Abstract
The TREX complex is involved in both transcription elongation and mRNA export and is recruited to nascent transcription complexes. We have examined Yra1p, Sub2p and Hpr1p recruitment to nine genes of varying lengths and transcription frequencies. All three proteins increase from the 5′ to the 3′ ends of the four intronless genes examined. A modified chromatin immunoprecipitation assay that includes an RNase step indicates that Sub2p is bound to nascent RNA, Yra1p is associated with both RNA and DNA, and Hpr1p is associated with DNA. Although Hpr1p is recruited similarly to both intronless and intron-containing genes, low Yra1p and Sub2p levels are present on a subset of intron-containing genes. The residual Yra1p and Sub2p recruitment is less RNA-associated, and this correlates with high levels of U1 SnRNP on these genes. These experiments support a model in which TREX is recruited via the transcription machinery and then Yra1p and Sub2p are transferred to the nascent RNA. On some intron-containing genes, retention and/or transfer of Yra1p and Sub2p to nascent RNA are inhibited.
Keywords: intron, RNA export, splicing, transcription, TREX
Introduction
The defining feature of the eukaryotic cell—the nuclear membrane—separates transcription and nuclear mRNA processing from most if not all translation. Within the nucleus, transcripts undergo multiple covalent processing events including the addition of a 7-methylguanosine triphosphate cap to the 5′ end, splicing to remove intervening noncoding regions, and cleavage and polyadenylation of the 3′ end. There are also noncovalent changes, which include the packaging of RNA into mRNP complexes. Some of these mRNP proteins are important for RNA export from the nucleus to the cytoplasm, including the yeast (Saccharomyces cerevisiae) proteins Yra1p and Sub2p (Aly/REF and UAP56 in vertebrates, respectively). It is hypothesized that Sub2p recruits Yra1p to nascent RNA (Luo et al, 2001; Strasser and Hurt, 2001), after which Sub2p is displaced from Yra1p by the mRNA export factor Mex67p/TAP (Strasser et al, 2000; Stutz et al, 2000; Zhou et al, 2000). Mex67p/TAP forms a heterodimer with Mtr2p/p15, which interacts directly with the nuclear pore to facilitate mRNP transport to the cytoplasm (Segref et al, 1997; Strasser et al, 2000).
mRNP packaging begins during transcription. An elongation complex called THO (Hpr1p, Tho2p, Mft1p, Thp2p) was biochemically purified with the mRNA export factors Sub2p and Yra1p as well as a previously unknown protein Tex1p (Chavez et al, 2000; Strasser et al, 2002). This complex was termed TREX for ‘transcription/export' (Strasser et al, 2002), as mutations in its components show phenotypes indicative of both transcription elongation and mRNA export defects. For example, mutations in Yra1p and Sub2p are defective in transcribing G-C-rich sequences and show elevated levels of transcription-dependent recombination (Chavez et al, 2000; Fan et al, 2001; Jimeno et al, 2002). In addition, cells deleted for THO components are deficient in mRNA export at 37 degrees (Schneiter et al, 1999; Libri et al, 2002; Strasser et al, 2002).
Chromatin immunoprecipitation (ChIP) experiments show that Hpr1p, Tho2p, Sub2p and Yra1p are recruited to actively transcribed genes. All three of these proteins show similar patterns of recruitment: they increase from the 5′ end to the 3′ end of the open reading frame (ORF) and dissociate from the site of transcription coincident with cleavage and polyadenylation (Lei et al, 2001; Strasser et al, 2002; Zenklusen et al, 2002; Kim et al, 2004). However, it is unclear from the ChIP assay whether these factors are associated with DNA, perhaps via the transcription machinery, or with nascent RNA. Yra1p contains an RNA-recognition motif (RRM) suggesting that it binds to RNA. Indeed, Yra1p interacts directly with RNA in vitro. However, this interaction is not mediated by the RRM but rather occurs via arginine-rich regions (Rodrigues et al, 2001; Zenklusen et al, 2001). Although not directly demonstrated, Sub2p may also have RNA-binding activity, as it is predicted to be a DEAD-box helicase (Sträßer and Hurt, 2000; Kistler and Guthrie, 2001; Zhang and Green, 2001). A purified complex containing the TREX components associates with RNA in vitro as well as DNA (Jimeno et al, 2002). Moreover, it is possible that the early steps of TREX recruitment are not reflected in the steady-state protein distribution, that is, the initial recruitment of TREX could be to DNA and then some components transferred to the nascent RNA.
TREX proteins including Yra1p and Sub2p have also been implicated in splicing. Sub2p was originally identified as the yeast homolog of the metazoan splicing factor UAP56 and is required to facilitate U2 snRNP recruitment (Kistler and Guthrie, 2001; Libri et al, 2001; Zhang and Green, 2001). Yra1p was recently purified with yeast U2 snRNP components, suggesting that Yra1p may also have a role in splicing (Wang and Rymond, 2003). In higher eukaryotes, TREX complex components were identified in four different spliceosome purifications (Neubauer et al, 1998; Hartmuth et al, 2002; Jurica et al, 2002; Rappsilber et al, 2002). Moreover, the Yra1p homolog Aly/REF is an integral component of the exon junction complex (EJC), which is deposited at the exon–exon junction during splicing (Le Hir et al, 2000). The Sub2p homolog UAP56 also associates with the EJC, but it is not an integral component of the complex (Reichert et al, 2002). These results suggest that Yra1p and Sub2p may act at the interface between splicing and mRNA export and could bind specifically to properly spliced mRNAs.
To learn more about recruitment of TREX components to active transcription sites, we have examined the recruitment of Yra1p, Sub2p and Hpr1p to a panel of nine different endogenous genes. By normalizing to PolII transcription, we were able to measure the levels of Yra1p, Sub2p and Hpr1p per PolII/nascent mRNA complex. Yra1p, Sub2p and Hpr1p are recruited similarly to a set of four different intronless genes. However, an assay developed to distinguish between RNA- and DNA- (transcription machinery) binding shows that these three proteins have different fates at the site of transcription. Sub2p is RNA-associated, which fits with the increase of this protein from the 5′ end to the 3′ end of genes. In contrast, the THO component Hpr1p is bound to the DNA, presumably via the transcriptional machinery. Yra1p shows yet a third pattern; the data suggest that only a fraction of the total Yra1p at the transcription site is associated with RNA. Our results suggest a two-step recruitment model, in which Yra1p and Sub2p are recruited via the TREX complex and are then transferred to the nascent RNA. Extending our analysis to five intron-containing genes indicates that Hpr1p is similarly recruited to intronless and intron-containing genes. However, Yra1p and Sub2p are poorly recruited to and retained on three out of five intron-containing genes. These and other results indicate that spliceosome assembly may interfere with the transfer of Yra1p and Sub2p to nascent RNA, suggesting a relationship between the cotranscriptional recruitment of export factors and pre-mRNA retention.
Results
TREX components increase from 5′ to 3′ on four different intronless genes
Most ChIP studies have concentrated on the recruitment of TREX components to a very limited set of genes, predominantly PMA1 (Lei et al, 2001; Lei and Silver, 2002; Strasser et al, 2002; Zenklusen et al, 2002). Moreover, most studies do not control for changes in transcription rate or polymerase density, which will obviously influence the level of TREX recruitment. We have therefore extended previous analyses to include three additional intronless genes of different lengths and expression levels (Table I), and we have normalized to RNA polymerase II (PolII) recruitment. To measure simultaneously TREX and PolII recruitment from the same chromatin preparation, we used a monoclonal antibody to PolII for ChIP (8WG16). Although this reagent has been reported to show a bias against highly phosphorylated forms of PolII (Thompson et al, 1989), ChIP data with 8WG16 are not substantially different from those with an anti-hemagglutinin (HA) antibody and an HA-tagged RNA polymerase subunit (Komarnitsky et al, 2000 and Figure 1B). Indeed, ChIPs using 8WG16 indicate that PolII recruitment to the four different intronless genes largely reflects previously published differences in transcription frequency (Figure 1C and Table I).
Table 1.
