Abstract
Dyneins and kinesins move in opposite directions on microtubules. The question of how the same-track microtubules are able to support movement in two directions remains unanswered due to the absence of details on dynein–microtubule interactions. To address this issue, we studied dynein–microtubule interactions using the tip of the microtubule-binding stalk, the dynein stalk head (DSH), which directly interacts with microtubules upon receiving conformational change from the ATPase domain. Biochemical and cryo-electron microscopic studies revealed that DSH bound to tubulin dimers with a periodicity of 80 Å, corresponding to the step size of dyneins. The DSH molecule was observed as a globular corn grain-like shape that bound the same region as kinesin. Biochemical crosslinking experiments and image analyses of the DSH–kinesin head–microtubule complex revealed competition between DSH and the kinesin head for microtubule binding. Our results demonstrate that dynein and kinesin share an overlapping microtubule-binding site, and imply that binding at this site has an essential role for these motor proteins.
Keywords: cryo-electron microscopy, dynein, kinesin, protein structure, tubulin
Introduction
Dyneins and kinesins are microtubule-based molecular motors that play important roles in various cellular processes, including axonal transport, chromosome segregation during mitosis, and flagellar assembly and motility (Vallee and Sheetz, 1996; Hirokawa, 1998; Vale, 2003). Dyneins use ATP to transport their cargo toward the minus end of microtubules, whereas kinesins generally move towards the plus end. Although dyneins and kinesins share the same track microtubules, the architecture of the two motor proteins is completely different, and relatively little is known about the mechanism of dynein motility.
Among the numerous polypeptides that compose the massive dynein complex (1000–2000 kDa), the heavy chain acts as the fundamental motor unit. The dynein heavy chain contains three domains: a globular head with ATPase activity, a cargo-binding stem and a microtubule-binding stalk (Figure 1A). The long, slender stem, which is located at the N-terminus of the heavy-chain sequence, binds to cargo and interacts with various intermediate and light chains. The amino-acid sequence of the stem varies among dyneins, allowing the motor to accept various cargoes (Tynan et al, 2000). The globular head, which comprises the C-terminal two-thirds of the heavy chain, has a ring-like arrangement of six subdomains, as observed by electron microscopy (EM) (Samso et al, 1998; Burgess et al, 2003). Recent sequence analyses have indicated that each subdomain is composed of an ∼220-residue AAA (ATPases Associated with various cellular Activities) superfamily of mechano-enzymes (Neuwald et al, 1999). The first four AAA domains (D1–D4, Figure 1A) include P-loop motifs that are well conserved among dyneins, with the first P-loop being absolutely conserved (Gibbons et al, 1991; Ogawa, 1991). In contrast, the last two AAA domains have no P-loops. Only D1 displays measurable ATPase activity, and the next three AAA domains (D2–D4) are thought to regulate dynein activity (Shiroguchi and Toyoshima, 2001). Recently, Burgess et al (2003) observed a change in the stem–stalk angle, depending on the nucleotide state of the head, which may represent the power stroke of dynein.
Figure 1.
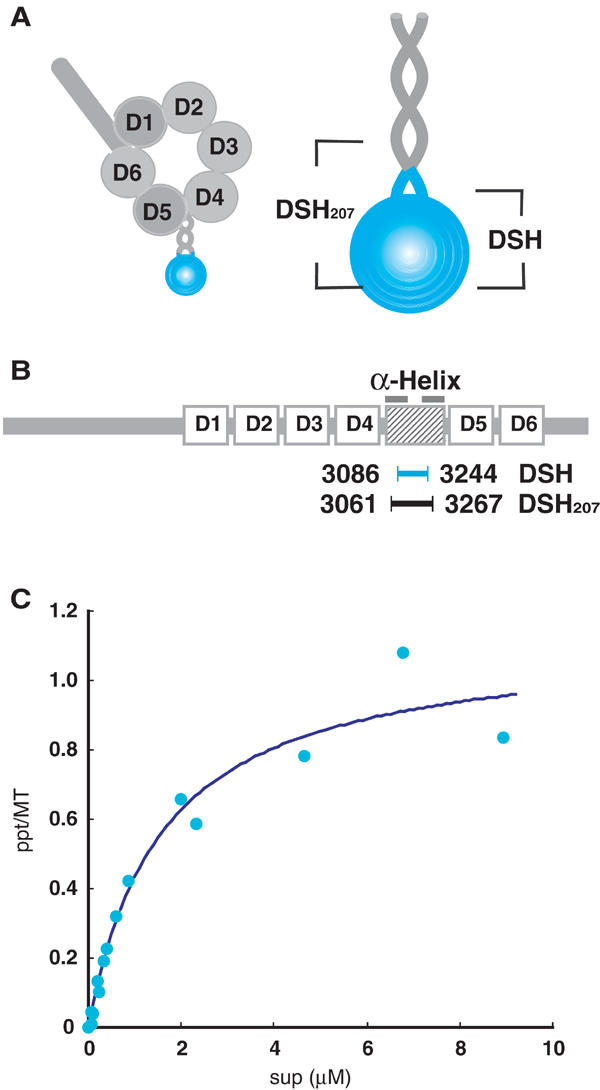
Characterization of the dynein stalk head (DSH). (A) Proposed structure of the dynein heavy chain and an enlarged view of the dynein stalk. The DSH sequence used in this study is colored blue. (B) Domain organization of the dynein heavy chain. D1–D6 represent six AAA domains. The two bars on the shaded box represent helices that form an antiparallel coiled-coil. DSH (blue) and DSH207 (black) are located between the helices. (C) Binding isotherms for DSH. The dissociation constant (Kd) and binding stoichiometry were calculated by fitting the data to rectangular hyperbolae. The dissociation constant (Kd) was 1.59 μM with a stoichiometry of 1.14.
The slender stalk protrudes from the head, and is topped with a small globular domain that allows dynein to bind microtubules in an ATP-sensitive manner. The stalk structure was first visualized by quick-freeze/deep-etch EM as a 10–15 nm ‘B-link' in the flagellar outer dynein arm (Goodenough and Heuser, 1982, 1984). A microtubule co-sedimentation assay using recombinant stalk proteins demonstrated that the amino-acid sequence encoding the stalk lies between the fourth and fifth AAA domains (Gee and Vallee, 1998). The stalk is composed of an antiparallel coiled-coil and an intervening segment of 125 residues. Alanine scanning of dynein led to the identification of three clusters of amino acids within the dynein stalk that are important for physical contact with microtubules (Koonce and Tikhonenko, 2000).
Although these studies qualitatively show that the stalk domain is responsible for microtubule binding, the binding/dissociation constants, binding stoichiometry and structural features of dynein–microtubule interactions are yet to be elucidated. Such quantitative studies have previously been difficult, due to nonspecific interactions occurring at the fragment ends of expressed dynein stalks (Gee et al, 1997), In addition, structural studies on dynein–microtubule interactions are of particular interest for comparison with the extensively studied kinesin–microtubule structures (Amos and Hirose, 1997; Mandelkow and Hoenger, 1999; Rice et al, 1999; Kikkawa et al, 2000). These comparison studies should confirm whether dyneins and kinesins bind to the same region, and provide evidence for commonality or difference between those oppositely directed motors.
