Abstract
Bach1 is a transcriptional repressor of heme oxygenase-1 and β-globin genes, both of which are known to be transcriptionally induced by heme. To test the hypothesis that heme regulates the activity of Bach1, we expressed wild type and mutated versions of Bach1 together with or without its heterodimer partner MafK in human 293T and GM02063 cells and examined their subcellular localization. Inhibition of heme synthesis enhanced the nuclear accumulation of Bach1, whereas treating cells with hemin resulted in nuclear exclusion of Bach1. While the cadmium-inducible nuclear export signal (NES) of Bach1 was dispensable for the heme response, a region containing two of the heme-binding motifs was found to be critical for the heme-induced nuclear exclusion. This region functioned as a heme-regulated NES dependent on the exporter Crm1. These results extend the regulatory roles for heme in protein sorting, and suggest that Bach1 transduces metabolic activity into gene expression.
Keywords: globin, heme, heme oxygenase, Maf, oxidative stress
Introduction
The coupling of metabolic activity and gene expression is fundamental in maintaining homeostasis. Heme is essential for life as a prosthetic group of many heme proteins in reactions involving molecular oxygen, electron transfer, and diatomic gases. In addition, heme binds to several transcription factors that regulate genes involved in oxygen utilization in lower eukaryotes and prokaryotes (Monson et al, 1992; Zhang and Guarente, 1995; Qi et al, 1999). In higher eukaryotes, heme binds to two mammalian transcription factors, Bach1 (Oyake et al, 1996; Ogawa et al, 2001) and NPAS2 (Dioum et al, 2002). The fact that Bach1 and NPAS2 have no apparent evolutionary or functional relationship suggests the involvement of heme in a diverse range of gene expression pathways. Heme, therefore, appears to play a key role as a signaling molecule in coupling metabolic activity and gene activity.
One of the targets of Bach1 is the heme oxygenase-1 (HO-1) gene. HO-1 is the rate-limiting enzyme involved in heme degradation, generating ferrous iron, carbon monoxide, and biliverdin, which is rapidly reduced to bilirubin. Carbon monoxide, biliverdin, and bilirubin possess antioxidant and anti-inflammatory activities in vivo (Otterbein et al, 2003). The cytoprotective roles of bilirubin and carbon monoxide suggest a role for HO-1 as an antioxidant defense enzyme that converts heme into antioxidants. While its role in lower eukaryotes is not clear, HO-1 is essential for higher eukaryotes in order to cope with various aspects of cellular stress and to regulate cellular iron metabolism (Poss and Tonegawa, 1997; Yachie et al, 1999; Shibahara et al, 2002). Importantly, the expression of HO-1 is induced by the substrate heme (Tenhunen et al, 1970; Shibahara et al, 1978; Alam et al, 1989). Thus, heme functions as both an inducer and substrate of the cytoprotective HO-1 system. Additionally, the transcription of ho-1 is robustly induced in mammalian cells by oxidative stress, various pro-inflammatory stimulants such as cytokines, heavy metals, heat shock, UV light, and LPS (Shibahara et al, 1985, 1987; Alam et al, 1989; Keyse and Tyrrell, 1989; Taketani et al, 1989). The induction of HO-1 by diverse stress conditions poses an important question regarding the mechanistic basis for the multifaceted responsibility that realizes cytoprotective functions of ho-1.
The inducible enhancers of ho-1 carry multiple stress responsive elements (StREs; Inamdar et al, 1996) that are closely related to Maf-recognition elements (MAREs; Kataoka et al, 1994). The heterodimers of Nrf2 and small Maf proteins, both belonging to the basic leucine zipper (bZip) class of transcription factors, activate ho-1 through binding to MAREs (Alam et al, 1999, 2000; Ishii et al, 2000; Kataoka et al, 2001; Sun et al, 2004). In contrast, heterodimers of Bach1 and the small Maf proteins such as MafK repress transcription (Sun et al, 2002, 2004). Gene-targeting experiments in mice revealed that loss of bach1 function is sufficient to uncouple ho-1 from stress-responsive control, and resulted in the constitutive expression of HO-1 (Sun et al, 2002). Importantly, Bach1 repressor activity is dominant over the activators such as Nrf2, effectively keeping ho-1 expression at low levels under normal conditions (Sun et al, 2002, 2004). These results suggest that Bach1 ‘poises' the ho-1 enhancers.
A very similar competitive regulation by Maf heterodimers operates on the β-globin gene. MAREs, originally referred to as NF-E2 sites, play critical roles in the enhancer function of the β-globin locus control region (LCR) (Ney et al, 1990a, 1990b). Biochemical and chromatin immunoprecipitation (ChIP) studies have shown that the heterodimer of small Maf/Bach1 binds to MAREs within the β-globin LCR in murine erythroid leukemia (MEL) cells prior to terminal differentiation (Igarashi et al, 1998; Brand et al, 2004). Following DMSO-induced terminal differentiation of MEL cells, NF-E2, a heterodimer of p45 and the small Maf proteins (Andrews et al, 1993a, 1993b; Igarashi et al, 1994), occupies MAREs to activate transcription (Brand et al, 2004). Importantly, heme induces transcription of the β-globin gene (Charnay and Maniatis, 1983).
Given that these repressing and activating transcription factors not only form heterodimers with the small Maf protein but also bind to the same enhancer sequences, it is critical to regulate their activities in response to various inducible conditions. The DNA-binding activity of Bach1 is inhibited in vitro and in vivo upon heme binding (Ogawa et al, 2001; Sun et al, 2004), suggesting a simple model in which the repression of these genes by Bach1 is alleviated by increased levels of heme. Here we demonstrate that heme induces the nuclear export of Bach1 through a novel heme-dependent nuclear export signal (NES) on Bach1, which involves two heme-binding heme regulatory motifs (HRMs). Our results suggest that heme regulates the DNA binding and subcellular localization of Bach1, clearly placing Bach1 as an important and interesting target of heme action. Furthermore, these observations expand the regulatory role of heme pertaining to protein sorting within a cell.
Results
Nuclear exclusion of Bach1 induced by heme
We recently reported that cadmium, a strong inducer of ho-1, stimulates the nuclear export of Bach1 (Suzuki et al, 2003). To explore the possibility that heme regulates the subcellular localization of Bach1, we examined the effect of hemin, ferric protoporphyrin IX, which is reduced to ferroprotoporphyrin IX (heme) within cells, on the subcellular distribution of Bach1. In an effort to mimic the situation in vivo, MafK and FLAG-tagged Bach1 expression plasmids were coexpressed in 293T human embryonic kidney cells, and the localization of Bach1 and MafK was analyzed by indirect immunofluorescence staining. Consistent with our previous observations (Suzuki et al, 2003), Bach1 accumulated in the nuclei in the presence of overexpressed MafK (Figure 1A). We then treated 293T cells with 10 μM hemin or with 10 μM cadmium chloride for 4 h. These conditions are known to trigger HO-1 induction. Bach1 accumulated in the cytoplasmic region and was absent from the nuclei when the cells were treated with cadmium, as reported previously (Suzuki et al, 2003). Interestingly, hemin treatment also resulted in the translocation of Bach1 from the nucleus to the cytoplasm (Figure 1A and B). In contrast, while we observed a weak but significant MafK staining in the cytoplasmic region upon hemin or cadmium treatment, the MafK signal intensity remained strong in the nuclei (Figure 1A). These results indicate that the subcellular localization of Bach1 is regulated in response to cadmium and increased levels of heme.
Figure 1.
