Abstract
Pathway specificity is poorly understood for mitogen-activated protein kinase (MAPK) cascades that control different outputs in response to different stimuli. In yeast, it is not known how the same MAPK cascade activates Kss1 MAPK to promote invasive growth (IG) and proliferation, and both Fus3 and Kss1 MAPKs to promote mating. Previous work has suggested that the Kss1 MAPK cascade is activated independently of the mating G protein (Ste4)–scaffold (Ste5) system during IG. Here we demonstrate that Ste4 and Ste5 activate Kss1 during IG and in response to multiple stimuli including butanol. Ste5 activates Kss1 by generating a pool of active MAPKKK (Ste11), whereas additional scaffolding is needed to activate Fus3. Scaffold-independent activation of Kss1 can occur at multiple steps in the pathway, whereas Fus3 is strictly dependent on the scaffold. Pathway specificity is linked to Kss1 immunity to a MAPK phosphatase that constitutively inhibits basal activation of Fus3 and blocks activation of the mating pathway. These findings reveal the versatility of scaffolds and how a single MAPK cascade mediates different outputs.
Keywords: Kss1, MAPK phosphatase, Msg5, Ste5 scaffold, Ste11
Introduction
Mitogen-activated protein kinase (MAPK) cascades form the cores of many receptor-linked signaling pathways in eukaryotes and respond to many external stimuli. The same MAPK cascade can be used by more than one signal transduction pathway to effect different responses, raising the question of how pathway specificity is achieved (Madhani and Fink, 1998; Tan and Kim, 1999). One level of pathway specificity arises from variations in the strength and duration of activation of a MAPK cascade by stimulus (Traverse et al, 1992; Marshall, 1995), which is influenced by the level of MAPK phosphatase (Bhalla et al, 2002).
Scaffold proteins also provide specificity (Yu et al, 1998; Burack and Shaw, 2000; Elion, 2001; Roy et al, 2002; Weston and Davis, 2002). In budding yeast, the Ste5 and Pbs2 scaffolds join different subsets of the same kinases to different plasma membrane anchors that sense different stimuli (Figure 1A; Choi et al, 1994; Whiteway et al, 1995; Maeda et al, 1995; Posas and Saito, 1997). Despite this overlap, each scaffold provides specificity based on the distinct phenotypes of ste5 and pbs2 null mutants. Ste5 binds the β subunit (Ste4) of a heterotrimeric G protein that is coupled to mating pheromone receptors, and joins MAPKKK Ste11 and MAPKK Ste7 with MAPK Fus3, which promotes mating in response to mating pheromone. Pbs2 binds Sho1, a plasma membrane sensor, and joins Ste11 with itself, a MAPKK, and MAPK Hog1, which promotes survival in high osmolarity. In each case, binding of scaffold to membrane-linked anchor is thought to facilitate activation of Ste11 by Ste20, a p21-activated kinase (PAK) that is anchored to Cdc42 GTPase in an active state. The scaffolds therefore specify two events, MAPKKK activation in response to stimulus and linkage of the activated MAPKKK to MAPKK and MAPK. This three-tier model of kinases with scaffold is also used in other pathways (Burack and Shaw, 2000; Morrison and Davis, 2003).
Figure 1.
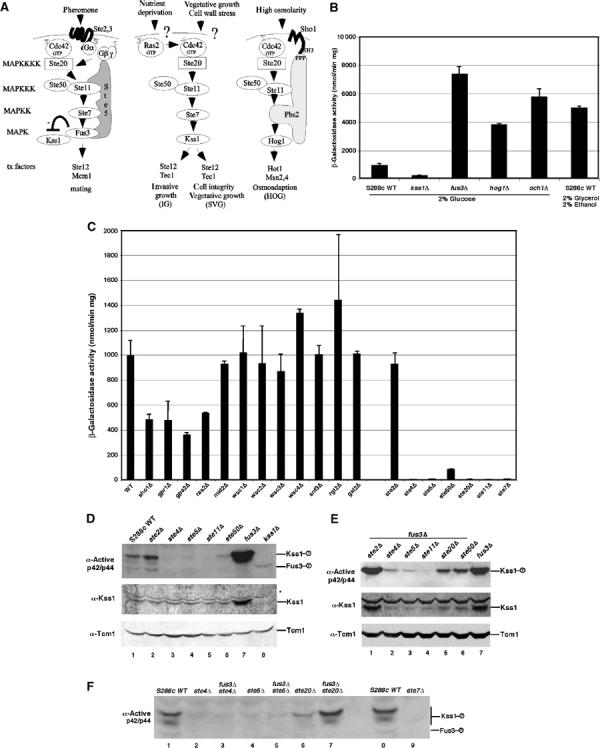
Activation of Kss1 during vegetative growth is dependent on the mating G protein β subunit (Ste4) and scaffold (Ste5), but not on upstream regulators of the high osmolarity growth (HOG), protein kinase A, protein kinase C, and sucrose nonfermenting pathways. (A) Cartoon of mating response, IG, SVG and HOG pathways in Saccharomyces cerevisiae. (B) FRE(Tec1)-lacZ reporter gene expression. β-Galactosidase assays were performed on mitotically dividing S288c strains harboring a pFRE(TEC1)-lacZ plasmid (Madhani and Fink, 1997a). Standard deviation is shown for triplicate samples. (C) Fre(Tec1)-LacZ activity in S288c strains with mutations in upstream regulators of a variety of protein kinase pathways. (D) Kss1 activity and protein in S288c strains with mating pathway mutations. (E) Kss1 activity and protein in S288c fus3Δ strains with mutations in the mating pathway. Immunoblot analysis was performed on whole-cell extracts prepared from vegetatively dividing cells. (F) Kss1 activity in fus3Δste and ste strains. The active form of Kss1 was detected with an anti-phospho p42/p44 peptide antibody and total Kss1 protein was detected with a polyclonal antibody. The asterisk indicates a protein that crossreacts nonspecifically with the Kss1 antibody. Tcm1 is a ribosomal protein that serves as a normalization control. Similar results were found in W303a. Depending on gel conditions, Kss1 migrates as a doublet when activated for unknown reasons.
The Ste20, Ste11, and Ste7 kinases also activate the Kss1 MAPK to promote invasive growth (IG) in haploid cells, pseudohyphal development (PD) in diploid cells, and cell integrity during vegetative growth (sterile vegetative growth (SVG)) (Figure 1A; Madhani and Fink, 1998; Lee and Elion, 1999; Pan et al, 2000). They also activate Kss1 during mating (Gartner et al, 1992; Cherkasova et al, 2001; Sabbagh et al, 2001). IG and mating pathway specificity is linked to the ability of Kss1 and Fus3 to recognize distinct substrates such as different transcription factors (Elion et al, 1993; Madhani and Fink, 1997a; Breitkreutz and Tyers, 2002). This configuration of a MAPKKK and MAPKK activating multiple MAPKs is a common feature of MAPK cascades, and includes human extracellular regulated kinases (Erks) and c-Jun N-terminal kinases (Jnks) (Widmann et al, 1999; Pearson et al, 2001).
It is not understood how Kss1 is selectively activated during proliferative responses. Both IG and PD are induced by activated Ras2 and by nutritional changes, which has led to the hypothesis that these conditions activate the Kss1 MAPK cascade (Gimeno et al, 1992; Roberts and Fink, 1994; Mosch et al, 1996; Pan et al, 2000). Kss1 is activated by mutations in Och1, which is a mannosyltransferase required for synthesis of mannoproteins (Lee and Elion, 1999) and may be upregulated by Sho1 (Cullen et al, 2000). Since genetic evidence places Ras2 and Sho1 upstream of Ste11 and Och1 in parallel with Ste11 (Figure 1A), these regulators are unlikely to be part of a mechanism that distinguishes between Kss1 and Fus3 activation. The mating receptor, G protein, and Ste5 scaffold are not required for IG (Roberts and Fink, 1994). These observations led to the hypothesis that specificity is provided by an alternative scaffold that selectively activates a Ste11, Ste7, and Kss1 MAPK cascade in response to Ras2 input (Roberts and Fink, 1994).
The question of whether or not Ste5 regulates Kss1 during IG is unresolved. During mating, Kss1 is activated by the same receptor–G protein–Ste5 scaffold system that activates Fus3 (Ma et al, 1995; Breitkreutz et al, 2001; Cherkasova and Elion, 2001; Sabbagh et al, 2001), but is subsequently attenuated by activated Fus3 (Sabbagh et al, 2001) (Figure 1A). Downregulation of Kss1 by Fus3 has been proposed to block erroneous crossactivation of the IG pathway during mating (Madhani et al, 1997b; Sabbagh et al, 2001). However, the fact that low levels of mating pheromone induce IG-like responses in a wild-type strain (Oehlen and Cross, 1998; Roberts et al, 2000; Erdman and Snyder, 2001) suggests that Ste5 could normally activate a Kss1-dedicated pathway as long as Fus3 activity is below a certain threshold. To date, a systematic analysis of positive regulators of basal Kss1 and Fus3 kinase activity has not been done, nor has it been established that conditions that induce IG lead to changes in Kss1 kinase activity.
