Abstract
Type 2 diabetes (T2D) is characterized by insulin resistance and increased hepatic glucose production, yet the molecular mechanisms underlying these abnormalities are poorly understood. MicroRNAs (miRs) are a class of small, noncoding RNAs that have been implicated in the regulation of human diseases, including T2D. miR-26a is known to play a critical role in tumorigenesis; however, its function in cellular metabolism remains unknown. Here, we determined that miR-26a regulates insulin signaling and metabolism of glucose and lipids. Compared with lean individuals, overweight humans had decreased expression of miR-26a in the liver. Moreover, miR-26 was downregulated in 2 obese mouse models compared with control animals. Global or liver-specific overexpression of miR-26a in mice fed a high-fat diet improved insulin sensitivity, decreased hepatic glucose production, and decreased fatty acid synthesis, thereby preventing obesity-induced metabolic complications. Conversely, silencing of endogenous miR-26a in conventional diet–fed mice impaired insulin sensitivity, enhanced glucose production, and increased fatty acid synthesis. miR-26a targeted several key regulators of hepatic metabolism and insulin signaling. These findings reveal miR-26a as a regulator of liver metabolism and suggest miR-26a should be further explored as a potential target for the treatment of T2D.
Keywords: Endocrinology, Genetics, Metabolism
Introduction
Type 2 diabetes (T2D) is characterized by insulin resistance and abnormally elevated hepatic glucose production (HGP). Although many genetic and physiological factors interact to produce and aggravate insulin resistance, a common underlying mechanism is dysregulated insulin signaling by insulin receptor substrates (IRS1 and IRS2) that are inappropriately phosphorylated on serine residues (1). Many insulin-regulated Ser/Thr kinases, such as S6K1, mTOR1, and GSK3β, inhibit the function of IRS proteins. For example, T2D patients have elevated expression and activity of GSK3β in smooth muscles; conversely, Gsk3b knockout mice exhibit increased glucose homeostasis and increased insulin sensitivity (2).
Lipids are associated with insulin signaling, and ectopic lipid accumulation is proposed to be the root cause of insulin resistance in liver and muscle (3, 4). Fatty acids are a major class of lipids in mammalian cells, and dysregulation of fatty acid metabolism is a key event responsible for insulin resistance. Acyl-CoA synthetase long-chain (ACSLs) family proteins are essential enzymes for cellular fatty acid metabolism that catalyze the initial step of acyl-CoA formation from long-chain fatty acids. ACSL3 and ACSL4, 2 ACSLs that are abundant in the liver, have been implicated in the development of hepatic insulin resistance (5, 6). In addition, ectopic lipid accumulation can activate PKC family members, which consequently affect cellular events through kinase activity (3). For example, PKCs, such as PKCδ and PKCθ, phosphorylate IRS proteins and impair insulin signaling upon sensing the accumulation of lipids.
Another hallmark of T2D is excessive HGP, which primarily results from sustained gluconeogenesis (7). The rate of gluconeogenesis is largely determined by 3 rate-limiting gluconeogenic enzymes: phosphoenolpyruvate carboxykinase (PCK1), fructose-1,6-bisphosphatase (FBP1), and glucose-6-phosphatase (G6PC). PCK1 catalyzes the first committed step in gluconeogenesis, and its transcription is regulated by several transcription factors involved in nutrient and hormone signals, notably glucagon and glucocorticoids, and the opposing hormone insulin (8). Silencing of hepatic PCK1 in db/db mice improves their control of glycemia, insulin sensitivity, and dyslipidemia, consistent with the function of PCK1 in gluconeogenesis (9). In addition to these gluconeogenic enzymes, the WNT-signaling effector TCF7L2 was recently identified as a critical regulator of HGP and glucose homeostasis (10). In mice, loss of Tcf7l2 through either global knockout or liver-specific inducible depletion is associated with hypoglycemia and reduced HGP. Conversely, overexpression of TCF7L2 in the liver leads to enhanced HGP and glucose intolerance.
Metabolic disease and malignancy are thought to share common biological mechanisms. Metabolic reprogramming is proposed to be a core hallmark of cancer (11). Likewise, many T2D-associated genes are involved in tumorigenesis or cell-cycle regulation (12, 13). MicroRNAs (miRs) are emerging as new regulators of metabolism and malignancy during development and disease (14–16). Many cancer-associated miRs, such as miR-122 (17), miR-103/107 (18), and miR-143 (19), have been identified as critical regulators of metabolic disease. More recently, it was shown that obesity-induced overexpression of miR-802 impairs glucose homeostasis and attenuates insulin sensitivity (20), although the function of this miR in tumorigenesis remains unclear. Therefore, it is of interest to investigate the potential role of cancerous miRs in metabolic disease.
