to local histone hyperacetylation, which in turn poises the promoter for rapid activation upon Pho4 nuclear translocation. Thus, the NuA4 complex plays an essential role in PHO5 induction before the chromatin remodeling step by presetting the promoter for activation.
Results
The NuA4 complex is essential for transcriptional induction of the PHO5 gene
We have previously shown that the level of PHO5 mRNA is reduced in several NuA4 mutants compared to wild-type strain (Boudreault et al (2003) and references therein). However, these observations were made only on basal non-induced levels of PHO5 transcription. In order to assess the role of NuA4 in the activation of the PHO5 gene, we analyzed by Northern blotting the rate of PHO5 mRNA accumulation after shift to low phosphate (−Pi) media. In this experiment, we used a temperature-sensitive (ts) mutant for Esa1, NuA4 catalytic HAT subunit, the isogenic wild-type strain, and compared the induction of PHO5 at permissive (RT) and restrictive temperature (37°C). We have previously demonstrated that this esa1 ts mutant (esa1-Δ414 (Clarke et al, 1999)) is deficient in NuA4 HAT activity at permissive temperature and that shifting to restrictive conditions completely abolishes the activity and disrupts the complex (Allard et al, 1999). In the wild-type strain, PHO5 mRNA levels are significantly induced after 2 h in low Pi medium and reached the maximum level after 5 h at the permissive temperature or 4 h at 37°C (Figure 1A and B). In the mutant strain no induction is observed at the restrictive temperature, indicating that Esa1 and/or the NuA4 complex is essential for induction of the PHO5 gene. Moreover, under permissive conditions the level of PHO5 mRNA is significantly reduced in the esa1 mutant compared to the wild type. No further induction was observed after longer incubation in low Pi media (data not shown). Almost identical results were also obtained with a different esa1 ts mutant (esa1-L254P (Clarke et al, 1999)) (data not shown). These observations demonstrate that wild-type Esa1 activity is required for induction of the PHO5 gene, and identify NuA4 as the first coactivator HAT complex essential for the transition from inactivated to activated state.
Figure 1.
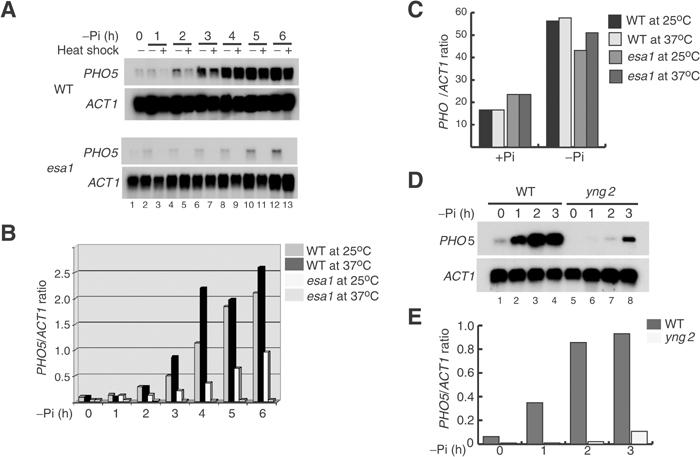
Esa1/NuA4 is essential for the transcriptional induction of the PHO5 gene. (A) Transcriptional induction of the PHO5 gene is severely impaired in the esa1 ts mutant at restrictive temperature. WT or esa1 ts mutant (esa1) were grown to mid-log phase in YPD at room temperature (25°C) and then transferred to phosphate-free medium (−Pi) for various times at (25°C) or heat shocked at (37°C). Extracted RNA was analyzed by Northern blots with PHO5 and ACT1 probes. (B) The histogram represents the PHO5/ACT1 ratio for each condition shown in (A) (quantified by phosphorimager). (C) Transcriptional induction of the PHO8 gene is NuA4 independent. As in (A), the WT or esa1 mutant strains were grown in YPD (+Pi) and shifted to (−Pi) medium for 1 h at 25 or 37°C. The Northern blot was hybridized with PHO8 and ACT1 probes and the histogram represents the PHO8/ACT1 ratio value (× 102). (D) Deletion of the YNG2 gene affects PHO5 induction. Strains QY202 (WT) and QY203 (yng2) were grown in YPD and transferred to phosphate-free medium (−Pi) for 1, 2 and 3 h. RNA samples were analyzed as in (A). (E) Northern blot signals from (D) were quantified as in (B).
To investigate whether this effect is specific to PHO5, we analyzed the induction of the coregulated PHO8 gene. After shifting the cell to low phosphate media, we observed an induction of the PHO8 gene in the wild-type and esa1 mutant strains at both permissive and restrictive temperatures (Figure 1C). These data demonstrate that Esa1 is not required for the induction of PHO8, indicating that its role is specific to PHO5 gene activation.
We have previously demonstrated that cells carrying a deletion of YNG2, which encodes another NuA4 subunit, contains a complex with reduced nucleosomal HAT activity and lowered basal expression of PHO5 (Nourani et al, 2001). We have also shown that Yng2 association with Esa1 greatly stimulates its activity towards chromatin substrates (Boudreault et al, 2003). In order to confirm that Esa1's role in PHO5 induction is through NuA4 complex HAT activity, we analyzed PHO5 transcript levels after phosphate starvation in yng2 mutant and isogenic wild-type strains (Figure 1C and D). PHO5 induction was detected after 1 h in the wild-type strain and reached a maximum level after 2 h. In contrast, the yng2 mutant strain showed weak induction only after 3 h of incubation in low Pi media. However, in contrast to the esa1 mutant, longer incubation under inducing conditions allows PHO5 mRNA to accumulate (likely due to the reduced but still present NuA4 HAT activity, data not shown). Nevertheless, this result indicates that nucleosomal acetylation by the NuA4 complex is the key to PHO5 transcription activation.
To address the possibility that NuA4 could indirectly affect PHO5 induction by disrupting signaling to the Pho4 activator, we analyzed the localization of a Pho4–GFP fusion protein in the esa1 and yng2 mutants used above. In phosphate-rich media, there is no clear localization of Pho4–GFP in either wild-type or mutant strains (Figure 2). After 3 h of phosphate starvation, the majority of cells show nuclear accumulation of the fusion protein in both the wild-type and mutant strains. These results demonstrate that NuA4 does not affect Pho4 nuclear translocation upon phosphate starvation, suggesting that the complex is required at subsequent steps in the PHO5 activation process.
Figure 2.
