Abstract
Bacterial meningitis is a serious infection of the CNS that results when blood-borne bacteria are able to cross the blood-brain barrier (BBB). Group B Streptococcus (GBS) is the leading cause of neonatal meningitis; however, the molecular mechanisms that regulate bacterial BBB disruption and penetration are not well understood. Here, we found that infection of human brain microvascular endothelial cells (hBMECs) with GBS and other meningeal pathogens results in the induction of host transcriptional repressor Snail1, which impedes expression of tight junction genes. Moreover, GBS infection also induced Snail1 expression in murine and zebrafish models. Tight junction components ZO-1, claudin 5, and occludin were decreased at both the transcript and protein levels in hBMECs following GBS infection, and this repression was dependent on Snail1 induction. Bacteria-independent Snail1 expression was sufficient to facilitate tight junction disruption, promoting BBB permeability to allow bacterial passage. GBS induction of Snail1 expression was dependent on the ERK1/2/MAPK signaling cascade and bacterial cell wall components. Finally, overexpression of a dominant-negative Snail1 homolog in zebrafish elevated transcription of tight junction protein–encoding genes and increased zebrafish survival in response to GBS challenge. Taken together, our data support a Snail1-dependent mechanism of BBB disruption and penetration by meningeal pathogens.
Keywords: Infectious disease, Microbiology, Vascular Biology
Introduction
Bacterial meningitis is the most common life-threatening infection of the CNS. To cause meningitis, blood-borne bacteria must penetrate the blood-brain barrier (BBB), which consists of an extremely specialized layer of brain microvascular endothelial cells (BMECs). The BBB separates the brain from circulating blood, and disruption of BBB integrity is a hallmark event in the pathophysiology of bacterial meningitis. This disruption is thought to result from the combined effect of bacterial entry and penetration of BMECs, direct cellular injury by bacterial cytotoxins, and/or activation of host inflammatory pathways that compromise BMEC barrier function (1).
Streptococcus agalactiae, or group B Streptococcus (GBS), is a Gram-positive bacterial pathogen that is an important cause of invasive disease in newborns and a subset of adults (2). Currently, GBS is the leading cause of meningitis in the neonate (3, 4). Despite advances in intensive care management and antibiotic therapy, mortality can approach 10%, with 25% to 50% of surviving neonates exhibiting permanent neurological sequelae, including cerebral palsy, mental retardation, blindness, deafness, and seizure (2). GBS possesses many virulence factors that may contribute to the interaction with brain endothelium, including lipoteichoic acid (LTA) (5), β-hemolysin/cytolysin (β-H/C) (6), pili (7, 8), serine-rich repeat (Srr) proteins (9, 10), and HvgA (11). Recently, we have demonstrated that the GBS pilus protein PilA and Srr-1 interact with components of the host extracellular matrix (ECM) to promote BBB interactions and the development of meningitis (8, 9). Other meningeal pathogens, such as Streptococcus pneumoniae (SPN), E. coli K1, Neisseria meningitidis, and Haemophilus influenza type B (HiB, also bind ECM components and ECM receptors (e.g., integrins and laminin receptor) to mediate bacterial-BBB interactions (12–15). Given that host ECM components and receptors preferentially localize to the basolateral surface of polarized BBB endothelium (16), we hypothesized that disruption of junctional protein complexes in brain endothelium is the first step leading to bacterial access to basally expressed receptors.
The BBB, composed primarily of a specialized layer of BMECs, separates the brain and its surrounding tissues from circulating blood, thereby maintaining CNS homeostasis (17). The brain endothelial cells are characterized by the presence of tight intercellular junctions that promote high transendothelial electrical resistance and therefore impede paracellular flux of macromolecules (18). In BMECs, tight junctions are composed of 4 types of integral membrane proteins: occludin, claudins, junctional adhesion molecules, and cell-selective adhesion molecules, all of which are linked through cytoplasmic proteins (zonala-occludin 1 [ZO-1], -2, -3, cingulin) to the actin cytoskeleton (19). Furthermore, tight junction integrity is important for the maintenance of apical and basal cell polarity (20). Here, we demonstrate for the first time to our knowledge that infection of brain endothelium with GBS and other meningeal pathogens induces expression of the host transcription factor Snail1 (SNAI1), a global repressor of tight junction gene expression (21, 22), resulting in increased bacterial BBB penetration. Our results suggest a novel mechanism for bacterial-BBB disruption and explain how bacterial pathogens access basally expressed host proteins to promote CNS entry.
Results
Induction of SNAI1 in brain endothelium during bacterial infection.
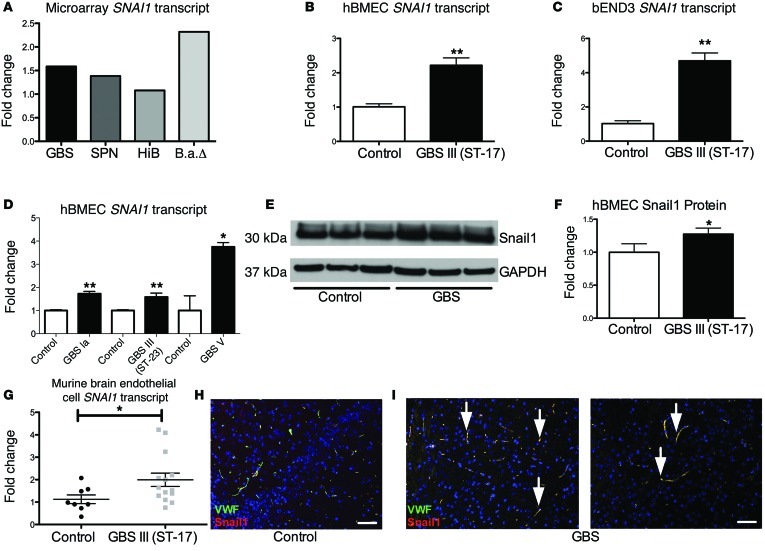
We first performed a comparative global transcriptional analysis of immortalized human BMECs (hBMECs) upon infection with bacteria associated with CNS disease, including GBS, S. pneumoniae, HiB, and Bacillus anthracis (B.a.), to examine host factors that are upregulated during infection. We have previously published a complete microarray dataset from B.a. (23) and GBS (6, 8) infection of hBMECs and a partial list of affected genes in hBMECs in response to SPN (24). Data analysis of these experiments using a statistical algorithm developed for high-density oligonucleotide arrays (25) revealed that infection with all pathogens, with the exception of HiB, resulted in significant induction of SNAI1 (Figure 1A). Snail1 is a global transcriptional repressor of tight junctions (22) and plays an important role in the epithelial-to-mesenchymal transition (EMT) during development (26). To confirm the microarray results, hBMECs and a murine brain endothelial cell line, bEND3, were infected with a hypervirulent GBS clinical isolate that is highly associated with meningitis (sequence type [ST] 17, serotype III). Quantitative PCR (qPCR) analysis revealed that the SNAI1 transcript was significantly increased in infected cells compared with uninfected control cells (Figure 1, B and C). SNAI1 induction occurred following infection with 3 different GBS clinical serotypes (Figure 1D), but not with nonpathogenic bacterial strains or S. aureus, which is not classically associated with meningitis (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI74159DS1). We further analyzed protein expression by Western blot analysis and found that Snail1 protein levels were significantly induced during GBS infection (Figure 1, E and F). To examine whether SNAI1 is induced by GBS in vivo, we used an established murine model of hematogenous meningitis (6). Mice were injected i.v. with WT GBS or vehicle control. At the time of death, brain endothelial cells were isolated and RNA was purified for qPCR analysis. SNAI1 transcripts were significantly increased in GBS-infected mice compared with levels in control mice (Figure 1G). We further examined the localization of Snail1 in brain tissue and observed that Snail1 colocalized with von Willebrand factor (VWF), further supporting the observation that Snail1 is expressed in endothelial cells during active infection (Figure 1, H and I). Taken together, these data suggest that Snail1 is induced in brain endothelial cells in vitro and in vivo in response to GBS infection.
Figure 1. GBS upregulates Snail1 in brain endothelium.