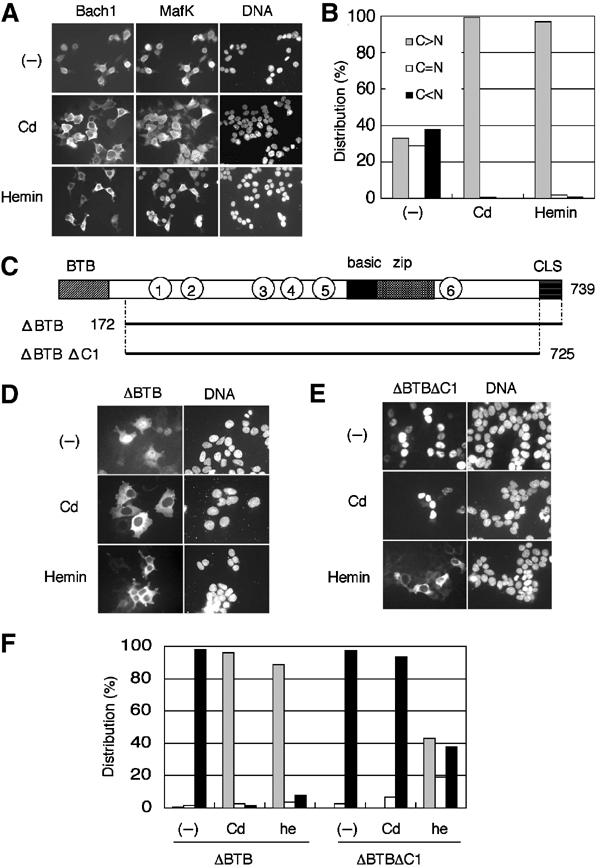
Cytoplasmic accumulation of Bach1 in response to heme. (A) 293T cells were cotransfected with FLAG-Bach1 and MafK expression plasmids. At the end of culturing, cells were treated with 10 μM CdCl2 (second row) or hemin (third row) for 4 h. Bach1 and MafK were detected using anti-FLAG and anti-MafK antibodies, respectively. Images for Bach1, MafK, and DNA are shown (left to right). (B) Subcellular localization of Bach1 was classified into three categories: C>N, cytoplasmic-dominant accumulation (gray bar); C=N, roughly equal distribution in cytoplasmic and nuclear compartments (white bar); and C<N, nuclear-dominant accumulation (black bar). Results of counting 200 cells are shown. (C) Domains of Bach1. BTB and CLS facilitate cytoplasmic accumulation, whereas the basic region facilitates nuclear accumulation (Suzuki et al, 2003). CP motifs are shown with circled numbers. Bach1 derivatives tagged with FLAG at the N-termini are shown below, with the positions of amino-acid residues marked. (D, E) 293T cells were transfected with Bach1ΔBTB or Bach1ΔBTBΔC1 expression plasmids and treated with cadmium (second row) or hemin (third row). Images are DNA (right) and Bach1 (left). (F) Subcellular distributions of Bach1ΔBTB or Bach1ΔBTBΔC1 in 293T cells were counted as in (B). Results of counting 200 cells are shown.
Mechanistic distinction between heme- and cadmium-induced effects on Bach1
Bach1 accumulates within the cytoplasmic region when overexpressed alone in a cell (Suzuki et al, 2003). Since MafK induces the nuclear accumulation of Bach1 (Suzuki et al, 2003; see Figure 1A), the Bach1–MafK interaction may be altered by increased levels of heme, resulting in the cytoplasmic accumulation of Bach1. To exclude this possibility, we utilized Bach1 lacking the BTB domain (Bach1ΔBTB; Figure 1C). The Bach1 BTB domain mediates oligomer formation, generating a multivalent DNA-binding complex (Igarashi et al, 1998; Yoshida et al, 1999). Since Bach1ΔBTB accumulates within the nuclei even in the absence of MafK coexpression (Suzuki et al, 2003; see Figure 1D), we reasoned that the heme effect on its subcellular localization could be measured independent of the heterodimeric interaction. As reported previously (Suzuki et al, 2003), Bach1ΔBTB translocates to the cytoplasm very efficiently when cells are treated with cadmium chloride (Figure 1D and F). Importantly, Bach1ΔBTB altered its localization to the cytoplasmic region following hemin treatment (Figure 1D and F). These results indicate that cadmium and heme regulate Bach1 localization independent of its interaction with MafK or other small Maf proteins.
In our previous report, we showed that cytoplasmic localization signal (CLS), a C-terminal short sequence conserved between Bach1 and Bach2 (Hoshino et al, 2000), is essential for the cadmium-induced nuclear export of Bach1 (Suzuki et al, 2003). To examine whether CLS was also involved in the heme-induced cytoplasmic relocalization of Bach1, we examined another FLAG-Bach1 derivative that lacked both CLS and BTB (Bach1ΔBTBΔC1; Figure 1C). While Bach1ΔBTBΔC1 did not alter its subcellular localization following the addition of cadmium chloride, Bach1ΔBTBΔC1 changed its subcellular localization following the addition of hemin and accumulated within the cytoplasmic region (Figure 1E and F). Similar results were also obtained in experiments comparing the distribution of full-length Bach1 and Bach1ΔC1 (data not shown). These results indicate that, while CLS is essential for the cadmium-induced nuclear export of Bach1, it is not essential for the relocalization of Bach1 in response to heme. Thus, heme and cadmium regulate the subcellular localization of Bach1 through distinct mechanisms.
Inhibition of heme synthesis induces the nuclear accumulation of Bach1
Every cell synthesizes heme. To test whether endogenous levels of heme influence the subcellular localization of Bach1, we examined the effect of succinylacetone (SA), an inhibitor of δ-aminolevulinic acid dehydratase, the second enzyme involved in the heme synthesis pathway. A Flag-Bach1 expression plasmid was transfected into 293T cells without the MafK expression plasmid, and the cells were treated with or without 5 mM SA for 24 h (Figure 2). In the absence of SA, Bach1 was mainly localized in the cytoplasm, as reported previously (Suzuki et al, 2003). When treated with SA, Bach1 accumulated within the nuclei, suggesting that heme levels within a cell can affect the nucleocytoplasmic distribution of Bach1. Interestingly, upon further incubation with 10 μM hemin for 4 h following SA treatment, the nuclear accumulation of Bach1 was abolished and a prominent cytoplasmic relocalization was observed (Figure 2), confirming that the effect of SA was due to decreased levels of heme within the cells. Besides SA, the ferric iron chelator DFO, which causes inhibition of heme synthesis, also induced the nuclear accumulation of Bach1 (Figure 2). These results indicate that endogenous levels of heme substantially affect the subcellular localization of Bach1. Taken together with the results in Figure 1, heme may stimulate the cytoplasmic retention or nuclear export of Bach1.
Figure 2.
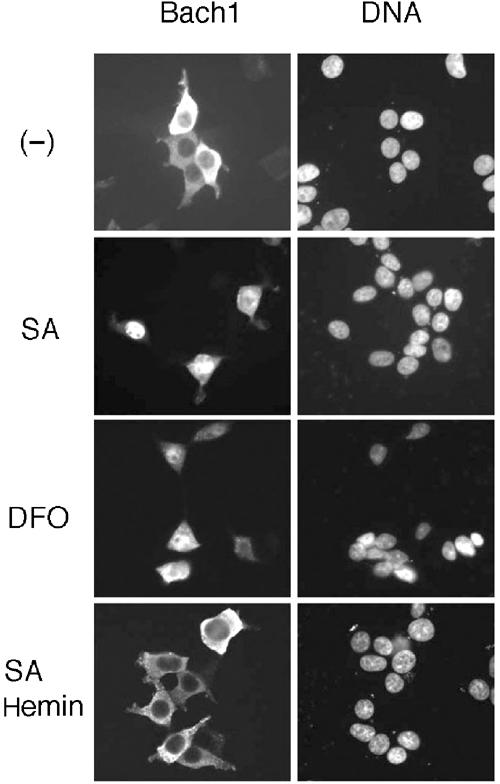
Effects of inhibitors of heme synthesis on Bach1 localization. 293T cells were transfected with FLAG-Bach1 expression plasmid. Cells were maintained in medium lacking serum in the absence or presence of 5 mM SA or 100 μM DFO.
Heme-binding motifs are critical for the cytoplasmic relocalization of Bach1
The Bach1–heme interaction is mediated by evolutionarily conserved multiple HRMs including the cysteine–proline (CP) dipeptide sequence in Bach1 (Ogawa et al, 2001). Among the six CP motifs shown in Figure 3A, previous experiments indicate that CP3–6 are involved in heme binding. In an effort to determine whether these CP motifs are involved in the nuclear exclusion of Bach1 in response to heme, we examined response of the FLAG-tagged Bach1mCP1–6, carrying Cys-to-Ala changes in all of the CP motifs (see Figure 3B), to hemin. Its proper expression was confirmed by immunoblotting analysis (Figure 3C). When expressed alone in 293T cells, wild-type Bach1 was localized in the cytoplasm under normal culture conditions, whereas Bach1mCP1–6 was clearly localized in the nucleus (Figure 3D). These observations provide evidence that some of the CP motifs were involved in the regulation of the subcellular localization of Bach1. When treated with cadmium, Bach1mCP1–6 was detected mainly in the cytoplasm and was excluded from the nucleus (Figure 3D). In contrast, Bach1mCP1–6 remained in the nuclei even following the addition of hemin to the medium. Thus, CP motifs are essential for the heme-induced exclusion of Bach1 from the nucleus. Furthermore, these results confirm that cadmium- and heme-induced nuclear exclusion is controlled by mechanisms that are distinct from each other.