To resolve the microtubule-binding interface of dynein, we prepared recombinant dynein stalk head (DSH) protein and analyzed its biochemical and structural properties by cryo-EM and helical three-dimensional (3D) image reconstruction. The DSH protein competed efficiently with intact cytoplasmic dynein for microtubule binding, showing that the microtubule-binding activity of recombinant DSH is similar to that of native dynein. A DSH–microtubule co-sedimentation assay revealed that one DSH protein bound to one tubulin dimer. The 80 Å periodicity derived from the cryo-EM image of the DSH–microtubule complex allowed us to determine the path length of dynein. The high-resolution 3D structure of DSH appeared as a globular corn grain-like shape. Biochemical crosslinking of a mixture of DSH, kinesin head (KH) and microtubules, followed by further cryo-EM analyses revealed that the two motors competed for a single binding region on a tubulin dimer. Together with the 3D structure of the DSH–microtubule complex, our data provide evidence that DSH and kinesin share an overlapping binding site on tubulin.
Results
Recombinant DSH is monomeric
Gee et al (1997) expressed a recombinant dynein stalk fragment in insect cells, but the fragments aggregated due to nonspecific interactions at their ends. To address this issue and to determine the strength and stoichiometry of DSH binding to microtubules, we generated a new construct expressing the globular region of the dynein stalk in Escherichia coli (DSH; Figure 1B). Purified recombinant DSH protein did not aggregate. Although its size as estimated by gel filtration (31 kDa; data not shown) was larger than the expected molecular weight of 19k, the estimated size is clearly below that of the corresponding dimer, indicating that recombinant DSH exists as a monomer. The larger size is probably due to the elongated shape of the molecule, as discussed below.
Successful expression of stable monomeric DSH protein allowed us to determine the strength and stoichiometry of DSH binding to microtubules. Varying concentrations of DSH were incubated and co-sedimented with microtubules. Quantification of microtubule-bound DSH disclosed a dissociation constant (Kd) of 1.59 μM, which is higher than the values previously obtained for kinesins in the presence of AMP-PNP (4 nM–0.28 μM) (Lockhart et al, 1995; Thormahlen et al, 1998; Kikkawa et al, 2000). At saturation, approximately 1.1 DSH molecules bound per tubulin dimer (Figure 1C).
DSH inhibited the binding of cytoplasmic dynein to microtubules
To confirm that the microtubule binding of DSH is functionally comparable to that of native dyneins, we analyzed the effect of DSH to the microtubule binding of cytoplasmic dynein. Here, we measured the landing rate, the frequency with which microtubules land on the dynein-covered glass surface (Figure 2). The landing assay is superior to conventional biochemical assays, because the assay is a direct visualization of the binding. In the absence of nucleotide and DSH, 66 microtubules/field landed, whereas the landing rate decreased to five microtubules/field with increasing amount of DSH. The decrease is explained by assuming that the DSH occupied the similar binding region as cytoplasmic dynein and decreased the available binding site. In fact, the degrees of inhibition (28, 60 and 93%) well correlated with the presumed occupancies of the microtubule by DSH (12, 32 and 62%, respectively). As a control, we also measured the landing rate in the absence of cytoplasmic dynein, and no microtubule landing was observed (data not shown). Therefore, this landing assay demonstrated that DSH inhibits the binding of cytoplasmic dynein to microtubules, and shows that the microtubule binding of DSH is comparable to that of cytoplasmic dyneins.
Figure 2.
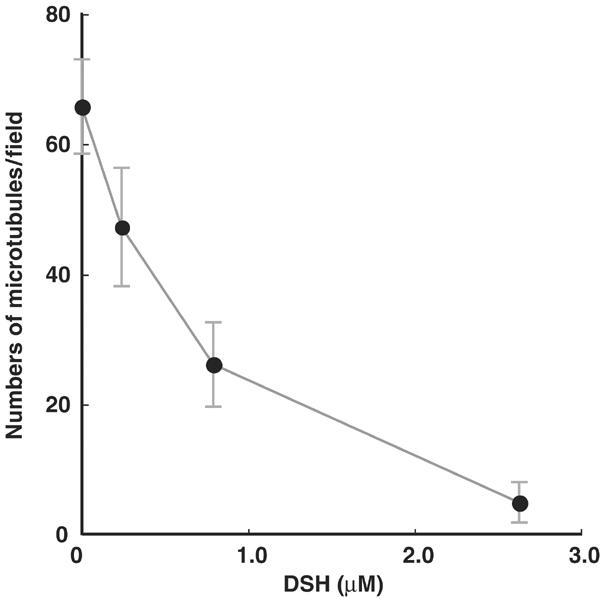
Numbers of microtubules landing as a function of DSH concentration. The numbers of microtubules attached to the surface covered with cytoplasmic dynein in the presence of different DSH concentrations. Each data point represents observations of 15 different fields (48 μm × 44 μm).
Cryo-EM of the DSH–microtubule complex
To elucidate the mechanism by which DSH interacts with microtubules at the molecular level, we analyzed the DSH–microtubule complex using cryo-EM. In the cryo-EM images, the globular heads of the DSH appeared as small dots on the microtubules (Figure 3A). Computed diffraction patterns of these images disclosed additional layer lines at around 80 Å (Figure 3C), which are generally absent from the diffraction patterns of microtubules, due to the similarity between α- and β-tubulin (Figure 3D) (Grimstone and Klug, 1966; Amos and Klug, 1974). The presence of 80 Å layer lines in DSH-bound microtubules indicates that DSH-specific binding sites occur on the microtubules with a longitudinal periodicity of 80 Å.
Figure 3.
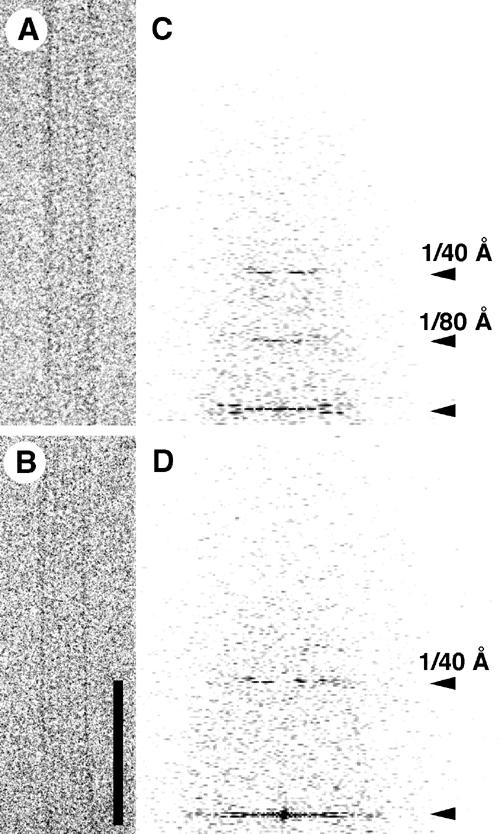
Cryo-EM images of a DSH–microtubule complex. Cryo-EM images (bar=800 Å) showing (A) a DSH–microtubule complex and (B) an undecorated microtubule. (C, D) Computed diffraction patterns of the images.