Upregulation of SNAI1 transcription as assessed by microarray analysis during infection with GBS, SPN, HiB, and B.a. ΔpX01 (B.aΔ) (A). Changes in mRNA transcripts in confluent hBMECs or bEND3 monolayers after a 5-hour infection with GBS at an MOI of 10) (B and C). Confluent hBMECs were infected for 5 hours with different serotypes of GBS at an MOI of 10 (type Ia and III) or an MOI of 0.1 (type V). RNA was extracted and SNAI1 transcripts analyzed (D). Changes in protein levels of confluent hBMEC monolayers after infection for 5 hours with GBS at an MOI of 10 (E and F). Mice (n = 8/group) were infected i.v. with GBS or vehicle control. Upon sacrifice, brain tissue was collected and endothelial cells isolated for RNA extraction and qPCR analysis (G). Brains of infected mice (n = 10) showed Snail1 colocalized with VWF factor (scale bar: 50 μm) (H and I). Experiments were performed at least 3 times in triplicate. For qPCR analysis, error bars represent the SEM of at least 3 biological replicates (B–D and G), or the SD for protein analysis of a representative experiment (F). Student’s t test was used to determine statistical significance. *P < 0.05; **P < 0.01.
GBS infection disrupts tight junctions in brain endothelium.
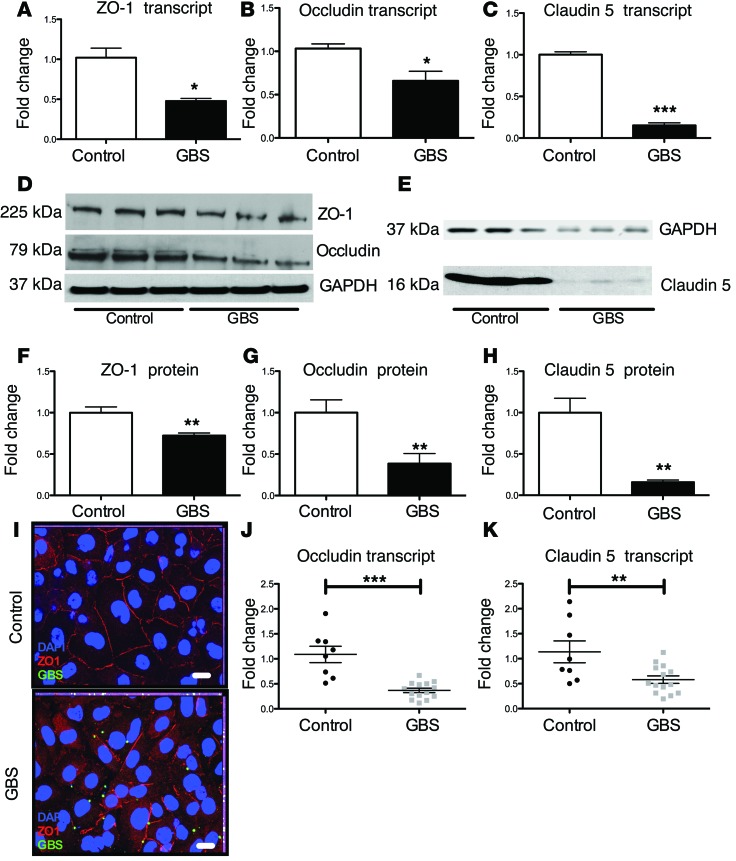
Snail1 is a global repressor of tight junctions, as Snail1 binds the promoter regions of the gene encoding occludin, genes in the claudin family, and genes of other adherence junction proteins (21, 22). Therefore, we sought to determine the effect of GBS infection on transcript and protein abundance levels of tight junction proteins in brain endothelium. GBS infection resulted in decreased transcript and protein levels of ZO-1, occludin, claudin 5 (Figure 2, A–H), and VE-cadherin (Supplemental Figure 1, B and C). Consistent with these results, GBS infection of hBMECs resulted in an overall reduction and disruption of ZO-1 staining at the intercellular junctions, as observed by immunofluorescence staining (Figure 2I). Similar results were observed when visualizing occludin (Supplemental Figure 1D) and claudin 5 during GBS infection (data not shown). Further, we observed no difference in cell monolayer density or cell viability, as measured by Trypan blue staining during infection (Supplemental Figure 1E), suggesting that the observed differences in tight junction staining were not a result of increased cell death. Finally, endothelial cells isolated from infected mice had significantly lower claudin 5 and occludin expression levels when compared with those in uninfected mice (Figure 2, J and K). These data demonstrate that GBS is able to disrupt the transcript and protein levels of these tight junction components in brain endothelial cells in vivo and in vitro.
Figure 2. GBS disrupts tight junctions of brain endothelium.
GBS decreased transcript abundance of zo-1 (A), occludin (B), and claudin 5 (C) in confluent hBMECs (A and B) or bEND3 (C) following GBS infection for 5 hours at an MOI of 10. qPCR experiments were performed at least 3 times in triplicate. Error bars represent the SEM of at least 3 biological replicates (A–C). Protein levels of ZO-1 and occludin in hBMECs (D, F, and G) or bEND3 (E and H) during GBS infection were determined by Western blot analysis. Lysates were probed for ZO-1, occludin, or claudin 5. Western blot analysis was normalized to GAPDH (D–H). Experiments were performed at least 3 times in triplicate. Error bars represent the SD of a representative experiment (F–H). hBMECs were stained for ZO-1 and visualized by immunofluorescence (scale bars: 20 μm) (I). Mice (n = 8/group) were infected i.v. with GBS or vehicle control. Upon sacrifice, brain tissue was collected and endothelial cells isolated for RNA extraction and qPCR analysis (J and K). Student’s t test was used for image analysis. *P < 0.05; **P < 0.01; ***P < 0.001.
Snail1 is necessary and sufficient to disrupt tight junctions.
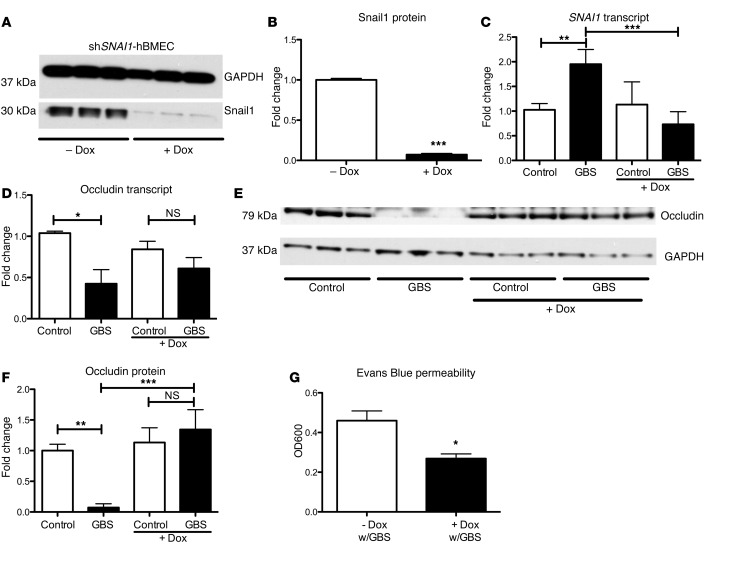
To examine whether increases in Snail1 contribute to the loss of tight junctions, we generated a doxycycline-inducible knockdown cell line that targets SNAI1 using the TRIPZ lentiviral vector system (see Experimental Procedures). The generated hBMEC line (shSNAI1-hBMEC) was treated with doxycycline to induce expression of shSNAI1, which resulted in a reduction of Snail1 (Figure 3, A and B). As expected, GBS did not induce SNAI1 in doxycycline-treated shSNAI1-hBMECs (Figure 3C). Additionally, whereas GBS infection resulted in decreased occludin transcript levels in shSNAI1-hBMECs, GBS infection led to a modest rescue of occludin in doxycycline-treated shSNAI1-hBMECs (Figure 3D). Furthermore, doxycycline treatment of cells resulted in a rescue of occludin (Figure 3, E and F). Finally, to examine whether SNAI1 knockdown increases barrier integrity, we used an Evans Blue Transwell migration assay as described previously (23, 27). In response to GBS infection, less Evans Blue migration was observed in doxycycline-treated shSNAI1-hBMECs (Figure 3G). No changes in tight junction proteins or Snail1 were observed in doxycycline-treated control hBMECs (Supplemental Figure 2A), nor did doxycycline inhibit GBS growth at the concentration used (Supplemental Figure 2B). These results suggest that Snail1 expression is necessary for GBS-mediated downregulation and disruption of tight junction proteins, which results in decreased endothelial barrier integrity.