Figure 3.
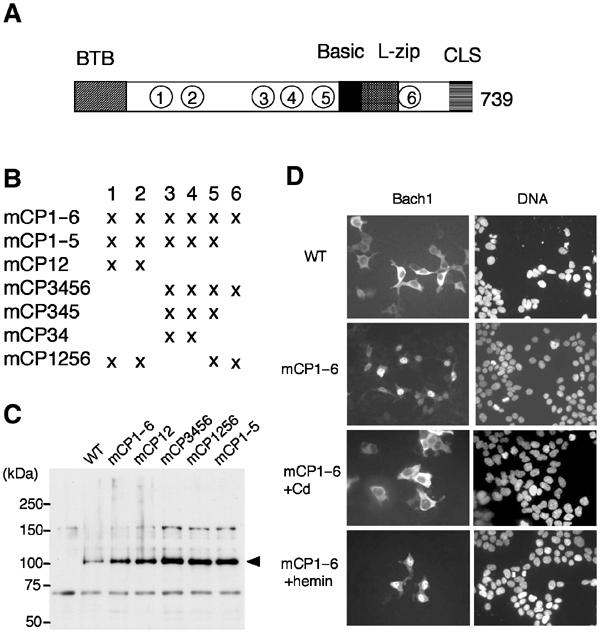
Involvement of heme-binding motifs in the heme response. (A, B) Schematic representation of Bach1 and Bach1 derivatives containing Cys-to-Ala mutations in the CP motifs 1–6 in various combinations. (C) 293T cells were transfected with FLAG-Bach1 or various FLAG-Bach1mCP expression plasmids. Extracts were analyzed by immunoblotting with anti-FLAG antibody. (D) 293T cells were transfected with FLAG-Bach1 or -Bach1mCP1–6, and treated with cadmium (third row) or hemin (fourth row).
To identify the critical CP motif involved in the heme-induced nuclear exclusion, we prepared a series of Bach1 derivatives carrying various combinations of mutations in CP motifs (Figure 3B). These Bach1 derivatives were epitope-tagged with FLAG and expressed in GM02063 human fibroblastic cells together with MafK to ensure their nuclear accumulation under normal culture conditions. As shown in the above experiment, wild-type Bach1 clearly demonstrated a heme response, while mCP1–6 failed to respond at all (Figure 4A). mCP12 responded to heme as the wild-type Bach1. In contrast, mCP3456 completely lacked a heme response. mCP34 also showed little response to heme. Importantly, mCP1256, which retains CP3 and 4, displayed a significant response to heme. A profile plot analysis of the signal intensities clearly showed heme-induced nuclear exclusion of Bach1 and mCP1256 but not of mCP34 (Figure 4B). These results indicate that, among the six CP motifs, CP3 and 4 are critical and sufficient for heme-induced nuclear exclusion.
Figure 4.
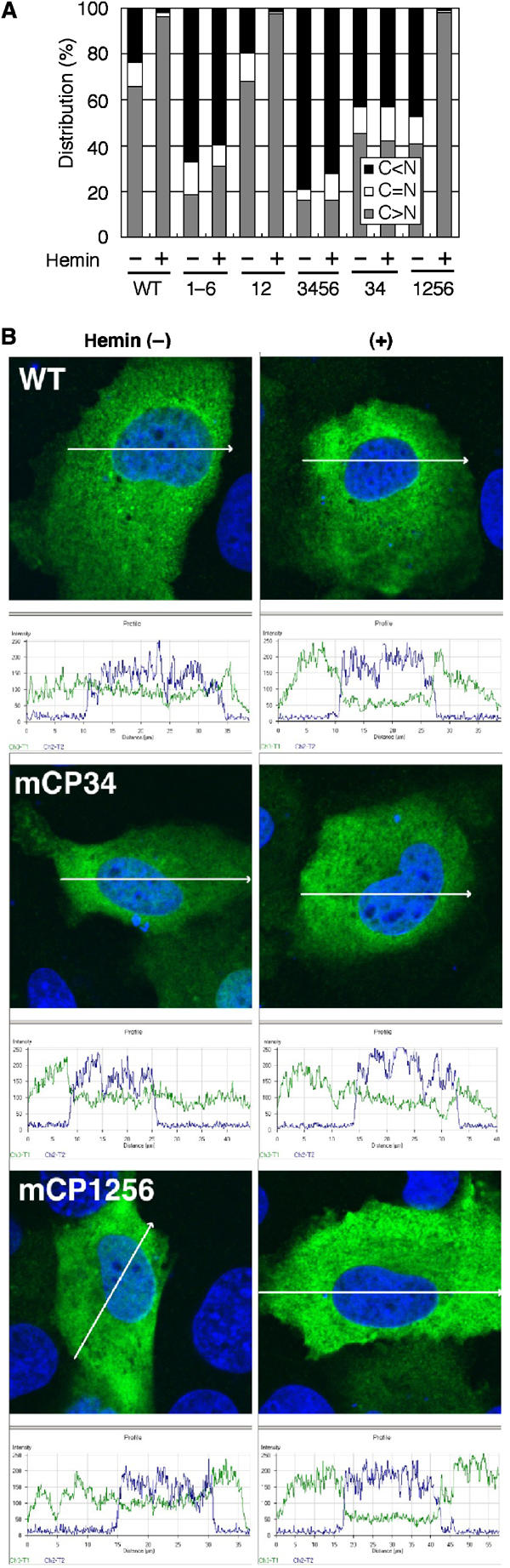
Delineation of CP motifs involved in the heme-induced nuclear exclusion of Bach1. (A) Human fibroblast GM02063 cells were transfected with FLAG-tagged Bach1 or Bach1mCP derivative expression plasmids, together with MafK expression plasmid. Cells were treated with or without 10 μM hemin at the end of culturing. The subcellular localization of Bach1 was classified into three categories as in Figure 1. Results of counting 400 cells are shown. (B) Profile plots of signal intensity along the arrows are shown in green (Bach1, mCP34, or mCP1256) or blue (DNA). Without hemin treatment, the majority of cells showed a significant nuclear signal of Bach1 in confocal optical sections.
Determination of a heme-dependent NES
Having determined that CP3 and 4 are critical in the heme-induced cytoplasmic relocalization of Bach1, we then investigated the region(s) that were necessary and sufficient in effecting heme-induced nuclear exclusion. We first expressed Bach1 fragment 417–587 possessing CP3–5 as a fusion with EGFP (Figure 5A). This fragment contained the basic region that functions as a nuclear import signal (Suzuki et al, 2003). The fusion protein GFP-B1(417–587) was localized in the nucleus under normal culture conditions (Figure 5B, upper panel). It was localized in the cytoplasm following treatment of the cells with hemin for 4 h (Figure 5B, lower panel). We then examined the contributions of individual CP motifs by changing each cysteine residue to alanine (mCP3–5) using GFP-B1(417–587). When expressed in 293T cells (Figure 5C), GFP-B1(417–587)mCP5 showed diffuse subcellular localization similar to the wild-type fusion protein in response to hemin treatment, indicating that CP5 is not necessary for the observed heme response. In contrast, GFP-B1(417–587)mCP3 was not relocalized to the cytoplasm even after hemin treatment. Compared with mCP3 and 5, GFP-B1(417–587)mCP4 displayed an intermediate response. In this case, we observed an increased signal intensity in the cytoplasmic region following hemin treatment; however, the extent was less than that for mCP5 or the wild-type reporter.
Figure 5.