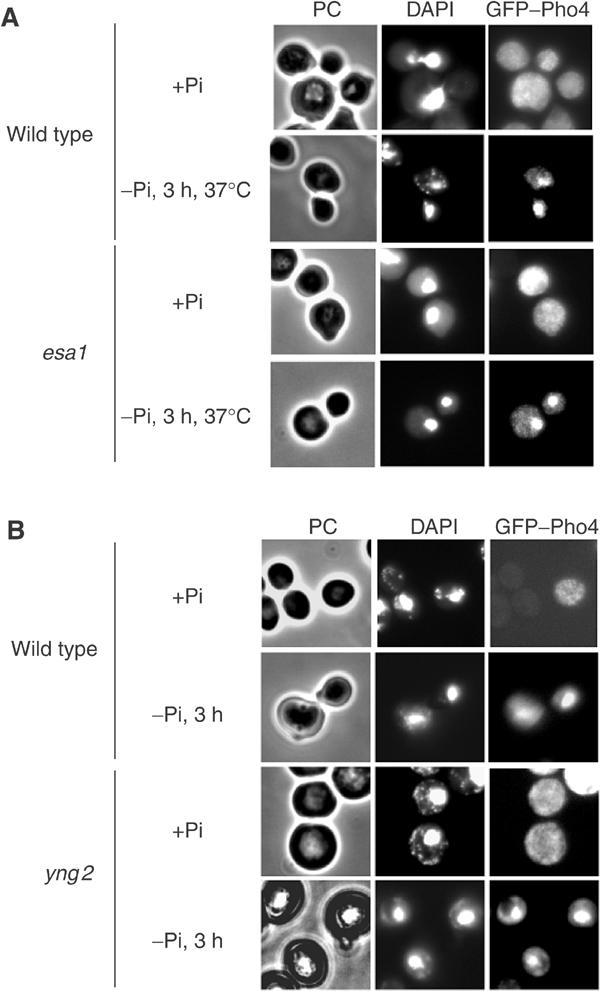
The Pho4 nuclear accumulation after phosphate starvation is not NuA4 dependent. (A) Esa1 does not affect Pho4 localization. The wild-type (WT) and esa1 ts mutant (esa1) expressing a GFP–Pho4 fusion protein were cultured to mid log phase in YPD (+Pi) and then shifted to phosphate-deprived medium (−Pi) for 3 h at the restrictive temperature (37°C). The GFP–Pho4 fusion protein was observed by fluorescence microcopy (GFP–Pho4 panel). DAPI-stained nuclei are shown in the central panel, and cells visualized by phase contrast (PC) in the left panel. (B) Yng2 is not involved in Pho4 nuclear accumulation. The WT and yng2 mutant expressing a GFP–Pho4 fusion protein were analyzed as in (A), but without heat shock.
The NuA4 complex is required for chromatin remodeling at the PHO5 promoter
The effect of esa1 or yng2 mutations on PHO5 transcriptional induction could be due to a defect at the subsequent chromatin remodeling step. To this end, we compared chromatin remodeling at the PHO5 promoter in both esa1 and yng2 mutants (Figure 3). Analyzing the accessibility of the ClaI restriction site located in the nucleosome −2 of the PHO5 promoter is a reliable and quantitative way to assess the remodeling event (Svaren et al, 1995). We isolated nuclei from esa1 mutant and wild-type (WT) strains after incubation in low Pi medium at the permissive and the restrictive temperature. In phosphate-rich medium, the ClaI site is poorly accessible in both wt and esa1 mutant strains (Figure 3A, lanes 1, 2, 5 and 6). At 3 h after shifting to low Pi medium at the restrictive temperature, we observed a clear three-fold increase of the ClaI accessibility in wild type, while the level of digestion remained low in the esa1 mutant (compare lanes 3, 4 with 7, 8). This indicates that Esa1 is required for the remodeling of the UAS2-containing nucleosome at the PHO5 promoter upon induction. Even with longer incubation under inducing conditions at the permissive temperature (where some PHO5 induction is detected in the esa1 mutant, Figure 1A), ClaI digestion remained significantly lower in the mutant strain (compare lanes 10 and 12). These results demonstrate a perfect correlation between the defect in transcription activation and inefficient chromatin remodeling at the PHO5 promoter in the esa1 mutant, indicating that normal levels of Esa1 activity are required for normal opening of this promoter. Similar results were obtained when yng2 mutants were tested for ClaI accessibility after 3 h of phosphate starvation (Figure 3B). Again, while the wild-type cells showed an increase in digestion under inducing conditions, accessibility remained unchanged in yng2 cells (compare lanes 1–4 and 5–8). This indicates that NuA4 complex activity is critical for remodeling of this nucleosome at the PHO5 promoter.
Figure 3.
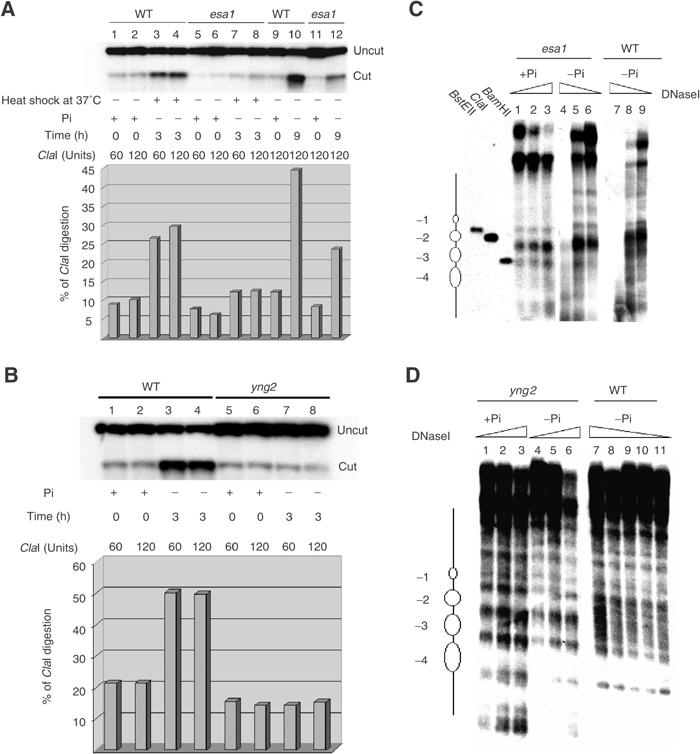
The NuA4 complex controls PHO5 promoter remodeling during phosphate starvation. (A) Esa1 is required for the PHO5 promoter transition upon phosphate deprivation. Nuclei were isolated from wild-type (WT) or esa1 ts mutant (esa1) grown in YPD (+Pi) and shifted to phosphate-poor medium (−Pi) for 3 h at 37°C or 9 h at 25°C. The nuclei were treated with 60 or 120 Units of ClaI for 30 min, DNA was then isolated and analyzed by Southern blot. The histogram represents the percentage of ClaI cleavage (quantified by phosphorimager). (B) Yng2 has a major impact on PHO5 promoter remodeling. Nuclei were isolated from wild type (WT) or YNG2 deleted strain (yng2) grown in YPD (+Pi) and shifted to phosphate-poor medium (−Pi) for 3 h. The ClaI cleavage assay was performed as in (A). (C) PHO5 chromatin is refractory to the remodeling in the esa1 mutant strain. The nuclei were isolated from the esa1 ts mutant grown in YPD (+Pi) or phosphate-deprived medium (−Pi) for 6 h (25°C), and also from wild-type cells grown 6 h in −Pi medium. In all, 100 mg of nuclei were digested by DNaseI (0.5, 1 or 2 U/ml) for 20 min. Isolated DNA was analyzed by Southern blot. Positioned nucleosomes at PHO5 promoter are represented on the left. (D) Nucleosomes are not remodeled in yng2 mutant cells after 3 h under inducing conditions. The nuclei were isolated from the yng2 mutant grown in YPD (+Pi) or phosphate-deprived medium (−Pi) for 3 h and also from wild type grown for 3 h in −Pi medium. DNaseI assays were carried out as in (C), except 0.25, 0.125, 0.0625, 0.031 or 0.015 U/ml of enzyme was used for the WT.