3D structure of the DSH–microtubule complex
The 15 Å resolution 3D structure of the DSH–microtubule complex was reconstructed by helical image analysis of cryo-EM images (Hirose et al, 1995; Kikkawa et al, 1995; Sosa et al, 1997). At this resolution, several structural features become discernible.
The interaction between DSH and microtubule is of particular interest. Surface rendering (Figures 4A, B and 7B) demonstrated that DSH is a corn grain-shaped structure attached to the ridge of a single microtubule protofilament, spanning between α- and β-tubulins (Figure 7B). The structure displays little direct interaction with adjoining protofilaments. At this resolution, DSH binding induced no significant structural changes in microtubules. The polarity of the microtubule was determined from anti-clockwise skewing of the protofilament, as observed from the plus end (Figure 4B) (Chretien et al, 1996). A side view of the complex revealed two major interactions between DSH and the microtubule, occurring on the left side of the protofilament (Figure 4A, arrowheads), which will be discussed in detail later. A top view showed DSH tilted in a clockwise direction relative to the axis of tubulin (Figure 4B, dashed lines).
Figure 4.
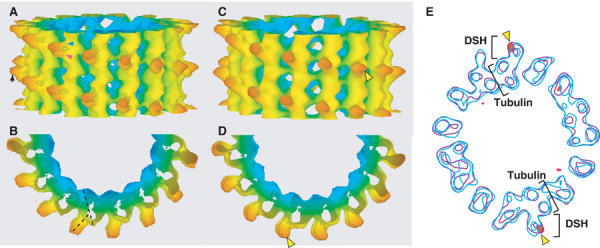
3D maps of DSH–microtubule complexes. (A) The 15 Å resolution map of the DSH–microtubule complex. The isosurface is colored according to the radius. In all figures, the plus end of the microtubule is oriented up, unless otherwise specified. Arrowheads show two major interactions between DSH and the microtubule. (B) View from the plus end of the microtubule. Axes of tubulin and DSH are depicted with dashed black lines. (C, D) The 3D 20 Å resolution map of the DSH207–microtubule complex. Yellow arrowheads show the positions where t-test detected a difference at a level of 99%. (E) Overlaid cross-sections of difference map between the DSH–microtubule and DSH207–microtubule complexes (red) and the density maps of DSH–microtubule (blue) and DSH207–microtubule complexes (purple). The contour of the difference map was chosen to enclose regions of significant difference.
Figure 7.
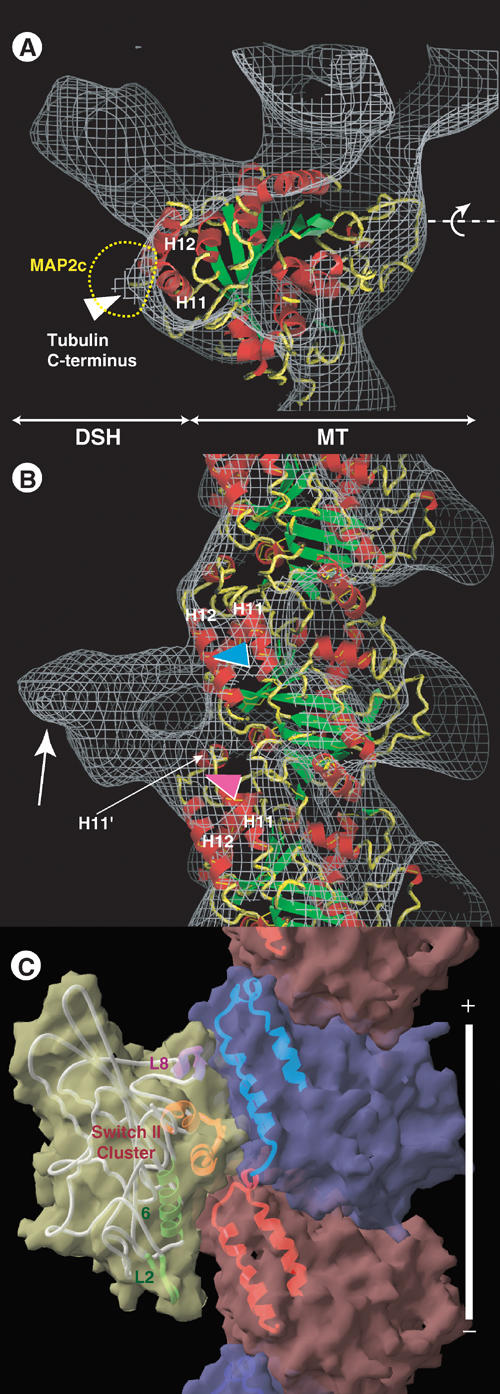
Docking of the tubulin structure onto the EM-derived structure of the DSH–microtubule. (A) Superimposition of the EM-derived map (gray chicken-wire surface) of the DSH–microtubule complex combined with the atomic model of tubulin (ribbon diagram). (B) Side view. Several elements of the motor and tubulin structure are specified to facilitate orientation. In (A), the dashed yellow line indicates the MAP2c/tau outer shape. (C) Side view of the atomic models of the KIF1A–microtubule complex in an ATP-like state.
Although the atomic model of DSH is not currently available, the DSH observed in this reconstruction is also intriguing. The DSH molecule was a globular corn grain-like shape and its long and short axes were approximately 45 and 30 Å in length, respectively (Figure 4A). The volume of the DSH molecule is about half that of the KH (∼40 kDa), which provided clear evidence that DSH is monomeric. The size is also consistent with those previously reported for the globular domains at the tip of the dynein stalk in quick-freeze deep-etch and rotary shadow EM images (Goodenough and Heuser, 1982, 1984; Gee et al, 1997). In those quick-freeze deep-etch EM images, the globular domain is connected to the large head domain by an antiparallel α-helical coiled-coil (Gee et al, 1997; Koonce, 1997). The DSH protein contains the amino-acid sequences that are predicted to form a coiled-coil structure of dynein stalk at both the N- and C-terminal ends. In our reconstruction, we observed an ∼10 Å projection pointing away from the microtubule, which was parallel to the DSH axis (Figures 4A and 7B, arrow). This projection may be the coiled-coil domain.