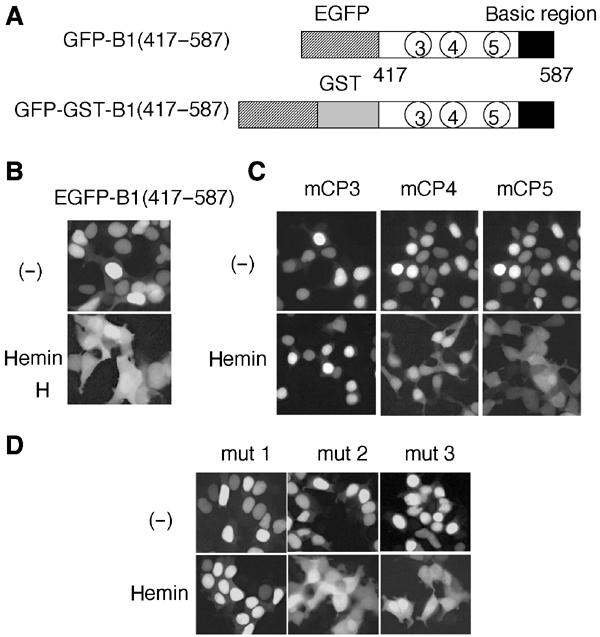
Delineation of heme-responsive CP motifs within Bach1. (A) Schematic representation of reporter proteins containing indicated regions of Bach1 fused with EGFP and GST. (B–D) GFP–B1(417–587) and its derivatives carrying indicated mutations were expressed in 293T cells. Hemin was added for the last 4 h before observation.
Cadmium induces the Crm1-dependent nuclear export of Bach1 (Suzuki et al, 2003). We then examined whether the heme effect involves Crm1-dependent nuclear export. To this end, we utilized a modified version of human Crm1, hCrm1-K1. hCrm1-K1 possesses a substitution of Ser for Cys at position 528, which is the binding site of LMB. As such, hCrm1-K1 is insensitive to LMB when expressed in mammalian cells (Kudo et al, 1999a; Hoshino et al, 2000). We examined the effects of hCrm1 or hCrm1-K1 on the subcellular localization of a Bach1 reporter protein. In this experiment, we expressed Bach1 417–587 as a fusion with EGFP and glutathione-S-transferase (GFP-GST-B1(417–587); see Figure 5A). It is known that increasing protein size often provides for clear distribution in the presence of both NLS and NES by inhibiting passive diffusion (Kudo et al, 1999b). As shown in Figure 6A, while GFP-GST-B1(417–587) accumulated within the nuclei in the presence of hCrm1 or hCrm1-K1 under the normal culture conditions, heme clearly induced cytoplasmic accumulation and exclusion from the nuclear region. This conditional nuclear export of GFP-GST-B1(417–587) was inhibited by treatment with LMB when coexpressed with the wild-type hCrm1. However, when cotransfected with hCrm1-K1, GFP-GST-B1(417–587) was localized within the cytoplasmic region and excluded from the nucleus even in the presence of LMB. We obtained similar results by expressing the full-length Bach1 and MafK together with hCrm1 or hCrm1-K1 (Figure 6B). To examine whether Bach1 binds to Crm1, a GST–Bach1 fusion protein (residues 417–645 containing CP3–5) was charged with hemin and then incubated with liver cell extracts. As shown in Figure 6C, a small but significant fraction of Crm1 bound to the GST–Bach1 fragment. When taken together, these results clearly established that heme activated the Crm1-dependent nuclear export of Bach1.
Figure 6.
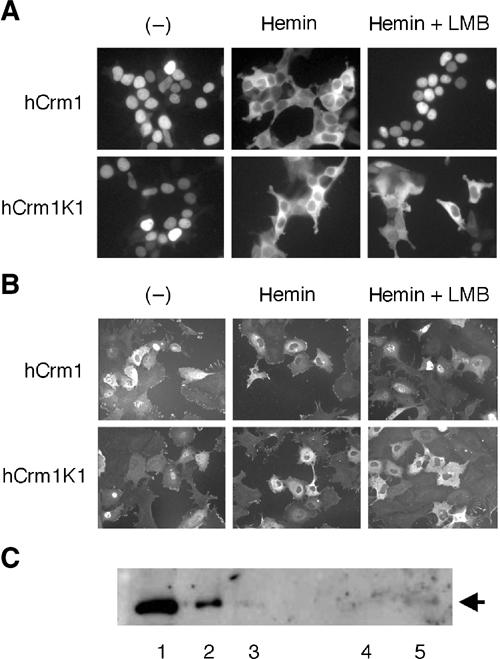
Involvement of Crm1 in the hemin-induced nuclear exclusion of Bach1. (A, B) Localization of GFP-GST-B1(417–587) in 293T cells (A) or Bach1 in GM02063 cells (B) in the presence of Crm1 or Crm1K1 was examined. Cells were treated with indicated reagents. (C) Glutathione beads (lane 4) or glutathione beads with GST–Bach1(417–645) (lane 5) were incubated with liver cell extracts and bound proteins were analyzed by immunoblotting analysis using anti-Crm1 antibodies. Fractions of input extracts were also examined (5.7, 1.9, or 0.6 μg in lanes 1–3, respectively).
One of the characteristic features of a typical NES is the cluster of essential hydrophobic amino acids represented by the LXXLXXLXL motif (Fischer et al, 1995; Wen et al, 1995). While the region between 417 and 587 lacks this type of cluster, there are several hydrophobic residues juxtaposing the critical CP3 and 4 regions (see Figure 7A, shown with lines above the sequence). In an effort to investigate the involvement of these hydrophobic residues in the heme-induced cytoplasmic localization, we carried out an alanine scanning mutagenesis of these hydrophobic residues. We replaced the hydrophobic residues with alanine using GFP-B1(417–587) (Figure 7A; mut 1, 2, and 3). Among the three clustered mutations examined here, only mut 1 completely abolished hemin-induced nuclear export (Figure 5D). The fragments represented by mut 2 or 3 showed considerable cytoplasmic accumulation when treated with hemin.
Figure 7.
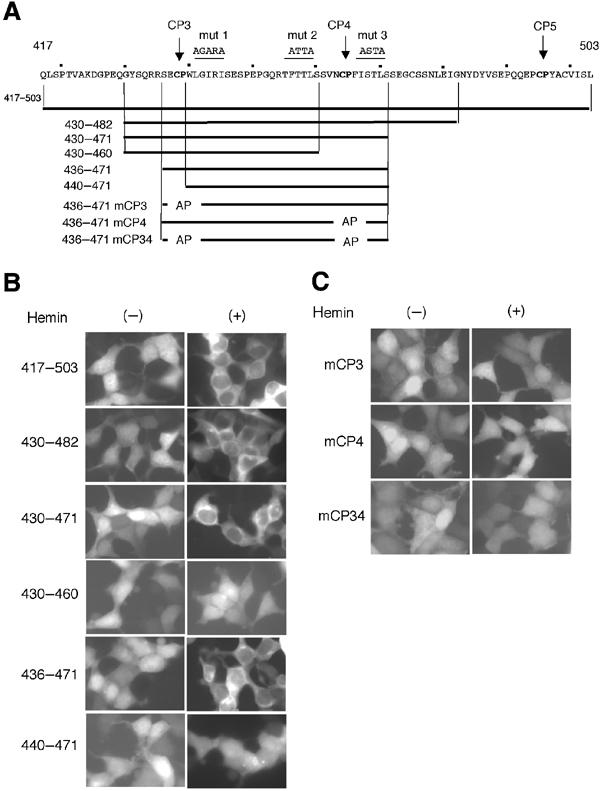
Demarcation of heme-regulated NES. (A) Amino-acid sequence (residues 417–503) containing CP3–5 motifs (bold) is shown. Mutations in hydrophobic residues are shown above the sequence (mut 1, 2, and 3). Subfragments examined as EGFP fusion proteins are shown below the sequence. (B, C) Indicated EGFP fusions were expressed in 293T cells and their distribution was observed in the absence or presence of hemin.