To further characterize the role of the NuA4 complex in the changes of nucleosomal organization at the PHO5 promoter upon activation, in vivo DNaseI protection assays were performed and DNA was analyzed by Southern blotting (Figure 3C and D). Nuclei were isolated from esa1 and yng2 mutants grown in +Pi (lane 1–3) or −Pi media (lanes 4–6) and digested with increasing amount of DNaseI. The positioned nucleosomes are clearly present on the PHO5 promoter in esa1 and yng2 mutants grown under non-inducing conditions. When esa1 mutant cells are grown in −Pi media, there is a minor change in the DNaseI profile over the PHO5 promoter, which encompasses the −2 and −3 nucleosomes (Figure 3C, lanes 4–6). This moderate change correlates with the reduced ClaI accessibility and a deficient induction of the esa1 mutant at a permissive temperature (see above). In contrast, the DNaseI digestion pattern shows an extensive hypersensitive region overlapping the −2 and −3 nucleosomes in the wild-type strain after incubation in −Pi media (Figure 3C, lanes 7–9). This indicates that the PHO5 promoter is fully remodeled under these conditions. Similar results were obtained with the yng2 mutant cells (Figure 3D). After 3 h of phosphate starvation, the pattern of DNaseI digestion remained unchanged, showing the presence of the four positioned nucleosomes (compare lanes 1–3 and 4–6). In contrast, the incubation of the wild-type strain under inducing conditions resulted in a specific loss of DNaseI protected regions corresponding to nucleosomes −2 and −3 (lanes 7–11). Altogether, these results indicate that the NuA4 complex affects the dynamic reconfiguration of PHO5 promoter chromatin structure, which is in fact responsible for the effect observed at the transcriptional level.
NuA4 is no longer required once the PHO5 gene is activated
We have demonstrated that the PHO5 gene cannot be induced in the absence of NuA4. To determine whether the continued presence of NuA4 is required after transcriptional induction of PHO5, we analyzed the effect of in vivo NuA4 disruption on PHO5-activated transcription. To this end, the esa1 temperature-sensitive mutant strain was grown at the permissive temperature in low phosphate media until maximum induction was reached and then shifted to the nonpermissive temperature. The results presented in Figure 4A show the induction of PHO5 mRNA levels when cells are grown at room temperature in low Pi (lanes 1–3). PHO5 transcript levels remained unchanged after cells were shifted to 37°C (lanes 4 and 5). Similar results were obtained when the heat shock was performed after 3 h under inducing conditions (data not shown). In comparison, returning the cells to phosphate-rich media provokes full repression of PHO5 transcription and disappearance of mRNA signal within an hour (data not shown). These observations strongly suggest that disrupting the NuA4 complex has no effect on transcription of the already induced PHO5 gene. To confirm this result, we analyzed the effect of NuA4 disruption in pho80 mutant cells, which exhibit constitutively derepressed PHO5 transcription. Wild-type, pho80 and pho80 esa1 double mutants were grown in rich media and subjected to heat shock for 1 or 2 h (Figure 4B). As expected, the deletion of the Pho80 cyclin-encoding gene resulted in a large increase of PHO5 signal (compare lanes 1, 4 and 7). This derepressed signal remains largely unchanged after 1 or 2 h of heat shock either in pho80 or pho80 esa1 double mutant cells (lanes 4–6 and 7–9). These results clearly demonstrate that NuA4 activity is not continuously required for PHO5 transcription. Thus, NuA4 is required for the initial transition from repressed to derepressed state, but has only a minor impact in maintaining transcriptionally active conditions.
Figure 4.
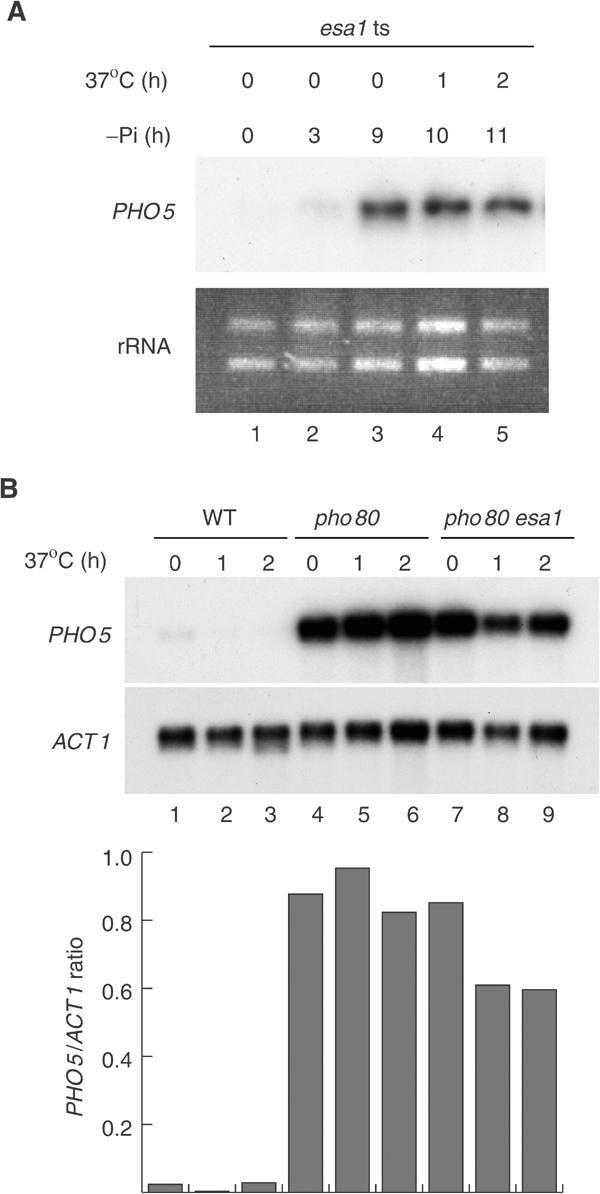
Esa1 is not required for PHO5 transcription after promoter remodeling. (A) Esa1 depletion after induction in phosphate-deprived medium does not affect PHO5 transcription. The esa1 ts mutant (esa1) was grown in YPD, transferred to phosphate-free medium (−Pi) for 9 h at 25°C and then heat shocked for 1 or 2 h at 37°C. PHO5 mRNA signals were analyzed by Northern blot. Ribosomal RNA (rRNA) is used as loading control. (B) Esa1 is not required for PHO5 transcription in the constitutively derepressed Pho80 mutant. The wild-type (WT), PHO80 deleted and pho80/esa1 ts mutants were grown at 25°C and then heat shocked for 1 or 2 h at 37°C. RNA samples were analyzed as in Figure 1.
The NuA4 complex is present at the PHO5 promoter in vivo under repressing conditions
Chromatin immunoprecipitation (ChIP) experiments under repressing conditions have shown that Esa1-dependent acetylated histone H4 isoforms are enriched at the PHO5 promoter (Vogelauer et al, 2000). We have obtained similar results with antibodies for the hyperacetytated forms of histone H4 and H2A, which are generated by Esa1/NuA4 in vivo and in vitro (data not shown (Boudreault et al, 2003)). If there is specific Esa1-dependent histone H4/H2A acetylation prior to gene induction, we would expect specific association of NuA4 at the PHO5 promoter region under repressing conditions. To test this possibility, ChIP analysis was performed to study the association of a TAP-tagged version of the Epl1 protein, an essential NuA4 subunit (Boudreault et al, 2003), to the PHO5 promoter. Crosslinked chromatin from isogenic strains containing either Epl1-TAP or Dot1-TAP as control was precipitated with IgG-sepharose beads and analyzed by PCR for the presence of PHO5, SSA4 and GAL7 promoter DNA regions. SSA4 served as a positive control and GAL7 as a negative control according to previously published data (Reid et al, 2000). As shown in Figure 5, we observed PHO5 and SSA4 promoter DNA specifically precipitated from Epl1-TAP chromatin (lanes 2 and 4) when compared to Dot1-TAP chromatin (lanes 1 and 3) and GAL7 signals (lanes 5–6). Similar results were obtained with other NuA4-tagged subunits (data not shown and see below). These data demonstrate a specific recruitment of the NuA4 complex to the PHO5 promoter under repressing conditions prior to gene activation. Surprisingly, we could not detect an increase in NuA4 at the PHO5 promoter upon gene induction. In fact, a significant drop of NuA4 binding was observed after 3 h under inducing conditions (Supplementary Figure S1). This correlates with the reported unchanged levels of histone H4 acetylation during PHO5 activation (Vogelauer et al, 2000; Reinke and Horz, 2003; Steger et al, 2003). ChIP experiments after 3 h under inducing conditions confirmed the previously reported loss of specific nucleosomes on the PHO5 promoter during activation and the unchanged levels of H4 acetylation on those remaining (Reinke and Horz, 2003) (Supplementary Figure S1).