Characteristics of the genes used in ChIP analysis
| ORF length (bp) | Transcriptional frequency (mRNAs/h)a | Intron length | Intron position | |
|---|---|---|---|---|
| PMA1 | 2757 | 95 | NA | NA |
| PGK1 | 1251 | 139 | NA | NA |
| ADH3 | 1128 | 22 | NA | NA |
| FBA1 | 1080 | 166 | NA | NA |
| ASC1 | 1233 | 124 | 273 | 538–810 |
| ACT1 | 1436 | 45.5 | 308 | 11–318 |
| RPL28 | 961 | 126.3 | 511 | 50–560 |
| DBP2 | 2643 | 61.5 | 1002 | 1274–2275 |
|
SEC27 |
2870 |
5 |
200 |
19–218 |
| NA: not applicable. | ||||
| a Data from Holstege et al (1998). | ||||
Figure 1.
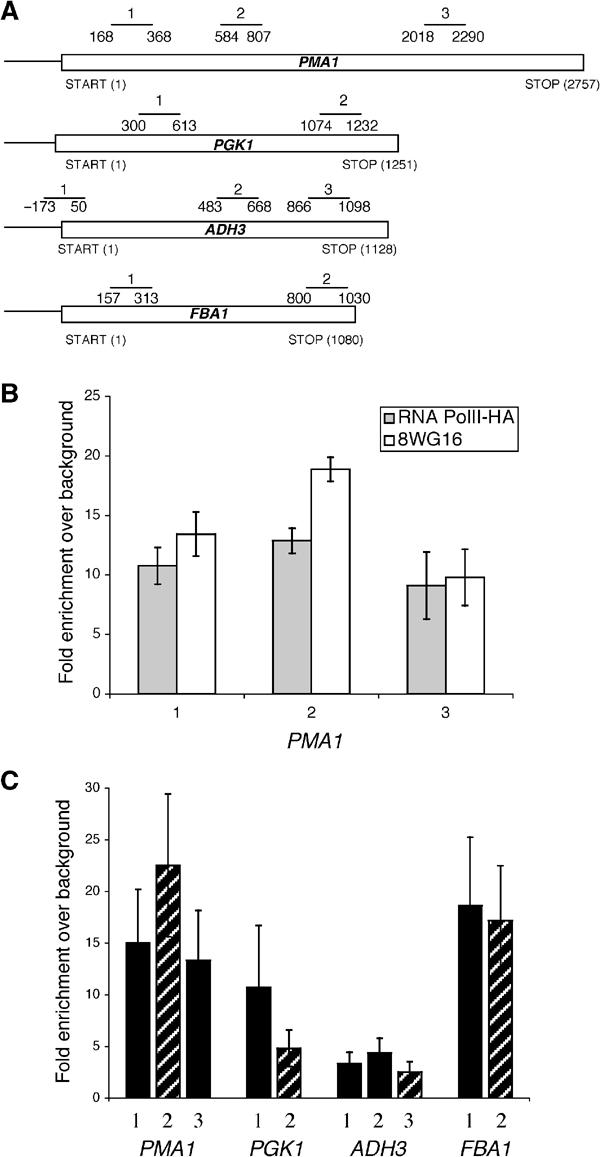
RNA PolII is differentially recruited to four intronless genes. (A) The schematic diagram shows the relative positions of the primer pairs used to analyze the recruitment to the intronless genes PMA1, PGK1, ADH3 and FBA1. (B) The amount of PolII detected via ChIP using a monoclonal antibody against PolII (8WG16; white bars) or an anti-HA antibody to recognize an HA-tagged RNA PolII (gray bars) is very similar. The position of the PMA1 primer pairs is indicated in (A). (C) The amount of PolII recruited to four different intronless genes was examined using ChIPs with the monoclonal PolII antibody (8WG16). The data are presented as the fold enrichment of PolII relative to the background binding and represent the average results from greater than 10 experiments. The striped bars indicate primer pairs that are approximately 1000 bp after transcription initiation.
To compare Yra1p, Sub2p and Hpr1p recruitment to this set of genes, we performed immunoprecipitations from the same chromatin preparation with antibodies specific to each epitope-tagged TREX component and with 8WG16 (see Materials and methods for details). As reported previously, Yra1p, Sub2p and Hpr1p show a very similar pattern of normalized recruitment to PMA1; all three proteins increase from the 5′ end to the 3′ end of the ORF (Figure 2). This same general pattern is observed on PGK1, ADH3 and FBA1. To accommodate mRNA length differences between multiple genes, we compared TREX factor recruitment at approximately 1000 bp after transcription initiation. Yra1p and Sub2p are relatively poorly recruited to FBA1 after approximately 1000 bp of transcription (compare striped bars, Figure 2A and B). Hpr1p is similarly recruited to PGK1, ADH3 and FBA1, but it is more strongly recruited to PMA1 (compare striped bars, Figure 2C). These statistically significant differences indicate that TREX components are differentially recruited to and stabilized on this set of genes.
Figure 2.
TREX recruitment to four different intronless genes. Single chromatin preparations from yeast strains expressing epitope-tagged Yra1p (KAY136), Sub2p (FSY1473) or Hpr1p (FSY1525) were immunoprecipitated using either antibodies recognizing PolII (8WG16) or the epitope tag. To determine the amount of TREX component recruited to the site of transcription per PolII/nascent RNA complex, the amount of TREX component recruited was divided by the amount of PolII recruited (see Materials and methods for details). The striped bars indicate which primer pairs are located approximately 1000 bp after transcription initiation. (A) Yra1p, (B) Sub2p and (C) Hpr1p. The data presented are the average of two independent experiments.
RNA-bound proteins are detectable via ChIP
The general 5′ to 3′ increase of Yra1p, Sub2p and Hpr1p suggests that these factors may associate with the nascent transcript. To demonstrate that ChIP can detect a protein–nascent mRNA interaction, a positive control was first constructed: a protein that only interacts with nascent RNA and not at all with the transcriptional machinery. To this end, we inserted two tandem MS2 stem loops into the 5′-UTR of a GAL1-GFP plasmid (see Materials and methods). This reporter gene was transformed into yeast along with a plasmid expressing a fusion protein containing the bacteriophage MS2 coat protein and three HA tags (MS2-HA). This system has been well characterized, even in yeast (Stripecke et al, 1994).
A ChIP experiment with standard anti-HA antibodies indicates that MS2-HA is recruited to pGAL1-MS2-GFP only when the gene is actively transcribed (Figure 3B; gray versus white bars). As an additional control, we established that there is no recruitment of MS2-HA to a GAL1-GFP transcript lacking MS2 stem loops, indicating that recruitment is MS2 stem loop mediated (data not shown). There is only a modest loss of signal at the 3′ end of the gene, suggesting that the nascent mRNA between the HA-tagged RNA binding protein and the 3′ DNA template remains intact and is not severely degraded.
Figure 3.
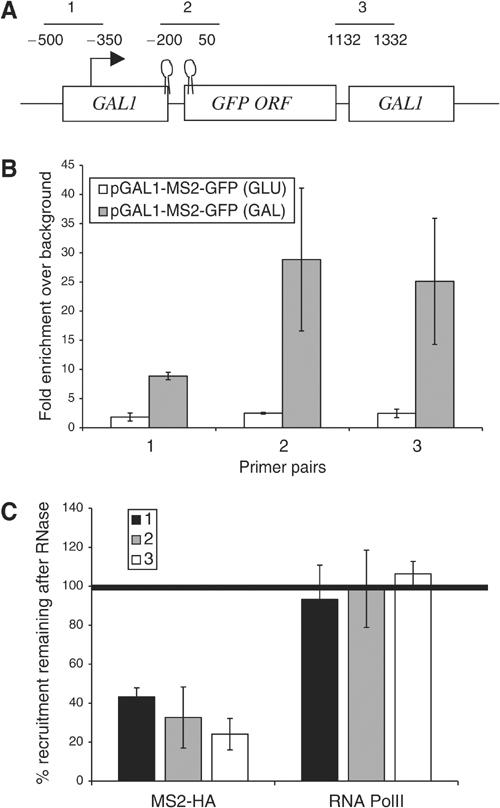
The recruitment of the MS2-HA fusion protein to the nascent transcription complex is sensitive to RNase. ChIPs were performed on yeast expressing an MS2-HA fusion protein and pGAL-MS2-GFP-pA-CEN (KAY406). (A) The diagram shows the features of the GAL1-MS2-GFP-pA construct and the position of three primer sets used in the ChIPs described in (B, C). (B) An MS2-HA fusion protein is cotranscriptionally recruited to a GFP mRNA containing two MS2 binding sites in the 5′-UTR (gray bars). The recruitment of MS2-HA is dependent on transcription; the MS2 fusion protein is not recruited when transcription from the GAL1 promoter is repressed by the addition of glucose (white bars). (C) An RNase treatment step was added to ChIPs with antibodies against either PolII or MS2-HA to determine whether RNase can distinguish between RNA- and DNA-bound proteins in a ChIP assay. The data are presented as the percentage of protein that remains after RNase treatment (see Materials and methods). Only between 20 and 40% of the MS2-HA recruited to GAL1-MS2-GFP remains after RNase treatment. The data presented are the average of two independent experiments.