In this study, we screened for upstream regulators of the Kss1 MAPK cascade and identified Ste4 and Ste5 as key positive regulators of Kss1 activation during vegetative and IG, with little contribution by Ras2 and Sho1. Ste4 and Ste5 also activated Fus3 during vegetative and IG. Pathway specificity was linked to constitutive inactivation of Fus3 by MAPK phosphatase Msg5. Our results support a model in which a single-core G protein–scaffold–kinase pathway responds to multiple stimuli and regulates two functionally distinct MAPKs, with a MAPK phosphatase determining the threshold for entry into two developmental choices. Interestingly, Ste5 provides different functions in activating the two MAPKs. Fus3 is dependent on multiple tethering functions of the scaffold and is likely to be activated on the scaffold. In contrast, Kss1 can be activated off of the scaffold by an active pool of Ste11 that is generated as a result of Ste5 recruitment to Ste4. This novel mechanism provides a means by which signal amplification might occur as a result of scaffold-induced activation.
Results
Ste4 and Ste5 are essential for basal activation of Kss1, while Ras2, Sho1, Gpr1, and Gpa2 are not
To identify positive regulators of the SVG pathway, we screened a library of yeast null mutants in the S288c strain background using the FRE (filamentation and invasion response element) (Tec1)-lacZ reporter gene (Madhani and Fink, 1997a). Control tests showed that the FRE(Tec1)-lacZ reporter was a sensitive monitor for both SVG and IG pathways under basal and induced conditions. FRE(Tec1)-lacZ expression was reduced by a kss1Δ mutation and increased by fus3Δ and hog1Δ mutations that abolish attenuating mechanisms (Figure 1B). Furthermore, FRE(Tec1)-lacZ expression was stimulated in an och1Δ strain with reduced cell integrity (Lee and Elion, 1999) and in low glucose conditions (i.e. 2% glycerol/2% ethanol) that induce IG (Cullen and Sprague, 2000). Initial screening showed that FRE(Tec1)-lacZ expression is not regulated by a wide variety of plasma membrane-linked receptors and other proteins known to sense environmental conditions (Figure 1C; data not shown). However, a reproducible 50% reduction in β-galactosidase activity was found in strains that lacked Ras2, Sho1, Gpa2 or Gpr1, indicating that these proteins weakly stimulate but are not essential for signaling through the Kss1 MAPK cascade during VG in rich medium. This finding was consistent with the observation that a Ras2A19V mutant did not stimulate the activation of Kss1 to an obvious extent (Supplementary Figure 1).
In contrast, FRE(Tec1)-lacZ expression was abolished in S288c strains that lack either the Gβ subunit Ste4 (ste4Δ) or the MAPK cascade scaffold Ste5 (ste5Δ), whereas mutation of the mating pheromone receptor Ste2 (ste2Δ) had no effect (Figure 1C). Similar results were found in W303 strains (data not shown). Ste4 and Ste5 were as critical as Ste20, Ste11, and Ste7, and more critical than Ste50, which positively regulates Ste11, and is essential for IG (Ramezani Rad et al, 1998). The requirement for Ste4 and Ste5 in basal activation of Kss1 was confirmed by immunoblot analysis of the level of active Kss1 protein in whole-cell extracts. Activated Kss1 was detected with an anti-MAPK antibody directed against dual phosphorylated forms of mammalian p42/p44 that crossreacts with activated Kss1 and Fus3, and compared to the total level of Kss1 protein with an anti-Kss1 antibody. The level of active Kss1 was nearly abolished in the ste4Δ and ste5Δ strains, but was slightly elevated in the ste2Δ mutant lacking the α factor mating pheromone receptor (Figure 1D).
It was possible that the requirement for Ste4 and Ste5 was indirectly due to negative regulation of Kss1 levels by inactive Fus3, which blocks accumulation of active Kss1 and Kss1 protein (Cherkasova et al, 2003). However, Ste4 and Ste5 were required to activate Kss1 in a fus3Δ strain, which has greatly elevated levels of active Kss1 and Kss1 protein as a result of the loss of attenuation by Fus3 (Figure 1E). Active Kss1 was barely detected in fus3Δste4Δ and fus3Δste5Δ double mutants compared to a large amount of active Kss1 in the fus3Δ single mutant and residual active Kss1 in ste20Δ fus3Δ and ste50Δfus3Δ double mutants (Figure 1E). The level of active Kss1 was as reduced in ste5Δfus3Δ and ste4Δfus3Δ double mutants as it was in ste5Δ and ste4Δ single mutants (Figure 1F). Therefore, signaling through the Kss1 MAPK cascade during proliferation is directly dependent on Ste4 and Ste5 and not due to a change in the status of Fus3.
Ste4 and Ste5 activate Kss1 under conditions that stimulate IG
A prediction of our findings was that Ste4 and Ste5 should be required for Kss1 to be activated during conditions that induce IG. No direct evidence exists for stimulus-induced effects on Kss1 activation during proliferation. We tested three conditions that induce IG: growth on solid agar plates containing 2% glucose (Roberts and Fink, 1994), glucose depletion (Cullen and Sprague, 2000), and 1-butanol induction, which induces morphological changes resembling, but not identical to, pseudohyphal growth (Lorenz et al, 2000a; Zeitlinger et al, 2003). Assessment of IG in S288c strains in which the naturally occurring mutation in the FLO8 gene was corrected with a FLO8 plasmid (Liu et al, 1996) showed that ste4Δ and ste5Δ strains were as deficient in invading the agar as flo11Δ and ste20Δ strains (Figure 2A and B). Kss1 activation was not detected in ste4Δ and ste5Δ mutants grown in liquid medium containing 2% glycerol/2% ethanol, although these conditions increased the level of active Kss1 in the control strain (Figure 2C). Moreover, although 1-butanol induced the activation of both Kss1 and Fus3 within 30 min in the S288c wild-type strain (Figure 2D), no activation or morphological changes occurred in the ste4Δ and ste5Δ strains. Similar results were found in W303 (data not shown). Thus, Ste4 and Ste5 are required for Kss1 to be activated by multiple stimuli that induce IG.
Figure 2.
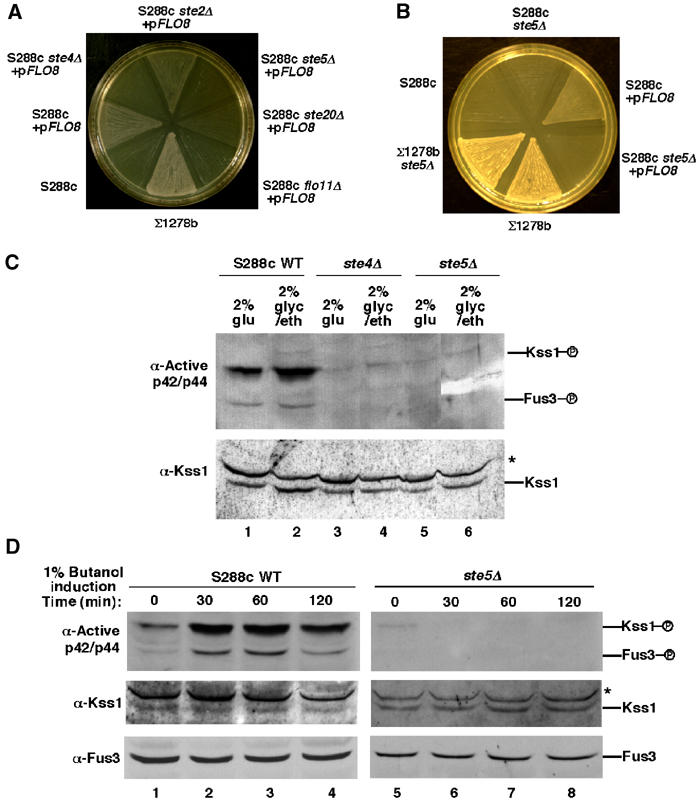
Activation of Kss1 during IG is dependent on Ste4 and Ste5. (A, B) IG of S288c strains harboring a FLO8 plasmid requires STE4 and STE5. (C) Enhancement of Kss1 activity as a result of growth in the absence of glucose requires STE4 and STE5. Immunoblot analysis of extracts made from cells grown in liquid medium containing 2% glucose or 2% glycerol/2% ethanol. (D) 1-Butanol activates Kss1 and Fus3 in the presence of Ste4 and Ste5. S288c strains growing in liquid medium containing 2% glucose were induced with 1% 1-butanol for the indicated times.