Figure 5.
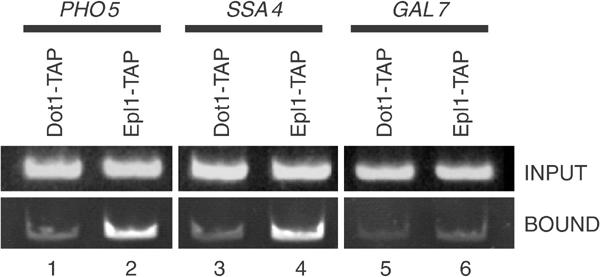
NuA4 is present at the PHO5 promoter in vivo. ChIP experiments were performed on formaldehyde crosslinked chromatin extracted from Epl1-TAP- or Dot1-TAP-expressing strains. The IgG-sepharose-bound and input materials were amplified by PCR with specific pairs of primers for PHO5, SSA4 and GAL7 promoters. Epl1-TAP, but not Dot1-TAP, is specifically found at PHO5 and SSA4 promoters, while absent from GAL7 promoter.
The NuA4 complex interacts with Pho2 homeodomain protein in vitro and in vivo
As PHO5 transition from repressed to activated state requires both Pho2 and Pho4 activators, either or both of these could be responsible for recruitment of NuA4 to the promoter. We tested a direct physical interaction by in vitro GST-pull-down using purified NuA4 complex and a series of GST-fusion proteins harboring different parts of Pho2 and the mapped activation domains Pho4, Rap1, Gcn4 and VP16 (Figure 6A and B). Gcn4 and VP16 fusions are used as positive controls (Utley et al, 1998). The N-terminal domain of Pho4 is essential for its ability to challenge the repressive chromatin of the PHO5 promoter (Svaren and Horz, 1997). Beads carrying equal amounts of GST fusions were incubated with the NuA4 complex, washed and assayed for nucleosomal HAT activity (Figure 6C). Importantly, Pho2 and Pho4 are not substrates for acetylation by NuA4 in vitro (data not shown), arguing against such a role for the complex in PHO5 regulation. As expected, Gcn4 and VP16 were able to pull down significant NuA4 HAT activity onto the beads (lanes 8–9). While GST alone, Rap1 and Pho2C failed to interact with NuA4, weak HAT activity was recovered on beads with the Pho4 and Pho2mid regions (lanes 2–4, 6 and 7). Strikingly, Pho2 N-terminal domain showed the highest affinity for the NuA4 complex, even when compared to Gcn4 and VP16 positive controls (lane 5). These results demonstrate a strong direct interaction between Pho2 N-terminal domain and the NuA4 complex in vitro. This interaction seems specific to NuA4 and does not involve Tra1, Epl1, Yng2 and Esa1 subunits as two other purified HAT complexes, SAGA and picNuA4 (Boudreault et al, 2003), are not similarly pulled down by Pho2 (Supplementary Figure S2). To determine whether this NuA4–Pho2 physical interaction also exists in the cell, we performed co-immunoprecipitation experiments using whole-cell extract from a TAP-tagged Pho2 strain expressing or not an HA-tagged version of the NuA4 subunit Epl1 (Figure 6D). Anti-HA Western blot on the material associated with the IgG-sepharose beads showed the presence of significant amounts of HA-Epl1 when both Pho2-TAP and HA-Epl1 are co-expressed (compare lanes 4–6). These in vivo data confirm direct physical interaction between NuA4 and Pho2. Furthermore, NuA4-Pho2 interaction was detected in vivo under normal growth conditions, that is, high phosphate, arguing that Pho2 is responsible for recruitment of the NuA4 complex at the PHO5 promoter when the gene is fully repressed.
Figure 6.
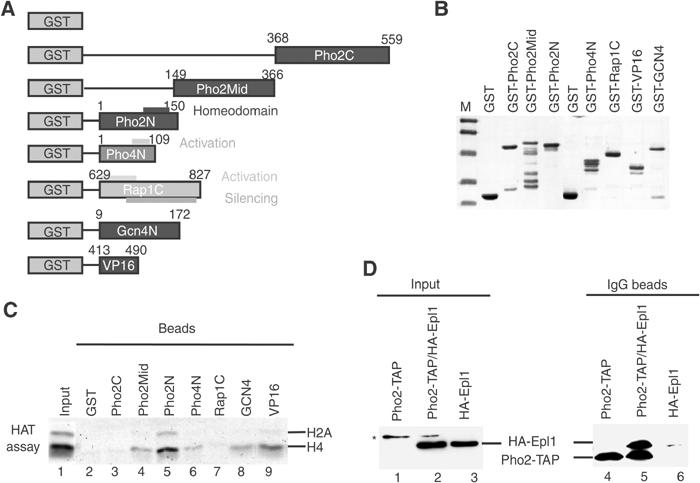
NuA4 interacts specifically with Pho2 in vitro and in vivo. (A) Diagram illustrating the GST fusion proteins used in this experiment. (B) Normalized amounts of the indicated fusion proteins were run in 12% SDS–PAGE and coomassie stained. Relative amounts of GST-Pho4N, -VP16 and -Gcn4 were increased two-fold in the pull-down assays. (C) Pho2 N-terminal region interacts with NuA4 in vitro. Equal amounts of input and beads from the GST pull-downs with purified NuA4 were assayed for nucleosomal HAT activity. (D) Coimmunoprecipitation of Pho2 with the NuA4 complex. Whole-cell extracts from cells expressing Pho2-TAP, HA-Epl1 or both proteins were incubated with IgG beads. The input and bead-associated material were analyzed by Western blots. Pho2-TAP is detected only on beads, not in input/cell extracts (asterisk marks a nonspecific band).
Pho2, but not Pho4, is responsible for recruitment of the NuA4 complex to PHO5 promoter region
To test the idea that Pho2 recruits NuA4 in vivo under repressing conditions, we analyzed the association of the NuA4 complex to the PHO5 promoter region in phosphate-rich media by ChIP assays. Formaldehyde crosslinked chromatin was extracted from a strain expressing the Epl1–13MYC fusion protein in a wild-type or a pho2Δ background (Figure 7A). Immunoprecipitated material demonstrates the presence of PHO5-specific DNA only when Epl1-13MYC was expressed in the wild-type context and assayed with anti-MYC antibody (lanes 5–8). No enrichment was observed with anti-HA antibody or at other loci. These data clearly demonstrate that Pho2 is required for recruitment of NuA4 complex to PHO5 promoter.
Figure 7.