We next modified the fixation conditions slightly and added an RNase treatment step to the ChIP protocol. (There is no PolII normalization used in this case; see Materials and methods for details.) Only 20–40% of the recruited MS2-HA remained after the samples were treated with RNase (Figure 3C). As expected of a protein bound to nascent RNA, the RNase sensitivity was more pronounced at the 3′ end of the gene. In contrast, the PolII signal did not decrease but actually remained constant or increased slightly with RNase treatment (Figure 3C). We often observe increases in PolII signal after RNase treatment (Figure 4), which can probably be attributed to an increase in the accessibility of the PolII epitope after digestion of nascent RNA and associated factors. This interpretation is consistent with the fact that RNase is often included in ChIP protocols to increase the signal of DNA-associated factors (Spencer et al, 2003). The striking difference between MS2-HA and PolII indicates that RNase sensitivity under these conditions can distinguish between RNA-bound and DNA-bound (transcription machinery-bound) factors.
Figure 4.
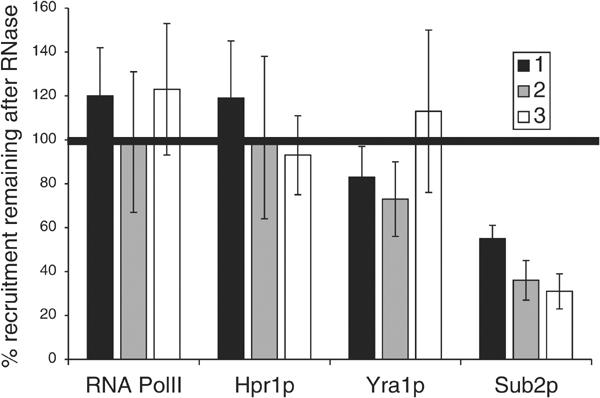
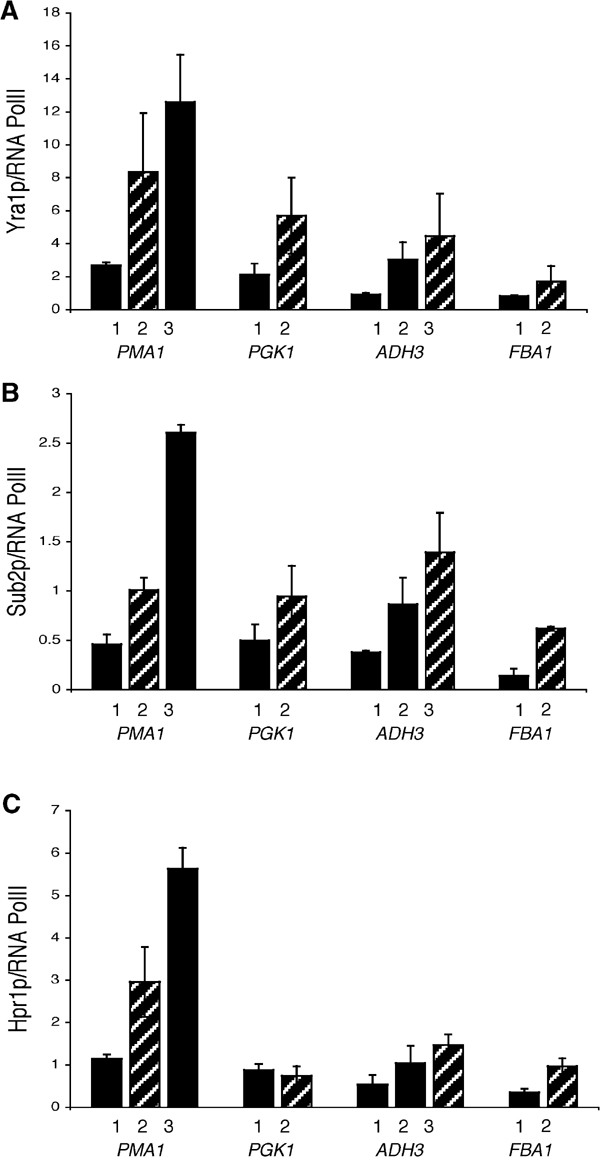
Sub2p, Yra1p and Hpr1p are differentially associated with the nascent mRNA. ChIPs with an RNase treatment step were performed on yeast strains expressing epitope-tagged Sub2p (FSY1473), Yra1p (KAY136) and Hpr1p (FSY1525). The methods used are identical to those described in Figure 3. The PMA1 primer pairs used are described in Figure 1. The RNase sensitivity of Sub2p is almost identical to MS-HA; only 20–40% of the Sub2p remains at the site of transcription after RNase treatment (n=4). In contrast, the RNase sensitivity of Hpr1p (n=4) resembles that of PolII (n=10); it is insensitive to RNase treatment. Yra1p shows partial RNase sensitivity (n=7): it is more RNase sensitive than PolII at both the 5′ and middle primer; however this difference is statistically significant only at the 5′ end of PMA1.
The amount of Sub2p recruited to actively transcribing genes decreases upon RNase treatment
We then applied the RNase assay to the recruitment of TREX components and PolII to the PMA1 gene. The RNase sensitivity of Sub2p recruitment is almost identical to that of the MS2-HA protein to the GAL1-MS2-GFP gene (compare Figure 4 with Figure 3C). This indicates that PMA1-bound Sub2p is predominantly nascent RNA-associated. In contrast to Sub2p, Hpr1p recruitment is essentially indistinguishable from that of PolII (Figure 4). Yra1p shows a third pattern (Figure 4). Although apparently RNase-insensitive, the difference between Yra1p and PolII RNase sensitivity is statistically significant at the 5′ (PP2) end of PMA1 (80% for Yra1p; 120% for PolII; Figure 4). The differences between Sub2p, Yra1p, Hpr1p and PolII RNase sensitivity are not due to some unique feature of the PMA1 gene. Similar results were obtained on PGK1 (data not shown and Figure 7).
Figure 7.
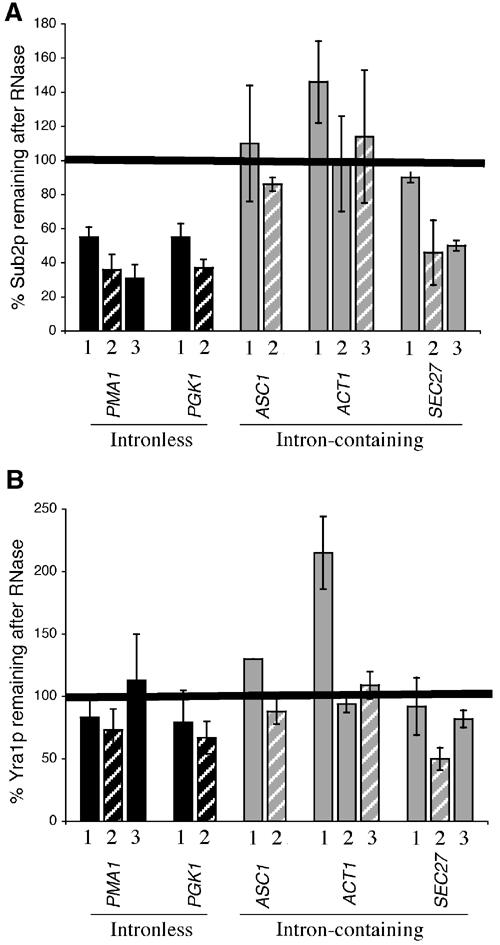
Poor cotranscriptional recruitment of Yra1p and Sub2p is correlated with a decrease in RNA association. Chromatin preparations from strains expressing epitope-tagged Yra1p (KAY136) or Sub2p (FSY1473) were incubated with or without RNase. The primer pairs used in these experiments are described in Figure 5A. (A) Sub2p and (B) Yra1p. The data presented are the average of two independent experiments.