Figure 3. Snail1 is necessary to disrupt brain endothelial tight junctions.
shRNA knockdown of Snail1 in hBMECs resulted in a significant reduction of Snail1 protein abundance (A and B). SNAI1 expression was no longer upregulated upon GBS infection with shRNA knockdown (C). Occludin transcript and protein levels, with knockdown of SNAI1 during GBS infection (D–F). shRNA hBMEC barrier function was greater as visualized by Evans Blue migration; dye was added to the upper chamber and quantified colorimetrically in the lower chamber (OD600) (G). Experiments were performed at least 3 times in triplicate; error bars represent the SEM of at least 3 biological replicates (C, D, and G), or the SD of a representative experiment (B and F). One-way ANOVA (C, D, and F) and Student’s t test (B and G) were used to analyze statistical significance. *P < 0.05; **P < 0.01; ***P < 0.001.
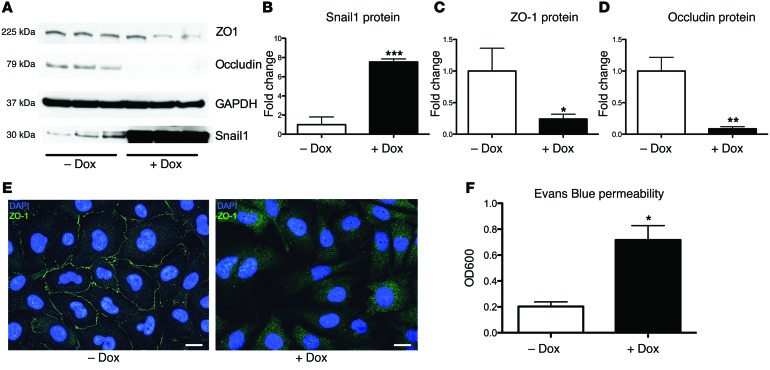
To determine whether Snail1 is sufficient to disrupt tight junctions in BBB endothelium, we constructed a doxycycline-inducible Snail1-overexpression cell line in hBMECs (hBMEC-TRE:SNAI1) using the lentiviral system described in Methods. When treated with doxycycline, protein levels of Snail1 were increased in hBMEC-SNAI1 compared with levels in the noninduced controls (Figure 4, A and B). Induction of Snail1 resulted in decreased levels of ZO-1, occludin (Figure 4, A, C, and D), and VE-cadherin (Supplemental Figure 2, C and D). Overexpression was also coupled with a loss in tight junction integrity as visualized by ZO-1 staining (Figure 4E). No changes in Snail1, ZO-1, or occludin were observed in a control cell line, which contained all the elements of the overexpression line except for the Snail1 ORF (hBMEC-TRE) (Supplemental Figure 2E). To determine the impact of Snail1 overexpression on hBMEC integrity and barrier function, we measured hBMEC permeability to Evans Blue dye as described previously (23, 27). Increased permeability to Evans Blue was observed in doxycycline-treated hBMEC-TRE:SNAI1 when compared with that in untreated cells (Figure 4F). The control cell line (hBMEC-TRE) did not exhibit any permeability (Supplemental Figure 2F). These data suggest that Snail1 expression is sufficient to cause the loss of tight junctions and subsequent barrier integrity in brain endothelium.
Figure 4. Snail1 is sufficient to disrupt brain endothelial tight junctions.
Overexpression of Snail1 resulted in a loss of tight junction proteins (A–D). Control cell line shows the expected cobblestone pattern, and overexpression of Snail1 resulted in a loss of tight junction staining (scale bars: 20 μm) (E). Snail1 overexpression increased hBMEC permeability as assessed by Evans Blue dye migration after induction with doxycycline; dye was added to the upper chamber and quantified colorimetrically in the lower chamber (OD600) (F). Experiments were performed at least 3 times in triplicate; error bars represent the SEM of at least 3 biological replicates (F), or the SD for protein analysis of a representative experiment (B–D). Student’s t test was used to determine significance. *P < 0.05; **P < 0.01; ***P < 0.001.
Contribution of bacterial factors and host signaling pathways to Snail1 induction.
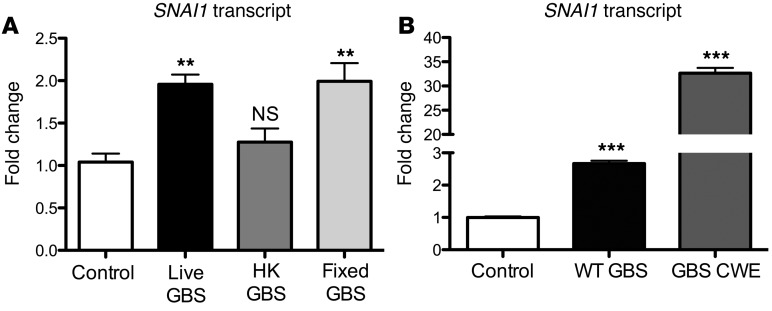
To determine whether live bacterial challenge was required for SNAI1 induction, we treated hBMECs with live, heat-killed (HK), or formalin-fixed GBS. Neither HK nor formalin-fixed GBS had viable bacteria present as determined by CFU enumeration on Todd Hewitt Agar (THA) plates. Interestingly, HK GBS did not induce SNAI1, however formalin-fixed GBS resulted in the induction of SNAI1 transcripts comparable to the levels seen in infection with live bacteria (Figure 5A). This result suggests that a GBS surface factor initiates SNAI1 transcription and that this surface factor is susceptible to heat denaturation. To further support this hypothesis, we prepared a previously described (28) cell wall extract from GBS that contained components attached to the bacterial cell surface. Strikingly, GBS cell wall extract greatly induced SNAI1 transcript levels when compared with those of the control (Figure 5B). Taken together, these data suggest that GBS expresses a surface factor that induces SNAI1 in hBMECs.
Figure 5. GBS factors contribute to SNAI1 expression.
Fixed GBS was able to induce SNAI1 expression at levels that were comparable to those in live GBS; however, HK GBS was not able to induce SNAI1 (A). Cell wall extracts (CWE) prepared from WT GBS were greatly able to induce SNAI1 (B). Experiments were performed at least 3 times in triplicate; error bars represent the SEM of at least 3 biological replicates. One-way ANOVA was used to determine significance. **P < 0.01; ***P < 0.001.
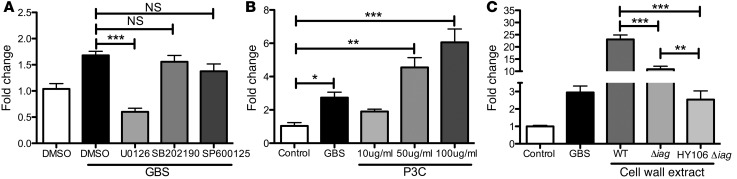
We have previously examined the global transcriptional profile of hBMECs following infection with B.a. and the toxin-deficient ΔpXO1 mutant (23). As shown in Figure 1A, the toxin-deficient mutant (B.a.Δ) induced the SNAI1 transcript, while the WT B.a. strain did not (23). We confirmed these microarray results by qPCR in hBMECs (data not shown). Interestingly, lethal toxin (LT), which is encoded on the pXO1 plasmid, cleaves components of the MAPK signaling pathway (29). Thus, we hypothesized that MAPK signaling pathways might contribute to Snail1 induction during bacterial infection, as B.a. does not induce SNAI1 in hBMECs, whereas B.a.Δ does induce SNAI1 transcripts. To further examine the signaling pathways leading to SNAI1 induction, we inhibited MEK1/2/ERK1/2, p38, and JNK to assess the contribution of each to SNAI1 upregulation. Inhibition of the MEK1/2/ERK1/2 components of the MAPK pathway with U0126 resulted in a significant reduction of SNAI1 transcripts in response to GBS infection, while inhibitors of p38 (SB202190) and JNK (SP600125) had no effect (Figure 6A). As TLR2 contributes to the pathogenesis of GBS meningitis (5) and downstream signaling of TLRs activate the MAPK pathway, we hypothesized that treatment of hBMECs with the TLR2 agonist Pam3CSK4 (P3C) would induce SNAI1 in hBMECs. We observed a dose-dependent increase in SNAI1 in response to P3C (Figure 6B). We have previously described a GBS mutant (Δiag) that lacks the ability to properly anchor (LTA) (6), a known TLR2 ligand (30). It is believed that the Δiag mutant still produces unanchored LTA that is shed by the bacteria (6). Furthermore, the polysaccharide capsule likely traps the shed LTA, as a double mutant in capsule and LTA anchoring results in less LTA associated with the bacteria (6). To test whether LTA could be responsible for SNAI1 induction in hBMECs, we prepared cell wall extracts from WT, Δiag, and the capsule HY106/Δiag double mutant and treated our hBMECs with these fractions. We observed that cell wall extract from the Δiag mutant resulted in significantly reduced levels of SNAI1 transcripts when compared with those of cell wall extracts prepared from WT GBS cell wall extract. Strikingly, the cell wall extracts prepared from the double mutant resulted in an even greater reduction of SNAI1 induction (Figure 6C). Taken together, these data suggest that bacterial LTA, TLR2 activation, and signaling through MEK1/2/ERK1/2 lead to SNAI1 induction.