Structure of a longer DSH
To determine the position of the coiled-coil domain, we made a longer DSH expression construct (DSH207), which included 207 aa in total with an extra 25 aa at both the N- and C-termini. The 3D structure of DSH207 appeared to be similar to that of DSH (Figure 4C and D), in that both molecules bound to the same position on the microtubule, and both stuck out at the same angle (Figure 4). However, the protrusion of DSH207 (Figure 4C and D, yellow arrowheads) was significantly longer than that of DSH as indicated by a Student's t-test at the 99% significance level (Figure 4E, shown with red on the contour maps). Although the protrusion was shorter than the expected 40 Å (based on the assumption that the extra 25 aa form an α-helical coiled-coil), this may be due to the flexibility of the stalk region. Also, the similarity of the two structures indicates that DSH and DSH207 bind to microtubule by common globular domain, and not by the N- or C-termini.
Biochemical competition between DSH and KH
In our 3D representation of DSH–microtubule binding (Figure 4A and B), DSH is located on the left side of the protofilament, spanning the two tubulin monomers with a pair of connections. Surprisingly, this configuration is similar to the binding between kinesins and microtubules. Thus, we next examined whether DSH and KH compete for the same binding site on tubulin.
Initially, we tested for competition between DSH and KH binding to microtubules, using the zero-length crosslinker 1-ethyl-3-[3-(dimethylamino)propyl]carbodiimide (EDC). When DSH and microtubules were incubated in the presence of EDC, a major crosslinked product (Figure 5B, lane 1, asterisk) comprising one DSH and one tubulin monomer (75 kDa) was observed. This 75 kDa band was stained with both anti-α-tubulin and anti-β-tubulin antibodies on a Western blot (data not shown), consistent with our cryo-EM observation that DSH spans the two tubulin monomers. In view of the finding that kinesin is also crosslinked to both α- and β-tubulins (Walker, 1995; Tucker and Goldstein, 1997), we assessed whether DSH and KH compete for crosslinking to tubulin. At increasing concentrations of KH (Figure 5B), the amount of the 75 kDa product (DSH+tubulin) decreased, concomitant with an increase in a new 97 kDa product (KH+tubulin) (Figure 5D). In the reverse experiment, where increasing amounts of DSH were added to constant amounts of KH–microtubule mixture, the 75 kDa product increased in favor of the 97 kDa product, which decreased (Figure 5C and E). These crosslinking data demonstrated that DSH and KH compete for microtubule binding, indicating that the two motors share an overlapping binding site.
Figure 5.
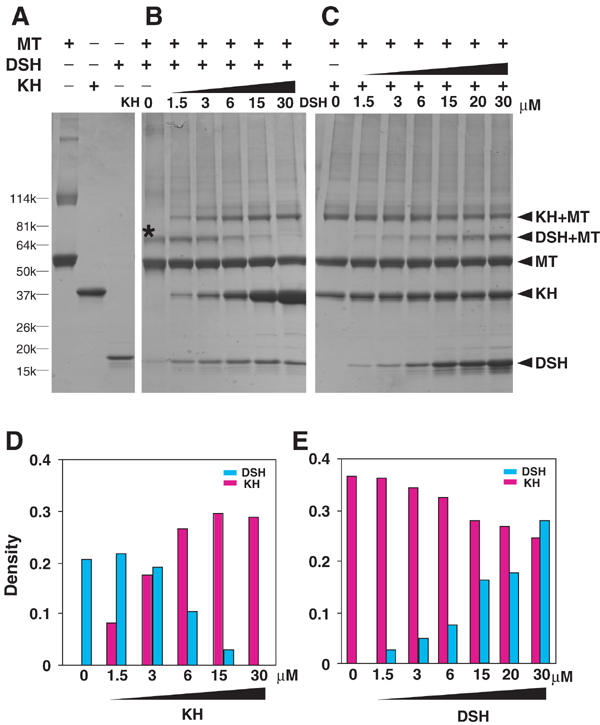
Competition between DSH and KH in the crosslinking reaction. (A) SDS–PAGE images of crosslinked combinations of microtubule, KH and DSH. Competition experiments between DSH and KH were performed (B) in the presence of increasing amounts of KH with 6 μM DSH and (C) in the presence of increasing amounts of DSH with 6 μM KH. A major crosslinked product of tubulin and DSH is marked with an asterisk (*). The molecular compositions of crosslinked products are shown on the right. MT denotes microtubule. (D, E) Densities of DSH–tubulin product and KH–tubulin product in (B, C), expressed in arbitrary units.
Relative positions of motors and tubulin dimers
We then re-examined our high-resolution structures to confirm that DSH and kinesin bind across the same tubulin subunit interface (Figure 6A), rather than occupying similar but different positions (Figure 6B). We differentiated one tubulin from another and determined the positions of the tubulin dimers relative to the motors. In the reciprocal space, an 80 Å layer line contained the signals differentiating between the two tubulins. Unfortunately, the 80 Å layer line signal from each individual microtubule image is below the background level (Figure 3D). However, we were able to improve the signal to above the background level by averaging data from a large number of molecules (see Materials and methods).
Figure 6.
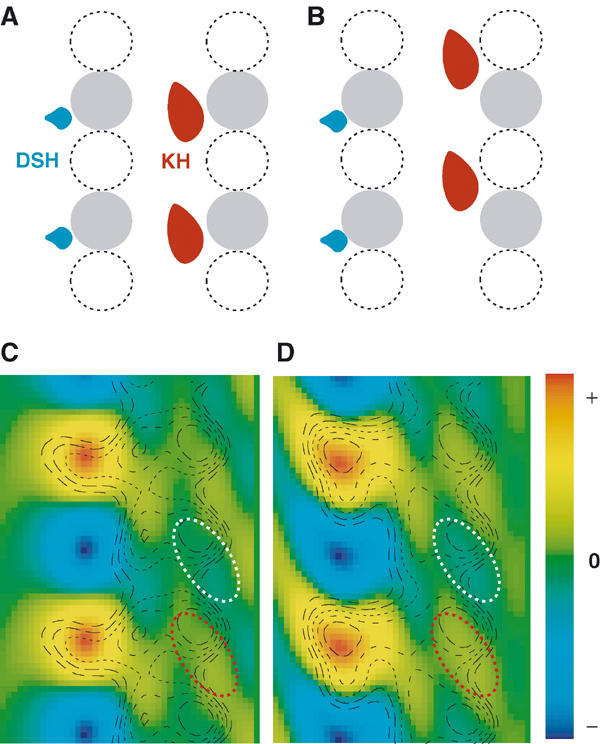
Relative positions of DSH, KH and tubulin dimers. (A, B) Two possible arrangements of the relative positions of DSH, KH and the tubulin dimer. One tubulin (e.g. β-tubulin) is shown in gray, while the other (e.g. α-tubulin) is shown with a dashed line. (C) DSH–microtubule complex: a section of the density map generated from the 80 Å layer line, shown as a color map. The radial projection map is overlaid as a contour plot to orient the density map. (D) KH–microtubule complex. White and red ellipses enclose lower and higher density stripes, respectively, in the microtubule.