The RNase insensitivity of Hpr1p suggests that it is primarily DNA- or transcription machinery-associated. The RNase sensitivity of Sub2p indicates that it is primarily associated with nascent transcripts and suggests that the 5′ to 3′ increase in Sub2p protein levels is due at least in part to the increasing number of binding sites on the growing nascent mRNA. Although the RNase sensitivity of Yra1p is marginal, the experiment was repeated seven times; the data indicate that there are small but significant differences between the RNase sensitivities of Yra1p and PolII. This suggests that the Yra1p population at the site of transcription may be divided between the nascent RNA and the DNA transcription machinery.
Yra1p and Sub2p are poorly recruited to intron-containing genes
The roles of Yra1p and Sub2p in splicing and the fact that they both show an association with nascent RNA inspired a comparison of normalized TREX factor recruitment between the four intronless genes and five intron-containing genes. ASC1, ACT1, RPL28, DBP2 and SEC27 are transcribed at varying rates and contain introns of different sizes and positions (Table I). As shown for the intronless genes, the intron-containing genes recruit different amounts of PolII, which also corresponds to previous studies of transcription rates (Figure 5B and Table I).
Figure 5.
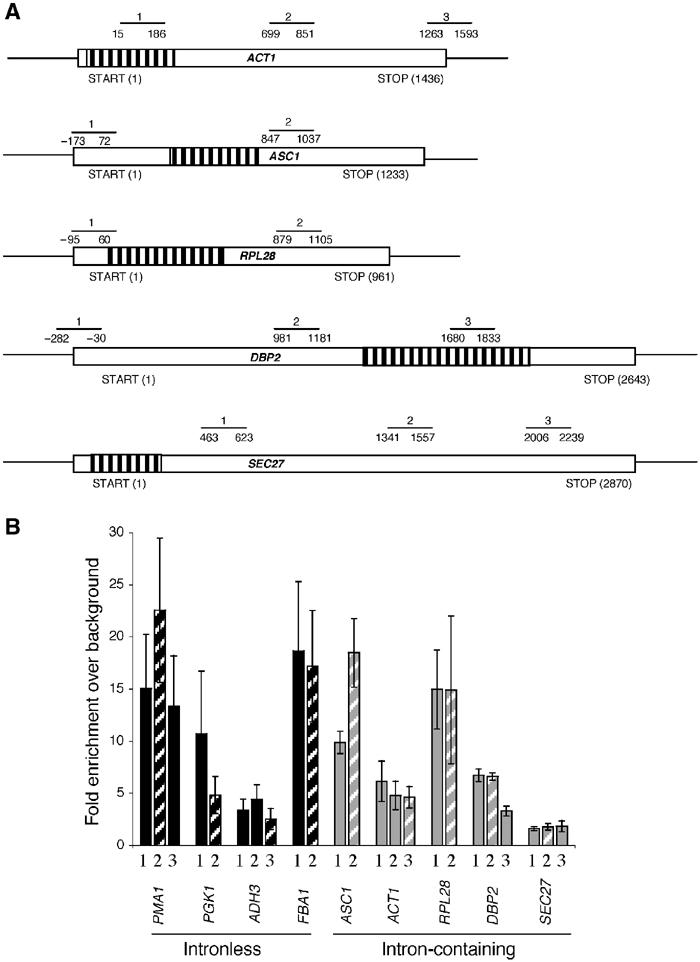
RNA PolII recruitment reflects the transcriptional frequency of nine intronless and intron-containing genes. (A) The schematic diagram shows the relative positions of the primer pairs used to examine the recruitment of PolII and TREX components to intron-containing genes. The striped regions indicate the position of the intron. (B) ChIPs with the monoclonal PolII antibody (8WG16) indicate that different amounts of PolII are recruited to four intronless (PMA1, PGK1, ADH3 and FBA1) and five intron-containing (ASC1, ACT1, RPL28, DBP2 and SEC27) genes. The striped bars indicate the amount of PolII recruitment approximately 1000 bp after transcription initiation. The data are presented as fold PolII enrichment relative to background binding and represent the average results from greater than 10 experiments (see Materials and methods for details).
Hpr1p is recruited similarly to eight out of the nine genes regardless of intron status (Figure 6A). The exception is the super-Hpr1p-recruiter PMA1 (see above). Yra1p shows two different recruitment patterns on intron-containing genes: it is poorly recruited to ASC1, ACT1 and RPL28 and well recruited to DBP2 and SEC27 (Figure 6B). Recruitment of Yra1p to these two genes is comparable to that observed on intronless genes. The differential recruitment of Yra1p to ACT1 and DBP2 agrees with a previous study (Lei and Silver, 2002). Sub2p shows a very similar recruitment pattern to Yra1p with one exception: Sub2p is also poorly recruited to DBP2 in our hands (Figure 6C), contrary to a previous report (Lei and Silver, 2002).
Figure 6.
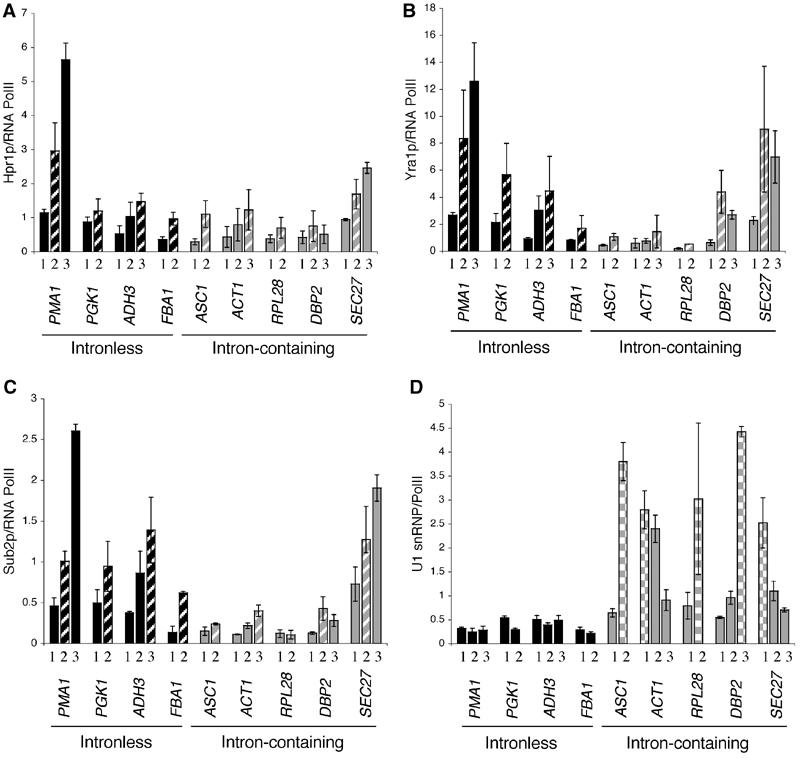
Yra1p and Sub2p are poorly recruited to a subset of intron-containing genes. ChIPs were performed on strains expressing epitope-tagged Yra1p (KAY136), Sub2p (FSY1473), Hpr1p (FSY1525) and the U1 SnRNP component, Prp42p (YKK25). The amount of Yra1p, Sub2p, Hpr1p and U1 snRNP recruited per PolII/nascent RNA complex is examined on five different intron-containing genes. The primer pairs used are described in Figure 5A and the striped bars (A–C) indicate primer pairs that are approximately 1000 bp after transcription initiation and the checkered bars (D) indicate the position of the intron. (A) Hpr1p, (B) Yra1p, (C) Sub2p and (D) U1 snRNP. PCR reactions for all nine genes were performed from the same ChIP samples; therefore, gene-to-gene differences in TREX and U1 snRNP recruitment cannot be attributed to experimental variation. The data presented are the average of two independent experiments.