The requirement for Ste4 and Ste5 in activation of Kss1 during IG in S288c contrasted Σ1278b, which does not require Ste4 or Ste5 for IG or SVG (Roberts and Fink, 1994; Lee and Elion, 1999). Σ1278b shares only a small amount of genetic lineage with S288c (Winzeler et al, 2003) and undergoes significantly more robust IG compared to S288c/FLO8 strains (Figure 2A) (Liu et al, 1996). A comparison of Kss1 activation in Σ1278b, S288c, and W303 showed that Σ1278b does not require Ste5 to activate Kss1 and has significantly more active Kss1 and total Kss1 protein compared to S288c and W303, and a significantly greater ability to stimulate the FRE(Tec1)-lacZ reporter gene (Supplementary Figure 2A and B). The dependence on Ste5 for Kss1 activation in S288c was found to correlate with lower levels of basal activation of Kss1 (Supplementary Figure 2C). The feral parent of S288c, EM93 (Mortimer and Johnston, 1986), had very low levels of active Kss1 (data not shown), suggesting that S288c and Σ1278b reflect differences in basal signaling among naturally occurring yeast strains.
Only the recruitment function of Ste5 that leads to Ste11 activation is required to activate Kss1
The strict dependence of Kss1 activation on Ste4 and Ste5 in S288c and W303 and the absence of the requirement for Ste5 under conditions of higher basal activation in Σ1278b raised the question of whether the G protein and Ste5 activate Kss1 and Fus3 by different mechanisms, since very little active Fus3 is detected in these strains. Ste5 activates Fus3 through multiple tethering functions, which can be divided into two key events: activation of Ste11 and activation of Fus3 by activated Ste11. Ste11 activation involves Ste5 binding to both Ste4 and Ste11 to facilitate phosphorylation of Ste11 by Ste20. Fus3 activation involves Ste5 binding to Ste11, Ste7, and Fus3, to facilitate activation of Ste7 and Fus3 (Choi et al, 1994; Inouye et al, 1997; Feng et al, 1998; Pryciak and Huntress, 1998; Bardwell et al 2000; Wang and Elion, 2003). To determine whether Ste5 activates Kss1 by a similar mechanism, we assessed whether Ste5 mutants with defects in binding to either Ste4 (Ste5C180A; Feng et al, 1998) or Ste11 (Ste5L482/485A; Wang and Elion, 2003) could activate Kss1. Neither Ste5C180A nor Ste5L482/485A supported the activation of Kss1 in either ste5Δ or ste5Δ fus3Δ strains, before or after induction with 1% butanol (Figure 3A). Therefore, Ste5 must be able to bind to both Ste4 and Ste11 in order to activate Kss1.
Figure 3.
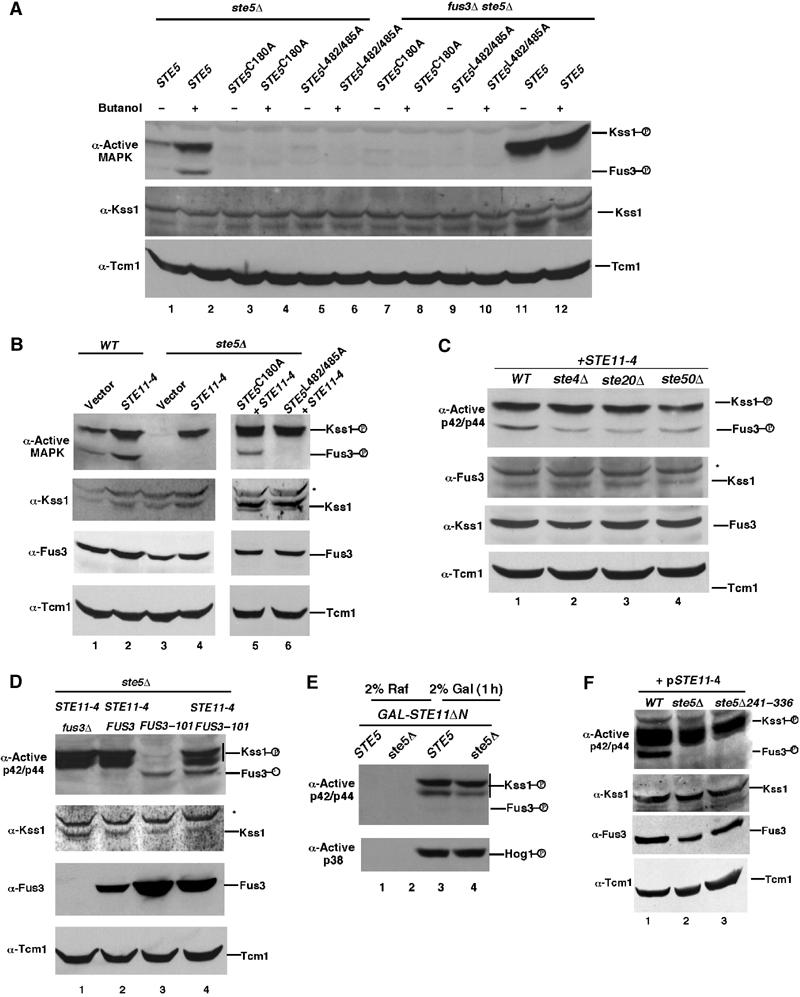
Kss1 only requires Ste5 to activate Ste11. (A) Kss1 activation is dependent on Ste5 binding to Ste4 and Ste11. Level of Kss1 activity in ste5Δ and ste5Δ fus3Δ strains expressing Ste5, Ste5C180A or Ste5L482/485A from centromeric plasmids. The ‘+' indicates 1 h induction with 1% 1-butanol. (B) Ste11-4 activates Kss1 in the absence of Ste5. Levels of active Kss1 and Fus3 and total Kss1 and Fus3 proteins in wild-type and ste5Δ cells expressing either a control vector or Ste11-4 from a centromeric vector. (C) Ste11-4 activates Kss1 and Fus3 in the absence of STE4, STE20, and STE50. (D) Ste5 is needed for Ste11-4 to activate Fus3-101. Active Kss1 and Fus3 in a fus3Δ ste5Δ strain expressing FUS3 or FUS3-101, with or without STE11-4. (E) Ste11ΔN activates Kss1, but not Fus3, in the absence of Ste5. Ste11ΔN was expressed from the GAL1 promoter. Cells were grown in 2% raffinose medium and then in 2% galactose medium for 1 h. The abundance of Kss1 and Fus3 is lower in 2% raffinose medium than in 2% galactose medium, resulting in very low basal activity. (F) Ste5Δ241–336 supports activation of Kss1, but not Fus3, by Ste11-4. Extracts were prepared from the S288c ste5Δ pSTE11-4 strain with CEN vectors expressing Ste5 (lane 1), Ste5Δ241–336 (lane 3) or a control vector (lane 2).
Two constitutively active derivatives of Ste11, Ste11-4 (Stevenson et al, 1992), and Ste11ΔN (Cairns et al, 1992), were used to determine whether Kss1 must be bound to Ste5 to be activated by Ste11. Ste11-4 has a T596 → I mutation in subdomain VII (596TDFG599) of the catalytic domain and retains its ability to bind to Ste5. Ste11ΔN lacks the autoinhibitory N-terminal regulatory domain that binds Ste5. Ste11-4 activated Kss1 with equal efficiency in wild-type, ste5Δ, ste5C180A, and ste5L482/485A strains (Figure 3B, lanes 1–6). This behavior was similar to that of Hog1, which was also activated by Ste11-4 in the presence and absence of Ste5 (data not shown). In contrast, Ste11-4 did not activate Fus3 when Ste5 was either not present or unable to bind to Ste11-4 (i.e. ste5Δ and ste5L482/485A; Figure 3B, lanes 4, 6), but could activate Fus3 in pathway mutants that permitted binding of Ste11-4 to Ste5, including ste5C180A (Figure 3B, lane 5) and ste4Δ, ste20Δ, and ste50Δ (Figure 3C). The inability to detect active Fus3 in the absence of Ste5 was not a detection problem based on parallel analysis with Fus3-101 (Figure 3D), which autophosphorylates weakly in the absence of Ste5 (Brill et al, 1994). Furthermore, similar results were found with Ste11ΔN, which activated Kss1 with equal efficiency in wild-type and ste5Δ strains, but did not activate Fus3 (Figure 3E).