Using the averaged data set, we generated density maps highlighting only the 80 Å layer lines (Figure 6B and C, colored maps), and superimposed radial projection maps as contour plots to orient the density maps. In these superimposed maps, positions of motors and tubulin dimers are clearly observed. On the left side, strong signals indicated the center of gravity of the motor molecule. On the right side, stripes on the inner side of the microtubule correspond to α- and β-tubulins, representing the high density of one tubulin monomer (Figure 6C and D, red ellipsoid) compared with the low density of the other (white ellipsoid).
As shown in Figure 6C, DSH is located at the same height as the high-density region in the inner side of each microtubule. This is also true for the KH–microtubule complex (Figure 6D). Comparing the positions of the tubulins relative to the motors allowed us to conclude that DSH and KH bind to the same tubulin subunit, rather than to a different one. Together with the recent study on KH–microtubule complex (Krebs et al, 2004), both motors bind to the intradimer interface. Thus, biochemical crosslinking experiments and these structural data confirm that DSH and KH compete for an overlapping binding site on tubulin dimers.
Docking tubulin atomic model onto the DSH–microtubule complex
Having concluded that DSH and KH share an overlapping binding site, the atomic model of tubulin (Lowe et al, 2001) was docked onto the DSH–microtubule complex, and the resulting structure was compared to that of the monomeric kinesin (KIF1A)–microtubule complex (Kikkawa et al, 2000). In a side view (Figure 7B), two major connections are observed between DSH and the microtubule. The upper connection is located toward the middle of H12 in the upper tubulin monomer (Figure 7B, blue arrowhead), while the lower connection is located near the pocket formed by the C-terminal part of the upper tubulin H12 and H11′ of the lower tubulin (Figure 7B, red arrowhead). These two binding sites are also utilized in tubulin–kinesin binding. The upper and lower connections correspond to the L8 and switch II clusters of kinesin, respectively (Figure 7C). In a top view (Figure 7A), DSH is seen bound to the left side of the protofilament, again similar to the binding behavior of kinesin. According to a recent report (Al-Bassam et al, 2002), the right side of the protofilament, including the tubulin C-terminus, is the main binding region for MAP2c and tau (Figure 7A, yellow dashed line).
Discussion
Interaction between dynein and microtubules
In this study, DSH was used to determine the biochemical and structural properties of the dynein microtubule-binding domain: the dissociation constant and the 80 Å periodicity, which are fundamental parameters for understanding dynein mechanism.
The binding competition experiment showed that DSH inhibits microtubules binding to cytoplasmic dynein. Thus, microtubule binding of DSH seems comparable to that of native dyneins. Our work includes the first quantitative estimation of the dynein–microtubule binding dissociation constant (Kd), yielding a value of 1.59 μM. This value is higher than the values for kinesins. This higher Kd, which indicates weaker binding, may be related to the smaller contact area between DSH and the microtubule (∼600 Å2), compared to the area between kinesin and microtubule (∼1200 Å2 for monomeric kinesin KIF1A in the ATP-like state) (Kikkawa et al, 2001).
Our biochemical and structural data also provided fundamental information to clarify the track used by dynein. We found that one DSH binds to one tubulin dimer with 80 Å periodicity. Structurally, a periodicity of 240 Å was previously observed in EM images of axonemal dynein stalks in situ (Goodenough and Heuser, 1982; Burgess et al, 1991; Burgess, 1995), and also in microtubules decorated with outer dynein arms in vitro. On the contrary, 80 Å periodicity was observed as the processive bead movement attached to 22S dynein (Hirakawa et al, 2000) and inner-arm dynein c (Sakakibara et al, 1999). Although the unit of the bead along the microtubule is 80 Å, the path length, which is the distance between consecutive binding sites as dynein moves along the microtubule (Howard, 2001), can be predicted as the integer multiples of 80 Å but less than 240 Å, together with the 240 Å periodical binding of axonemal dynein. Thus, our use of DSH, which is small enough to avoid steric hindrance when binding to the microtubule, allowed us to define a unitary path length of 80 Å.
Dynein and kinesin bind to an overlapping binding site
Here, we provide two independent lines of evidence indicating that DSH and kinesin share an overlapping binding site: biochemical competition and the relative positions of motors and tubulin dimers. At 15 Å resolution, the binding site can be localized to about 10 residues; both dynein and kinesin bind to the H11 (403–409) and the C-terminal part of H12 (420–430). Thus, it is interesting to consider the functional implications of a shared binding site.
In a cellular context, the surprising result that anterograde and retrograde motors share the binding site raises the question as to how dynein passes kinesin when they encounter each other. We observed that kinesins in ATP state have lower Kd values than DSH, indicating stronger binding to microtubules. Based on this observation, we offer a simple kinesin priority model. In this model, a kinesin has the ‘right-of-way' and remains on its protofilament track when it encounters dynein. This is supported by the observations that kinesin binds more tightly to the microtubule than individual dynein stalk, and follows the protofilament axis (Ray et al, 1993). When this ‘traffic jam' occurs, dynein may detour to a neighboring protofilament, which allows it to pass kinesin. This is supported by reports that dynein exhibits greater lateral movement among microtubule protofilaments (Vale and Toyoshima, 1988; Wang et al, 1995).
It is also of great interest to speculate how dynein and kinesin evolved to bind to the same region on tubulin, which comprises only 10–15% of the outer surface area. This is particularly surprising given that dynein and kinesin belong to different enzyme classes. Dynein is a member of the AAA ATPase protein family, while kinesin, together with myosin, possibly originates from a common ancestral GTP-binding protein (Kull et al, 1996). Additionally, there are no apparent amino-acid sequence similarities between the microtubule-binding domains of the DSH and KH proteins. Therefore, it is unlikely that the two motors are derived from a common ancestral protein. Rather, the two motors likely evolved independently from different ancestral proteins, and the common binding site is an example of convergent evolution. Ancestral proteins may have evolved to utilize the site on tubulin, because this binding site may play an essential role for the motility. One possible explanation for this convergent evolution is that both dynein and kinesin recognize polymerized tubulin by identifying conformational changes. Since the binding region is located at the interface between tubulin monomers, motors may distinguish between tubulin polymerization/depolymerization states based on the binding site conformation. Although the power stroke of the motor has attracted attention, our finding, the shared binding site between kinesin and dynein, underlines potential importance of motor–microtubule interaction in the motility.
Motors and MAPs bind to different regions
We observed that the motor-binding region for kinesin and dynein is on the left side of the protofilament, distinct from the binding domain of microtubule-associated proteins (MAPs) located on the right side. This is in contrast to two previous reports that MAP2 and cytoplasmic dynein both bind to the C-terminus of tubulin (Paschal et al, 1989; Hagiwara et al, 1994). In both reports, native MAP2 and dynein were used in a competition assay. As these native proteins are large (cytoplasmic dynein is 500 Å long and MAP2 is 1500 Å long), it is likely that MAP2 excludes dynein–microtubule binding by steric inhibition.