In an attempt to understand why Sub2p and Yra1p are differentially recruited to these intron-containing genes, we examined the recruitment of U1 snRNP component, Prp42p (Figure 6D). As shown previously, U1 snRNP is well recruited to intron-containing genes and poorly if at all to non-intron-containing genes (Kotovic et al, 2003). However, only ASC1, ACT1 and RPL28 associate with U1 snRNP across their entire ORFs. (The first primer pairs of ASC1 and RPL28 are in the promoters.) In contrast, DBP2 and SEC27 have relatively low levels of U1 snRNP, before and after the introns, respectively. These results indicate that there is an inverse correlation between U1 snRNP and Sub2p/Yra1p recruitment to intron-containing genes. It suggests a model in which spliceosome assembly interferes with Yra1p and Sub2p recruitment, transfer or stabilization on some intron-containing RNAs (see Discussion).
Sub2p and Yra1p are less RNA-associated on intron-containing genes
To further address this hypothesis, we examined the RNase sensitivity of Yra1p and Sub2p on intron-containing genes (Figure 7). In contrast to the intronless genes PMA1 and PGK1, Sub2p is relatively insensitive to RNase only on those intron-containing genes that show low Sub2p recruitment, for example, ASC1 and ACT1 (Figure 7A). Consistent with this correlation, the 3′ end of the intron-containing SEC27 gene recruits Sub2p well and shows ‘intronless' levels of Sub2p RNase sensitivity. The data suggest that low levels of Sub2p on ASC1 and ACT1 remain associated with the DNA or with the transcription machinery.
Owing to the marginal RNase sensitivity of Yra1p, it was not possible to detect large changes in the RNase sensitivity of Yra1p on intron-containing genes. However, there are two statistically significant differences. Yra1p shows an increased RNase insensitivity at the 5′ ends of ACT1 and ASC1 (Figure 7B). As these primer pairs are before or within the introns, the result suggests that Yra1p like Sub2p may not be efficiently transferred to nascent mRNA on a subset of intron-containing genes. In addition, Yra1p is more sensitive to RNase on SEC27, that is, an intron-containing gene that recruits high levels of Yra1p.
Yra1p recruitment increases upon intron removal
If the low Yra1p levels on a subset of intron-containing genes are due to spliceosome assembly, then intron removal should restore Yra1p recruitment and increase its RNA association. To test this hypothesis, we constructed two strains, with an intronless ASC1 (−intron; SALY56) or a wild-type ASC1 (+intron; SALY59) integrated at the ASC1 genomic locus, and compared Yra1p recruitment to both ASC1 and PMA1. Higher levels of Yra1p were recruited to intronless ASC1 (Figure 8A), whereas there was no difference in Yra1p recruitment to PMA1 in the two strains (data not shown). To account for the difference in length of the two ASC1 transcripts, we plotted the data as a function of distance from the ATG. A similar but smaller increase in Yra1p recruitment is observed when the intron is removed from ACT1 (data not shown; see Discussion). As predicted, the increase in Yra1p recruitment on intronless ASC1 was accompanied by an increase in RNase sensitivity (Figure 9B). This suggests that the presence of an intron in ASC1 inhibits Yra1p transfer to the nascent RNA.
Figure 8.
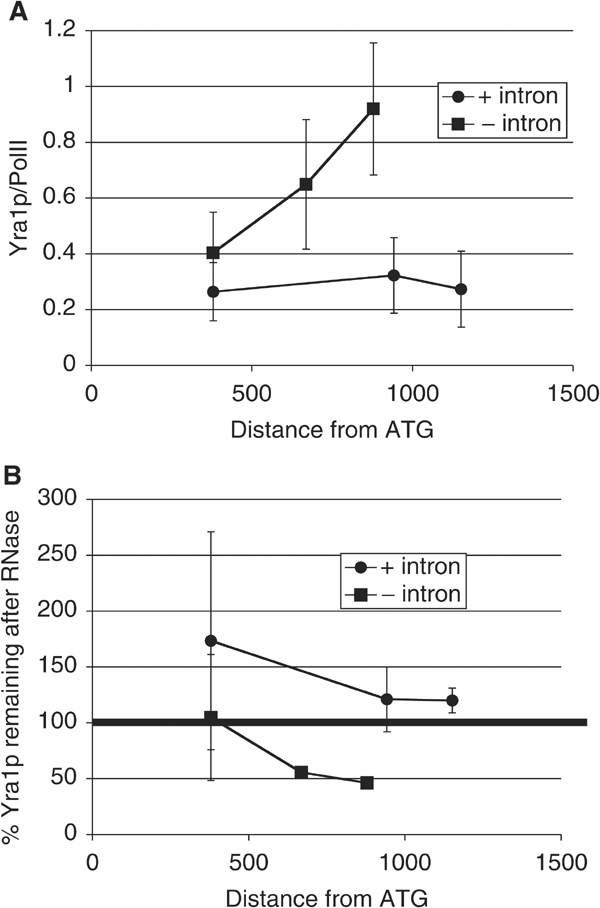
Removal of the intron from ASC1 restores Yra1p levels and its RNA association. ChIPs were performed using epitope-tagged Yra1p strains expressing either ASC1+intron (SALY56) or ASC1−intron (SALY59). (A) The amount of Yra1p recruited to ASC1 increases when the intron is removed; compare squares (−intron) with circles (+intron). The data are presented relative to the distance from the ATG to account for the differences in transcript length between the two ASC1 constructs. (B) Yra1p becomes more RNA associated on ASC1 when the intron is removed (compare squares (−intron) with circles (+intron).
Figure 9.
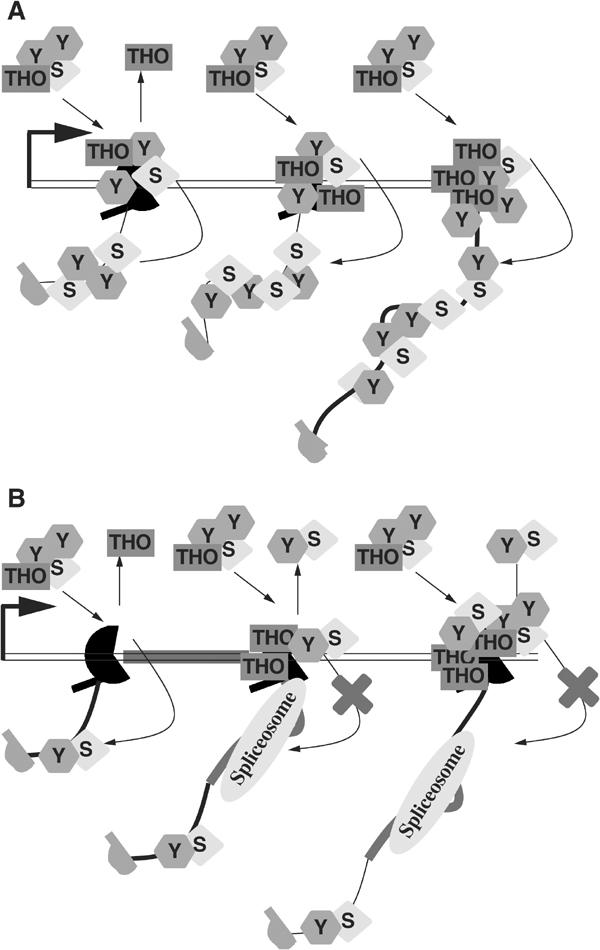
Model for TREX component recruitment to intronless and intron-containing genes. (A) Our data indicate that once the TREX complex is recruited to the site of transcription, presumably via the transcription machinery, the complex dissociates. The THO complex remains associated with the DNA or transcription machinery. Sub2p is immediately and efficiently transferred to the nascent RNA. The Yra1p divides into two pools: one remains associated with the DNA and the other is transferred to the RNA. (B) On intron-containing genes, the TREX complex is recruited normally but the transfer of Sub2p and Yra1p to the RNA may be hindered by the spliceosome. As a result, less Sub2p and Yra1p are retained at the site of transcription (S=Sub2p; Y=Yra1p).