These findings suggested that Kss1 does not need to bind to Ste5 to be activated by Ste11, whereas Fus3 must be bound. We tested this interpretation with a Ste5Δ241-336 mutant that lacks the binding site for Fus3 and Kss1 (Choi et al, 1994). Ste5Δ241-336 could support activation of Kss1 by Ste11-4; however, Fus3 was not activated (Figure 3F). Thus, Ste11 activates Fus3 while bound to Ste5, whereas Kss1 does not have to be bound to Ste5 to be activated. This finding implies that Ste7 activates Fus3 within the Ste5 complex but activates Kss1 independently of Ste5.
Kss1 dependence on the Ste5 scaffold is bypassed by increasing the levels of Ste11 and Ste7
Overexpression of Ste11 inhibits Fus3 activation unless Ste5 is also increased, which is consistent with Fus3 activation requiring a scaffold complex that contains Ste11 (Choi et al, 1994). We further tested the possibility that a scaffold is not required for Kss1 to be activated by Ste11, by determining whether increased levels of Ste11 or Ste7 would stimulate Kss1 activation in the absence of Ste5. Overexpression of either Ste11 or Ste7 increased the level of active Kss1 in a ste5Δ strain, with a nearly complete bypass when Ste11 and Ste7 were co-overexpressed (Figure 4A). Co-overexpression of Kss1 with Ste11 and Ste7 resulted in a further increase in active Kss1 (Figure 4B). In contrast, the level of active Fus3 did not increase under any of these conditions (Figure 4A and B). These results are consistent with active Ste11 and Ste7 being able to activate Kss1 independently of Ste5.
Figure 4.
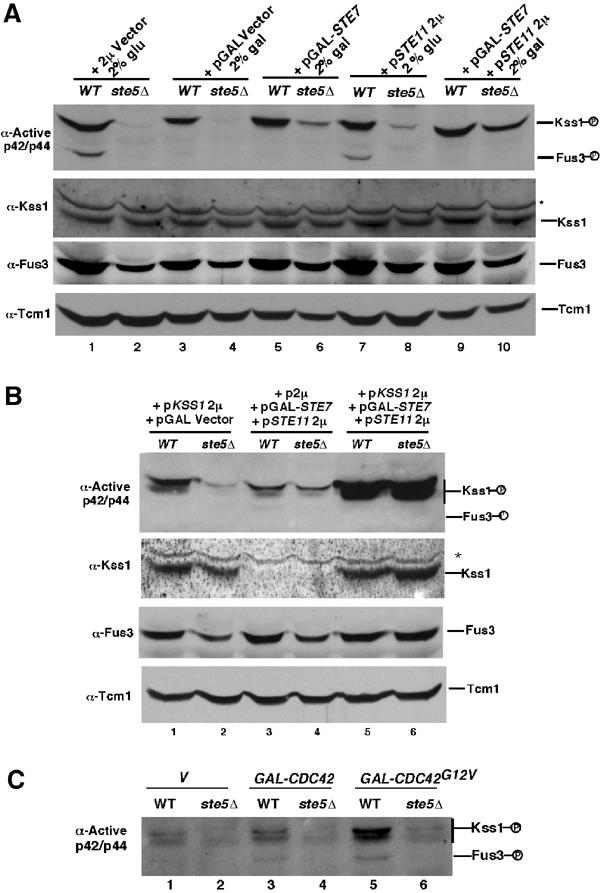
Fus3, but not Kss1, must bind to Ste5 to be activated by Ste11. (A) Overexpression of either Ste11 or Ste7 bypasses the need for Ste5 in basal activation of Kss1. Wild-type and ste5Δ strains harbor a vector expressing STE11 from its native promoter (pSTE11 2 μ) on a multicopy plasmid or STE7 from the GAL1 promoter (GAL-STE7) on a centromeric plasmid. (B) Co-overexpression of Ste7, Ste11, and Kss1 completely bypasses the need for Ste5 to activate Kss1. Relative levels of active Kss1 and total Kss1 protein in wild-type and ste5Δ cells overexpressing Kss1, Ste7, and/or Ste11. The strong intensity of the Kss1 band in strains expressing KSS1 2 μ obscures the weak band of endogenous Kss1. (C) Cdc42G12V does not stimulate Kss1 activity in a ste5Δ strain. GAL1p-CDC42G12V was expressed for 1 h in medium containing 2% galactose.
Msg5 phosphatase inhibits constitutive activation of Fus3
It was not clear how the same G protein–scaffold system selectively activates one MAPK during proliferation and two MAPKs during mating, since the only difference between these two states is pheromone activation of a receptor that is coupled to the G protein (Figure 1A). For example, the expression of Cdc42G12V increases the level of active Kss1 with negligible effect on Fus3, even though the stimulation of Kss1 requires Ste5 (Figure 4C). We tested whether the G protein–scaffold system activates Kss1 more efficiently than Fus3 by comparing the relative fold induction of Kss1 and Fus3 as a function of brief (15 min) exposure to physiological concentrations of mating pheromone (α factor) and increasing the level of Ste5 in the absence of pheromone stimulation. The fold increase in Fus3 activation was greater than Kss1, with the preference for Fus3 greatest at highest α factor concentration (Figure 5A and B). Increasing the level of Ste5 or expressing a hyperactive allele of Ste5, Ste5hyp1 (Hasson et al, 1994) also caused a greater fold increase in the activity of Fus3 compared to Kss1 (Figure 5C and D). Therefore, the G protein–scaffold system is unlikely to activate Kss1 more efficiently than Fus3.
Figure 5.
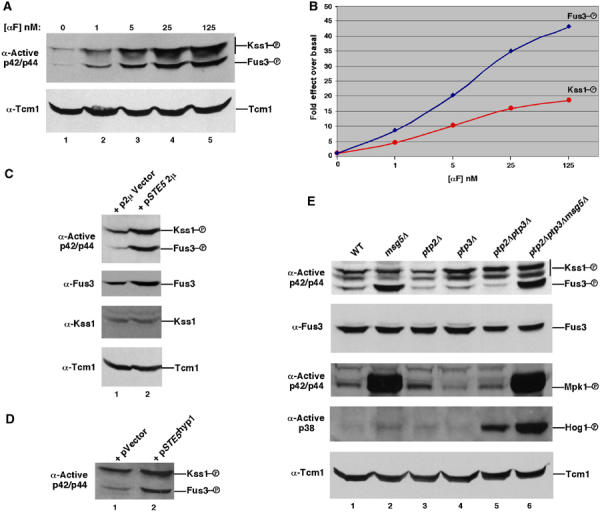
Fus3 is efficiently activated during vegetative growth, but is inhibited by Msg5 MAPK phosphatase. (A) Activation of Kss1 and Fus3 by mating pheromone. Vegetatively growing cells were treated with α factor for 15 min. Parallel immunoblotting showed that the levels of Fus3 and Kss1 remained the same under these conditions (data not shown). (B) Fold effect of activation of Fus3 by α factor is greater than that of Kss1 based on densitometric analysis. The level of active MAPK relative to total MAPK protein was determined by densitometry for each lane and the fold increase as a consequence of α factor addition was plotted. Quantitation was done with the Scion image 1.62c densitometry program of the public domain software NIH image (available at http://rsb.info.nih.gov/nih-image/) using multiple exposures of immunoblots. (C) Effect of increasing Ste5 levels on Fus3 and Kss1 activity. (D) Effect of gain-of-function STE5hyp1 mutant on Fus3 and Kss1. (E) Level of active Kss1, Fus3, Mpk1, and Hog1 in msg5Δ, ptp2Δ, and ptp3Δ MAPK phosphatase mutants. Kss1, Fus3, and Mpk1 were detected with anti-p42/p44 phosphopeptide antibody. Hog1 with anti-active p38 antibody. The positions of Mpk1 and Hog1 were deduced from mpk1 and hog1 mutants. The level of Kss1 protein did not change in the mutants compared to wild type (data not shown).
An alternative explanation for pathway specificity is that Fus3 and Kss1 are both basally activated by the G protein–scaffold system, but subsequent inactivation of Fus3 leaves Kss1 to effect responses. Fus3 is immediately inactivated upon mating pheromone withdrawal, consistent with it being subject to constitutive inactivation (Choi et al, 1999). To test this hypothesis, we compared the effect of mutations in the three major MAPK phosphatases (Msg5, Ptp2 and Ptp3) on the activity of Fus3 and Kss1 in addition to the two other MAPKs expressed in haploids, Hog1 and Mpk1. Msg5 is a dual-specificity MAPK phosphatase (Doi et al, 1994) that is thought to inactivate Fus3 during recovery from pheromone-induced G1 arrest (Doi et al, 1994), and also regulate Mpk1 (Martin et al, 2000) and Kss1 (Davenport et al, 2001). Ptp2 and Ptp3 are phospho-tyrosine phosphatases (Watanabe et al, 1995) that have been linked to Fus3, Kss1 (Zhan et al, 1997) and Hog1 (Wurgler-Murphy et al, 1997; Mattison et al, 1999).