Figure 6. Contribution of MAPK signaling in SNAI1 expression.
hBMECs were treated with either DMSO vehicle control or a specific MAPK pathway inhibitor. U0126 had the greatest effect on inhibition of GBS-induced SNAI1 expression (A). Treatment of hBMECs with the TLR2 agonist P3C resulted in a dose-dependent increase in SNAI1 expression (B). Treatment of hBMECs with extracts from Δiag or HY106 Δiag resulted in significantly lower SNAI1 expression levels (C). Experiments were performed at least 3 times in triplicate; error bars represent the SEM of at least 3 biological replicates. One-way ANOVA was used to determine significance. *P < 0.05; **P < 0.01; ***P < 0.001.
Contribution of Snail1 to GBS-BBB penetration in vivo.
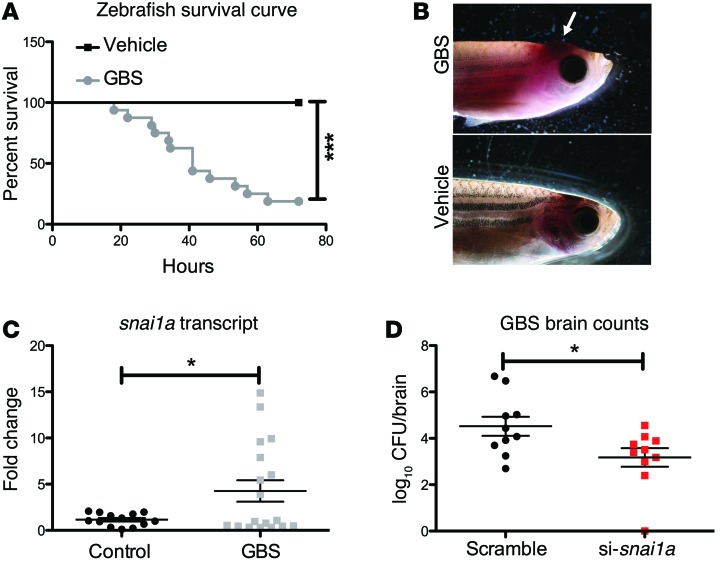
As the Snail1-KO mice are embryonically lethal (31), we developed an in vivo model of GBS infection using Danio rerio (zebrafish) adults. Adult zebrafish have been used to model Streptococcus iniae and, recently, GBS (32, 33). When zebrafish were injected with WT GBS or a PBS vehicle control, we observed up to 90% mortality in GBS-infected fish (Figure 7A) and high bacterial loads in the blood and brain (Supplemental Figure 3, A and B). We also observed aberrant swimming behavior in the infected fish (Supplemental Figure 3, C and D), corroborating previous reports (32). Furthermore, GBS-infected fish exhibited cerebral swelling and edema typical of CNS infection (Figure 7B). We next sought to determine whether the zebrafish SNAI1 homolog snai1a is upregulated during GBS infection. We isolated RNA from the brains of infected zebrafish, and snai1a transcripts were measured by qPCR. As shown in Figure 7C, GBS significantly upregulated snai1a in the brain tissue of infected zebrafish. To examine the role of snai1a during GBS infection in zebrafish, we transiently knocked down snai1a using siRNA duplexes. Injection of siRNA decreased brain snai1a transcripts (Supplemental Figure 3E) and resulted in a significant decrease in the amount of GBS recovered from brain tissue of infected fish (Figure 7D), while no differences in bacterial levels recovered from the blood were observed (Supplemental Figure 3F). These data suggest that snai1a plays a role in promoting BBB integrity, as decreased expression attenuated GBS passage to the brain in adult zebrafish.
Figure 7. Contribution of Snail1 to GBS-BBB penetration in vivo.
Kaplan-Meier survival curve for zebrafish (n = 9/group) infected with GBS (A). Representative images of GBS-infected zebrafish showing cerebral swelling and edema compared with noninfected controls (B). GBS infection increased snai1a transcript levels in brain homogenates of zebrafish following GBS infection (n = 19) compared with those in noninfected zebrafish (n = 12) (C). Knockdown of snai1a using siRNA duplexes (n = 10/treatment group) did not change GBS bloodstream survival, but attenuated bacterial penetration into the brain (D). Representative data from 1 of 2 independent experiments are shown. Student’s t and log-rank tests were used to determine significance. *P < 0.05; ***P < 0.001.
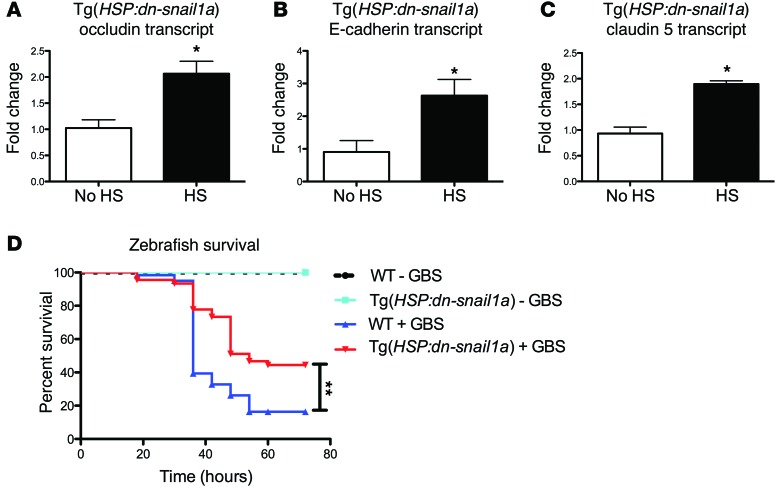
To further examine the role of snai1a in vivo, we developed a larval model of zebrafish GBS infection (34). We generated Tg(HSP:dn-snai1a) animals that function as a dominant-negative inhibitor of endogenous snai1a. Heat-shocked HSP:dn-snai1a larvae 3 days post fertilization (dpf) had increased transcript levels of occludin, claudin 5, and E-cadherin compared with those in larvae that were not heat shocked (Figure 8, A–C). Heat shocking WT larvae did not increase occludin, claudin 5, or E-cadherin levels (Supplemental Figure 4, A–C). To examine whether increasing expression of tight junctions could result in increased survival in response to GBS challenge, we infected heat-shocked HSP:dn-snai1a and WT larvae at 3 dpf and infected them 5 hours after heat shocking. At 72 hours after infection, only 10% of WT larvae survived, whereas about 40% of the HSP:dn-snai1a larvae were still viable (Figure 8D). These data suggest that inhibition of snai1a in vivo can result in lower mortality in response to GBS infection in zebrafish larvae.
Figure 8. Inhibition of Snail1 impacts expression of junctional components and reduces GBS virulence in zebrafish larvae.
Transgenic zebrafish expressing an inducible dominant-negative snai1a, Tg(HSP:dn-snai1a), upregulated occludin, E-cadherin, and claudin 5 following heat shock (HS) (n = 9) when compared with controls with no heat shock (no HS) (n = 4) 6 hours after heat shock (A–C). Kaplan-Meier survival curve of WT or Tg(HSP:dn-snai1a) transgenic zebrafish following heat shock and GBS challenge (n = at least 10/group). Transgenic zebrafish exhibited significantly lower mortality rates following GBS challenge compared with those of WT zebrafish (D). Data represent 3 independent experiments combined. Student’s t and log-rank tests were used to determine significance. *P < 0.05; **P < 0.01.
Discussion
The initiation of bacterial meningitis occurs when blood-borne bacteria penetrate the BBB, gaining access to the CNS. The exact mechanisms of BBB traversal vary among meningeal pathogens (35). However, the majority of bacteria associated with meningitis possess the ability to bind to host factors such as ECM components and receptors, which preferentially localize to the basolateral surface of the BBB (1). It is not completely understood how blood-borne meningeal pathogens engage basolateral receptors in the presence of a functional BBB. Tight junctions promote barrier integrity, creating an impassable barrier to diffusing receptors, thus separating basolateral and apical membranes and maintaining cell polarity (36). Disruption of tight junctions results in the loss of cell polarity (37). Several studies have demonstrated that meningeal pathogens degrade tight junction proteins or remodel the junctional complexes. For example Neisseria meningitidis induces specific cleavage of occludin through the release of host MMP8, resulting in endothelial cell detachment and increased paracellular permeability (38). E. coli K1 initiates a signaling process that leads to dissociation of β-catenins from cadherins and results in disruption of barrier integrity (39). Recent studies have also shown that the type IV pili of N. meningitidis lead to recruitment of the PAR3/PAR6/PKCζ polarity complex and delocalization of junctional proteins, which results in anatomical gaps that are used by the bacteria to penetrate into the CNS (40, 41). In this study, we similarly show that GBS is able to disrupt tight junctions in brain endothelium and that this process is facilitated by the Snail1 host transcription factor.