Based on the fact that motors and MAPs bind to different regions, we propose that the left side of the protofilament provides a foothold for the motors, whereas the right side, including the tubulin C-terminal flexible domain (E hook), is a site for MAP binding and regulation of motor processivity (Figure 8). This hypothesis is compatible with reports that ATPase activity and processivity of dynein were reduced by cleaving the E hook with subtilisin (Paschal et al, 1989; Wang and Sheetz, 2000), and that MAP binding to the right side of the protofilament decreases the run length for both kinesin and dynein (Seitz et al, 2002). In order to test this hypothesis, future studies, such as mutational analyses of tubulin, will be required.
Figure 8.
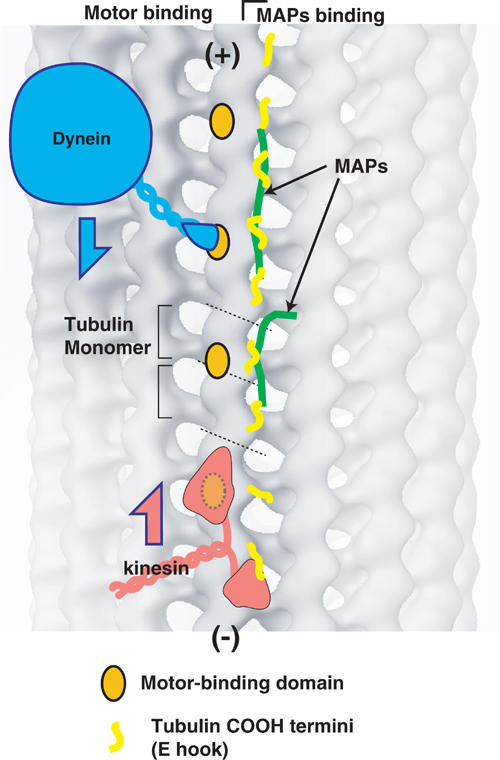
Model for describing the mechanism by which microtubules support molecular motor motility. Binding sites for both dynein and kinesin are presented as orange spots. The C-termini of tubulin molecules, which represent the main binding site for MAP, are depicted with yellow lines. Both dynein and kinesin move between these motor-binding sites with a step size of 8 nm. The C-termini (E hooks) of tubulin on the right of the protofilament function as guides for processivity of the motors. MAP proteins and post-translational modification of tubulin may regulate motor processivity by altering the flexibility and charges of the E hooks.
Materials and methods
Expression and purification of proteins
The DSH and DSH207 expression vectors were constructed by inserting a DNA fragment encoding residues D3086–K3244 and M3061–K3267, respectively, of the Saccharomyces cervisiae cytoplasmic dynein into the Nde1–Xho1 sites of pET17b (Novagen). The resulting constructs were transformed into BL21(DE3) cells for expression. Cells were pre-cultured in x3YT medium supplemented with ampicillin (0.1 mg/ml) and 0.5% glucose for 12 h at 37°C, and transferred to glucose-free medium until OD600=0.6. After 4 h of induction with 0.8 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 23°C, cells were centrifuged and resuspended in buffer A (10 mM Pipes-Na, pH 7, 1 mM MgCl2, 0.5 mM EDTA and protease inhibitors) and lysed twice using a Microfluidizer M-110L (Microfluidics). The soluble protein fraction was mixed with SP-sepharose equilibrated with buffer A, and eluted with 0.3 M NaCl. Peak fractions were diluted to lower the ionic strength and loaded onto Resource S columns (Amersham Biosciences). A linear NaCl gradient (0–0.5 M) was applied to elute the proteins. Purified DSH or DSH207 was stored in liquid nitrogen for later use. Porcine or bovine tubulin was purified as described previously (Shelanski et al, 1973). KH protein, which includes amino acids 4–332 of the Drosophila kinesin heavy chain (Yang et al, 1989) with a 6xHis tag at the C-terminus, was expressed and purified as previously reported (Kikkawa et al, 1994).
Co-sedimentation assays
Co-sedimentation assays were performed in buffer A with 50 mM NaCl and 20 μM Taxol. DSH (0.18–21 μM) was mixed with 3.5 μM microtubules and incubated for 10 min at 25°C. Samples were centrifuged at 45 000 g for 20 min at 25°C. After removing the supernatant, pellets were rinsed and resuspended in buffer A with 50 mM NaCl. Supernatants and pellets were analyzed by SDS–PAGE on either 15 or 4–20% gradient polyacrylamide gels. Coomassie blue-stained gels were scanned and analyzed using the NIH image 1.62.
Microtubule landing assay on the cytoplasmic dynein-coated surface
Cytoplasmic dynein was prepared from porcine brain (Bingham et al, 1998). The activity of the dynein was confirmed by the microtubule gliding in the presence of ATP. The landing assay (deCastro et al, 1999) was performed in assay buffer (25 mM K-acetate, 10 mM Pipes-K, pH 7, 4 mM MgSO4, 1 mM EGTA and 1 mM DTT) with the following sequence: (1) 5 mg/ml bovine serum albumin for 5 min; (2) 40 μg/ml cytoplasmic dynein for 3 min; (3) 0.5 mg/ml protein A for 2 min; (4) washed twice with the assay buffer; (5) 0.15 μM of Taxol-stabilized microtubules were sheared to 5–10 μm in length by pipetting, and then mixed with DSH (0, 0.24, 0.79 and 2.63 μM) in the assay buffer with 10 mM NaCl for 3 min; and (6) twice with the assay buffer to wash out unbound microtubules. The chamber was observed with dark-field optical microscope (Toba et al, 2004), and microtubules that landed onto the glass surface were counted in 15 randomly selected fields (48 μm × 44 μm).
Biochemical crosslinking
In all, 6 μM DSH with increasing amounts of KH (1.5–30 μM), or 6 μM KH with increasing amounts of DSH (1.5–30 μM) were mixed with 3 μM microtubules in buffer A supplemented with 0.1 M NaCl, 1 mM 5-adenylylimidodiphosphate (AMPPNP) and 5 mM EDC. The samples were incubated for 30 min at 25°C, SDS–PAGE sample buffer was added to quench the reaction, and the samples were electrophoresed in a 4–20% gradient gel. For analysis of the composition of the crosslinked product, the samples were immunoblotted with DM1A (for α-tubulin) and H-235 (for β-tubulin) antibodies, followed by staining with the appropriate peroxidase-coupled secondary antibodies.
Cryo-electron microscopy
Microtubules were polymerized in 7% DMSO–PEM buffer (100 mM Pipes, pH 6.8, 1 mM EGTA, 1 mM MgCl2, 1 mM GTP and 7% DMSO) and adsorbed to a holey carbon grid. For the DSH–microtubule and DSH207–microtubule complexes, 5 μl DSH (0.5 mg/ml) in PEM buffer was mixed with adsorbed microtubules on a grid for 30 s. Grids were blotted and plunged into liquid ethane. Images of the DSH–microtubule and DSH207–microtubule complexes were observed under low-dose conditions with JEM-2010F or JEM-2100F electron microscopes at a nominal magnification of 40 000 at 17 000–20 000 and 16 000–25 000 Å defocus, respectively.