To narrow down the heme-regulated NES sequence, we fused various fragments around CP3–5 motifs with EGFP (Figure 7A), expressed them in 293T cells, and compared their localization in the absence or presence of hemin. These fusion proteins were localized in both the nucleus and cytoplasm in the absence of hemin (Figure 7B, left panels). Since these fragments lacked the basic region of Bach1 that functions as an NLS (Suzuki et al, 2003) and they are small in size, we assume that these GFP fusions entered the nucleus through passive diffusion. When treated with hemin, all of them with CP3 and 4 showed clear nuclear exclusion (Figure 7B, right panels), indicating that the minimal hemin-responsive region encompassed amino-acid residues 436–471. Deletions of either CP3 or 4 (430–460 or 440–471) completely abolished hemin-induced nuclear exclusion. To confirm that CP3 and 4 were important for the hemin response, we mutated these CP motifs on the shortest fragment (Figure 7A). As shown in Figure 7C, mutations in the CP motifs completely abolished hemin-induced nuclear exclusion. When taken together with the results presented in Figures 5 and 6, these results indicate that the region between residues 436 and 471 functions as a heme-dependent NES and that the CP3 and 4 motifs are both involved in this function.
The heme-dependent NES binds heme in vitro
To examine whether the heme-dependent NES actually binds to heme directly, we prepared a recombinant GST fusion protein carrying residues 417–480 of Bach1 (see Figure 7A) and conducted a heme-binding assay. When hemin was titrated with increasing amounts of the fusion protein, the absorption spectrum of free hemin was changed to a new spectrum, which exhibited the Soret band at 371 nm (Figure 8, top left panel). As the fusion protein itself does not have any appreciable absorption between 700 and 350 nm, the spectral changes of hemin shown in Figure 8 are primarily due to alteration in the electronic structure and coordination state of the heme iron caused by the interaction between the hemin iron and the protein fragments. The titration curve (Figure 8, top right panel) showed that hemin bound to the protein with almost 1:1 stoichiometry. The spectrum with Soret band at 370 nm is not commonly seen in ferric heme proteins, such as hemoglobins and cytochromes, but is reminiscent of hemin complexes with p-nitrobenzenethiolate and a heme-responsive element peptide with a CP motif (Tang et al, 1976; Zhang and Guarente, 1995). In these model systems, the hemin iron is coordinated by a thiolate ligand, and we infer that the axial ligand of the hemin complex with the GST–Bach1 fragment fusion protein is a thiolate as well. We conclude that hemin binds to a cystein residue in NES to form a 1:1 complex.
Figure 8.
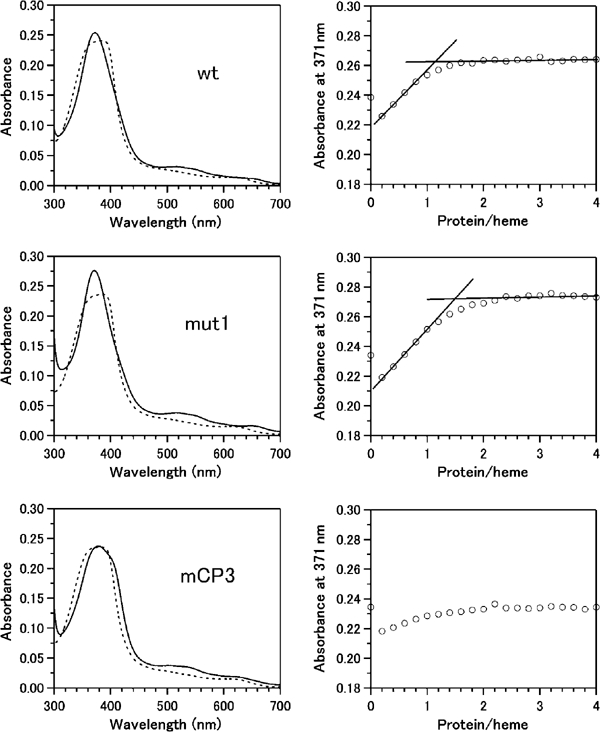
Heme binding to the GST–Bach1 fragment fusion protein. Absorption spectra of hemin in the absence (dotted lines) and presence (solid lines) of the GST–Bach1(417–480) fragments at [Bach1]/[heme] ratio of 3 are shown on the left. Titration curves of hemin with increasing amounts of the fusion protein represented as absorbance at 371 nm as a function of the molar concentration ratios of the protein to heme are shown on the right. Lines are drawn as a visual aid for stoichiometic heme binding and do not carry any analytical meanings. From top to bottom panels, the wild-type, mut 1, and mCP3 GST–Bach1 fragments. A minor spectral change observed for mCP3 (the bottom panels) is probably due to nonspecific heme interaction with the fusion protein.
To gain further insight into the relationship between the hemin binding and the heme-dependent nuclear export properties of Bach1, we focused on some of the mutations that abolished the heme-dependent nuclear export (see Figure 7A) and compared their effect upon hemin binding in vitro. When the mut 1 mutations were introduced in the context of 417–480, the Bach1 fragment with the mut 1 mutations still showed a spectrum and titration curve (Figure 8, middle panels) similar to those of the wild-type fragment, clearly excluding the possibility that these mutations affected the heme-binding activity. In contrast, mCP3AP mutation and mCP34AP double mutations did not show a Soret band at 371 nm (Figure 8, bottom panel, and data not shown), indicating that these mutations abolished heme–Bach1 interaction. We conclude that at least CP3 in NES is critically involved in the interaction with heme.
Regulation of transcription by heme
In an effort to investigate the physiological significance of the heme-induced export of Bach1, we compared the effects of wild-type Bach1 and the Bach1 mCP derivatives on a ho-1 reporter gene expression system (Figure 9). The reporter plasmid pHO15luc contains the upstream 15 kb DNA of ho-1 and is repressed by Bach1 (Alam et al, 2000; Sun et al, 2002). The reporter and Bach1 expression plasmids were cotransfected. Bach1mCP derivatives repressed reporter expression more efficiently than wild-type Bach1, suggesting again that Bach1 was negatively regulated by endogenous levels of heme. In the presence of heme, wild-type Bach1 completely lost repressor activity. In contrast, Bach1mCP1–6 continued to repress reporter gene expression, indicating that the CP motifs were indeed involved in the heme response. Bach1mCP1256 did not show effective repression in the presence of heme, indicating that CP3 and 4 were indeed important for the heme response in the reporter assays. However, CP3 and 4 were not the only motifs governing the heme sensitivity since Bach1mCP34, which was insensitive to heme in the subcellular localization assays, also failed to repress transcription. Thus, the properties of Bach1mCP34 clearly dissociate heme effects on nuclear export from transcription repression. While mCP12 did not repress reporter gene in the presence of heme, mCP3456 effectively repressed the activity in the presence of heme, suggesting critical roles for CP3–6. mCP12345 showed an intermediate response to heme, suggesting the involvement of CP6 in another layer of regulation by heme. These results taken together indicate that CP3–6 are critical for the regulation of Bach1 activity by heme. CP5 and/or 6 may affect the activity of Bach1 in terms of activities other than nuclear export. Since heme inhibits the DNA-binding activity of Bach1, CP5 and/or 6 may be involved in this regulation.
Figure 9.
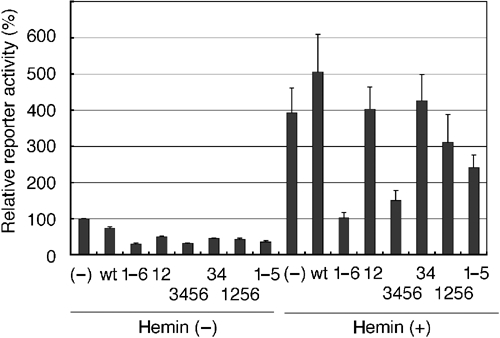
Effect of heme upon Bach1 in transcription reporter assays. 293T cells were transfected with the reporter pHO15luc together with or without FLAG-tagged Bach1 or a battery of Bach1mCP derivative expression plasmids. Cells were treated with 10 μM hemin for 4 h at the end of culturing. Relative reporter gene activities are shown from three experiments.