The miR-26 family, which is composed of miR-26a and miR-26b, plays crucial roles in tumorigenesis by targeting critical regulators involved in development, cell cycle, and differentiation (21–24). In particular, miR-26a acts as a potent tumor suppressor in the liver, the central organ involved in maintaining glucose and lipid homeostasis. Despite the significance of miR-26a in cancer, its function in metabolism and metabolic disease is unknown.
In this report, we show that miR-26a prevents obesity-induced insulin resistance, excessive lipid synthesis, and elevated HGP. miR-26a targets genes encoding factors involved in insulin signaling (GSK3β, PKCδ, and PKCθ), fatty acid metabolism (ACSL3 and ACSL4), and gluconeogenesis (PCK1 and TCF7L2). Taken together, these findings reveal critical roles for miR-26a in liver metabolism and suggest it has potential as a therapeutic target for improving insulin sensitivity and inhibiting lipogenesis and gluconeogenesis, thereby preventing the development of T2D in humans with diet-induced obesity (DIO).
Results
miR-26a expression is reduced in obese mice and humans.
Given the biological importance of miR-26 in hepatocarcinogenesis (21, 22, 24) and the critical role of the liver in controlling glucose and lipid metabolism, we speculated that miR-26 might have a role in T2D. miR-26a transcripts were much more abundant than miR-26b transcripts in the liver (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI75438DS1), consistent with recent reports (21, 25). Therefore, we chose miR-26a for further analysis. Gene ontology analysis on miR-26a target genes revealed a significant enrichment of regulators involved in metabolic processes (Supplemental Figure 2), supporting the idea that miR-26a may have a role in metabolic disease.
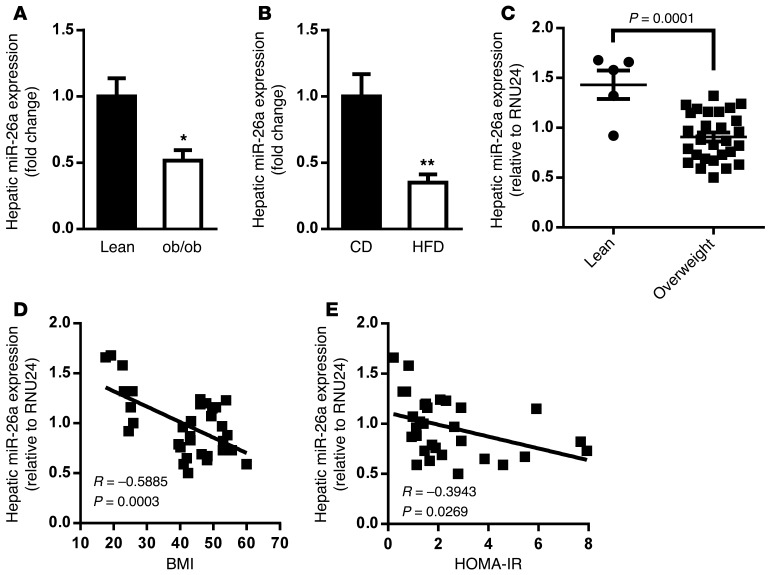
We first examined miR-26a expression in the genetic obese leptin-deficient (ob/ob) mouse model. Quantitative reverse-transcriptase PCR (QRT-PCR) analysis showed that miR-26a expression was slightly less in various obesity-associated organs of ob/ob mice than in lean mice (Supplemental Figure 3). Interestingly, miR-26a expression was significantly downregulated in the livers of ob/ob mice compared with control lean mice (Figure 1A). This reduction of hepatic miR-26a was recapitulated in a DIO mouse model, for which WT mice were fed a high-fat diet (HFD) for 16 weeks as compared with mice fed a standard chow diet (CD) (Figure 1B).
Figure 1. miR-26a expression is reduced in obese mice and humans.