Snail1 is a zinc finger transcription factor that has been extensively studied in the context of EMT and cancer metastasis (31, 42–44). EMT is involved in normal embryonic development and repair of epithelial injury and is associated with changes in claudin expression and regulation. Induction of Snail1 induces EMT and downregulates tight junction proteins including claudins and occludin (21, 45). Snail1 binds to an E-box motif of promoters, repressing transcription (21, 46). However, Snail1 can also downregulate claudin 1 and ZO-1, without affecting their transcription (22). Notably, Snail1 expression can also disrupt apical/basal polarity complexes in epithelial cells (47). Here, we demonstrate that infection with multiple pathogens associated with meningitis results in the induction of SNAI1, which we demonstrate to be necessary and sufficient to disrupt tight junctions in brain endothelium. Our results are consistent with a recent study describing a role for Snail1 in bacterial transit across epithelial cells (48). Notably, TLR2/4 and MAPK p38 signaling induced Snail1 expression in the nasopharynx. Similarly, our results suggest that in brain endothelium, Snail1 induction is initiated by TLR2 and involves the ERK pathway of the MAPK signaling cascade.
MAPK signaling pathways modulate the expression of tight junction proteins and alter the molecular composition within tight junction complexes (49). Three MAPK pathways have been well characterized. First, the ERKs, which are activated by several extracellular stimuli, include growth factors, cytokines, and oxidative stress. Second, the JNKs and third, p38, respond to different types of cellular stress and inflammatory mediators (50). Activation of ERK1/2 results in increased ZO-1 degradation during traumatic brain injury (51), decreased ZO-1 and claudin 5 expression in brain endothelium during HIV infection (52, 53), and reduced occludin and claudin 5 expression in BBB endothelium during ischemic stroke (54). Whether Snail1 is similarly involved in tight junction disruption under these conditions remains to be determined. Host factors and cytokines can also influence tight junction turnover and claudin expression, presumably in part to accommodate neutrophil migration across epithelial and endothelial barriers (55). Interestingly, we did not observe an increase in SNAI1 transcript levels in hBMECs upon addition of TNF-α or IL-1β (data not shown).
Using a murine model of hematogenous meningitis, we observed a significant increase in SNAI1 transcripts in brain tissue during GBS infection when compared with noninfected controls. Further experiments with murine models are technically challenging, as SNAI1-null alleles are embryonically lethal at E8.5 (31, 56, 57). To examine a functional role for Snail1 during infection in vivo, we developed and used adult and larval zebrafish models for Streptococcal infection as previously described (32, 33). As the zebrafish Snail1 homolog Snail1a is also required for EMT and neural crest development (58), this model was more amenable to the use of siRNA silencing to globally knock down expression of Snail1a. We showed that siRNA-mediated reduction of snai1a transcription resulted in decreased brain penetration of GBS in adult zebrafish. Finally, to further examine a functional role for Snail1a in vivo, we generated and characterized an inducible Snail1a dominant-negative transgenic zebrafish. This animal showed increased survival when compared with WT animals, suggesting that inhibition of Snail1a is sufficient to protect zebrafish larvae from GBS infection. To our knowledge, this is the first characterization of the role of Snail1 in the pathogenesis of bacterial infection in vivo. Our results also suggest that GBS LTA may induce SNAI1 expression. Future studies will seek to further determine the mechanism of LTA activation and whether other LTA-associated proteins play a role. The subsequent loss of tight junctions may represent the critical first step that enables bacterial pathogens to engage basolaterally expressed host receptors and promote BBB disruption and progression to meningitis.
Methods
Bacterial strains and cell lines.
The Streptococcus agalactiae (GBS) highly virulent clinical isolate COH1 (serotype III, ST-17) was primarily used for the experiments (59), along with other GBS serotypes, NCTC10/84 (V) (60), 515 (Ia) (61), and NEM316 (serotype III, ST-23) (62). Other bacteria were used and propagated as previously described: SPN (24), HiB (63), and the B.a. Sterne strain ΔpXO1 (23). The GBS mutants used include COH1 Δiag, NCTC10/84 ΔpilA, and NCTC10/84 ΔcylE (5–7). Cell wall extracts were prepared as previously described (28). Briefly, GBS pellets were resuspended in 20% sucrose, 20 mM Tris-HCl, and 10 mM MgCl2 treated with protease inhibitor and mutanolysin for 2 hours at 37°C, followed by centrifugation, and the supernatant was collected as cell wall extract. Immortalized hBMECs (a gift of Kwang Sik Kim and Monique Stins, Johns Hopkins University, Baltimore, Maryland, USA) were cultured in RPMI 1640 containing 10% FBS, 10% NuSerum, and 1% nonessential amino acids as described previously (5, 6, 64). Immortalized bEND3 cells were purchased from ATCC and cultured to ATCC specifications in DMEM-F12 containing 10% FBS.
Infection assays.
hBMECs or bEND3 were grown to confluency on collagenized 24-well plates (Corning). GBS or other bacteria were then incubated with hBMECs for 4 to 5 hours. Following infection and/or treatment with inhibitors, cell lysates were collected for protein and RNA, or fixed for staining according to the procedures described below. For MAPK inhibitor assays, 30 minutes prior to infection, hBMECs were treated with the inhibitors U0126 (Reagents Direct), SB202190 (Reagents Direct), or SP600125 (Reagents Direct) — all at 20 μM. After inhibitor was added, hBMECs were infected with WT GBS at an MOI of 10 and incubated for 5 hours. HK or formalin-fixed GBS were prepared by growing GBS to OD600 0.4 in Todd-Hewitt broth (THB), followed by centrifugation and resuspension of the pellet in PBS and treatment at 95°C for 10 minutes, or treatment of GBS with 4% paraformaldehyde for 15 minutes. Bacteria were then washed and used to treat hBMECs at an estimated desired MOI. RNA lysates were collected and prepared as described below, and qPCR was performed on samples to determine Snail1 expression levels.
Microarray analysis and qPCR.
Bacterial infection, RNA isolation, cDNA preparation, microarray analysis, and qPCR were performed as described previously (8, 23). Heat-killing experiments were performed as described previously (65). For qPCR, the primer sequences used have been published for human SNAI1 (66), human ZO-1 (TJP1) (67), human OCLN (68), and human GAPDH (8); murine Cldn5, Snai1, and Gapdh (69), and murine Ocln (forward: 5′-GATGCAGGTCTGCAGGAGTATAA-3′; reverse: 5′-ATCCTTAATTGGAGTGTTCAGCC-3′); and zebrafish occludin (oclna), gapdh (70), and E-cadherin (cdh1) (71). Fold change was calculated by the ΔΔCT method.
Protein analysis.
Confluent cell monolayers were infected with GBS. Cells were lysed using RIPA buffer (Thermo Scientific) containing 100 mM NaF, 1 mM PMSF, and protease inhibitor cocktail (Calbiochem). Lysates were probed with antibodies against GAPDH (1:150,000; catalog MAB374; EMD Millipore); ZO-1 (1:1,000; catalog 617300; Invitrogen); occludin (1:1,000; catalog 331500; Invitrogen); claudin 5 (1:1,000; catalog ABT45; EMD Millipore); pan-cadherin (1:1,000; catalog 4068; Cell Signaling Technology); and Snail1 (1:1,000; catalog ab85931; Abcam). Appropriate HRP secondary antibodies were used to detect primary antibodies, and blots were visualized using SuperSignal West Pico Chemiluminescent Substrate ECL (Thermo Scientific) and radiographic film (Denville Scientific Inc.). For florescence microscopy following infection, hBMECs were fixed with 4% PFA and permeabilized with 0.1% Triton X-100. Junction proteins and GBS were detected using anti–Z0-1 (1:100; catalog 617300; Invitrogen) and anti-GBS (1:50; catalog BM5557P; Acris) antibodies, followed by Cy3- or FITC-conjugated secondary antibodies. Coverslips were mounted using DAPI containing VECTASHIELD (Vector Laboratories) and visualized with a Zeiss florescence microscope.