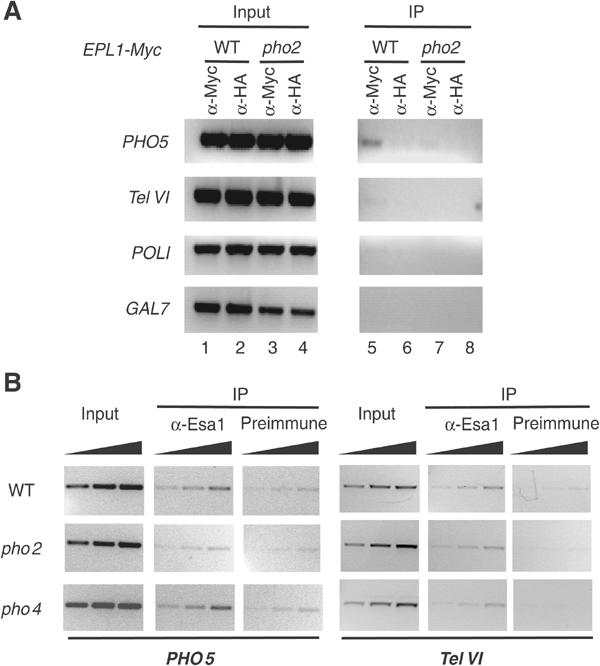
NuA4 specific recruitment to the PHO5 promoter is Pho2 dependent. (A) Recruitment of NuA4 to the PHO5 promoter is lost in pho2 mutant cells. ChIPs with indicated antibodies were performed on formaldehyde cross-linked chromatin extracted from wild-type or pho2-deleted strains both expressing the Epl1-13Myc-tagged protein. The presence of the PHO5, TelVI (ORF YFR057w near telomere region), POLI and GAL7 promoter DNA in both the immunoprecipitated and input material was assayed by PCR. (B) NuA4 recruitment is dependent on Pho2 but not Pho4. Formaldehyde crosslinked chromatin was extracted from wild-type, pho2 and pho4 strains grown in phosphate-rich medium. ChIPs with anti-Esa1 and preimmune sera were performed and increasing amounts (0.5 ×, 1 × and 2 ×) of input and bound material were used in PCR reactions to amplify either PHO5 promoter region (lanes 1–9) or TelVI (lanes 10–18) as control locus.
It has been assumed that Pho2 presence on the PHO5 promoter always occurs through cooperative binding with Pho4. To elucidate whether the presence of NuA4 on PHO5 was dependent on Pho2–4 cooperation, we performed ChIP analysis in Δpho2 and Δpho4 strains (Figure 7B). Using anti-Esa1 and preimmune serum, we confirmed that NuA4 binding the PHO5 promoter is dependent on Pho2 (compare wt to pho2 panels). The signal detected at another locus (or with preimmune serum) was not affected by PHO2 deletion. On the other hand, deletion of PHO4 had no effect on NuA4 at the PHO5 promoter (pho4 panels). These data indicate that NuA4 is recruited to PHO5 under repressing conditions by the homeodomain protein Pho2 in the absence of its Pho4 partner.
To confirm directly the presence of Pho2 at the PHO5 promoter under repressing conditions, we performed ChIPs using an anti-Pho2 polyclonal serum (Bhoite et al, 2002) or an HA-tagged Pho2-expressing strain. In both cases, we detected the presence of Pho2 at the PHO5 promoter with different chromatin preparations. The results obtained with anti-Pho2 show a 2.5-fold enrichment of Pho2 at the PHO5 promoter (Figure 8A). As for NuA4, the detected presence of Pho2 was tested for dependence on cooperative binding with Pho4. Pho2 presence at the PHO5 promoter under repressing conditions is not affected by deletion of the PHO4 gene (Figure 8B), again indicating that Pho2 recruits NuA4 independently of Pho4. Recruitment of NuA4 requires Pho2 to interact with an activator-binding domain in the HAT complex. Recently, we produced a strain (epl1(1–380)) in which the nucleosomal acetyltransferase core of NuA4 (Esa1/Epl1/Yng2) is physically dissociated from the rest of the complex (Boudreault et al, 2003). This picNuA4 complex cannot be recruited by activators like Pho2 in vitro, while it can provide global non-targeted H4/H2A acetylation in vivo (Boudreault et al, 2003; Supplementary Figure S2). Thus, if NuA4 must be specifically recruited by Pho2 to allow PHO5 gene activation, it should not be targeted in the epl1(1–380) strain. Indeed, the PHO5 gene cannot be efficiently derepressed in these mutant cells (Figure 8C, compare lanes 1–4 to 5–8). This also shows that the global HAT action of Esa1/picNuA4 still present in these cells is not sufficient to allow PHO5 activation.
Figure 8.
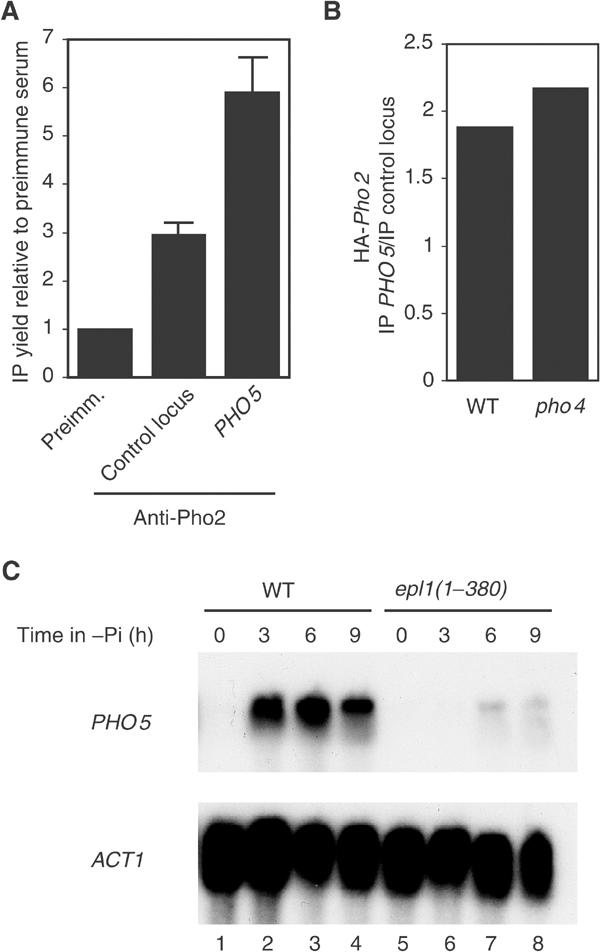
Pho2 is present at the PHO5 promoter under repressing conditions and recruitment of Esa1 activity, not its global HAT action, is required for PHO5 activation. (A) ChIP experiment showing the specific enrichment of Pho2 at the PHO5 promoter in vivo in phosphate-rich media. Signals were quantified by real-time PCR from two different IPs with polyclonal anti-Pho2 serum. A non-coding region of chromosome V is used as negative control and data are presented as a ratio of IP yield (bound/input) with anti-Pho2 versus preimmune serum at each locus. Similar results were obtained with two other chromatin preparations and in three experiments using HA-tagged Pho2 to perform the IP. (B) Similar ChIP experiments in wild type and PHO4 deleted background. HA-Pho2-expressing strains were used with non-tagged strains as controls. PHO5 and control locus IP signals (bound/input) in the nontagged strains were subtracted from the values in the HA-tagged strains. The HA-Pho2 enrichment is then presented as a ratio of PHO5 IP versus control locus IP. PHO4 deletion had no effect on Pho2 presence at PHO5, a result reproduced with four independent chromatin samples with anti-HA and anti-Pho2 serum. (C) A recruitable form of Esa1 HAT, not its global action, is required for PHO5 gene activation. A strain expressing a truncated version of Epl1, epl1(1–380), contains only a globally acting nontargeted form of Esa1 within the Piccolo NuA4 complex (Boudreault et al, 2003). Wild-type (WT) and epl1(1–380) mutant cells were incubated for the indicated period of time in phosphate-free media and analyzed by Northern blot as in Figure 1A.