Discussion
We have examined the cotranscriptional recruitment of three TREX proteins to nine different yeast genes. Yra1p, Sub2p and Hpr1p all show a striking 5′ to 3′ increase in recruitment on almost all these genes. However, an RNase assay distinguishes between the three proteins. Only Sub2p appears cleanly bound to nascent RNA, with no evidence for an association with DNA or with the transcription machinery. Hpr1p, in contrast, is directly or indirectly bound to DNA. Another THO complex component, Tho2p, shows the same complete RNase insensitivity as Hpr1p (data not shown). Yra1p is in a third category and apparently contacts DNA as well as the nascent RNA. The data suggest that Yra1p and Sub2p are recruited to the site of transcription as part of the TREX complex and then Sub2p and to a lesser extent Yra1p are transferred to the nascent RNA. Hpr1p is recruited similarly to both intronless and intron-containing genes. In contrast, both Yra1p and Sub2p accumulate poorly on a substantial subset of intron-containing genes. Taken together, we suggest that successful transfer of Yra1p and Sub2p from the transcription machinery to the nascent RNA may be a critical step in linking transcription, splicing and export (Figure 9).
The simplest explanation for the striking 5′ to 3′ enrichment of all TREX complex components is that the entire complex is recruited by or transferred to the nascent RNA. This possibility is consistent with the fact that Yra1p, Sub2p and Hpr1p all dissociate from the site of transcription after polyadenylation has occurred (Kim et al, 2004). A problem, however, is the striking distinction between the RNase sensitivity of Sub2p and Hpr1p. Moreover, Sub2p relies on Hpr1p for nascent recruitment, that is, Sub2p recruitment to PMA1 is substantially decreased in an hrp1Δ strain both with and without normalization to PolII (KC Abruzzi and M Rosbash, preliminary results; Zenklusen et al, 2002, respectively). This suggests that Sub2p is recruited via the TREX complex to DNA or to the transcription machinery and is then transferred to the nascent RNA. An equally plausible possibility is that TREX is recruited to nascent mRNA and Hpr1p is subsequently transferred to the DNA. However, we would then expect Hpr1p to be decreased on the intron-containing genes. The data argue in favor of the ‘DNA and then transfer to RNA' interpretation.
The 5′ to 3′ increase in Sub2p signal is probably due to the longer nascent RNAs associated with the 3′ ends of genes. However, the same increase in the recruitment of Yra1p and especially Hpr1p requires another explanation, as it is different from other nascent proteins such as transcription and polyadenylation factors (Licatalosi et al, 2002; Ahn et al, 2004; Kim et al, 2004). Although changes in epitope availability cannot be excluded, there is probably more Yra1p, Hpr1p and other THO components at the 3′ end of genes, or they bind more tightly in these regions (lower off-rate). Many changes occur as polymerase transcribes across a gene, including changes in PolII phosphorylation (Komarnitsky et al, 2000), the recruitment of factors important for 3′-end formation (Licatalosi et al, 2002; Ahn et al, 2004; Kim et al, 2004) and DNA supercoiling (Huertas and Aguilera, 2003). It is possible that one or more of these 3′-end events could stimulate TREX binding or retention.
The complete transfer of Sub2p to RNA is consistent with an analysis of giant Balbiani ring nascent mRNPs, namely, that the Sub2p ortholog coats the entire mRNP uniformly (Kiesler et al, 2002). The Yra1p ortholog has a different distribution (Kiesler et al, 2002), consistent with the differences we observe in RNase sensitivity. Nonetheless, Yra1p may also be completely transferred to the nascent RNA. The intermediate RNase sensitivity may reflect some residual affinity for the transcription machinery, which is captured during the in vivo crosslinking procedure. Another possibility is that Yra1p is incompletely transferred to the RNA, leaving some protein bound to DNA or to the transcription machinery. This might indicate that there is less Yra1p than Sub2p bound to nascent RNA. Alternatively, Yra1p and Sub2p could still be stochiometric on nascent RNA, which implies a prior suprastochiometric recruitment of Yra1p to the transcription machinery. This could be due either to a suprastochiometric association of Yra1p relative to Sub2p within TREX or to an additional mode of Yra1p recruitment, followed by stochiometric and coupled transfer of Sub2p and Yra1p to nascent RNA (Figure 9A). Indeed, coupled transfer accommodates the known association of Sub2p with Yra1p, and the similarity in Sub2p and Yra1p recruitment is striking—their differences in RNase sensitivity notwithstanding.
Another example of Yra1p and Sub2p co-regulation is the low levels of Yra1p and Sub2p on ACT1, ASC1 and RPL28. These intron-containing genes are expressed at different rates, range between 1000 and 1400 bp in length, and contain introns either at the 5′ end or middle of the genes. As this effect is paralleled by lower RNase sensitivity and high U1 snRNP recruitment, it suggests that cotranscriptional spliceosome assembly inhibits transfer to, or retention on, nascent pre-mRNA (Figure 9B). This may be due to passive occlusion (the spliceosome occupies pre-mRNA sites that would otherwise receive Sub2p and Yra1p), or it may reflect some more active inhibition of transfer. These models are supported by results showing that the removal of the ASC1 intron increases Yra1p recruitment and Yra1p association with the nascent RNA (Figure 8).
Despite the increase in Yra1p recruitment to the ASC1 cDNA, our results suggest that the inhibition of Yra1p and Sub2p recruitment may be complicated. When the intron is removed from ACT1, the increase in Yra1p recruitment is less striking (data not shown). Similar results are observed for Sub2p recruitment to the ASC1 cDNA; Sub2p increases only marginally and the differences are not significant (data not shown), suggesting that the splicing inhibition of Yra1p and Sub2p recruitment and transfer is gene specific and complicated. This may be related to the diverse effects of splicing on transcription and polyadenylation in yeast as well as metazoa (Brinster et al, 1988; Niwa et al, 1990; Palmiter et al, 1991; Dye and Proudfoot, 1999; Lacy-Hulbert et al, 2001; Furger et al, 2002).
In contrast to ASC1, ACT1 and RPL28, the SEC27 and DBP2 intron-containing genes recruit and retain high levels of Yra1p and/or Sub2p. Our data suggest that the nascent transcripts of these long and unusual intron-containing genes contain substantial stretches of RNA free of spliceosomes. Although U1 snRNP recruitment is robust in the region adjacent to the 5′-intron of SEC27, U1 snRNP is undetectable in the middle and 3′ end of SEC27. Consistent with this observation, Sub2p is less associated with the RNA only at the 5′ end of SEC27. In the case of DBP2, no U1 snRNP is recruited prior to the 3′-intron. This suggests that DBP2 resembles an intronless gene until the intron emerges 1273 bp after transcription initiation and that Yra1p binds normally prior to intron emergence. Previously published experiments showed that mutations in early and late splicing factors decrease and increase Yra1p recruitment on DBP2, respectively (Lei and Silver, 2002). It is difficult to interpret these results in the light of our U1 snRNP data, but perhaps they are due to complicated effects of splicing on transcriptional recruitment.
The low levels of Sub2p and Yra1p on some intron-containing genes was surprising, given the relationship of these proteins to splicing (e.g., Kistler and Guthrie, 2001; Libri et al, 2001; Zhang and Green, 2001). Although our data do not preclude a role in splicing, they do argue against the presence of an exon junction complex (EJC) in yeast, at least as a major means of recruiting Yra1 and Sub2p to most intron-containing genes. The earlier study examining the recruitment of Yra1p and Sub2p to DBP2 and ACT1 explained the low recruitment of Yra1p and Sub2p to ACT1 by invoking an EJC-like mechanism, that is, by proposing that Yra1p requires a long 5′-exon to bind to intron-containing RNAs (Lei and Silver, 2002). However, our data show that 5′-exon length does not correlate positively with Yra1p and Sub2p recruitment: Yra1p and Sub2p are poorly recruited to the long 5′-exon-containing gene ASC1 and well recruited to the short 5′-exon-containing gene SEC27. Given the features of the five intron-containing genes we have examined, we suspect that low recruitment will be the more common result for the entire set of yeast intron-containing genes.
Most unspliced pre-mRNAs are poorly exported to the cytoplasm in yeast (Legrain and Rosbash, 1989; Rutz and Seraphin, 2000). A recent study indicates that nuclear Mlp1p may be a central player in this retention process: it recognizes U1 snRNP-bound pre-mRNAs at the nuclear periphery and helps prevent their export (Galy et al, 2004). Our data suggest that yeast may have a second line of defense against the nuclear export of pre-mRNAs. mRNP export factors may not be adequately loaded onto pre-mRNA during early spliceosome formation. This may inhibit their efficient export from the nucleus.
Materials and methods
Strains and media
All yeast strains used in this study are from the W303 background and are derivatives of Y368 except where noted (Table II). We used standard methods for yeast manipulations (Guthrie and Fink, 1991).