The msg5Δ mutation caused a large increase in the levels of active Fus3 and Mpk1 during vegetative growth, with no obvious effect on the amount of active Kss1 or active Hog1 (Figure 5E, lanes 1 and 2). In contrast, the ptp2Δ and ptp3Δ mutations, alone or in combination, had little effect on the level of active Fus3 or Mpk1, whereas the level of active Kss1 increased somewhat and that of Hog1 increased greatly (lanes 3–6). The increase in Fus3 activity in the msg5Δ strain is not the result of activation of Mpk1, because Mpk1 does not positively regulate mating pathway activation (Lee and Elion, 1999). Furthermore, the greater increase in Fus3 activity compared to Kss1 may be an underestimate, because the anti-active MAPK antibody is directed against a region in p42/p44 that is more conserved in Kss1 than in Fus3 (Materials and methods). Therefore, the G protein–scaffold system efficiently activates Fus3 in addition to Kss1 during vegetative growth, with the relatively greater pool of active Kss1 the result of selective inactivation of Fus3 by Msg5.
Differential input by Ste5 and Msg5 is critical for pathway specificity
We determined whether Msg5 regulates pathway specificity by analyzing the phenotype of the msg5Δ mutant, using reporter genes and cell morphology as monitors. Loss of Msg5 constitutively activated the mating pathway, based on an increased level of active Fus3 (Figure 5E, lanes 1 and 2) and expression of FUS1-lacZ and FUS1-GFP reporter genes (Figure 6A), cell enlargement and morphological changes resembling shmoo formation in many cells (Figure 6A) and slower growth rate (data not shown). In contrast, IG was inhibited based on decreased expression of FRE(Tec1)-lacZ (Figure 6B), which correlated with a decreased level of active Kss1 (Figure 5E). This result is consistent with the inhibitory effect of Fus3 on IG and Kss1 activation as a result of its activation by pheromone (Sabbagh et al, 2001). Therefore, Msg5 prevents misactivation of the mating pathway and enforces the Kss1 fate.
Figure 6.
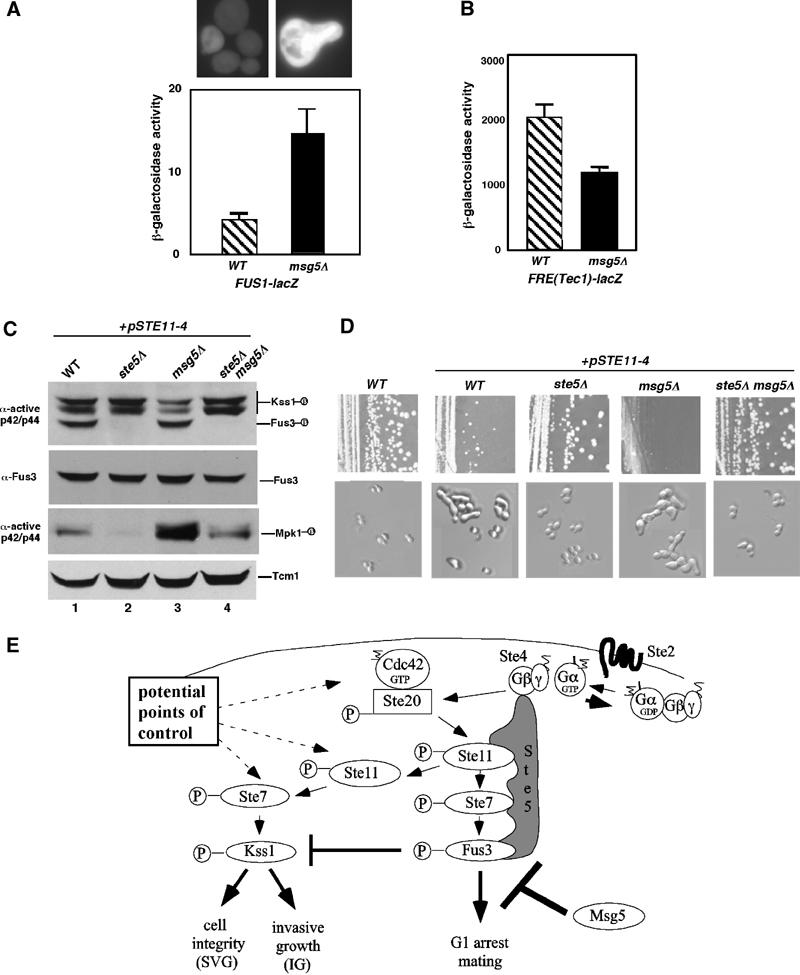
Differential regulation of Kss1 and Fus3 by Ste5 and Msg5 is required for pathway specificity. (A) Constitutive mating pathway activation in msg5Δ strain based on FUS1-lacZ, FUS1-GFP, and cell morphology. (B) Decreased FRE(Tec1)-lacZ expression in a msg5Δ strain. (C) Level of active Kss1, Fus3, and Msg5 in STE11-4 strains with ste5Δ and/or msg5Δ mutations. (D) Growth and morphology of STE11-4 strains with ste5Δ and/or msg5Δ mutations. Strains were streaked for single colonies on a SC-selective plate and incubated at room temperature. Nomarski images were taken of the cells. (E) Model for how a single G protein–scaffold and MAPK phosphatase system regulates IG and mating pathways.
The combined effect of Msg5 and Ste5 on pathway specificity during MAPK activation was further analyzed in STE11-4 strains. The msg5Δ mutation, which increases the level of active Fus3 and Mpk1 compared to Kss1 (Figure 6C), also increased the degree of morphological changes resembling shmoos and caused a nearly complete cell cycle arrest compared to the STE11-4 strain (Figure 6D). These phenotypes are consistent with the increased level of active Fus3 compared to Kss1, which likely results from coincident attenuation by Fus3. The introduction of a ste5Δ mutation into the STE11-4 and STE11-4 msg5Δ strains abolished activation of Fus3 equivalently (Figure 6C) and restored efficient growth (Figure 6D) and a round budding morphology (Figure 6D). Although the ste5Δ mutation had no effect on Hog1 (data not shown), it did reduce the level of active Mpk1 (Figure 6C), which is activated as a consequence of morphological changes induced by mating pheromone (Zarzov et al, 1996; Buehrer and Errede, 1997). Thus, dual control of Fus3 and Kss1 by the G protein–Ste5 scaffold system, together with MAPK phosphatase selectivity, allows for proper differentiation and survival.
Discussion
Receptor-independent recruitment of a scaffold to G protein anchor stimulates a proliferative MAPK
Previous work defined the Ste20 → Ste11 → Ste7 → Fus3/Kss1 MAPK cascade as a mediator of mating, IG, and cell integrity, with specificity at the level of the MAPKs (Madhani and Fink, 1998; Breitkreutz and Tyers, 2002). However, it has not been clear how different inputs into the same pathway selectively activate one MAPK (Kss1) to promote IG and both MAPKs (Fus3 and Kss1) to induce mating. Our genetic screen unexpectedly reveals that the Kss1 MAPK cascade utilizes the mating G protein and the Ste5 scaffold to signal during vegetative growth and IG, but not the receptor that is activated by mating pheromone. The dependence of Kss1 activation on the RING-H2 domain of Ste5 indicates that Kss1 is activated as a result of Ste5 recruitment to Ste4 in the absence of mating pheromone. These findings lead to a simplified view in which a single G protein-coupled scaffold system regulates one MAPK cascade under a variety of conditions and in the absence of an obvious external stimulus (Figure 6E). Inputs by other sensors of environment such as Ras2 and Sho1 may stimulate the Kss1 MAPK cascade, but to a much lesser degree than Ste4 and Ste5. Flexibility is achieved by a simpler role of Ste5 in Kss1 activation. While it is not possible to test the validity of release of active Ste11 from Ste5 in the physiological context of invasively growing cells, this interpretation is strongly supported by the behavior of specific Ste11 and Ste5 mutants, and the fact that activation by 1-butanol, activated Cdc42, and overexpressed Ste11 and Ste7 each cause preferential activation of Kss1 compared to Fus3.