NuA4 action is critical to allow Pho4 binding to the PHO5 promoter
As NuA4 is recruited to the PHO5 promoter prior to Pho4 nuclear translocation under inducing conditions, and as NuA4, like Pho4, is required for chromatin remodeling and transcription activation, we analyzed Pho4 binding to the PHO5 promoter in the absence of NuA4 after phosphate starvation. When normal cells are placed under inducing conditions, we observe a rapid specific enrichment of Pho4 at the PHO5 promoter, which follows its nuclear translocation (Figure 9A, WT −Pi versus +Pi). In contrast, the esa1 mutant strain did not show such enrichment after 3 h under inducing conditions at the nonpermissive temperature, even though Pho4 is clearly nuclear in these cells (esa1 −Pi versus +Pi). When the cells are placed in phosphate-free media for a longer period of time (6 h), Pho4 is very highly enriched at PHO5 in normal cells, while esa1 mutant cells exhibit at least 10-fold less Pho4 at the PHO5 promoter (Figure 9B). These data demonstrate that prior action of NuA4 is critical to allow Pho4 binding to the PHO5 promoter upon nuclear translocation. This also explains why NuA4 is required for chromatin remodeling and transcription activation as both depend on Pho4 binding.
Figure 9.
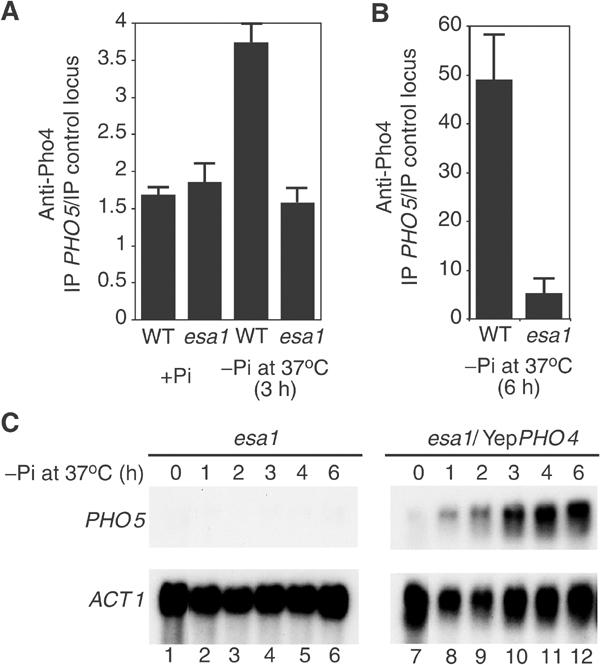
NuA4 action is necessary to allow Pho4 binding to the PHO5 promoter, but is dispensable upon overexpression of Pho4. (A) Pho4 is enriched at the PHO5 promoter in wild-type but not esa1 mutant cells under inducing conditions. ChIP experiment using anti-Pho4 and preimmune sera with wild-type or esa1 ts cells grown in phosphate-rich (+Pi) media at 25°C or placed for 3 h in phosphate-free (−Pi) media at 37°C. IP ratios were calculated by real-time PCR reactions and presented as in Figure 8B using preimmune serum signals as subtracted backgrounds. (B) A similar ChIP experiment was performed with wild-type and mutant cells incubated for 6 h under inducing conditions at 37°C. Under these conditions, the presence of Pho4 at the PHO5 promoter is 10-fold higher in wild type than in mutant cells. (C) Overexpression of Pho4 bypasses both Pho2 and NuA4 requirements for PHO5 activation. esa1 ts mutant cells were transformed with a high copy vector that drives overexpression of the Pho4 transactivator. Cells were incubated in low phosphate media at the non-permissive temperature for the indicated periods of time and analyzed by Northern blot.
To test our model depicting an essential specific link between Pho2 and NuA4 to allow PHO5 promoter acetylation and efficient Pho4 binding, we used a system in which Pho2 is dispensable for derepression. It was shown that when Pho4 is overexpressed in vivo, Pho2 is no longer necessary for PHO5 gene activation (Fascher et al, 1990). Bypassing Pho2 requirement should also make NuA4 dispensable for activation. We overexpressed Pho4 in an esa1 ts strain and analyzed PHO5 induction at the nonpermissive temperature (Figure 9C). Clearly, overexpression of Pho4 activates the PHO5 gene in esa1 mutant cells even at 37° (compare lanes 1–6 to 7–12). Thus, these data indicate that NuA4 requirement for PHO5 activation is specifically linked to Pho2 function, supporting the model of specific and direct local recruitment by the activator. It also indicates that a major function of NuA4 at the PHO5 promoter is indeed to facilitate Pho4 binding upon its nuclear translocation. The NuA4 defect can be overcome by overexpression of Pho4, which drives binding to the promoter in the absence of the complex, bypassing the need for presetting by NuA4.
Discussion
In addition to the DNA-binding factors, Pho2 and Pho4, two well-studied mechanisms including the remodeling of nucleosomes by ATP-dependent complexes such as SWI/SNF or INO80, and the covalent modifications of histone tails by activities such as Gcn5-containing complexes, are involved in PHO5 activation. However, a number of studies suggest that none of these coactivator activities is independently fully essential for PHO5 transcriptional induction (see Introduction and references therein). In our work, we provide clear evidence that the NuA4 HAT complex is absolutely required for derepression of the PHO5 gene in response to phosphate starvation. Indeed, when phosphate deprivation is carried out in parallel with NuA4 complex depletion, the PHO5 gene remains fully repressed. The role of histone acetylation in PHO5 derepression is not a new observation. The deletion or mutation of the histone H4 tail decreases either the rate of derepression or the level of induction (Durrin et al, 1991; Barbaric et al, 2001). However, these studies showed that, while H4 acetylation was important for the rate and level of induction, it was not essential for PHO5 activation. The absolute requirement of Esa1/NuA4 could be explained by the fact that it acetylates both H4 and H2A N-terminal tails in vivo (Boudreault et al, 2003). Furthermore, it was previously shown that histone H2A tail is important for the repression of basal transcription of the PHO5 gene (Lenfant et al, 1996). Thus, simultaneous mutations of H4 and H2A tails in the same cell could disable PHO5 induction as NuA4 mutants do. On the other hand, NuA4 might play an additional role at the PHO5 promoter not related to acetylation of histone H4/H2A tails. For instance, NuA4 may acetylate different unknown non-histone substrates or simply co-activate transcription independently of acetylation.
It is important to compare the involvement of Esa1/NuA4 versus Gcn5/SAGA HAT complexes in PHO5 activation. While Gcn5 is only required for the normal rate of chromatin remodeling and activation (Barbaric et al, 2001), Esa1 is fully required for these events (Figure 3). While Gcn5 affects PHO5 transcription in a constitutively derepressed state (pho80 mutant; Gregory et al, 1998), Esa1 has no effect in such a context and is dispensable once the gene is activated (Figure 4). While SAGA is found at the PHO5 promoter only after phosphate starvation in a Pho4-dependent manner (Barbaric et al, 2003), NuA4 is recruited by Pho2 before activation (under repressing conditions) in a Pho4-independent manner (Figures 5, 6, 7 and 8). While histone H3 acetylation increases over the promoter during PHO5 activation (before nucleosomes unfold), histone H4/H2A acetylation remains largely unchanged (Reinke and Horz, 2003; supplementary Figure S1). These data indicate that SAGA plays a role at a later step during the activation process, while NuA4 action is essential and occurs prior to Pho4 translocation to the nucleus.