Table 2.
Strains and plasmids
| Strain/plasmid | Genotype | References |
|---|---|---|
| Strains | ||
| FSY1473 | MATa, ura3, lys2, trp1, ade, his3, leu2, sub2∷HIS3 plus pFS2625 | Zenklusen et al (2002) |
| FSY1525 | MATa, ura3, lys2, trp1, his3, ade, leu2, sub2∷HIS3, Hpr1-ProtA plus pSUB2-URA3-CEN | Zenklusen et al (2002) |
| KAY136 | MATa, yra1∷HIS3, ade2, his3, leu2, trp1, ura3 plus pKA14 | This study |
| KAY406 | FY368 plus pMS2-HA-LEU2-CEN & pGAL-MS2-GFP-TRP1-CEN | This study |
| YKK25 | MATa, ura3, leu2, trp1, his3, can1-100, psi ADE+ GAL+ prp42∷HA-PRP42-KanR | Kotovic et al (2003) |
| SALY56 | MATa, yra1∷HIS3, ASC1∷ASC1-URA3, ade2, his3, leu2, trp1, ura3 plus pKA14 | This study |
| SALY59 | MATa, yra1∷HIS3, ASC1∷ASC1-intron-URA3, ade2, his3, leu2, trp1, ura3 plus pKA14 | This study |
| YSB749 | MATa, ura3-52, leu2-3, 112, trp1-1, his3200, ade2-1, RPB3-(HA)3∷LEU2 | Komarnitsky et al (2000) |
| Plasmids | ||
| pASC9 | ASC1-URA3-2μ | Furger et al (2002) |
| pD16 | ASC1-intron-URA3-2μ | Furger et al (2002) |
| pFS2625 | SUB2-PROTA-LEU2-CEN | Zenklusen et al (2002) |
| pFS2233 | HA-YRA1(+intron)-TRP1-CEN | Zenklusen et al (2001) |
| pKA14 | HA-YRA1(+intron)-LEU2-CEN | This study |
| pSAL1 | ASC1pro-ASC1-ASC1-3′UTR-URA3-2μ | This study |
| pSAL2 | ASC1pro-ASC1-intron-ASC1-3′UTR-URA3-2μ | This study |
| ASC1pro-BamHI-SalI-ASC1-3′UTR-URA3-2μ | This study | |
| TRP1-2μ | Libri et al (2002) | |
| GAL1-TRP1-2μ | This study | |
| GAL1-(MS2)-GFP-pA-TRP1-CEN | This study | |
| TDH3-LEU2-CEN | This study | |
| MS2-HA-LEU2-CEN | This study |
Plasmid and strain construction
To construct a LEU2-marked plasmid expressing YRA1 with a triple HA epitope tag, pFS2233 (Zenklusen et al, 2001) was cut with BamH1 and the resulting fragment was ligated into BamH1-digested pRS315 (Sikorski and Hieter, 1989) to produce pKA14. To construct pGAL1-TRP1-2μ, the GAL1 locus was PCR amplified using overlap PCR to replace the GAL1 ORF with BamHI/SalI cloning sites. This PCR product was cloned using BglII/XhoI into BamHI/SalI of TRP1-2μ (Libri et al, 2002). To construct pGAL1-GFP-TRP1-2μ, the green fluorescent protein (GFP) was PCR amplified from plasmid pJK19-1 (a gift from P Silver) and cloned as a BamHI/SalI fragment into GAL1-TRP1-2μ. To construct pGAL1-(MS2)-GFP-pA-TRP1-CEN, the GAL1 promoter, GFP ORF, and GAL1 3′ region were PCR amplified from pGAL1-GFP-pA-TRP1-2μ using overlap PCR to introduce two MS2 stem loops (CGTACACCATCAGGGTACG CGAGCTAGCCCATCGCGTACACCATCAGGGTACG) into the GAL1 5′ untranslated region. This PCR product was cloned using BglII/XhoI into BamHI/SalI of pRS314. The MS2 stem loop sequence was described Jurica et al (2002). TDH3-LEU2-CEN was constructed by PCR amplifying the TDH3 locus using overlap PCR in which BamHI/SalI cloning sites replace the TDH3 ORF. This PCR product was cloned using BglII/XhoI into BamHI/SalI of pRS315. To construct pMS2-HA-LEU2-CEN, the bacteriophage MS2 coat protein ORF was PCR amplified from MS2-MBP (a gift from M Jurica and M Moore; Jurica et al, 2002) to introduce sequences encoding an N-terminal SV40 nuclear localization signal and three C-terminal HA tags. This PCR product was cloned using BamHI/XhoI into BamHI/SalI of pTDH3-LEU2-CEN. To construct pASC1-BamHI-SalI-ASC1, the ASC1 locus was PCR amplified using overlap PCR to replace the ASC1 ORF with BamHI/SalI cloning sites. This PCR product was digested with BglII/XhoI and ligated into BamHI/SalI-digested pRS426 to create pASC1-BamHI-SalI-ASC1. The ASC1 ORFs, with or without the intron, were amplified from plasmids pASC9 and pD16 (gifts from N Proudfoot; Furger et al, 2002), respectively. The resulting PCR products were digested with BamHI/SalI and ligated into pASC1-BamHI-SalI-ASC1 to generate pSAL1 (+intron) and pSAL2 (−intron). pSAL1 and pSAL2 were linearized using AhdI, amplified with primers SALO48 and SALO49, and the resulting product was transformed into yeast (KAY136) to generate SALY56 and SALY59.
Chromatin immunoprecipitations
ChIPs were performed using methods previously described (Komarnitsky et al, 2000) with the following exceptions. The crosslinking time was reduced from 20 to 5 min when RNase treatment was performed. Yeast cells were lysed using a Mini-Bead Beater 8 (BioSpec Products). All centrifugation steps were performed in a refrigerated microcentrifuge at maximum speed. Samples were sonicated for six cycles of a 20 s pulse followed by a 20 s pause using a 550 Sonic Dismembranator (Fisher Scientific). This sonication treatment is sufficient to shear the chromatin to an average size of between 500 and 1000 bp. Monoclonal antibodies against total RNA PolII (8WG16) were purchased from Covance. The anti-HA monoclonal (12CA5) was purchased from Roche. Each of these antibodies was preincubated with protein A–Sepharose 4B beads (Zymed). For protein-A-tagged factors, IgG Sepharose beads (Amersham Pharmacia) were used.
The amount of DNA in the ‘input' and ‘IP' samples was quantitated using real-time PCR (Rotorgene by Corbett Research). To verify that the real-time PCR machine gave accurate and reliable results, we did a side-by-side comparison of real-time PCR and radioactive PCR followed by phosphor imaging. We found that these two techniques gave nearly identical results (data not shown). The real-time PCR reactions contained 0.5 U of platinum Taq polymerase (Invitrogen), 0.1 mM dNTP mixture (Invitrogen), 0.25 μM each primer, 1.5 mM MgCl2 and 1 × platinum Taq PCR buffer (20 mM Tris–HCl (pH 8.4), 500 mM KCl) and 0.2 × concentration of Sybr Green (Molecular Probes). PCR was performed under the following conditions: 90 s at 95°C followed by 45 cycles of 10 s at 94°C, 30 s at 55°C and 45 s at 72°C. Each ‘input' and ‘IP' sample was analyzed in triplicate using real-time PCR, and the resulting values were averaged to determine the concentration of each sample. The primers used in this study were chosen using Primer3 from the Whitehead Institute (Rozen and Skaletsky, 2000) and are described in Figures 1A and 5A, and Supplementary Table I. The PMA1 and intergenic primers (Intergenic V-1 and V-2) were previously described (Komarnitsky et al, 2000).
The calculations and normalizations for each ChIP were carried out as previously described (Komarnitsky et al, 2000; Ahn et al, 2004; Kim et al, 2004). Briefly, the enrichment of the protein of interest (i.e., Yra1p) above background was calculated by dividing (IP signal for gene-specific primer pair/IP signal for intergenic primer pair) by the (input signal for gene-specific primer pair/input signal for intergenic primer pair). The ‘intergenic' primer pair is in a nontranscribed region of chromosome five and should account for any nonspecific DNA binding. A fold enrichment of 1 indicates that the amount of DNA immunoprecipitated by the protein of interest is no higher than would be expected from nonspecific DNA binding.