Image analysis
Microtubules without seams (14, 15 and 16 protofilaments/2-start helix) were screened based on the moiré pattern of the image (Kikkawa et al, 2000). Selected micrographs were digitized with a LeafScan 45 (Scitex Ltd) at a pixel size of 2.5 Å. Images were analyzed using the helical symmetry of the complex. To correct the distortions of curved microtubules, each tube was straightened by fitting the axis to a cubic spline curve and interpolating the tube onto a straight line. The straightened tube was divided into single repeats and positional parameters were refined by comparing Fourier data with the reference data set, as described in previous reports (Beroukhim and Unwin, 1997; Kikkawa et al, 2000). Defocus levels were determined using CTFFIND3 (Mindell and Grigorieff, 2003). Individual data sets were corrected for contrast transfer function, aligned to the same origin and averaged. The 3D density map obtained from 14 or 16 protofilaments was transformed into 15 protofilaments layer-line data, and merged as previously described (Yonekura and Toyoshima, 2000). Final maps were calculated by combining the best 93 and 28 data sets representing ∼30 000 and ∼10 000 asymmetric units for the DSH–microtubule and DSH207–microtubule complexes, respectively. The effective resolution for the DSH–microtubule complex (15 Å) was determined by Fourier shell correlation with 0.5 criteria. The atomic model of tubulin (Nogales et al, 1998; Lowe et al, 2001) was docked as previously reported (Kikkawa et al, 2000, 2001) and manually adjusted using O (Jones et al, 1991). Most of the data analyses were carried out using the newly developed Ruby-Helix scripting system, which we will describe in detail elsewhere. The layer line data have been deposited into the EMD database.
Relative positions of motors and tubulin dimers
The following analyses were performed using data with a resolution of 25 Å. The density map was generated by Fourier–Bessel transformation and Fourier synthesis from an 80 Å layer line (n,l)=(−2,19). Similarly, a radial density projection was calculated using layer lines (n,l)=(0,0), (−2,19), (−4,38), (−6,57), (−8,76) (Figure 6). The KH–microtubule complex map was generated from previously published data (Kikkawa et al, 2000). In order to estimate the signal-to-noise ratio of the 80 Å layer line, we compared density maps generated from two independent data sets. The height differences of the high-density stripes were less than 8 Å, suggesting that the signal-to-noise ratio was high enough to allow us to judge accurately the relative positions of the motors and tubulin dimers.
Acknowledgments
We thank Drs RGB Anderson, WJ Snell and ST Brady for discussions and critical reading of the manuscript. We also thank K Yonekura, C Toyoshima and N Grigorieff for providing programs for image analysis. This work was supported, in part, by a Howard Hughes Medical Institute Award to the University of Texas (Southwestern Medical School) as well as grants from MEXT (Japan) to NH and YYT and from JST (Japan) to YYT.
References
- Al-Bassam J, Ozer RS, Safer D, Halpain S, Milligan RA (2002) MAP2 and tau bind longitudinally along the outer ridges of microtubule protofilaments. J Cell Biol 157: 1187–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos L, Klug A (1974) Arrangement of subunits in flagellar microtubules. J Cell Sci 14: 523–549 [DOI] [PubMed] [Google Scholar]
- Amos LA, Hirose K (1997) The structure of microtubule–motor complexes. Curr Opin Cell Biol 9: 4–11 [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Unwin N (1997) Distortion correction of tubular crystals: improvements in the acetylcholine receptor structure. Ultramicroscopy 70: 57–81 [DOI] [PubMed] [Google Scholar]
- Bingham JB, King SJ, Schroer TA (1998) Purification of dynactin and dynein from brain tissue. Methods Enzymol 298: 171–184 [DOI] [PubMed] [Google Scholar]
- Burgess SA (1995) Rigor and relaxed outer dynein arms in replicas of cryofixed motile flagella. J Mol Biol 250: 52–63 [DOI] [PubMed] [Google Scholar]
- Burgess SA, Dover SD, Woolley DM (1991) Architecture of the outer arm dynein ATPase in an avian sperm flagellum, with further evidence for the B-link. J Cell Sci 98: 17–26 [DOI] [PubMed] [Google Scholar]
- Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K (2003) Dynein structure and power stroke. Nature 421: 715–718 [DOI] [PubMed] [Google Scholar]
- Chretien D, Kenney JM, Fuller SD, Wade RH (1996) Determination of microtubule polarity by cryo-electron microscopy. Structure 4: 1031–1040 [DOI] [PubMed] [Google Scholar]
- deCastro MJ, Ho CH, Stewart RJ (1999) Motility of dimeric ncd on a metal-chelating surfactant: evidence that ncd is not processive. Biochemistry 38: 5076–5081 [DOI] [PubMed] [Google Scholar]
- Gee M, Vallee R (1998) The role of the dynein stalk in cytoplasmic and flagellar motility. Eur Biophys J 27: 466–473 [DOI] [PubMed] [Google Scholar]
- Gee M, Heuser JE, Vallee RB (1997) An extended microtubule-binding structure within the dynein motor domain. Nature 390: 636–639 [DOI] [PubMed] [Google Scholar]
- Gibbons IR, Gibbons BH, Mocz G, Asai DJ (1991) Multiple nucleotide-binding sites in the sequence of dynein beta heavy chain. Nature 352: 640–643 [DOI] [PubMed] [Google Scholar]
- Goodenough U, Heuser J (1984) Structural comparison of purified dynein proteins with in situ dynein arms. J Mol Biol 180: 1083–1118 [DOI] [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE (1982) Substructure of the outer dynein arm. J Cell Biol 95: 798–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimstone AV, Klug A (1966) Observations on the substructure of flagellar fibres. J Cell Sci 1: 351–362 [DOI] [PubMed] [Google Scholar]
- Hagiwara H, Yorifuji H, Sato-Yoshitake R, Hirokawa N (1994) Competition between motor molecules (kinesin and cytoplasmic dynein) and fibrous microtubule-associated proteins in binding to microtubules. J Biol Chem 269: 3581–3589 [PubMed] [Google Scholar]
- Hirakawa E, Higuchi H, Toyoshima YY (2000) Processive movement of single 22S dynein molecules occurs only at low ATP concentrations. Proc Natl Acad Sci USA 97: 2533–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N (1998) Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 279: 519–526 [DOI] [PubMed] [Google Scholar]
- Hirose K, Lockhart A, Cross RA, Amos LA (1995) Nucleotide-dependent angular change in kinesin motor domain bound to tubulin. Nature 376: 277–279 [DOI] [PubMed] [Google Scholar]
- Howard J (2001) Mechanics of Motor Proteins and the Cytoskelton. Sinauer: Sunderland, MA, ISBN 0878933344 [Google Scholar]