(A) Expression of miR-26a in livers of WT lean and ob/ob mice (n = 4–6). (B) Expression of miR-26a in livers of WT DIO mice fed a CD or a HFD (n = 5). (C) QRT-PCR analysis of human miR-26a expression in livers of lean (n = 5) and obese (n = 28, BMI > 25) individuals. (D) Correlation between hepatic miR-26a levels and BMI in a human cohort (n = 33). (E) Correlation between hepatic miR-26a levels and HOMA-IR in a human cohort (n = 33). miR-26a expression is normalized to 5S rRNA in mice and RNU24 in humans. Data are shown as mean ± SEM. *P < 0.05; **P < 0.01, 2-tailed Student’s t test.
We saw similar results for liver expression of miR-26a in a cohort of humans. miR-26a levels were markedly reduced in overweight (BMI > 25) compared with lean individuals (Figure 1C), and hepatic miR-26a expression significantly correlated with BMI (Figure 1D). Furthermore, the subjects’ homeostatic model assessment index of insulin resistance (HOMA-IR) negatively correlated with hepatic miR-26a expression (Figure 1E), indicating an association of miR-26a with insulin resistance. Expression of CTDSPL and CTDSP2, the host genes for miR–26a-1 and miR–26a-2, respectively, was unchanged in both the genetic and HFD-induced obesity mouse models (Supplemental Figure 4), indicating that miR-26a itself might be regulated posttranscriptionally, as reported for other miRs (26).
Improved glucose homeostasis and insulin sensitivity in miR-26a Tg mice.
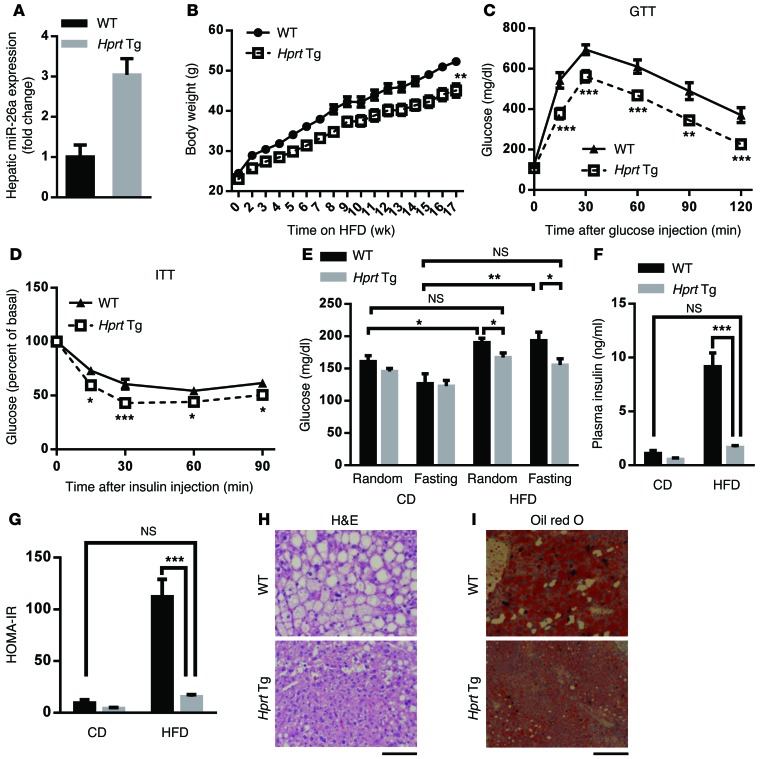
We investigated the function of miR-26a in vivo by using our recently established miR-26a Tg mouse line (27). Briefly, a genomic DNA fragment encoding the Mir26a1 locus, preceded by the synthetic CAG promoter and a loxP-flanked Neo-STOP cassette, was inserted into the Rosa26 locus to generate Tg(CAG-Neo-STOPfl-Mir26a1) mice. We then crossed Tg(CAG-Neo-STOPfl-Mir26a1) mice with Hprt-Cre Tg mice, which express CRE recombinase during the very early cleavage stage of embryogenesis, resulting in miR-26a1 induction in HPRT-expressing cells (herein referred to as Hprt-Mir26a Tg mice). miR-26a expression in livers of Hprt-Mir26a Tg mice was about 3-fold greater than that of WT controls (Figure 2A). This increased expression of miR-26a in Hprt-Mir26a Tg mice was comparable to the amount of downregulation of miR-26a seen in ob/ob or DIO mice (Figure 1, A and B), suggesting Hprt-Mir26a Tg mice provide an excellent model for characterizing the effects of miR-26a restoration on obesity-associated metabolic syndrome.
Figure 2. Global overexpression of miR-26a prevents obesity-induced insulin resistance, glucose tolerance, and lipid accumulation.