We have demonstrated that NuA4 is not only required for PHO5 transcriptional induction but also for chromatin remodeling over the promoter region (Figure 3). Our results clearly suggest that the essential requirement of NuA4 is related to the remodeling step and that it is specific to PHO5 as the same effect is not observed on the coregulated PHO8 gene (Figure 1). It was then surprising to find NuA4 at the PHO5 promoter under repressing conditions prior to activation (Figure 5). However, it was already known that PHO5 promoter region was enriched in Esa1-dependent hyperacetylated histone H4 under the repressive conditions (Vogelauer et al, 2000). This was often attributed to the global nontargeted action of Esa1. On the contrary, here we clearly show that the presence of Esa1 is dependent on both a DNA-bound transcription factor and the recruitable integrity of the NuA4 complex (Figures 7 and 8). Targeted hyperacetylation of nucleosomal histones under repressive conditions could prepare the promoter and potentiate a rapid remodeling step, followed by transcriptional induction in response to external stimulus.
How does NuA4-dependent hyperacetylation pre-set a promoter for rapid chromatin remodeling and activation? One possibility is that the DNA is rendered more accessible in hyperacetylated chromatin, which would allow efficient binding of Pho4 upon its translocation to the nucleus. Pho4 could then recruit partners to initiate the remodeling step and subsequent transcription. Indeed, Pho2 was implicated in Pho4 recruitment at the critical UAS1, while it plays a coactivator role at UAS2 (Barbaric et al, 1998). Accordingly, we show that Pho4 is clearly unable to bind PHO5 promoter in the absence of NuA4 action (Figure 9A and B). The absence of Pho4 binding also explains NuA4 requirement for chromatin remodeling. In agreement with a role in presetting the promoter for factor binding, Pho4 overexpression bypasses Pho2 requirement for its binding and makes NuA4 dispensable for PHO5 activation (Figure 9C) (Fascher et al, 1990). In addition, acetylation of specific lysines by NuA4 could be a mark for the binding of proteins or complexes required for the subsequent chromatin remodeling step. Indeed, histone acetylation by either SAGA or NuA4 HAT complexes was shown to stabilize SWI/SNF and SAGA binding to a promoter in vitro through bromodomain interactions (Hassan et al, 2002).
It is interesting to compare the co-regulated PHO5 and PHO8 genes. While PHO5 requires Esa1/NuA4 activity (this study), PHO8 needs Gcn5/SAGA (Gregory et al, 1999). Both genes require the Pho4 activator. Pho2 functions also on both genes, but is only essential for PHO5 induction (Munsterkotter et al, 2000). In agreement with a specific link between Pho2 and NuA4, Esa1 is also dispensable for PHO8 induction (Figure 1). Pho4 is able to bind PHO8 promoter in the absence of SAGA and SWI/SNF (Gregory et al, 1999), supporting our findings that specific NuA4-dependent acetylation of PHO5 under repressing conditions is required to allow efficient binding of Pho4 upon its translocation (Figure 9).
Our identification of transcription factor Pho2 as a specific direct target of the NuA4 complex is important, as this is the first natural NuA4 recruiter identified to date. Preliminary data support the Pho2-NuA4 functional link for transcription activation of other genes in vivo (A Auger and J Côté, unpublished data). While Pho2 was previously considered to always require cooperation with Pho4 for binding at PHO5, we clearly show the association of Pho2 under repressing conditions in the absence of Pho4 (Figures 6, 7 and 8). Furthermore, Pho2 was shown to bind by itself PHO5 promoter sites with high affinity in vitro (Barbaric et al, 1998; Terrell et al, 2002). The region of Pho2 responsible for NuA4 interaction maps away from the recently characterized transcription activation domain at the C-terminus (Bhoite et al, 2002). This could be explained by the different roles of Pho2 at UAS1 and UAS2 (Barbaric et al, 1998). Pho2 action at UAS1 is the key to Pho4 recruitment/binding, which in turn is essential for binding and remodeling at UAS2. Pho2 action at UAS2 is mainly for transactivation with Pho4. We suggest that the Pho2 homeodomain-containing region responsible for NuA4 recruitment is critical at UAS1, while the C-terminal transactivation domain plays a NuA4-independent role at UAS2. Accordingly, chromatin remodeling is required for Pho4 binding to UAS2, not UAS1 (Steger et al, 2003).
In conclusion, our data demonstrate that the NuA4 complex is recruited to the PHO5 promoter by the homeodomain factor Pho2 under repressing conditions. This recruitment creates a region of histone hyperacetylation that poises the promoter for activation. This pre-setting by NuA4 is essential to allow Pho4 binding after its nuclear translocation upon phosphate starvation, subsequent chromatin remodeling over the promoter region and transcription activation. Such presetting mechanism by homeodomain factor–HAT complex interactions may be conserved as the bicoid-related factor Pitx2 was shown to serve as a competence factor required for ordered recruitment of specific co-activator complexes, including the NuA4 highly related Tip60 complex, to the Cyclin D2 promoter (Kioussi et al, 2002).
Materials and methods
Yeast strains and plasmids
The esa1-Δ414 (LPY3123), esa1-L254P (LPY3500, used in Figure 9) and yng2Δ (QY203) mutant and isogenic wild-type strains were described (Clarke et al, 1999; Nourani et al, 2001). The QY303 strain was produced by cloning the esa1-Δ414 ts allele of LPY3123 into pFL36 (ARS/CEN/LEU2 vector with ESA1 promoter) to cover the ESA1 deletion in the BY4742 background. The haploid esa1-ts Pho80Δ strain was isolated after sporulation of the diploid strain obtained by crossing QY303 with pho80 deletion strain (BY4741 background). The 3xHA-Epl1 expression vector, epl1(1–380) mutant and TAP-tagged Epl1-expressing strains were described (Boudreault et al, 2003). The Epl1 protein was also tagged in vivo at the C-terminus with 13 Myc epitopes by PCR targeting to the EPL1 locus in the pho2 deletion mutant strain and isogenic wild type (BY4741 background). The Pho2 protein was similarly tagged with 3 HA epitopes in wild-type and pho4 mutant strains. The yeast EY0873 strain expressing Pho2 tagged at the C-terminus with the TEV-ZZ (named here TAP-tag for simplicity) cassette; the Pho4–GFP expression vector and the purified Pho4 antibodies were a generous gift from Dave Steger/Erin O'Shea. The YEP-PHO4 overexpression vector and anti-Pho2 serum were described (Fascher et al, 1990; Bhoite et al, 2002). Yeast strains were grown in minimal medium, YPD and induced in the phosphate-free synthetic medium prepared as described (Almer and Horz, 1986).
Northern blot, fluorescence microscopy and chromatin analysis
Yeast RNA was isolated and analyzed by Northern blots as described (Nourani et al, 2001). The PHO5, PHO8 and ACT1 probes used were ORFs, except for PHO5 in Figure 1, which was from the 5′ untranslated region (to show that the signal was not from other related PHO genes). For fluorescence miscroscopy, cells were fixed 5 min in cold methanol, washed once with cold acetone, twice with 1 × PBS and then resuspended in the same buffer. The DNA was stained with 60 mM DAPI. Chromatin structure mapping of the PHO5 promoter region with nucleases has been described (Svaren et al, 1995).