- Jones T, Zou J, Cowan S, Kjelgaard M (1991) Improved methods of building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kikkawa M, Ishikawa T, Nakata T, Wakabayashi T, Hirokawa N (1994) Direct visualization of the microtubule lattice seam both in vitro and in vivo. J Cell Biol 127: 1965–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa M, Ishikawa T, Wakabayashi T, Hirokawa N (1995) Three-dimensional structure of the kinesin head–microtubule complex. Nature 376: 274–277 [DOI] [PubMed] [Google Scholar]
- Kikkawa M, Okada Y, Hirokawa N (2000) 15 Å resolution model of the monomeric kinesin motor, KIF1A. Cell 100: 241–252 [DOI] [PubMed] [Google Scholar]
- Kikkawa M, Sablin EP, Okada Y, Yajima H, Fletterick RJ, Hirokawa N (2001) Switch-based mechanism of kinesin motors. Nature 411: 439–445 [DOI] [PubMed] [Google Scholar]
- Koonce MP (1997) Identification of a microtubule-binding domain in a cytoplasmic dynein heavy chain. J Biol Chem 272: 19714–19718 [DOI] [PubMed] [Google Scholar]
- Koonce MP, Tikhonenko I (2000) Functional elements within the dynein microtubule-binding domain. Mol Biol Cell 11: 523–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs A, Goldie KN, Hoenger A (2004) Complex formation with kinesin motor domains affects the structure of microtubules. J Mol Biol 335: 139–153 [DOI] [PubMed] [Google Scholar]
- Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD (1996) Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature 380: 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart A, Crevel IM, Cross RA (1995) Kinesin and ncd bind through a single head to microtubules and compete for a shared MT binding site. J Mol Biol 249: 763–771 [DOI] [PubMed] [Google Scholar]
- Lowe J, Li H, Downing KH, Nogales E (2001) Refined structure of alpha beta-tubulin at 3.5 Å resolution. J Mol Biol 313: 1045–1057 [DOI] [PubMed] [Google Scholar]
- Mandelkow E, Hoenger A (1999) Structures of kinesin and kinesin–microtubule interactions. Curr Opin Cell Biol 11: 34–44 [DOI] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142: 334–347 [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV (1999) AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res 9: 27–43 [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH (1998) Structure of the alpha beta tubulin dimer by electron crystallography. Nature 391: 199–203 [DOI] [PubMed] [Google Scholar]
- Ogawa K (1991) Four ATP-binding sites in the midregion of the beta heavy chain of dynein. Nature 352: 643–645 [DOI] [PubMed] [Google Scholar]
- Paschal BM, Obar RA, Vallee RB (1989) Interaction of brain cytoplasmic dynein and MAP2 with a common sequence at the C terminus of tubulin. Nature 342: 569–572 [DOI] [PubMed] [Google Scholar]
- Ray S, Meyhofer E, Milligan R, Howard J (1993) Kinesin follows the microtubule's protofilament axis. J Cell Biol 121: 1083–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, Pate E, Cooke R, Taylor EW, Milligan RA, Vale RD (1999) A structural change in the kinesin motor protein that drives motility. Nature 402: 778–784 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Kojima H, Sakai Y, Katayama E, Oiwa K (1999) Inner-arm dynein c of Chlamydomonas flagella is a single-headed processive motor. Nature 400: 586–590 [DOI] [PubMed] [Google Scholar]
- Samso M, Radermacher M, Frank J, Koonce MP (1998) Structural characterization of a dynein motor domain. J Mol Biol 276: 927–937 [DOI] [PubMed] [Google Scholar]
- Seitz A, Kojima H, Oiwa K, Mandelkow EM, Song YH, Mandelkow E (2002) Single-molecule investigation of the interference between kinesin, tau and MAP2c. EMBO J 21: 4896–4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelanski M, Gaskin F, Cantor C (1973) Microtubule assembly in the absence of added nucleotides. Proc Natl Acad Sci USA 70: 765–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroguchi K, Toyoshima YY (2001) Regulation of monomeric dynein activity by ATP and ADP concentrations. Cell Motil Cytoskeleton 49: 189–199 [DOI] [PubMed] [Google Scholar]
- Sosa H, Dias DP, Hoenger A, Whittaker M, Wilson-Kubalek E, Sablin E, Fletterick RJ, Vale RD, Milligan RA (1997) A model for the microtubule–Ncd motor protein complex obtained by cryo-electron microscopy and image analysis. Cell 90: 217–224 [DOI] [PubMed] [Google Scholar]
- Thormahlen M, Marx A, Muller SA, Song Y, Mandelkow EM, Aebi U, Mandelkow E (1998) Interaction of monomeric and dimeric kinesin with microtubules. J Mol Biol 275: 795–809 [DOI] [PubMed] [Google Scholar]
- Toba S, Gibson TM, Shiroguchi K, Toyoshima YY, Asai DJ (2004) Properties of the full-length heavy chains of Tetrahymena ciliary outer arm dynein separated by urea treatment. Cell Motil Cytoskeleton 58: 30–38 [DOI] [PubMed] [Google Scholar]
- Tucker C, Goldstein LS (1997) Probing the kinesin–microtubule interaction. J Biol Chem 272: 9481–9488 [DOI] [PubMed] [Google Scholar]
- Tynan SH, Gee MA, Vallee RB (2000) Distinct but overlapping sites within the cytoplasmic dynein heavy chain for dimerization and for intermediate chain and light intermediate chain binding. J Biol Chem 275: 32769–32774 [DOI] [PubMed] [Google Scholar]
- Vale RD (2003) The molecular motor toolbox for intracellular transport. Cell 112: 467–480 [DOI] [PubMed] [Google Scholar]
- Vale RD, Toyoshima YY (1988) Rotation and translocation of microtubules in vitro induced by dyneins from Tetrahymena cilia. Cell 52: 459–469 [DOI] [PubMed] [Google Scholar]
- Vallee RB, Sheetz MP (1996) Targeting of motor proteins. Science 271: 1539–1544 [DOI] [PubMed] [Google Scholar]
- Walker RA (1995) ncd and kinesin motor domains interact with both alpha- and beta-tubulin. Proc Natl Acad Sci USA 92: 5960–5964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Khan S, Sheetz MP (1995) Single cytoplasmic dynein molecule movements: characterization and comparison with kinesin. Biophys J 69: 2011–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Sheetz MP (2000) The C-terminus of tubulin increases cytoplasmic dynein and kinesin processivity. Biophys J 78: 1955–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Laymon R, Goldstein L (1989) A three-domain structure of kinesin heavy chain revealed by DNA sequence and microtubule binding analyses. Cell 56: 879–889 [DOI] [PubMed] [Google Scholar]
- Yonekura K, Toyoshima C (2000) Structure determination of tubular crystals of membrane proteins. II. Averaging of tubular crystals of different helical classes. Ultramicroscopy 84: 15–28 [DOI] [PubMed] [Google Scholar]