(A) QRT-PCR analysis of miR-26a expression in liver of Hprt-Mir26a Tg (Hprt Tg) mice and WT littermate controls (n = 4). (B–I) Mice were fed a HFD beginning at 6 to 8 weeks of age. Measurements were performed during the course of the HFD, as shown below. (B) Total BW (n = 10–13). (C) GTT, performed after 17 weeks of HFD (n = 7–8). (D) ITT, performed after 18 weeks of HFD (n = 8). (E and F) Blood glucose (E) and insulin (F) levels of mice that were fed with either CD or HFD for 18 weeks. Random or fasting conditions are noted (n = 6–10). (G) HOMA-IR, as calculated from fasting blood glucose and insulin levels (n = 5–8). (H and I) Representative H&E staining (H) and oil red O staining (I) of paraffin sections from livers of Tg and WT mice fed a HFD (n = 6). Scale bars: 100 μm. Data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005, 2-tailed ANOVA (B) and 2-tailed Student’s t test (C–G).
HPRT-Tg and WT mice of 8 to 12 weeks of age fed a CD had no significant differences in body weight (BW), organ weight, glucose disposal, or insulin sensitivity (Supplemental Figure 5). However, when 6- to 8-week-old Hprt-Mir26a Tg and WT mice were fed a HFD, Hprt-Mir26a Tg mice had significantly lower BWs than WT controls (Figure 2B). Furthermore, Hprt-Mir26a Tg mice had significantly better glucose tolerance and insulin sensitivity than did WT controls, as analyzed by the glucose tolerance test (GTT) and the insulin tolerance test (ITT), respectively (Figure 2, C and D). Hprt-Mir26a Tg mice fed a HFD showed significantly lower random and fasting blood-glucose levels as well as insulin levels, although no differences were detected between age-matched Hprt-Mir26a Tg mice and WT controls fed a CD (Figure 2, E and F). Consistent with this, the HOMA-IR was dramatically lower in Hprt-Mir26a Tg mice (Figure 2G). Notably, glucose levels, insulin levels, and HOMA-IR in Hprt-Mir26a Tg mice fed a HFD were comparable to those of age-matched WT controls fed a CD (Figure 2, E–G), indicating that miR-26a restoration is sufficient to increase insulin sensitivity and rescue features of obesity-associated metabolic syndrome.
Histological analyses further supported this idea and revealed that Hprt-Mir26a Tg mice had significantly less obesity-induced hepatic steatosis and lipid accumulation (Figure 2, H and I). Hprt-Mir26a Tg mice also had smaller adipocytes in visceral adipose tissue (VAT) and subcutaneous adipose tissue (SAT) than WT controls (Supplemental Figure 6A), further supporting an improvement of insulin sensitivity in those mice. No obvious histological differences were detected in other obesity-associated tissues, including brown adipose tissue, heart, and muscle (Supplemental Figure 6B). Taken together, these results suggest a causal role of miR-26a in obesity-associated insulin resistance, lipid accumulation, and glucose tolerance.
Liver-specific miR-26a Tg mice have improved glucose homeostasis and insulin action.
Given that the liver is the major organ controlling glucose and lipid metabolism and plays an important role in insulin resistance, we hypothesized that hepatic overexpression of miR-26a may be the primary contributor to the metabolic features seen in Hprt-Mir26a Tg mice. We therefore generated liver-specific miR-26a Tg mice (hereafter referred to as Alb-Mir26a Tg) by crossing Tg(CAG-Neo-STOPfl-Mir26a1) mice (WT controls) with Alb-Cre Tg mice that expressed CRE recombinase selectively in hepatocytes (28). miR-26a was specifically upregulated in the hepatocytes of Alb-Mir26a Tg mice (Supplemental Figure 7A). Alb-Mir26a Tg mice were viable, fertile, born at the expected frequency, and morphologically indistinguishable from their WT control littermates. When fed a CD, Alb-Mir26a Tg and WT mice showed no significant differences in BW, glucose disposal, or insulin sensitivity (Supplemental Figure 7, B–D).