Stimulation of the Kss1 MAPK cascade can occur at multiple points in the pathway
The results suggest that external and internal inputs could influence signaling through the Kss1/Fus3 MAPK cascade at multiple steps; for example, by increasing the pool of active Ste20, the pool of free Gβγ dimer, the level of Ste5 or the pool of active Ste11 or Ste7 (Figure 6E). Regulatory inputs that increase the pool of active Ste11 or Ste7 are predicted to be sufficient to stimulate Kss1 with no effect on Fus3, providing the potential for further flexibility in how the two MAPKs may be regulated. Furthermore, the absence of a requirement for Ste11 and Kss1 to bind to Ste5 for Kss1 to be activated suggests that the Ste5-independent signaling in Σ1278b could occur without an alternative scaffold. The significantly higher level of basal activation of Kss1 in Σ1278b raises the interesting possibility that the level of basal activation of the Kss1 pathway is a key determinant of whether a strain undergoes IG or PD. In contrast, the strict dependence of Fus3 for all of the tethering functions of Ste5 demonstrates that its activation mechanism is more dependent on direct contribution by the scaffold, less flexible, and more dependent on external pheromone stimulation.
MAPK phosphatase specificity limits pathway activation to Kss1 during growth
Previous work has shown that MAPK phosphatase can provide signaling flexibility by influencing the strength and duration of the effect of a given stimulus. Here, we show that constitutive inhibition of a MAPK by a specific MAPK phosphatase, Msg5, can determine the pathway fate. Although Msg5 was known to be important for pheromone-mediated inhibition of Fus3 (Doi et al, 1994), its involvement in basal activation of Fus3 was not realized. Multiple lines of evidence support a role for Msg5 in pathway specificity, including a striking increase in the level of active Fus3 in the msg5Δ strain and phenotypes associated with mating differentiation. In addition, there is a decrease in active Kss1 and expression of the FRE(Tec1)-LacZ reporter gene, as predicted for an increased pool of active Fus3. These findings lead to the surprising conclusion that Ste5 normally activates Fus3 under a variety of growth conditions in the absence of pheromone stimulation, possibly to an even greater extent than Kss1. Msg5 constitutively inhibits Fus3, with no obvious effect on Kss1, allowing selective accumulation of active Kss1. The constitutive inhibition of Fus3 by Msg5 stabilizes signaling through Kss1 under conditions meant to promote growth by preventing activation of mating pathway outputs and downregulation of Kss1 by active Fus3. This interpretation is consistent with the observation that low levels of mating pheromone stimulate IG rather than mating differentiation.
Of the four MAPKs expressed in haploid cells, Kss1 is unique in that it is not dramatically affected by mutations in Msg5, Ptp2 or Ptp3 phosphatases, suggesting that it has evolved to be a default MAPK that is responsive to small changes in stimulus. Further work is needed to determine the molecular basis of this specificity. Our findings lead us to speculate that the level of Fus3 activation determines pathway specificity, with the level of stimulus required to achieve a pool of active Fus3 being set by the level of Msg5. When the level of Fus3 inhibition exceeds the level of pathway activation, such as under conditions of either no or weak stimulation, only Kss1 should accumulate. This appears to be the case in glucose and glycerol/ethanol. Stronger stimuli such as 1-butanol and pheromone lead to the accumulation of active Fus3; however, at low levels of stimulation when the pool of active Fus3 is below a critical threshold (e.g. in 1-butanol and very low pheromone), cells are expected to divide and grow invasively rather than mate.
Versatility of a MAPK scaffold in signaling
Previous work on Ste5 led to the hypothesis that it functions by joining together all the three kinases of a MAPK cascade (MAPKKK, MAPKK, MAPK) to allow MAPK activation on the scaffold. Similar models have been made for scaffolds in other pathways, including Pbs2, the JIPs, KSR and β-arrestin. Our analysis confirms that the MAPK can be activated on the scaffold. In addition, it shows that a MAPK cascade scaffold protein can be versatile by providing distinct functions that are differentially utilized by different MAPKs that share common upstream activators (Figure 6E). The recruitment function of Ste5 (defined here as binding of Ste5 to Ste4) serves both Kss1 and Fus3 by facilitating the activation of associated Ste11 by active Ste20 that is bound to Cdc42. However, the tethering function of Ste5 (defined here as the binding of multiple kinases in the MAPK cascade) is only essential for Fus3 activation. This conclusion agrees with the original biochemical evidence that Ste5 facilitates the activation of Fus3 as a multi-kinase complex and the observation that Fus3 of highest specific activity is associated with the scaffold (Kranz et al, 1994; Choi et al, 1994, 1999). The conclusion that the active form of Ste11 can dissociate from Ste5 and activate a pool of Ste7 and Kss1 that is not bound to scaffold is supported by the observation that a plasma membrane-localized form of Ste5 does not recruit Ste11 or Kss1 to the plasma membrane, although it is able to recruit Ste7 and Fus3 (van Drogen et al, 2001). Further work is needed to ascertain whether the pool of active Ste11 that is released from Ste5 is the same pool that phosphorylates Ste7 on the scaffold, and whether Ste11 is modified while on the scaffold.
It is noteworthy that the kinase that is not entrained to the scaffold is not sensitive to inhibition by Msg5, whereas the kinase that is strictly dependent on scaffold is subject to inhibition. The absence of a requirement for Ste11 to be bound to Ste5 during activation of Kss1, combined with a possible immunity to inhibition by phosphatase, may provide a means of amplifying the initial signal at multiple loci in the cell in response to small changes in stimulation. In contrast, the greater dependence of Fus3 on the scaffold may be tied to its functions that are not shared by Kss1 and are critical for mating differentiation, that is, cell cycle arrest, polarized growth and cell fusion, which may involve a localization that is with or near Ste5 and potentially less subject to phosphatase inhibition. These possibilities are interesting to consider in light of the propensity of Msg5 to have access to Fus3 (Kholodenko et al, 2000). The fact that increasing the level of activating kinases in a cell can bypass the need for the recruitment function of a pathway scaffold supports the view that a key role of scaffolds is to enhance kinase–kinase interactions that are limited by diffusion. In addition, it reveals a new level of difficulty in assigning the appropriate pathway links for scaffold proteins in vivo.
Materials and methods
Strains, plasmids, media, microbiological techniques
Yeast strains and plasmids are listed in Table I. Yeast strains are from three backgrounds, S288c (Research Genetics), Σ1278b (Roberts and Fink, 1994) or W303a (Elion lab). S288c deletion strains (Winzeler et al, 1999) were obtained from Research Genetics, Inc. Strains used for α factor induction were bar1Δ (sst1Δ) and lacked a protease that degrades α factor (Ciejek and Thorner, 1979). Standard microbiological and molecular biological techniques were used to transform Escherichia coli, prepare plasmid DNA, and transform yeast.
Table 1.