Lentiviral vectors and virus production.
hBMECs were cotransfected at 50% to 60% confluency with 3 μg TRIPZ shRNA vector (RHS5087; Thermo Scientific), as well as the following components from individual vectors 0.4 μg GAG-POL, 0.4 μg Tat, 0.4 μg REV, 1.5 μg VPR, 0.8 μg of VSVg. Media were replaced with DMEM, 10% FBS, 1% MEM, and penicillin-streptomycin. Viral supernatant was collected 48 hours after transfection and filtered through 0.2 μM mixed cellulose ester (MCE) filters (catalog 09-719A; Fisherbrand, Fisher Scientific). Viral supernatants were stored at –80°C until transduction. Lentiviral supernatent (2 ml) collected after 48 hours of transfection was added to hBMECs at 70% to 80% confluency and incubated with polybrene. hBMECs were spin-infected by centrifugation at 1,500 g for 80 minutes at 30°C. Seventy-two hours after infection, 1 μg/ml doxycycline was added to induce TurboRFP (Sigma)and shRNA expression.
Creation of hBMEC Snail1 and shSnail1 cell lines.
The inducible vector pH-TRE.i.mCherry was constructed using pH-TRE as described previously (72). The IRES-mCherry cassette from pBMN.i.mCherry was inserted into pH-TRE downstream of the multiple cloning site (provided by Gary Nolan, Stanford University, Stanford, California, USA, and Clontech). pBMN-i-rtTA was constructed by removing rtTA from the Tet-On vector (Clontech) with EcoRI/BamHI and cloning it into pBMN-i-Lyt2. Human Snail1 was cloned from cDNA extracted from hBMECs. Primers were used to amplify the human Snail1 ORF (73), and the product was inserted into pH-TRE.i.mCherry, creating pH-TRE.Snail1.i.mCherry. The lentiviral vectors were produced as previously described (74). The resulting viral supernatant was mixed with polybrene and added to hBMECs. Cells were stained with PE-conjugated anti-Lyt2 antibodies (BD Biosciences) and sorted on a BD FACSAria Flow Cytometer (SDSU FACS Core Facility). This cell population was expanded and infected with pH-TRE.Snail1.i.mCherry. Dually infected cells were treated with doxycycline (1 μg/ml) and sorted for mCherry fluorescence to create hBMEC-SNAI1. The TRIPZ short hairpin lentiviral vector (Thermo Scientific) was used. Short hairpins directed against the human SNAI1 transcripts were purchased (GE Healthcare Dharmacon). Lentiviruses were created and naive hBMECs were spin-infected as described above. Cells were induced with doxycycline (1 μg/ml) and sorted by hCherry fluorescence to create the shSNAI1 hBMEC line.
Transwell permeability assay.
Briefly, the Snail1-overexpressing hBMEC line was seeded onto the apical side of collagen-coated polytetrafluoroethylene 0.4-μM pore-size membranes (Corning) and grown for 6 to 7 days to form intact monolayers. Snail1 expression was induced using 1 μg/ml doxycycline. Assessment of barrier integrity was performed as described previously (27).
Murine model of hematogenous meningitis.
We have previously described a mouse model of GBS meningitis (5, 8). Briefly, 6- to 8-week-old male CD-1 mice were injected i.v. with 6 × 107 to 7 × 107 CFU WT GBS or Dulbecco’s PBS (DPBS) vehicle control. At the time points of morbidity or death, the mice were euthanized, and brain tissue was collected for histology or endothelial cell isolation. Endothelial cells were isolated using the MACS CD31 endothelial cell isolation kit (Miltenyi Biotec).
Adult zebrafish model of GBS infection.
Six- to nine-month-old adult WT AB Zebrafish (Danio rerio) were anesthetized with 0.16% tricaine solution. Fish were placed upside down, supported with a moist sponge, and injected i.p. with 7 × 107 to 1 × 108 CFU COH1 GBS as previously described (32). Fish were euthanized when aberrant behavior was observed. Upon sacrifice, blood and brain samples were collected and diluted in PBS for bacterial enumeration. siRNA-knockdown duplexes were designed using the IDT RNAi program (Integrated DNA Technologies). RNA duplexes of si-Snai1a (5′-CCAGCAGUCAGCAAUGACUUCCCAG-3′), and scrambled (5′-ACCGACCGCCUUGUACCAACGAG-3′) were injected i.p. 24 hours before GBS infection. Fish were sacrificed 12 hours after infection, and tissues were collected for bacterial enumeration.
Zebrafish transgenesis.
Tg(HSP:dn-snai1a)sd37 were generated using Tol2-mediated transgenesis via the multisite Gateway cloning system. The final pDEST construct contained a 5′ zebrafish hsp70 promoter, truncated snai1a (300–783 bp) with an N terminus FLAG tag, and a 5′ polyadenylation signal. The following primers were used to amplify the truncated snai1a gene from zebrafish cDNA for cloning into the pDONR221 Gateway vector: forward, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTTATGGATTACAAGGATGACGACGATAAGGGTGGCAGTGGAGGGCAGAGAGAGGACAAGAGCG-3′; reverse, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTCTATTGGACATTGGCCGTGGAGGG-3′. Zebrafish embryos were microinjected at the 1-cell stage with a 25 pg final pDEST construct along with 50 pg Tol2 transposase RNA. Injected F0 adults were mated with AB strain zebrafish, and F1 offspring were screened by PCR to assess germline integration of the Tol2 construct. The primers used were FLAG forward, 5′-GATTACAAGGATGACGACGATAAG-3′ and Snail1a reverse, 5′-CTATTGGACATTGGCCGTGGAGGG-3′. F1 fish were used to generate larvae for all experiments described herein.
Larval zebrafish model of GBS infection.
Infection assays were performed as described previously (34). Briefly, zebrafish larvae at 3 dpf were anesthetized with tricane and placed on a 2% agar plate for stabilization. Diluted GBS (1 nl, MOI of 100) was injected into the caudal vein of the zebrafish with a FemtoJet microinjector (Eppendorf). Phenol red (3%) was added to the GBS dilution to visualize the injection. Fish were monitored every 6 hours. Infection experiments were performed in a blinded fashion. After death, fish were genotyped using the primers listed above and the experiment was deconvoluted.
Statistics.
GraphPad Prism version 5.0a (GraphPad Software) was used for statistical analysis. A 2-tailed Student’s t test and 1-way ANOVA with Bonferroni’s multiple comparisons post test were used where appropriate. A log-rank test was used to determine significance with survival curves. Data are represented as the mean ± SEM. Statistical significance was accepted at a P value of less than 0.05.
Study approval.
All mouse work was approved by the Office of Laboratory Animal Care of SDSU and conducted under accepted veterinary standards. All zebrafish were raised and maintained under UCSD IACUC guidelines.
Supplementary Material
Acknowledgments
We are grateful to Melody Neely for helpful suggestions, Roman Sasik for assistance with microarray data analysis, and the SDSU FACS Core Facility for expertise with flow cytometry. Microarray analysis was performed at the UCSD Biogem Core Facility (Gary Hardiman, Director). B.J. Kim acknowledges support from the San Diego chapter of the Achievement Rewards for College Scientists Foundation (ARCS) and the Kyoto Foundation. This work was also supported by fellowships from the Rees-Stealy Research Foundation/SDSU Heart Institute and the American Heart Association (14PRE18690037 to B.J. Kim), the NIH (R25-GM058906, to A. Bermudez and E. Reyes), and the National Institute of Neurological Disorders and Stroke (NINDS)/NIH (R01-NS051247, to K.S. Doran).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(6):2473–2483. doi:10.1172/JCI74159.
Anirban Banerjee’s present address is: Department of Biosciences and Bioengineering, IIT-Bombay, Mumbai, India.