Protein enrichment was normalized to PolII recruitment to account for differences in transcription frequency both across genes and between different genes. For these experiments, immunoprecipitations of the factor of interest (i.e., Yra1p) and PolII were preformed from the same chromatin preparation. The fold enrichment over background was determined individually for each protein (see description above), and the fold enrichment for the factor of interest (i.e., Yra1p) was divided by the fold enrichment for RNA PolII (i.e., Yra1p/PolII). When comparing the recruitment of TREX components on nine different genes (Figure 6), all of the PCR was performed from the same sample. Therefore, the gene-to-gene differences in TREX component and U1 snRNP recruitment cannot be attributed to experimental variation.
When an RNase treatment step was added to the ChIP protocol, chromatin from the same experiment was treated with either 7.5 U of RNase A and 300 U of RNase T1 (RNase A/T1 Cocktail; Ambion) or an equivalent volume of RNase storage buffer (10 mM Hepes pH 7.2, 20 mM NaCl, 0.1% Triton X-100, 1 mM EDTA and 50% glycerol v/v). After incubating at room temperature for 30 min, immunoprecipitations were performed as described above. To calculate the ‘percentage of factor remaining after RNase treatment', we divided the fold enrichment for the RNase-treated sample by the fold-enrichment for the non-RNase-treated sample and multiplied the result by 100. We did not normalize to PolII in the RNase experiments.
Supplementary Material
Supplemental Table I: Sequence of Primers
Acknowledgments
We thank the Buratowski, Silver and Struhl labs for their assistance in setting up the ChIP procedure, and S Buratowski, K Neugebauer, N Proudfoot, P Silver and F Stutz for yeast strains and plasmids. We appreciate the technical assistance of K Dower, H Felton, T McCarthy and L Kosker and are grateful to Elisa Izaurralde, Francoise Stutz, Torben Heick Jensen and members of the Rosbash Lab for valuable discussions and comments on this manuscript. KCA was supported by a postdoctoral fellowship from the NIH. This work was supported in part by a grant to MR from the NIH (grant GM23549).
References
- Ahn SH, Kim M, Buratowski S (2004) Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 13: 67–76 [DOI] [PubMed] [Google Scholar]
- Brinster RL, Allen JM, Behringer RR, Gelinas RE, Palmiter RD (1988) Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci USA 85: 836–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez S, Beilharz T, Rondon AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A (2000) A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J 19: 5824–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye MJ, Proudfoot NJ (1999) Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol Cell 3: 371–378 [DOI] [PubMed] [Google Scholar]
- Fan HY, Merker RJ, Klein HL (2001) High-copy-number expression of Sub2p, a member of the RNA helicase superfamily, suppresses hpr1-mediated genomic instability. Mol Cell Biol 21: 5459–5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furger A, O'Sullivan JM, Binnie A, Lee BA, Proudfoot NJ (2002) Promoter proximal splice sites enhance transcription. Genes Dev 16: 2792–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U (2004) Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116: 63–73 [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR (1991) Guide to yeast genetics and molecular biology. Methods Enzymol 194: 389–398 [PubMed] [Google Scholar]
- Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, Luhrmann R (2002) Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc Natl Acad Sci USA 99: 16719–16724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728 [DOI] [PubMed] [Google Scholar]
- Huertas P, Aguilera A (2003) Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12: 711–721 [DOI] [PubMed] [Google Scholar]
- Jimeno S, Rondon AG, Luna R, Aguilera A (2002) The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J 21: 3526–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ (2002) Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8: 426–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesler E, Miralles F, Visa N (2002) HEL/UAP56 binds cotranscriptionally to the Balbiani ring pre-mRNA in an intron-independent manner and accompanies the BR mRNP to the nuclear pore. Curr Biol 12: 859–862 [DOI] [PubMed] [Google Scholar]
- Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S (2004) Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J 23: 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler AL, Guthrie C (2001) Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev 15: 42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotovic KM, Lockshon D, Boric L, Neugebauer KM (2003) Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol Cell Biol 23: 5768–5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy-Hulbert A, Thomas R, Li XP, Lilley CE, Coffin RS, Roes J (2001) Interruption of coding sequences by heterologous introns can enhance the functional expression of recombinant genes. Gene Therapy 8: 649–653 [DOI] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ (2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J 19: 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrain P, Rosbash M (1989) Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57: 573–583 [DOI] [PubMed] [Google Scholar]
- Lei EP, Krebber H, Silver PA (2001) Messenger RNAs are recruited for nuclear export during transcription. Genes Dev 15: 1771–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei EP, Silver PA (2002) Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes Dev 16: 2761–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D, Dower K, Boulay J, Thomsen R, Rosbash M, Jensen TH (2002) Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol Cell Biol 22: 8254–8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D, Graziani N, Saguez C, Boulay J (2001) Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes Dev 15: 36–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL (2002) Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell 9: 1101–1111 [DOI] [PubMed] [Google Scholar]
- Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R (2001) Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413: 644–647 [DOI] [PubMed] [Google Scholar]
- Neubauer G, King A, Rappsilber J, Calvio C, Watson M, Ajuh P, Sleeman J, Lamond A, Mann M (1998) Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat Genet 20: 46–50 [DOI] [PubMed] [Google Scholar]
- Niwa M, Rose SD, Berget SM (1990) In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev 4: 1552–1559 [DOI] [PubMed] [Google Scholar]
- Palmiter RD, Sandgren EP, Avarbock MR, Allen DD, Brinster RL (1991) Heterologous introns can enhance expression of transgenes in mice. Proc Natl Acad Sci USA 88: 478–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M (2002) Large-scale proteomic analysis of the human spliceosome. Genome Res 12: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert V, Le Hir H, Jurica MS, Moore M (2002) 5′ exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes Dev 16: 2778–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JP, Rode M, Gatfield D, Blencowe BJ, Carmo-Fonseca M, Izaurralde E (2001) REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc Natl Acad Sci USA 98: 1030–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Rutz B, Seraphin B (2000) A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J 19: 1873–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R, Guerra CE, Lampl M, Gogg G, Kohlwein SD, Klein HL (1999) The Saccharomyces cerevisiae hyperrecombination mutant hpr1Delta is synthetically lethal with two conditional alleles of the acetyl coenzyme A carboxylase gene and causes a defect in nuclear export of polyadenylated RNA. Mol Cell Biol 19: 3415–3422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E (1997) Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J 16: 3256–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer VA, Sun JM, Li L, Davie JR (2003) Chromatin immunoprecipitation: a tool for studying histone acetylation and transcription factor binding. Methods 31: 67–75 [DOI] [PubMed] [Google Scholar]
- Strasser K, Bassler J, Hurt E (2000) Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J Cell Biol 150: 695–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Hurt E (2001) Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413: 648–652 [DOI] [PubMed] [Google Scholar]
- Sträßer K, Hurt E (2000) Yra1p, a conserved nuclear RNA binding protein, interacts directly with Mex67 and is required for mRNA export. EMBO J 19: 410–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, Hurt E (2002) TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308 [DOI] [PubMed] [Google Scholar]
- Stripecke R, Oliveira CC, McCarthy JE, Hentze MW (1994) Proteins binding to 5′ untranslated region sites: a general mechanism for translational regulation of mRNAs in human and yeast cells. Mol Cell Biol 14: 5898–5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F, Bachi A, Doerks T, Braun I, Seraphin B, Wilm M, Bork P, Izaurralde E (2000) REF, an evolutionarily conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA export. RNA 6: 638–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson NE, Steinberg TH, Aronson DB, Burgess RR (1989) Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J Biol Chem 264: 11511–11520 [PubMed] [Google Scholar]
- Wang Q, Rymond BC (2003) Rds3p is required for stable U2 snRNP recruitment to the splicing apparatus. Mol Cell Biol 23: 7339–7349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Vinciguerra P, Strahm Y, Stutz F (2001) The yeast hnRNP-Like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol Cell Biol 21: 4219–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Vinciguerra P, Wyss JC, Stutz F (2002) Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol Cell Biol 22: 8241–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Green MR (2001) Identification and characterization of yUAP/Sub2p, a yeast homolog of the essential human pre-mRNA splicing factor hUAP56. Genes Dev 15: 30–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R (2000) The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407: 401–405 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table I: Sequence of Primers