Heme inhibits DNA binding of Bach1 in vivo
To address whether the DNA-binding activity of Bach1 is regulated independently of CP3 and 4, we carried out ChIP assays. We transfected the ho-1 reporter plasmid together with Bach1 and MafK expression plasmids into GM02063 cells, and carried out ChIP using anti-Bach1 antibodies (Figure 10A). Both Bach1 and Bach1mCP34 clearly bound to the E2 enhancer region in the absence of hemin. While Bach1mCP34 showed less binding to the enhancer in ChIP as compared with the wild-type Bach1, the reason for this observation is not clear at present. Their binding was markedly lost upon hemin treatment (Figure 10A). The effect of hemin was judged specific to Bach1, since coexpressed MafK remained bound to the enhancer in the presence of hemin (Figure 10B). Furthermore, when Nrf2 was coexpressed with MafK, its binding to E2 was not inhibited by hemin treatment (Figure 10B). These results showed that heme inhibited the enhancer occupancy of Bach1 in a cell. Importantly, CP3 and 4, and hence the heme-dependent nuclear export of Bach1, were not essential for the regulation of enhancer binding.
Figure 10.
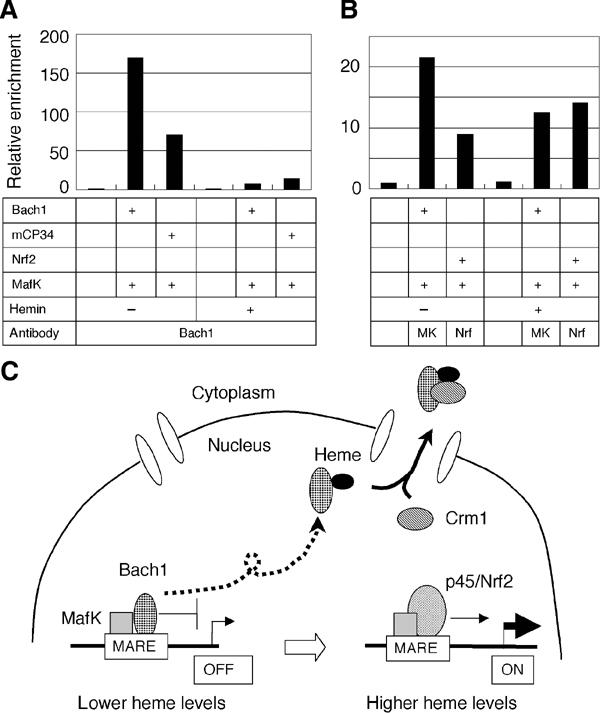
Regulation of DNA binding by heme in vivo. (A) GM02063 cells were transfected with the ho-1 reporter plasmid and indicated Bach1 and MafK expression plasmids. Cells were treated with or without hemin, and binding of Bach1 to the ho-1 E2 enhancer was examined by ChIP. (B) Binding of MafK or Nrf2 to the ho-1 E2 enhancer was examined as in (A). (C) Model describing the regulation of ho-1 or other target genes by Bach1 and heme. Besides MafK, other Maf-related factors may also serve as partners for Bach1. Bach1 occupies MARE enhancers to repress transcription under normal conditions. An increase in heme levels alleviates Bach1-mediated repression through inhibition of its DNA-binding activity and subsequent Crm1-dependent nuclear export, making MAREs available for activating Maf complexes including Nrf2 or p45 NF-E2.
Discussion
To our knowledge, this study is the first report showing that heme regulates transcription by affecting the nucleocytoplasmic shuttling of nuclear protein. This conclusion is based on several observations. Most importantly, treatment of cells with SA resulted in nuclear accumulation of Bach1, indicating that endogenous levels of heme specified by the activities of heme synthesis pathway can substantially affect the subcellular localization of Bach1 (Figure 2). This regulation involved the heme-binding motifs CP3 and 4 in addition to the hydrophobic amino-acid residues juxtaposing CP3 (Figure 5). By several criteria, this region functions as a heme-regulated NES. When the region containing CP3 and 4 was fused with EGFP, the chimeric protein showed heme-induced nuclear exclusion (Figure 7). Furthermore, heme facilitates the Crm1-dependent nuclear export of Bach1 (Figure 6). Importantly, a short fragment containing CP3 and 4 bound heme in vitro (Figure 8). The simplest model is that this region is involved in a heme-regulated interaction with Crm1 that mediates nuclear export. While Crm1 bound to Bach1 in GST pull-down assays (Figure 6), we still do not know whether the interaction is direct or not. To test this hypothesis, we are currently searching for proteins that bind to this region in a heme-regulated manner. In any case, the available data indicate that heme regulates gene expression in part by changing the subcellular localization of Bach1. When taken together with our previous results (Ogawa et al, 2001; Sun et al, 2002), the model argues that Bach1 binds to MARE to repress transcription when heme levels are low. Increased levels of heme displace Bach1 from enhancers and stimulate its relocalization to the cytoplasm, making MARE accessible to activators such as NF-E2 p45 or Nrf2 (Figure 10C). In principle, this would allow a simple and tight coordination of heme metabolism and gene expression.
Indeed, hemin induces departure of Bach1 and subsequent binding of Nrf2 on the endogenous ho-1 enhancers in NIH3T3 cells (Sun et al, 2004). A very similar system operates in the regulation of β-globin genes. Prior to differentiation of MEL cells, MafK/Bach1 heterodimers recruit corepressor complexes including NuRD, which generate a transcriptionally repressive chromatin structure within the β-globin locus (Brand et al, 2004). Upon erythroid differentiation, an exchange of MafK-binding partners from Bach1 to p45 NF-E2 leads to the formation of the activator complex on the LCR, resulting in the activation of β-globin gene expression (Brand et al, 2004). Since heme is synthesized at high levels during erythroid differentiation, the replacement of Bach1 by the activator p45 may be initiated by heme. Indeed, ChIP assays revealed that exogenous hemin treatment displaced Bach1 from the LCR in MEL cells (Sun et al, 2004). Taken together with the results described here, heme may displace Bach1 by inhibiting its DNA-binding activity as well as by inducing its nuclear export, shifting the equilibrium balance toward p45 binding during erythroid differentiation. In this view, synthesis of heme is coupled with transcription of β-globin mRNA by the heme response of Bach1. This type of regulation, together with the heme-dependent regulation of globin mRNA translation (Han et al, 2001), may be important in avoiding toxicity due to the pro-oxidant nature of free heme during erythroid differentiation, since such a mechanistic link furthers the incorporation of heme into hemoglobin.
There is an interesting conceptual precedent in terms of heme-regulated protein sorting within a cell. δ-Aminolevulinic acid sythase (ALAS) is the rate-limiting mitochondrial enzyme involved in the heme synthesis pathway encoded by the nuclear DNA. The mitochondrial targeting signal of ALAS contains several CP motifs. Heme inhibits the transport of ALAS into mitochondria depending on the CP motifs (Lathrop, 1993), generating a feedback mechanism on heme synthesis. While structural analyses have yet to be reported for either Bach1 or ALAS, heme may induce a structural change in the Bach1 NES or ALAS mitochondrial targeting signal to regulate their activities.
The observations reported here have clearly demonstrated that Bach1 is regulated by heme in a cell, thus placing heme as a signaling molecule in gene expression in higher eukaryotes. The regulation of Bach1 by heme may be important for the stress response in general. When cells are injured, they release their contents, resulting in a local accumulation of heme (Wagener et al, 2003). The heme that is released has been suggested to play a signaling role in the inflammatory process (Wagener et al, 2003). It may be taken up by surrounding cells and then bind to target molecules such as Bach1. Although this study focused on Bach1, the regulatory role of heme in transcription factors may not be restricted to Bach1. For example, NPAS2, a regulator of the circadian clock, is a heme-binding protein (Dioum et al, 2002). Considering its critical roles in diverse metabolic pathways as a prosthetic group, heme may regulate other genes, as well, to transduce metabolic activity into changes in gene expression. The heme–Bach1 regulatory pathway deserves further investigation given its clinical significances in two aspects. First, de-regulation of heme metabolism has been implicated in mitochondrial and neural decay (Atamna et al, 2002). Second, HO-1 plays a critical cytoprotective role in diverse clinical settings such as ischemic reperfusion and transplantation (Otterbein et al, 2003). Gene responses under such conditions may also involve the heme–Bach1 pathways.