References
- 1.van Sorge NM, Doran KS. Defense at the border: the blood-brain barrier versus bacterial foreigners. Future Microbiol. 2012;7(3):383–394. doi: 10.2217/fmb.12.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edwards MS, Baker CJ. Group B streptococcal infections in elderly adults. Clin Infect Dis. 2005;41(6):839–847. doi: 10.1086/432804. [DOI] [PubMed] [Google Scholar]
- 3.Brouwer MC, Tunkel AR, van de Beek D. Epidemiology, diagnosis, and antimicrobial treatment of acute bacterial meningitis. Clin Microbiol Rev. 2010;23(3):467–492. doi: 10.1128/CMR.00070-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thigpen MC, et al. Bacterial meningitis in the United States, 1998–2007. N Engl J Med. 2011;364(21):2016–2025. doi: 10.1056/NEJMoa1005384. [DOI] [PubMed] [Google Scholar]
- 5.Doran KS, et al. Blood-brain barrier invasion by group B Streptococcus depends upon proper cell-surface anchoring of lipoteichoic acid. J Clin Invest. 2005;115(9):2499–2507. doi: 10.1172/JCI23829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doran KS, Liu GY, Nizet V. Group B streptococcal beta-hemolysin/cytolysin activates neutrophil signaling pathways in brain endothelium and contributes to development of meningitis. J Clin Invest. 2003;112(5):736–744. doi: 10.1172/JCI17335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maisey HC, Hensler M, Nizet V, Doran KS. Group B streptococcal pilus proteins contribute to adherence to and invasion of brain microvascular endothelial cells. J Bacteriol. 2007;189(4):1464–1467. doi: 10.1128/JB.01153-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerjee A, et al. Bacterial Pili exploit integrin machinery to promote immune activation and efficient blood-brain barrier penetration. Nat Commun. 2011;2:462. doi: 10.1038/ncomms1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seo HS, Mu R, Kim BJ, Doran KS, Sullam PM. Binding of glycoprotein Srr1 of Streptococcus agalactiae to fibrinogen promotes attachment to brain endothelium and the development of meningitis. PLoS Pathog. 2012;8(10):e1002947. doi: 10.1371/journal.ppat.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Sorge NM, Quach D, Gurney MA, Sullam PM, Nizet V, Doran KS. The group B streptococcal serine-rich repeat 1 glycoprotein mediates penetration of the blood-brain barrier. J Infect Dis. 2009;199(10):1479–1487. doi: 10.1086/598217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tazi A, et al. The surface protein HvgA mediates group B streptococcus hypervirulence and meningeal tropism in neonates. J Exp Med. 2010;207(11):2313–2322. doi: 10.1084/jem.20092594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim KJ, Chung JW, Kim KS. 67-kDa laminin receptor promotes internalization of cytotoxic necrotizing factor 1-expressing Escherichia coli K1 into human brain microvascular endothelial cells. J Biol Chem. 2005;280(2):1360–1368. doi: 10.1074/jbc.M410176200. [DOI] [PubMed] [Google Scholar]
- 13.Chung JW, et al. 37-kDa laminin receptor precursor modulates cytotoxic necrotizing factor 1-mediated RhoA activation and bacterial uptake. J Biol Chem. 2003;278(19):16857–16862. doi: 10.1074/jbc.M301028200. [DOI] [PubMed] [Google Scholar]
- 14.Orihuela CJ, et al. Laminin receptor initiates bacterial contact with the blood brain barrier in experimental meningitis models. J Clin Invest. 2009;119(6):1638–1646. doi: 10.1172/JCI36759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Unkmeir A, et al. Fibronectin mediates Opc-dependent internalization of Neisseria meningitidis in human brain microvascular endothelial cells. Mol Microbiol. 2002;46(4):933–946. doi: 10.1046/j.1365-2958.2002.03222.x. [DOI] [PubMed] [Google Scholar]
- 16.Bosman FT. Integrins: cell adhesives and modulators of cell function. Histochem J. 1993;25(7):469–477. doi: 10.1007/BF00159282. [DOI] [PubMed] [Google Scholar]
- 17.Betz AL. An overview of the multiple functions of the blood-brain barrier. NIDA Res Monogr. 1992;120:54–72. [PubMed] [Google Scholar]
- 18.Ballabh P, Braun A, Nedergaard M. The blood-brain barrier: an overview: structure, regulation, and clinical implications. Neurobiol Dis. 2004;16(1):1–13. doi: 10.1016/j.nbd.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 19.Dejana E, Tournier-Lasserve E, Weinstein BM. The control of vascular integrity by endothelial cell junctions: molecular basis and pathological implications. Dev Cell. 2009;16(2):209–221. doi: 10.1016/j.devcel.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Schneeberger EE, Lynch RD. Structure, function, and regulation of cellular tight junctions. Am J Physiol. 1992;262(6):L647–L661. doi: 10.1152/ajplung.1992.262.6.L647. [DOI] [PubMed] [Google Scholar]
- 21.Ikenouchi J, Matsuda M, Furuse M, Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci. 2003;116(pt 10):1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- 22.Ohkubo T, Ozawa M. The transcription factor Snail downregulates the tight junction components independently of E-cadherin downregulation. J Cell Sci. 2004;117(pt 9):1675–1685. doi: 10.1242/jcs.01004. [DOI] [PubMed] [Google Scholar]
- 23.van Sorge NM, et al. Anthrax toxins inhibit neutrophil signaling pathways in brain endothelium and contribute to the pathogenesis of meningitis. PLoS One. 2008;3(8):e2964. doi: 10.1371/journal.pone.0002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerjee A, Van Sorge NM, Sheen TR, Uchiyama S, Mitchell TJ, Doran KS. Activation of brain endothelium by pneumococcal neuraminidase NanA promotes bacterial internalization. Cell Microbiol. 2010;12(11):1576–1588. doi: 10.1111/j.1462-5822.2010.01490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasik R, Calvo E, Corbeil J. Statistical analysis of high-density oligonucleotide arrays: a multiplicative noise model. Bioinformatics. 2002;18(12):1633–1640. doi: 10.1093/bioinformatics/18.12.1633. [DOI] [PubMed] [Google Scholar]
- 26.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3(3):155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 27.Lembo A, et al. Regulation of CovR expression in Group B Streptococcus impacts blood-brain barrier penetration. Mol Microbiol. 2010;77(2):431–443. doi: 10.1111/j.1365-2958.2010.07215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kling DE, Madoff LC, Michel JL. Subcellular fractionation of group B Streptococcus. Biotechniques. 1999;27(1):24–26. doi: 10.2144/99271bm03. [DOI] [PubMed] [Google Scholar]
- 29.Bromberg-White J, Lee CS, Duesbery N. Consequences and utility of the zinc-dependent metalloprotease activity of anthrax lethal toxin. Toxins (Basel). 2010;2(5):1038–1053. doi: 10.3390/toxins2051038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning CJ. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. J Biol Chem. 1999;274(25):17406–17409. doi: 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- 31.Murray SA, Carver EA, Gridley T. Generation of a Snail1 (Snai1) conditional null allele. Genesis. 2006;44(1):7–11. doi: 10.1002/gene.20178. [DOI] [PubMed] [Google Scholar]
- 32.Patterson H, et al. Adult zebrafish model of bacterial meningitis in Streptococcus agalactiae infection. Dev Comp Immunol. 2012;38(3):447–455. doi: 10.1016/j.dci.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Phelps HA, Runft DL, Neely MN. Adult zebrafish model of streptococcal infection. Curr Protoc Microbiol. 2009;Chapter 9:Unit 9D.1. doi: 10.1002/9780471729259.mc09d01s13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim BJ, Hancock BM, Del Cid N, Bermudez A, Traver D, Doran KS. Streptococcus agalactiae infection in zebrafish larvae. Microb Pathog. 2015;79:57–60. doi: 10.1016/j.micpath.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim KS. Mechanisms of microbial traversal of the blood-brain barrier. Nat Rev Microbiol. 2008;6(8):625–634. doi: 10.1038/nrmicro1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matter K, Balda MS. Holey barrier: claudins and the regulation of brain endothelial permeability. J Cell Biol. 2003;161(3):459–460. doi: 10.1083/jcb.200304039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin K, Fogg VC, Margolis B. Tight junctions and cell polarity. Annu Rev Cell Dev Biol. 2006;22:207–235. doi: 10.1146/annurev.cellbio.22.010305.104219. [DOI] [PubMed] [Google Scholar]
- 38.Schubert-Unkmeir A, Konrad C, Slanina H, Czapek F, Hebling S, Frosch M. Neisseria meningitidis induces brain microvascular endothelial cell detachment from the matrix and cleavage of occludin: a role for MMP-8. PLoS Pathog. 2010;6(4):e1000874. doi: 10.1371/journal.ppat.1000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sukumaran SK, Prasadarao NV. Escherichia coli K1 invasion increases human brain microvascular endothelial cell monolayer permeability by disassembling vascular-endothelial cadherins at tight junctions. J Infect Dis. 2003;188(9):1295–1309. doi: 10.1086/379042. [DOI] [PubMed] [Google Scholar]