Chromatin immunoprecipitations
ChIPs were performed essentially as described (Hecht et al, 1999; Nourani et al, 2001). However, for precipitation using TAP-tagged proteins (Figure 5), 10 μl of IgG sepharose was used in 300 μl (approximately 1–2 mg of chromatin proteins) of sheared chromatin (0.2–1 kb). For the ChIPs in Figure 7, 10 μl of monoclonal (Myc or HA) antibodies and 4 μl of anti-Esa1 polyclonal antibody (Allard et al, 1999) were used for approximately 1–2 mg of chromatin proteins in an immunoprecipitation volume of 400 μl. After O/N incubation at 4°C, 20 μl of protein A beads were added and incubation continued for 90 min at room temperature. DNA was eluted from beads, precipitated and PCR reactions were carried out in a final volume of 50 μl by using 1–4% of immunoprecipitated material and 0.02–0.1% of input material. For ChIPs with anti-Esa1, an extra crosslinking step with 10 mM DMA was added before the actual treatment with formaldehyde. For the ChIPs in Figures 8 and 9, 1 μl of anti-Pho2 or anti-Pho4 antibodies were used and immunoprecipitations were quantified by real-time PCR (triplicates) using LightCycler (Roche). A noncoding region of chromosome V (positions 9716–9863) was used as control and the linear range of amplification was verified by serial dilutions for each pair of primers. Each experiment was repeated at the chromatin immunoprecipitation and PCR steps.
GST-pull-down and coimmunoprecipitation assays
The activation regions of various proteins as well as three sections of Pho2 were fused to GST (pGEX-4T3) and purified from Escherichia coli on glutathione sepharose following standard procedures. Protein concentrations were normalized by coomassie staining on gels and equivalent amounts were used in GST pull-down assays with purified NuA4 followed by nucleosomal HAT reactions essentially as described (Utley et al, 1998). The co-immunoprecipitation protocol was described in Nourani et al (2001). In all, 10 mg of total proteins was incubated for 2 h at 4°C with 50 μl of IgG beads in a final volume of 500 μl. Input (0.3%) and bound material (22.5%) were analyzed by Western blot.
Supplementary Material
Figure S1
Figure S2
Acknowledgments
We are indebted to Fred Winston for allowing A Nourani to perform experiments required for revisions of this paper while in his laboratory. We also thank Joelle Brodeur and Luc Gaudreau for performing ChIP experiments not included in the final version of this paper. We are grateful to Astride Clarke and Lorraine Pillus for the esa1 ts mutant strains, Dave Steger and Erin O'Shea for the PHO2-TEV-ZZ strain, the Pho4–GFP expression vector and Pho4 antiserum, David Stillman for Pho2 antiserum, Wolfram Horz for the YEP-Pho4 expression vector and Nicolas Lacoste for the Dot1-TAP strain. We thank Dave Steger, Mike Kladde, Colin Goding and Wolfram Horz for advice and useful discussions. This work was supported by a grant from the Canadian Institutes of Health Research (CIHR) to JC. AN and RTU were CIHR post-doctoral fellows. JC is a CIHR Investigator.
References
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Côté J (1999) NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J 18: 5108–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almer A, Horz W (1986) Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J 5: 2681–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaric S, Munsterkotter M, Goding C, Horz W (1998) Cooperative Pho2-Pho4 interactions at the PHO5 promoter are critical for binding of Pho4 to UASp1 and for efficient transactivation by Pho4 at UASp2. Mol Cell Biol 18: 2629–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaric S, Reinke H, Horz W (2003) Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol Cell Biol 23: 3468–3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaric S, Walker J, Schmid A, Svejstrup JQ, Horz W (2001) Increasing the rate of chromatin remodeling and gene activation—a novel role for the histone acetyltransferase Gcn5. EMBO J 20: 4944–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoite LT, Allen JM, Garcia E, Thomas LR, Gregory ID, Voth WP, Whelihan K, Rolfes RJ, Stillman DJ (2002) Mutations in the pho2 (bas2) transcription factor that differentially affect activation with its partner proteins bas1, pho4, and swi5. J Biol Chem 277: 37612–37618 [DOI] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD (2003) Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell 11: 1587–1598 [DOI] [PubMed] [Google Scholar]
- Boudreault AA, Cronier D, Selleck W, Lacoste N, Utley RT, Allard S, Savard J, Lane WS, Tan S, Côté J (2003) Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev 17: 1415–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AS, Lowell JE, Jacobson SJ, Pillus L (1999) Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol 19: 2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrin LK, Mann RK, Kayne PS, Grunstein M (1991) Yeast histone H4 N-terminal sequence is required for promoter activation in vivo. Cell 65: 1023–1031 [DOI] [PubMed] [Google Scholar]
- Fascher K-D, Schmitz J, Horz W (1990) Role of Trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J 9: 2523–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PD, Schmid A, Zavari M, Lui L, Berger SL, Horz W (1998) Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol Cell 1: 495–505 [DOI] [PubMed] [Google Scholar]
- Gregory PD, Schmid A, Zavari M, Munsterkotter M, Horz W (1999) Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J 18: 6407–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL (2002) Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111: 369–379 [DOI] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M (1999) Mapping DNA interaction sites of chromosomal proteins. Crosslinking studies in yeast. Methods Mol Biol 119: 469–479 [DOI] [PubMed] [Google Scholar]
- Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, Ohgi KA, Lin C, Gleiberman A, Wang J, Brault V, Ruiz-Lozano P, Nguyen HD, Kemler R, Glass CK, Wynshaw-Boris A, Rosenfeld MG (2002) Identification of a Wnt/Dvl/beta-Catenin → Pitx2 pathway mediating cell-type-specific proliferation during development. Cell 111: 673–685 [DOI] [PubMed] [Google Scholar]
- Lenfant F, Mann RK, Thomsen B, Ling X, Grunstein M (1996) All four core histone N-termini contain sequences required for the repression of basal transcription in yeast. EMBO J 15: 3974–3985 [PMC free article] [PubMed] [Google Scholar]
- Munsterkotter M, Barbaric S, Horz W (2000) Transcriptional regulation of the yeast PHO8 promoter in comparison to the coregulated PHO5 promoter. J Biol Chem 275: 22678–22685 [DOI] [PubMed] [Google Scholar]
- Nourani A, Doyon Y, Utley RT, Allard S, Lane WS, Côté J (2001) Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol Cell Biol 21: 7629–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JL, Iyer VR, Brown PO, Struhl K (2000) Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell 6: 1297–1307 [DOI] [PubMed] [Google Scholar]
- Reinke H, Horz W (2003) Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell 11: 1599–1607 [DOI] [PubMed] [Google Scholar]
- Santisteban MS, Kalashnikova T, Smith MM (2000) Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103: 411–422 [DOI] [PubMed] [Google Scholar]
- Steger DJ, Haswell ES, Miller AL, Wente SR, O'Shea EK (2003) Regulation of chromatin remodeling by inositol polyphosphates. Science 299: 114–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svaren J, Horz W (1997) Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem Sci 22: 93–97 [DOI] [PubMed] [Google Scholar]
- Svaren J, Venter U, Horz W (1995) In vivo analysis of nucleosome structure and transcription factor binding in Saccharomyces cerevisiae. Meth Molec Genet 6: 153–167 [Google Scholar]
- Terrell AR, Wongwisansri S, Pilon JL, Laybourn PJ (2002) Reconstitution of Nucleosome Positioning, Remodeling, Histone Acetylation, and Transcriptional Activation on the PHO5 Promoter. J Biol Chem 277: 31038–31047 [DOI] [PubMed] [Google Scholar]
- Utley RT, Ikeda K, Grant PA, Côté J, Steger DJ, Eberharter A, John S, Workman JL (1998) Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394: 498–502 [DOI] [PubMed] [Google Scholar]
- Vogelauer M, Wu J, Suka N, Grunstein M (2000) Global histone acetylation and deacetylation in yeast. Nature 408: 495–498 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2