Materials and methods
Plasmids
Expression plasmids for FLAG-tagged Bach1, FLAG-Bach1mCP1–6 (with all of the CP motifs mutated to AP), Bach1ΔBTB, and Bach1ΔBTBΔC1 were described previously (Suzuki et al, 2003). Sequences of the primers used in plasmid construction are available upon request. pEGFPGST, an expression plasmid for a fusion protein of EGFP and GST, was constructed as follows. A bacterial GST DNA was amplified using primers and pGEX6P1 (Pharmacia) as a template, and then inserted between the BglII and HindIII sites of pEGFP-C1 (Clontech). A Bach1 cDNA fragment (nucleotides 1422–1934 on the mouse bach1 cDNA (Oyake et al, 1996)), encompassing residues 417–587, was amplified using primers and inserted between the EcoRI and KpnI sites of pEGFPGST, resulting in pEGFPGSTB1(1422–1934). The same Bach1 cDNA fragment encoding the wild type or mutations in CP3AP, 4AP, or 5AP (Ogawa et al, 2001) was inserted between the EcoRI and KpnI sites of pEGFPC1, resulting in pEGFPB1(1422–1934), pEGFPB1(1422–1934mCP3), pEGFPB1(1422–1934mCP4), and pEGFPB1(1422–1934mCP5), respectively. A Bach1 cDNA fragment (nucleotides 1422–1682) encompassing residues 417–503 was amplified using primers and then inserted between the EcoRI and KpnI sites of pEGFPC1, resulting in pEGFPB1(1422–1682). Shorter fusion protein expression plasmids were constructed similarly using EcoRI- and KpnI-containing primer sets. The mutations in the hydrophobic residues around CP3 and 4 were created using PCR and ligation methods using pEGFPB1(1422–1934). cDNAs encoding Bach1mCP1–6, mCP1–5, mCP3456, 345, 34, and 1256 were created by site-directed mutagenesis using Bach1 cDNA cloned in the pAlter1 vector (Promega) as described by the manufacturer. After verifying mutations by DNA sequencing, cDNAs encompassing initiation methionine to stop codons were amplified using primers, digested with BclI and HindIII, and were inserted into the BamHI–HindIII sites of pcDNA3.1FLAG (Muto et al, 2002).
Immunocytochemistry and GFP observation
293T cells or GM02063 human fibroblastic cells were transfected with various Bach1 expression plasmids and cultured for 24 h. To examine the effects of reagents, transfected cells were treated with 20 ng/ml leptomycin B or 10–40 μM hemin for the indicated periods at the end of incubation. Reactions with FLAG antibodies and fluorescein-conjugated anti-mouse IgG antibodies were described previously (Suzuki et al, 2003). Subcellular localization of GFP fusion proteins was examined after fixation in 4% paraformaldehyde. Images were taken with a Leica epifluorescence microscope equipped with a charge-coupled device camera controlled by Qfluoro software (Leica). Profile plot analysis was carried out using ZEISS LSM510 confocal laser scanning microscope. Adobe Photoshop was used for the presentation of the images.
Crm1-binding assay
GST–Bach1(417–645) was purified and immobilized on glutathione beads as described previously (Ogawa et al, 2001). The column was washed with a PBS buffer containing 10 μM hemin and loaded with mouse liver cell extracts. After extensive washing with PBS, bound proteins were eluted with glutathione and analyzed in immunoblotting using anti-Crm1 antibodies.
Spectoroscopic analysis of heme binding
Escherichia coli BL21(DE3) cells transformed with the expression constructs for the GST–Bach1 fusion proteins were grown in TB media at 37°C until A600 nm reached 0.8. After the induction with 1 mM IPTG, cultures were incubated at 37°C for an additional 2 h. Cells were harvested by centrifugation and resuspended in PBS buffer (phosphate-buffer saline) containing 1 mM PMSF and 1 mM EDTA. The cell suspensions were sonicated on ice and centrifuged at 70 500 g for 30 min. Soluble protein fractions were loaded onto a glutathione-sepharose 4B column equilibrated with PBS buffer. After washing the column with PBS buffer, the GST–Bach1 fusion proteins were eluted with 50 mM Tris–HCl buffer, pH 8.0, containing 200 μM reduced glutathione. Glutathione was removed from the protein preparations by passing a column of Sephadex G25 equilibrated with 0.1 M phosphate buffer, pH 7.0.
In all, 500 μM of the GST–Bach1 fusion protein in 4-μl increments was added to 4 μM hemin in 2.5 ml of 0.1 M phosphate buffer, pH 7.0. After each addition of the protein, samples were incubated at 20°C for 10 min, and the absorption spectrum was recorded by a Perkin-Elmer Lambda 45 spectrophotometer at 20°C. By plotting the absorbance at 371 nm against the amount of the protein, titration curves were constructed.
Antibodies and ChIP
Polyclonal antiserum against Bach1 (A1–6) was generated by immunizing rabbits with a GST fusion protein of mouse Bach1 (amino-acid residues 624–739). Mouse HO-1 gene reporter plasmid pHO15luc (Alam et al, 2000) and various combinations of expression plasmids were transfected using Fugene6 (Roche) to GM02063 cells. After 36 h, cells were treated with or without hemin for another 2 h and processed for ChIP as described previously (Sun et al, 2004) using anti-Bach1 (A1–6), anti-Nrf2 (Santa Cruz Biotechnology, sc-13032), or anti-MafK (Igarashi et al, 1995) antibodies. Amounts of precipitated DNA were analyzed in duplicate by real-time PCR (LightCycler and SYBR Green 1, Roche). Relative enrichment means specific antibody signal normalized to respective input signal.
Acknowledgments
We thank Makoto Kobayashi (Tsukuba) for various suggestions and critical comments on the article. This work was supported by Grants-in-aid from the Ministry of Education, Science, Sport and Culture of Japan.
References
- Alam J, Shibahara S, Smith A (1989) Transcriptional activation of the heme oxygenase gene by heme and cadmium in mouse hepatoma cells. J Biol Chem 264: 6371–6375 [PubMed] [Google Scholar]
- Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL (1999) Nrf2, a Cap‘n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem 274: 26071–26078 [DOI] [PubMed] [Google Scholar]
- Alam J, Wicks C, Stewart D, Gong P, Touchard C, Otterbein S, Choi AMK, Burow ME, Tou J (2000) Mechanism of heme oxygenase-1 gene activation by cadmium in MCF-7 mammary epithelial cells. Role of p38 kinase and Nrf2 transcription factor. J Biol Chem 275: 27694–27702 [DOI] [PubMed] [Google Scholar]
- Andrews NC, Erdjument-Bromage H, Davidson MB, Tempst P, Orkin SH (1993a) Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature 362: 722–728 [DOI] [PubMed] [Google Scholar]
- Andrews NC, Kotkow KJ, Ney PA, Erdjument-Bromage H, Tempst P, Orkin SH (1993b) The ubiquitous subunit of erythroid transcription factor NF-E2 is a small basic-leucine zipper protein related to the v-maf oncogene. Proc Natl Acad Sci USA 90: 11488–11492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atamna H, Killilea DW, Killilea AN, Ames BN (2002) Heme deficiency may be a factor in the mitochondrial and neuronal decay of aging. Proc Natl Acad Sci USA 99: 14807–14812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M, Ranish JA, Kummer NT, Hamilton J, Igarashi K, Francastel C, Chi TH, Crabree GR, Aebersold R, Groudine M (2004) Dynamic changes in transcription factors complexes during erythroid differentiation revealed by quantitative proteomics. Nat Struct Mol Biol 11: 73–80 [DOI] [PubMed] [Google Scholar]
- Charnay P, Maniatis T (1983) Transcriptional regulation of globin gene expression in the human erythroid cell line K562. Science 220: 1281–1283 [DOI] [PubMed] [Google Scholar]
- Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez M, McKnight SL (2002) NPAS2: a gas-responsive transcription factor. Science 298: 2385–2387 [DOI] [PubMed] [Google Scholar]
- Fischer U, Huber J, Mattaj IW, Luhrmann R (1995) The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82: 475–483 [DOI] [PubMed] [Google Scholar]