- 40.Coureuil M, et al. Meningococcal type IV pili recruit the polarity complex to cross the brain endothelium. Science. 2009;325(5936):83–87. doi: 10.1126/science.1173196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coureuil M, et al. Meningococcus Hijacks a β2-adrenoceptor/beta-Arrestin pathway to cross brain microvasculature endothelium. Cell. 2010;143(7):1149–1160. doi: 10.1016/j.cell.2010.11.035. [DOI] [PubMed] [Google Scholar]
- 42.Blanco MJ, et al. Snail1a and Snail1b cooperate in the anterior migration of the axial mesendoderm in the zebrafish embryo. Development. 2007;134(22):4073–4081. doi: 10.1242/dev.006858. [DOI] [PubMed] [Google Scholar]
- 43.Peiro S, et al. Snail1 transcriptional repressor binds to its own promoter and controls its expression. Nucleic Acids Res. 2006;34(7):2077–2084. doi: 10.1093/nar/gkl141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scanlon CS, Van Tubergen EA, Inglehart RC, D’Silva NJ. Biomarkers of epithelial-mesenchymal transition in squamous cell carcinoma. J Dent Res. 2013;92(2):114–121. doi: 10.1177/0022034512467352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carrozzino F, et al. Inducible expression of Snail selectively increases paracellular ion permeability and differentially modulates tight junction proteins. Am J Physiol Cell Physiol. 2005;289(4):C1002–C1014. doi: 10.1152/ajpcell.00175.2005. [DOI] [PubMed] [Google Scholar]
- 46.Martinez-Estrada OM, et al. The transcription factors Slug and Snail act as repressors of Claudin-1 expression in epithelial cells. Biochem J. 2006;394:449–457. doi: 10.1042/BJ20050591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whiteman EL, Liu CJ, Fearon ER, Margolis B. The transcription factor snail represses Crumbs3 expression and disrupts apico-basal polarity complexes. Oncogene. 2008;27(27):3875–3879. doi: 10.1038/onc.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clarke TB, Francella N, Huegel A, Weiser JN. Invasive bacterial pathogens exploit TLR-mediated downregulation of tight junction components to facilitate translocation across the epithelium. Cell Host Microbe. 2011;9(5):404–414. doi: 10.1016/j.chom.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gonzalez-Mariscal L, Tapia R, Chamorro D. Crosstalk of tight junction components with signaling pathways. Biochim Biophys Acta. 2008;1778(3):729–756. doi: 10.1016/j.bbamem.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 50.Chen Z, et al. MAP kinases. Chem Rev. 2001;101(8):2449–2476. doi: 10.1021/cr000241p. [DOI] [PubMed] [Google Scholar]
- 51.Mori T, Wang X, Aoki T, Lo EH. Downregulation of matrix metalloproteinase-9 and attenuation of edema via inhibition of ERK mitogen activated protein kinase in traumatic brain injury. J Neurotrauma. 2002;19(11):1411–1419. doi: 10.1089/089771502320914642. [DOI] [PubMed] [Google Scholar]
- 52.Andras IE, et al. Signaling mechanisms of HIV-1 Tat-induced alterations of claudin-5 expression in brain endothelial cells. J Cereb Blood Flow Metab. 2005;25(9):1159–1170. doi: 10.1038/sj.jcbfm.9600115. [DOI] [PubMed] [Google Scholar]
- 53.Pu H, et al. HIV-1 Tat protein-induced alterations of ZO-1 expression are mediated by redox-regulated ERK 1/2 activation. J Cereb Blood Flow Metab. 2005;25(10):1325–1335. doi: 10.1038/sj.jcbfm.9600125. [DOI] [PubMed] [Google Scholar]
- 54.Shin JA, Kim YA, Jeong SI, Lee KE, Kim HS, Park EM. Extracellular signal-regulated kinase1/2-dependent changes in tight junctions after ischemic preconditioning contributes to tolerance induction after ischemic stroke. Brain Struct Funct. 2015;220(1):13–26. doi: 10.1007/s00429-013-0632-5. [DOI] [PubMed] [Google Scholar]
- 55.Walsh SV, Hopkins AM, Nusrat A. Modulation of tight junction structure and function by cytokines. Adv Drug Deliv Rev. 2000;41(3):303–313. doi: 10.1016/S0169-409X(00)00048-X. [DOI] [PubMed] [Google Scholar]
- 56.Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21(23):8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lomeli H, Starling C, Gridley T. Epiblast-specific Snai1 deletion results in embryonic lethality due to multiple vascular defects. BMC Res Notes. 2009;2:22. doi: 10.1186/1756-0500-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119(4):1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- 59. Wessels MR, Benedi V-J, Kasper DL, Heggen LM, Rubens CE. Type III capsule and virulence of group B Streptococci. In: Dunny GM, PP Cleary PP, McKay LL, eds. Genetics and molecular biology of Streptococci, Lactococci, and Enterococci. Washhington, DC, USA: American Society for Microbiology; 1991:219–223. [Google Scholar]
- 60.Wilkinson HW. Nontypable group B streptococci isolated from human sources. J Clin Microbiol. 1977;6(2):183–184. doi: 10.1128/jcm.6.2.183-184.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wessels MR, et al. Stimulation of protective antibodies against type Ia and Ib group B streptococci by a type Ia polysaccharide-tetanus toxoid conjugate vaccine. Infect Immun. 1993;61(11):4760–4766. doi: 10.1128/iai.61.11.4760-4766.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glaser P, et al. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol Microbiol. 2002;45(6):1499–1513. doi: 10.1046/j.1365-2958.2002.03126.x. [DOI] [PubMed] [Google Scholar]
- 63.Crisel RM, Baker RS, Dorman DE. Capsular polymer of Haemophilus influenzae, type b. I. Structural characterization of the capsular polymer of strain Eagan. J Biol Chem. 1975;250(13):4926–4930. [PubMed] [Google Scholar]
- 64.Nizet V, et al. Invasion of brain microvascular endothelial cells by group B streptococci. Infect Immun. 1997;65(12):5074–5081. doi: 10.1128/iai.65.12.5074-5081.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Equils O, Moffatt-Blue C, Ishikawa TO, Simmons CF, Ilievski V, Hirsch E. Pretreatment with pancaspase inhibitor (Z-VAD-FMK) delays but does not prevent intraperitoneal heat-killed group B Streptococcus-induced preterm delivery in a pregnant mouse model. Infect Dis Obstet Gynecol. 2009;2009:749432. doi: 10.1155/2009/749432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han SP, et al. SNAI1 is Involved in the proliferation and migration of glioblastoma cells. Cell Mol Neurobiol. 2011;31(3):489–496. doi: 10.1007/s10571-010-9643-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fink C, et al. Altered expression of ZO-1 and ZO-2 in Sertoli cells and loss of blood-testis barrier integrity in testicular carcinoma in situ. Neoplasia. 2006;8(12):1019–1027. doi: 10.1593/neo.06559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benedicto I, et al. The tight junction-associated protein occludin is required for a postbinding step in hepatitis C virus entry and infection. J Virol. 2009;83(16):8012–8020. doi: 10.1128/JVI.00038-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rowe RG, et al. Mesenchymal cells reactivate Snail1 expression to drive three-dimensional invasion programs. J Cell Biol. 2009;184(3):399–408. doi: 10.1083/jcb.200810113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clelland ES, Kelly SP. Tight junction proteins in zebrafish ovarian follicles: stage specific mRNA abundance and response to 17β-estradiol, human chorionic gonadotropin, and maturation inducing hormone. Gen Comp Endocrinol. 2010;168(3):388–400. doi: 10.1016/j.ygcen.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 71.Song S, Eckerle S, Onichtchouk D, Marrs JA, Nitschke R, Driever W. Pou5f1-dependent EGF expression controls E-cadherin endocytosis, cell adhesion, and zebrafish epiboly movements. Dev Cell. 2013;24(5):486–501. doi: 10.1016/j.devcel.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hilton BJ, Wolkowicz R. An assay to monitor HIV-1 protease activity for the identification of novel inhibitors in T-cells. PLoS One. 2010;5(6):e10940. doi: 10.1371/journal.pone.0010940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li W, Liu C, Tang Y, Li H, Zhou F, Lv S. Overexpression of Snail accelerates adriamycin induction of multidrug resistance in breast cancer cells. Asian Pac J Cancer Prev. 2011;12(10):2575–2580. [PubMed] [Google Scholar]
- 74.Kafri T, van Praag H, Ouyang L, Gage FH, Verma IM. A packaging cell line for lentivirus vectors. J Virol. 1999;73(1):576–584. doi: 10.1128/jvi.73.1.576-584.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.