Yeast strains and plasmids used in this study
| Strain name | Relevant genotype | Source or reference |
|---|---|---|
| Isogenic to S288c | ||
| BY4741 | leu2 ura3 his3 met15 | Research Genetics |
| RG3042 | fus3Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG6981 | kss1Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG2724 | hog1Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG4406 | och1Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG6116 | sho1Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG3731 | gpr1Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG152 | gpa2Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG2978 | ras2Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG5241 | mid2Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG1784 | wsc1Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG1161 | wsc2Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG6255 | wsc3Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG935 | wsc4Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG7201 | snf3Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG3836 | rgt2Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG2692 | gal2Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG5645 | ste2Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG2468 | ste4Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG4038 | ste5Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG3439 | ste50Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG5271 | ste11Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG956 | ste20Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG3857 | ste7Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG1408 | sst1Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| RG5953 | flo11Δ::KANr leu2 ura3 his3 met15 | Research Genetics |
| DSY28 | fus3Δ::KANr ste2Δ::KANr | This study |
| JAY458 | fus3Δ::KANr ste4Δ::KANr | This study |
| JAY408 | fus3Δ::KANr ste5Δ::KANr | This study |
| DSY19 | fus3Δ::KANr ste11Δ::KANr | This study |
| DSY34 | fus3Δ::KANr ste20Δ::KANr | This study |
| JAY493 | fus3Δ::KANr ste50Δ::KANr | This study |
| Isogenic to W303a | ||
| EY957 | sst1Δ ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 GAL+ | Elion et al (1993) |
| Y568 | trp1-1 his3-11,15 leu2-3,112 ura3-1 | Zhan et al (1997) |
| Y572 | ptp2Δ::LEU2 trp1-1 his3-11,15 leu2-3,112 ura3-1 | Zhan et al (1997) |
| Y567 | ptp3Δ::URA3 trp1-1 his3-11,15 leu2-3,112 ura3-1 | Zhan et al (1997) |
| Y578 | ptp2Δ::LEU2 ptp3Δ::URA3 trp1-1 his3-11,15 leu2-3,112 ura3-1 | Zhan et al (1997) |
| Y170 | msg5Δ::KANr trp1-1 his3-11,15 leu2-3,112 ura3-1 | Zhan et al (1997) |
| Y173 | msg5Δ::KANr ptp2Δ::LEU2 ptp3Δ::URA3 trp1-1 his3-11,15 leu2-3,112 ura3-1 | Zhan et al (1997) |
| DSY62 | ste5Δ::TRP1 trp1-1 his3-11,15 leu2-3,112 ura3-1 | This study |
| DSY63 | ste5Δ::TRP1 msg5Δ::KANr trp1-1 his3-11,15 leu2-3,112 ura3-1 | This study |
| AFY49 | ste11Δ::ura3 ste5Δ::TRP1 | Flotho and Elion (submitted) |
| DSY116 | ste11Δ::ura3 ste5Δ::TRP1 fus3-8::ADE2 | This study |
| Isogenic to Σ1278b | ||
| L5528 | his3::hisG ura3-52 | Roberts et al (1994) |
| L5554 | ste5Δ::HIS3+ | Roberts et al (1994) |
| Plasmid name |
Description |
Source or reference |
| B1817 | HIS3 CEN | Elion lab collection |
| B1819 | LEU2 CEN | Elion lab collection |
| B1820 | URA3 CEN | Elion lab collection |
| Yep13 | LEU2 2μ | Elion lab collection |
| pDAD-2 | pGAL1,10 URA3 2μ | Elion lab collection |
| pSKM49 | STE5-MYC9 CEN LEU2 | Elion lab collection |
| pYBS138 | STE5 URA3 CEN | Kranz et al (1994) |
| pJB224 | STE5 LEU2 2μ | Kranz et al (1994) |
| pCL-S5C180A-M9 | ste5C180A-MYC9 CEN LEU2 | A Flotho (unpublished) |
| pDJ174 | STE5hyp1 URA3 2μ | Hasson et al (1994) |
| pDH27 | 3'ste5(-509 to –13)::TRP1::5'ste5(+2395 to +2702) | Leberer et al (1993) |
| pBN3 | STE11 URA3 2μ | Elion lab collection |
| p11-4 | STE11-4 HIS3 CEN | H Madhani |
| pYGL-11ΔN | pGAL1-STE11ΔN LEU2 CEN | Cairns et al (1992) |
| pJBK-008 | STE2 URA3 CEN | Kanopka and Hartwell (1989) |
| Yep213-STE4 | STE4 LEU2 2μ | Elion lab collection |
| pSTE20-5 | STE20 URA3 CEN | Leberer et al (1992) |
| pMG5 | STE50 URA3 HIS3 CEN | MR Rad |
| pXT1 | KSS1 LEU2 2μ | Courchesne et al (1989) |
| pYEE81 | FUS3 URA3 CEN | Elion et al (1990) |
| pJB237 | FUS3-101 LEU2 CEN | Brill et al (1994) |
| pKCS5 | pGAL1-STE7-MYC HIS3 CEN | Choi et al (1994) |
| YCpR2V | RAS2A19V URA3 | Gimeno et al (1992) |
| pHL1 | FLO8 URA3 CEN | Liu et al (1996) |
| B641 | pGAL1,10-HO URA3 LEU2 CEN | Elion lab collection |
| BHM275 | FRE(TEC1)::LACZ URA3 2μ | Madhani and Fink (1997) |
| FB80 | MATα | Winston lab collection |
| pGAL1-42 | pGAL1-CDC42 LEU2 CEN | D Johnson |
| pGAL1-VAL12 | pGAL1-CDC42G12V LEU2 CEN | D Johnson |
| pYW-S5Δ241-336 | STE5(Δ241-336) URA3 CEN | Elion lab collection |
Immunoblotting
Primary antibodies used in this study were anti-phospho p44/42 MAPK (Cell Signaling Techology, Inc, #9101), anti-Kss1 polyclonal anti-serum (Santa Cruz), anti-Fus3 polyclonal antiserum (Elion et al, 1993), and anti-Tcm1 monoclonal antibody (gift from J Warner, Albert Einstein College of Medicine). Secondary antibodies used were rabbit anti-goat Ig-HRP, goat anti-mouse Ig-HRP and goat-anti-rabbit Ig-HRP. The p42/p44 anti-active MAPK antibody is directed against a region in ERK1 that is more conserved in Kss1 than Fus3 (C Salvadore, personal communication, Cell Signaling Technologies).
Additional details can be found in Supplementary data.
Supplementary Material
Supplemental materials and methods
Acknowledgments
We thank Annette Flotho for many helpful scientific discussions. We are especially grateful to K Struhl and C Baisden for access to their Research Genetics collection of yeast mutants. We thank A Flotho, GR Fink, and D Jenness for plasmids and strains and G Fink, H Madhani, M Tyers, and M Brandriss for stimulating discussions. This research was funded by an American Heart Association grant (01501176N) and National Institutes of Health grant (GM46962) to EAE. Salary support for JA was provided by the American Heart Association grant 01501176N and a post-doctoral fellowship from STINT (The Swedish Foundation for International Cooperation in Research and Higher Education), together with funds from the Swedish Wenner–Gren foundations.
References
- Bardwell AJ, Flatauer LJ, Matsukuma K, Thorner J, Bardwell L (2000) A conserved docking site in MEKs mediates high-affinity binding to MAP kinases and cooperates with a scaffold protein to enhance signal transmission. J Biol Chem 276: 10374–10386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla US, Ram PT, Iyengar R (2002) MAP kinase phosphatase as a locus of flexibility in a mitogen-activated protein kinase signaling network. Science 297: 1018–1023 [DOI] [PubMed] [Google Scholar]
- Breitkreutz A, Tyers M (2002) MAPK signaling specificity: it takes two to tango. Trends Cell Biol 12: 254–257 [DOI] [PubMed] [Google Scholar]
- Breitkreutz A, Boucher L, tyers M. (2001) MAPK specificity in the yeast pheromone response independent of transcriptional activation. Curr Biol 11: 1266–1271 [DOI] [PubMed] [Google Scholar]
- Brill JA, Elion EA, Fink GR (1994) A role for autophosphorylation revealed by activated alleles of FUS3, the yeast MAP kinase homolog. Mol Biol Cell 5: 297–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehrer BM, Errede B (1997) Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. Mol Cell Biol 17: 6517–6525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burack WR, Shaw AS (2000) Signal transduction: hanging on a scaffold. Curr Opin Cell Biol 12: 211–216 [DOI] [PubMed] [Google Scholar]
- Cairns BR, Ramer SW, Kornberg RD (1992) Order of action of components in the yeast pheromone response pathway revealed with a dominant allele of the STE11 kinase and the multiple phosphorylation of the STE7 kinase. Genes Dev 6: 1305–1318 [DOI] [PubMed] [Google Scholar]
- Cherkasova V, Elion EA (2001) far4, far5, and far6 define three genes required for efficient activation of MAPKs Fus3 and Kss1 and accumulation of glycogen. Curr Genet 40: 13–26 [DOI] [PubMed] [Google Scholar]
- Cherkasova VA, McCully R, Wang Y, Hinnebusch A, Elion EA (2003) A novel functional link between MAP kinase cascades and the Ras/cAMP pathway that regulates survival. Curr Biol 13: 1220–1226 [DOI] [PubMed] [Google Scholar]
- Choi KY, Kranz JE, Mahanty SK, Elion EA (1999) Characterization of Fus3 localization. Active Fus3 localizes in complexes of varying size and specific activity. Mol Biol Cell 10: 1553–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KY, Satterberg B, Lyons DM, Elion EA (1994) Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell 78: 499–512 [DOI] [PubMed] [Google Scholar]
- Ciejek E, Thorner J (1979) Recovery of S. cerevisiae a cells from G1 arrest by alpha factor pheromone requires endopeptidase action. Cell 18: 623–635 [DOI] [PubMed] [Google Scholar]
- Cook JG, Bardwell L, Thorner J (1997) Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature 390: 85–88 [DOI] [PubMed] [Google Scholar]
- Cullen PJ, Schultz J, Horecka J, Stevenson BJ, Jigami Y, Sprague GF Jr (2000) Defects in protein glycosylation cause SHO1-dependent activation of a STE12 signaling pathway in yeast. Genetics 155: 1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Sprague GF Jr (2000) Glucose depletion causes haploid invasive growth in yeast. Proc Natl Acad Sci USA 97: 13619–13624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport KD, Williams KE, Ullmann BD, Gustin MC (2001) Activation of the Saccharomyces cerevisiae filamentation/invasion pathway by osmotic stress in high-osmolarity glycogen pathway mutants. Genetics 153: 1091–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K, Gartner A, Ammerer G, Errede B, Shinkawa H, Sugimoto K, Matsumoto K (1994) MSG5, a novel protein phosphatase promotes adaptation to pheromone response in S. cerevisiae. EMBO J 13: 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion EA (2001) The Ste5p scaffold. J Cell Sci 114: 3967–3978 [DOI] [PubMed] [Google Scholar]