- Han AP, Yu C, Lu L, Fujiwara Y, Browne C, Chin G, Fleming M, Leboulch P, Orkin SH, Chen JJ (2001) Heme-regulated eIF2alpha (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J 20: 6909–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino H, Kobayashi A, Yoshida M, Kudo N, Oyake T, Motohashi H, Hayashi N, Yamamoto M, Igarashi K (2000) Oxidative stress abolishes leptomycin B-sensitive nuclear export of transcription repressor Bach2 that counteracts activation of Maf recognition element. J Biol Chem 275: 15370–15376 [DOI] [PubMed] [Google Scholar]
- Igarashi K, Hoshino H, Muto A, Suwabe N, Nishikawa S, Nakauchi H, Yamamoto M (1998) Multivalent DNA binding complex generated by small Maf and Bach1 as a possible biochemical basis for β-globin locus control region complex. J Biol Chem 273: 11783–11790 [DOI] [PubMed] [Google Scholar]
- Igarashi K, Itoh K, Hayashi N, Nishizawa M, Yamamoto M (1995) Conditional expression of the ubiquitous transcription factor MafK induces erythroleukemia cell differentiation. Proc Natl Acad Sci USA 92: 7445–7449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K, Kataoka K, Itoh K, Hayashi N, Nishizawa M, Yamamoto M (1994) Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature 367: 568–572 [DOI] [PubMed] [Google Scholar]
- Inamdar NM, Ahn YI, Alam J (1996) The heme-responsive element of the mouse heme oxygenase-1 gene is an extended AP-1 binding site that resembles the recognition sequences for MAF and NF-E2 transcription factors. Biochem Biophys Res Commun 221: 570–576 [DOI] [PubMed] [Google Scholar]
- Ishii T, Itoh K, Takahashi S, Sato H, Yanagawa T, Katoh Y, Bannai S, Yamamoto M (2000) Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J Biol Chem 275: 16023–16029 [DOI] [PubMed] [Google Scholar]
- Kataoka K, Handa H, Nishizawa M (2001) Induction of cellular antioxidative stress genes through heterodimeric transcription factor Nrf2/small Maf by antirheumatic gold(I) compounds. J Biol Chem 276: 34074–34081 [DOI] [PubMed] [Google Scholar]
- Kataoka K, Noda M, Nishizawa M (1994) Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol Cell Biol 14: 700–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyse SM, Tyrrell RM (1989) Heme oxygenase is the major 32-kDa stress protein induced in human skin fibroblasts by UVA radiation, hydrogen peroxide, and sodium arsenite. Proc Natl Acad Sci USA 86: 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S (1999a) Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cystein residue in the central conserved region. Proc Natl Acad Sci USA 96: 9112–9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Taoka H, Toda T, Yoshida M, Horinouchi S (1999b) A novel nuclear export signal sensitive to oxidative stress in the fission yeast transcription factor Pap1. J Biol Chem 274: 15151–15158 [DOI] [PubMed] [Google Scholar]
- Lathrop JT, Timko MP (1993) Regulation by heme of mitochondrial protein transport through a conserved aminoacid motif. Science 259: 522–525 [DOI] [PubMed] [Google Scholar]
- Monson EK, Weinstein M, Ditta GS, Helinski DR (1992) The FixL protein of Rhizobium meliloti can be separated into a heme-binding oxygen-sensing domain and a functional C-terminal kinase domain. Proc Natl Acad Sci USA 89: 4280–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, Tashiro S, Tsuchiya H, Kume A, Kanno M, Ito E, Yamamoto M, Igarashi K (2002) Activation of Maf/AP-1 repressor Bach2 promotes apoptosis and its interaction with PML nuclear bodies. J Biol Chem 277: 20724–20733 [DOI] [PubMed] [Google Scholar]
- Ney PA, Sorrentino BP, Lowrey CH, Nienhuis AW (1990a) Inducibility of the HS II enhancer depends on binding of an erythroid specific nuclear protein. Nucleic Acids Res 18: 6011–6017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ney PA, Sorrentino BP, McDonagh KT, Nienhuis AW (1990b) Tandem AP-1-binding sites within the human b-globin dominant control region functions as an inducible enhancer in erythroid cells. Genes Dev 4: 993–1006 [DOI] [PubMed] [Google Scholar]
- Ogawa K, Sun J, Taketani S, Nakajima O, Nishitani C, Sassa S, Hayashi N, Yamamoto M, Shibahara S, Fujita H, Igarashi K (2001) Heme mediates de-repression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J 20: 2835–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterbein LE, Soares MP, Yamashita K, Bach FH (2003) Heme oxygenase-1: unleashing the protective properties of heme. Trends Immunol 24: 449–455 [DOI] [PubMed] [Google Scholar]
- Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, Igarashi K (1996) Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol 16: 6083–6095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD, Tonegawa S (1997) Reduced stress defense in heme oxygenase 1-deficient cells. Proc Natl Acad Sci USA 94: 10925–10930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Hamza I, O'Brian MR (1999) Heme is an effector molecule for iron-dependent degradation of the bacterial iron response regulator (Irr) protein. Proc Natl Acad Sci USA 96: 13056–13061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara S, Kitamuro T, Takahashi K (2002) Heme degradation and human diseases: diversity in the soul of life. Antioxidants Redox Signaling 4: 593–602 [DOI] [PubMed] [Google Scholar]
- Shibahara S, Muller R, Taguchi H, Yoshida T (1985) Cloning and expression of cDNA for rat heme oxygenase. Proc Natl Acad Sci USA 82: 7865–7869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara S, Muller RM, Taguchi H (1987) Transcriptional control of rat heme oxygenase by heat shock. J Biol Chem 262: 12889–12892 [PubMed] [Google Scholar]
- Shibahara S, Yoshida T, Kikuchi G (1978) Induction of heme oxygenase by hemin in cultured pig alveolar macrophages. Arch Biochem Biophys 188: 243–250 [DOI] [PubMed] [Google Scholar]
- Sun J, Brand M, Zenke Y, Tashiro S, Groudine M, Igarashi K (2004) Heme regulates the dynamic exchange of Bach1 and NF-E2-related factors in the Maf transcription network. Proc Natl Acad Sci USA 101: 1461–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Hoshino H, Takaku K, Nakajima O, Muto A, Suzuki H, Tashiro S, Takahashi S, Shibahara S, Alam J, Taketo MM, Yamamoto M, Igarashi K (2002) Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J 21: 5216–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Tashiro S, Sun J, Doi H, Satomi S, Igarashi K (2003) Cadmium induces nuclear export of Bach1, a transcriptional repressor of heme oxygenase-1 gene. J Biol Chem 278: 49246–49253 [DOI] [PubMed] [Google Scholar]
- Taketani S, Kohno H, Yoshinaga T, Tokunaga R (1989) The human 32-kDa stress protein induced by exposure to arsenite and cadmium ions is heme oxygenase. FEBS Lett 245: 173–176 [DOI] [PubMed] [Google Scholar]
- Tang SC, Koch S, Papaefthymiou GC, Foner S, Frankel RB, Ibers JA, Holm RH (1976) Axial ligation modes in iron(III) Porphyrins. Models for the oxidized reaction states of cytochrome P-450 enzymes and the molecular structure of iron(III) protoporphyrin IX dimethyl ester p-nitrobenzenethiolate. J Am Chem Soc 98: 2114–2434 [DOI] [PubMed] [Google Scholar]
- Tenhunen R, Marver HS, Schmid R (1970) The enzymatic catabolism of hemoglobin: stimulation of microsomal heme oxygenase by hemin. J Lab Clin Med 75: 410–421 [PubMed] [Google Scholar]
- Wagener FA, van Beurden HE, von den Hoff JW, Adema GJ, Figdor CG (2003) The heme–heme oxygenase system: a molecular switch in wound healing. Blood 102: 521–528 [DOI] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell 82: 463–473 [DOI] [PubMed] [Google Scholar]
- Yachie A, Niida Y, Wata T, Igarashi N, Kaneda H, Toma T, Ohta K, Kasahara Y, Koizumi S (1999) Oxidative stress causes enhanced endothelial cell injury in human heme oxygenase-1 deficiency. J Clin Investig 103: 129–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida C, Tokumasu F, Hohmura KI, Bungert J, Hayashi N, Nagasawa T, Engel JD, Yamamoto M, Takeyasu K, Igarashi K (1999) Long range interaction of cis-DNA elements mediated by architectural transcription factor Bach1. Genes Cells 4: 643–655 [DOI] [PubMed] [Google Scholar]
- Zhang L, Guarente L (1995) Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J 14: 313–320 [DOI] [PMC free article] [PubMed] [Google Scholar]