- Elion EA, Satterberg B, Kranz JE (1993) FUS3 phosphorylates multiple components of the mating signal transduction cascade: evidence for STE12 and FAR1. Mol Biol Cell 4: 495–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman S, Snyder M (2001) A filamentous growth response mediated by the yeast mating pathway. Genetics 159: 919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Song LY, Kincaid E, Mahanty SK, Elion EA (1998) Functional binding between Gbeta and the LIM domain of Ste5 is required to activate the MEKK Ste11. Curr Biol 8: 267–278 [DOI] [PubMed] [Google Scholar]
- Fink GR, Madhani HD (1998) The riddle of MAP kinase signaling specificity. Trends Cell Biol 8: 348–353 [DOI] [PubMed] [Google Scholar]
- Gartner A, Nasmyth K, Ammerer G (1992) Signal transduction in Saccharomyces cerevisiae requires tyrosine and threonine phosphorylation of FUS3 and KSS1. Genes Dev 6: 1280–1292 [DOI] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Fink GR (1992) Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68: 1077–1090 [DOI] [PubMed] [Google Scholar]
- Hasson MS, Blinder D, Thorner J, Jenness DD (1994) Mutational activation of the STE5 gene product bypasses the requirement for G protein beta and gamma subunits in the yeast pheromone response pathway. Mol Cell Biol 14: 1054–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye C, Dhillon N, Thorner J (1997) Ste5 RING-H2 domain: role in Ste4-promoted oligomerization for yeast pheromone signaling. Science 278: 103–106 [DOI] [PubMed] [Google Scholar]
- Kholodenko BN, Brown GC, Hoek JB (2000) Diffusion control of protein phosphorylation in signal transduction. Biochem J 350: 901–907 [PMC free article] [PubMed] [Google Scholar]
- Kranz JE, Satterberg B, Elion EA (1994) The MAP kinase Fus3 associates with and phosphorylates the upstream signaling component Ste5. Genes Dev 8: 313–327 [DOI] [PubMed] [Google Scholar]
- Lee BN, Elion EA (1999) The MAPKKK Ste11 regulates vegetative growth through a kinase cascade of shared signaling components. Proc Natl Acad Sci USA 96: 12679–12684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Styles CA, Fink GR (1996) Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics 144: 967–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Cutler NS, Heitman J (2000a) Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae. Mol Biol Cell 11: 183–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Cook JG, Thorner J (1995) Phosphorylation and localization of Kss1, a MAP kinase of the Saccharomyces cerevisiae pheromone response pathway. Mol Biol Cell 6: 889–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Fink GR (1997a) Combinatorial control required for the specificity of yeast MAPK signaling. Science 275: 1314–1317 [DOI] [PubMed] [Google Scholar]
- Madhani HD, Fink GR (1998) The riddle of MAP kinase specificity. Trends Genet 14: 151–155 [DOI] [PubMed] [Google Scholar]
- Madhani HD, Styles CA, Fink GR (1997b) MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91: 673–684 [DOI] [PubMed] [Google Scholar]
- Maeda T, Takekawa M, Saito H (1995) Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269: 554–558 [DOI] [PubMed] [Google Scholar]
- Marshall CJ (1995) Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80: 179–185 [DOI] [PubMed] [Google Scholar]
- Martin H, Rodriguez-Pachon JM, Ruiz C, Nombela C, Molina M (2000) Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J Biol Chem 275: 1511–1519 [DOI] [PubMed] [Google Scholar]
- Mattison CP, Spencer SS, Kresge KA, Lee J, Ota IM (1999) Differential regulation of the cell wall integrity mitogen-activated protein kinase pathway in budding yeast by the protein tyrosine phosphatases Ptp2 and Ptp3. Mol Cell Biol 19: 7651–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DK, Davis RJ (2003) Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu Rev Cell Dev Biol 19: 91–118 [DOI] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR (1986) Genealogy of principal strains of the yeast genetic stock center. Genetics 113: 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosch HU, Roberts RL, Fink GR (1996) Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 93: 5352–5356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehlen L, Cross FR (1998) The mating factor response pathway regulates transcription of TEC1, a gene involved in pseudohyphal differentiation of Saccharomyces cerevisiae. FEBS Lett 429: 83–88 [DOI] [PubMed] [Google Scholar]
- Pan X, Harashima T, Heitman J (2000) Signal transduction cascades regulating pseudohyphal differentiation of Saccharomyces cerevisiae. Curr Opin Microbiol 3: 567–572 [DOI] [PubMed] [Google Scholar]
- Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH (2001) Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev 22: 153–183 [DOI] [PubMed] [Google Scholar]
- Posas F, Saito H (1997) Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276: 1702–1705 [DOI] [PubMed] [Google Scholar]
- Pryciak PM, Huntress FA (1998) Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gbetagamma complex underlies activation of the yeast pheromone response pathway. Genes Dev 12: 2684–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramezani Rad M, Jansen G, Buhring F, Hollenberg CP (1998) Ste50p is involved in regulating filamentous growth in the yeast Saccharomyces cerevisiae and associates with Ste11p. Mol Gen Genet 259: 29–38 [DOI] [PubMed] [Google Scholar]
- Roberts RL, Fink GR (1994) Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev 8: 2974–2985 [DOI] [PubMed] [Google Scholar]
- Roberts CJ, Nelson B, Marton JJ, Stoughton R, Meyer MR, Bennett HA, He YD, Dai H, Walker WL, Hughes TR, Tyers M, Boone C, Friend SH (2000) Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287: 873–880 [DOI] [PubMed] [Google Scholar]
- Roy F, Laberge G, Douziech M, Ferland-McCollough D, Therrien M (2002) KSR is a scaffold required for activation of the ERK/MAPK module. Genes Dev 16: 427–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbagh W Jr, Flatauer LJ, Bardwell AJ, Bardwell L (2001) Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol Cell 8: 683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson BJ, Rhodes N, Errede B, Sprague GF Jr (1992) Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev 6: 1293–1304 [DOI] [PubMed] [Google Scholar]
- Tan PB, Kim SK (1999) Signaling specificity: the RTK/RAS/MAP kinase pathway in metazoans. Trends Genet 15: 145–149 [DOI] [PubMed] [Google Scholar]
- Traverse S, Gomez N, Paterson H, Marshall C, Cohen P (1992) Sustained activation of the mitogen-activated protein (MAP) kinase cascade may be required for differentiation of PC12 cells: comparison of the effects of nerve growth factor and epidermal growth factor. Biochem J 288: 351–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Drogen F, Stucke VM, Jorritsma G, Peter M (2001) MAP kinase dynamics in response to pheromones in budding yeast. Nat Cell Biol 3: 1051–1059 [DOI] [PubMed] [Google Scholar]
- Wang Y, Elion EA (2003) Nuclear export and plasma membrane recruitment of the Ste5 scaffold are coordinated with oligomerization and association with signal transduction components. Mol Biol Cell 14: 2543–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Irie K, Matsumoto K (1995) Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol 15: 5740–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston CR, Davis RJ (2002) The JNK signal transduction pathway. Curr Opin Genet 12: 14–21 [DOI] [PubMed] [Google Scholar]
- Whiteway MS, Wu C, Leeuw T, Clark K, Fourest-Lieuvin A, Thomas DY, Leberer E (1995) Association of the yeast pheromone response G protein beta gamma subunits with the MAP kinase scaffold Ste5p. Science 269: 1572–1575 [DOI] [PubMed] [Google Scholar]
- Widmann C, Gibson S, Jarpe MB, Johnson GL (1999) Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol Rev 79: 143–180 [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Castillo-Davis CI, Oshiro G, Liang D, Richards DR, Zhou Y, Hartl DL (2003) Genetic diversity in yeast assessed with whole-genome oligonucleotide arrays. Genetics 163: 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Whelen Dow S, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Stroms RK, Véronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW (1999) Functional characterization of the Saccharomyces cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Wurgler-Murphy SM, Maeda T, Witten EA, Saito H (1997) Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol Cell Biol 17: 1289–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Fantl WJ, Harrowe G, Williams LT (1998) Regulation of the MAP kinase pathway by mammalian Ksr through direct interaction with MEK and ERK. Curr Biol 8: 56–64 [DOI] [PubMed] [Google Scholar]
- Zarzov P, Mazzoni C, Mann C (1996) SLT2(MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO J 15: 83–91 [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Simon I, Harbison CT, Hannett NM, Volkert TL, Fink GR, Young RA (2003) Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113: 395–404 [DOI] [PubMed] [Google Scholar]
- Zhan XL, Deschenes RJ, Guan KL (1997) Differential regulation of FUS3 MAP kinase by tyrosine-specific phosphatases PTP2/PTP3 and dual-specificity phosphatase MSG5 in Saccharomyces cerevisiae. Genes Dev 11: 1690–1702 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods