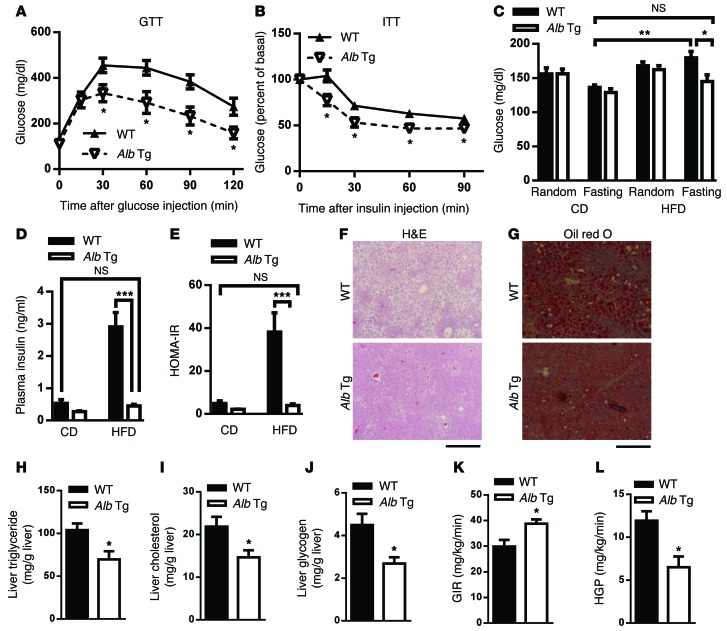
We also fed mice of both genotypes HFDs for 14 weeks. On a HFD, Alb-Mir26a Tg mice had BWs similar to those of WT controls (Supplemental Figure 8A), suggesting that hepatocyte-specific overexpression of miR-26a is not sufficient to prevent obesity. GTT analysis revealed that Alb-Mir26a Tg mice had significantly better glucose tolerance than did WT controls (Figure 3A). Insulin levels for mice of both genotypes were similar during GTT analysis (Supplemental Figure 8B), suggesting that improved glucose metabolism in Alb-Mir26a Tg mice results from improved insulin sensitivity, not increased insulin secretion. ITT analysis revealed that insulin resistance was also significantly ameliorated in Alb-Mir26a Tg mice (Figure 3B). On HFDs, fasting glucose and insulin levels, as well as HOMA-IR, were significantly lower in Alb-Mir26a Tg mice than in WT mice, although no significant differences in these measurements were seen between both genotypes fed a CD (Figure 3, C–E). Strikingly, the levels of these parameters in Alb-Mir26a Tg mice fed a HFD were comparable to those of WT controls fed a CD, which recapitulated the phenotype of Hprt-Mir26a Tg mice (Figure 2, C–G).
Figure 3. Hepatic overexpression of miR-26a prevents obesity-induced insulin resistance, glucose tolerance, and lipid accumulation.
Mice were fed a HFD beginning at 6 to 8 weeks of age. Measurements were performed during the course of the HFD, as shown below. (A) GTT, after 14 weeks of HFD (n = 4–5). (B) ITT, after 15 weeks of HFD (n = 6–9). (C and D) Blood glucose (C) and insulin (D) levels of mice kept either on CD or HFD for 15 weeks. Random or fasting conditions are noted (n = 6–9). (E) HOMA-IR, as calculated from fasting blood glucose and insulin levels (n = 6–9). (F and G) Representative H&E staining (F) and oil red O staining (G) of paraffin sections from livers of Alb-Mir26a Tg (Alb Tg) and WT mice fed HFD (n = 6). Scale bars: 100 μm. (H to J) Hepatic triglyceride (H), hepatic cholesterol (I) and fasted hepatic glycogen (J) were measured (n = 6). (K and L) Hyperinsulinemic-euglycemic clamp, after HFD. GIR (K) and HGP (L) were assessed during clamp (n = 4–6). Data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005, 2-tailed Student’s t test.
H&E analyses of liver tissues revealed that Alb-Mir26a Tg mice had less obesity-induced hepatic steatosis, reduced lipid accumulation, and smaller adipocytes in VAT and SAT (Figure 3, F and G, and Supplemental Figure 8C). In contrast, we did not detect any obvious histological and morphological differences in other obesity-associated tissues of Alb-Mir26a Tg mice (Supplemental Figure 8D). Consistent with this observation, the hepatic triglyceride levels were significantly lower in Alb-Mir26a Tg mice (Figure 3H). Hepatic cholesterol and glycogen levels in Alb-Mir26a Tg mice were also reduced (Figure 3, I and J).
To critically determine whether miR-26a overexpression improves insulin sensitivity and decreases HGP, Alb-Mir26a Tg mice and WT controls fed a HFD were subjected to hyperinsulinemic euglycemic clamp. Alb-Mir26a Tg mice had a significant increase in glucose infusion rate (GIR) (Figure 3K), demonstrating improved insulin sensitivity. HGP was markedly repressed in Alb-Mir26a Tg mice (Figure 3L). The difference in GIR could be mainly attributed to the insulin reduction of HGP because glucose disposal rate (GDR) remains similar in both groups (Supplemental Figure 9). These data indicate that miR-26a could improve insulin sensitivity mainly through the suppression of de novo glucose production.
Taken together, these results recapitulate the metabolic phenotype of Hprt-Mir26a Tg mice, suggesting that hepatic overexpression of miR-26a is sufficient to abolish DIO-induced insulin resistance and to rescue glucose homeostasis.
miR-26a increases insulin-stimulated AKT activation.
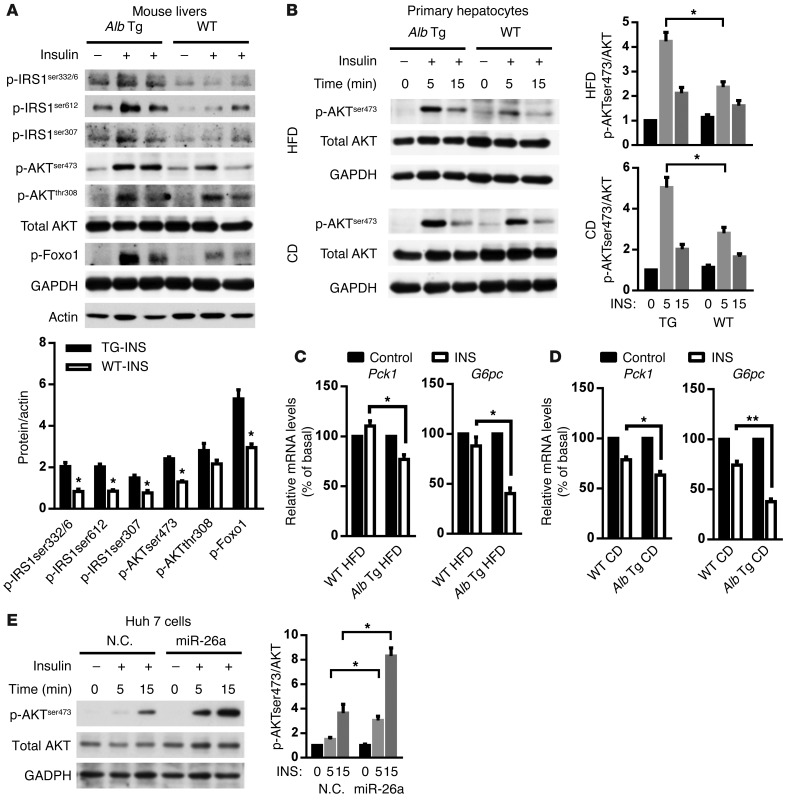
As shown above, both Hprt- and Alb-Mir26a Tg mice are resistant to obesity-induced insulin resistance. To further explore this, we directly assessed the effect of miR-26a on insulin signaling in mouse livers stimulated with insulin. Consistent with higher insulin sensitivity, Hprt- and Alb-Mir26a mice had greater responsiveness to insulin stimulation compared with obese WT control mice (Figure 4A and Supplemental Figure 10). A cell-intrinsic improvement in the ability of insulin to stimulate AKT activity was confirmed in primary hepatocytes from Alb-Mir26a Tg mice fed either a HFD or a CD (Figure 4B). In line with increased insulin signaling, overexpression of miR-26a in mice fed either a HFD or a CD increased the suppressive effects of insulin on gluconeogenesis, as demonstrated by reduced expression of Pck1 and G6pc (Figure 4, C and D).
Figure 4. miR-26a stimulates insulin signaling.
(A) Insulin signaling in livers of Alb-Mir26a Tg mice and WT littermate controls infused with insulin (0.25 U/kg) through the portal vein and fed a HFD for 16 weeks. Results are representative of 2 separate experiments. ImageJ quantification of the phospho-protein/AKT ratio is shown. (B and C) AKT phosphorylation in primary hepatocytes isolated from Alb-Mir26a Tg mice and WT littermate controls fed a HFD for 18 weeks (upper panels) or fed a CD (lower panels) and stimulated with insulin (10 nM) for the indicated times. Results are representative of 3 separate experiments. ImageJ quantification of the pAKT/AKT ratio is shown. (C) Expression of Pck1 and G6pc in primary hepatocytes isolated from Alb-Mir26a Tg mice and WT littermate controls fed a HFD for 18 weeks and stimulated with insulin (10 nM) for 6 hours (n = 3). (D) Expression of Pck1 and G6pc in primary hepatocytes isolated from Alb-Mir26a Tg mice and WT littermate controls fed a CD and stimulated with insulin (10 nM) for 6 hours (n = 3). (E) AKT phosphorylation in Huh7 cells transfected by miR-26a and stimulated with insulin (100 nM) for the indicated times. Results are representative of 3 separate experiments. ImageJ quantification of the pAKT/AKT ratio is shown. Data are shown as mean ± SEM. *P < 0.05; **P < 0.01, 2-tailed Student’s t test.
We further assessed the effect of miR-26a on insulin-stimulated AKT activity in cultured human Huh7 hepatoma cells. Huh7 cells transfected with miR-26a (28-fold increase in expression) had increased AKT activity after insulin stimulation (Figure 4E). Together, these results demonstrate that miR-26a improves insulin sensitivity.
miR-26a regulates fatty acid metabolism.
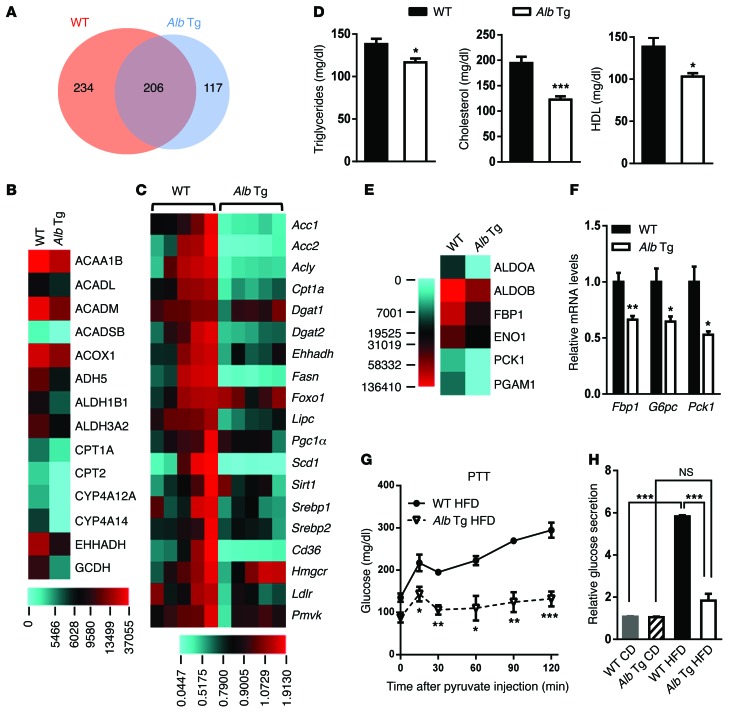
To elucidate mechanisms underlying the increased insulin sensitivity and improved glucose tolerance caused by miR-26a overexpression, we performed a large-scale proteomic analysis using quantitative mass spectrometry (MS). Total proteins were extracted from the livers of WT and Alb-Mir26a Tg mice fed a HFD for 16 weeks, cleaved into peptides with trypsin, and analyzed by ultra-performance liquid chromatography–electrospray tandem MS (UPLC-ESI-MS/MS). These experiments identified 557 proteins, of which 351 were differentially expressed between WT and Alb-Mir26a Tg mice (>1.5-fold) (Figure 5A and Supplemental Table 1). Gene ontology analysis of these dysregulated hepatic proteins revealed significant enrichment of regulators that participate in signal transduction and metabolism (Supplemental Figure 11 and Supplemental Table 2), supporting a role of miR-26a in metabolism. A total of 234 proteins were downregulated (>1.5-fold) in livers of Alb-Mir26a mice when compared with WT control mice, whereas 117 proteins were upregulated (>1.5-fold) (Supplemental Table 1). Pathway analyses on the 234 proteins downregulated in Alb-Mir26a mice revealed that a majority of these proteins are involved in metabolic processes (Supplemental Figure 12). Among the pathways analyzed, fatty acid metabolism ranked as the most significant dysregulated pathway, with 14 proteins having decreased expression in Alb-Mir26a Tg mice (Figure 5B and Supplemental Figure 12).
Figure 5. miR-26a regulates fatty acid metabolism and gluconeogenesis.
(A) Venn diagram comparing hepatic proteins in WT and Alb-Mir26a mice fed a HFD for 16 weeks, as identified by proteomic analysis. The numbers of proteins that are similarly or differentially expressed are shown. (B) Heat map of protein levels of genes involved in fatty acid metabolism, as indicated in Supplemental Figure 12. (C) Heat map of mRNA levels of genes involved in lipid and cholesterol metabolism. (D) Plasma triglycerides, cholesterol, and HDL levels in WT and Alb-Mir26a Tg mice fed a HFD. (E) Heat map of protein levels of genes involved in glycolysis/gluconeogenesis. (F) Expression of gluconeogenic genes in livers of Alb-Mir26a Tg and WT mice fed a HFD for 16 weeks. (G) PTT analysis of Alb-Mir26a Tg and WT mice fed a HFD for 16 weeks. (H) Glucose production in primary hepatocytes isolated from Alb-Mir26a Tg and WT mice fed either a CD or a HFD for 16 weeks. Results are normalized to the level in hepatocytes isolated from WT mice fed a CD. Data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005, 2-tailed Student’s t test.
We then evaluated mRNA levels of genes involved in fatty acid metabolism in livers of WT and Alb-Mir26a Tg mice fed a HFD. Consistent with the decreased protein levels, levels of Ehhadh and Cpt1a mRNAs were reduced in Alb-Mir26a Tg mice (Figure 5C). Moreover, expression of several key genes that regulate fatty acid synthesis and oxidation, including Acc1, Acc2, Acly, Dgat2, Fasn, Lipc, Scd1, and Srebf1, was also markedly downregulated in Alb-Mir26a Tg mice. This suggests that both hepatic fatty acid synthesis and oxidation are decreased in Alb-Mir26a Tg mice. Additionally, expression of Cd36 and Ldlr, 2 important regulators of cholesterol metabolism, was also decreased in Alb-Mir26a Tg mice. In WT mice, the 16-week HFD significantly induced hepatic expression of critical regulators of fatty acid metabolism, such as Acc1, Acly, Ehhadh, Fasn, and Scd1 (Supplemental Figure 13A). Notably, these increases did not occur in Alb-Mir26a Tg mice, suggesting miR-26a has a substantial role in maintaining normal fatty acid metabolism under diabetic conditions.
Because miR-26a regulates the hepatic gene-expression profile for lipid and cholesterol metabolism (Figure 5C and Supplemental Figure 13A), we examined the physiological outcome of miR-26a overexpression in mice. Plasma triglycerides, cholesterol, and HDL concentrations were significantly lower in Alb-Mir26a Tg mice than in WT mice (Figure 5D). Taken together, these results demonstrate that hepatic overexpression of miR-26a is sufficient to prevent obesity-induced abnormal metabolism of hepatic lipids and cholesterol.
miR-26a regulates gluconeogenesis.
In addition to elevated lipid metabolism, WT mice had significant upregulation of glycolysis/gluconeogenesis (Supplemental Figure 12). Levels of 9 proteins, as detected by proteomic analysis, were higher in WT mice than in Alb-Mir26a Tg mice (Figure 5, B and E). Increased HGP in T2D primarily results from gluconeogenesis, which is largely controlled by 3 rate-limiting enzymes: FBP1, G6PC, and PCK1. QRT-PCR analysis revealed that expression of the genes encoding these 3 genes was significantly reduced in Alb-Mir26a Tg mice (Figure 5F). Accordingly, Alb-Mir26a Tg mice had decreased glucose production during an intraperitoneal pyruvate tolerance test (PTT) (Figure 5G). These data demonstrate that hepatic overexpression of miR-26a inhibits obesity-induced HGP in vivo.
To determine whether the effect of miR-26a is cell autonomous, we performed a glucose production assay on primary hepatocytes isolated from WT and Alb-Mir26a Tg mice fed either a CD or a HFD. Overexpression of miR-26a did not affect cellular glucose production in hepatocytes from mice fed a CD, but it significantly decreased glucose production in hepatocytes from mice fed a HFD (Figure 5H and Supplemental Figure 13B). Although the HFD resulted in about a 5-fold increase in glucose production in hepatocytes from WT mice, the increase was much less in hepatocytes from Alb-Mir26a Tg mice. Strikingly, the glucose levels in hepatocytes from miR-26a Tg mice fed a HFD were comparable to those of cells from mice fed a CD, suggesting that overexpression of miR-26a in hepatocytes is sufficient to prevent obesity-induced HGP.
We then determined the effect of miR-26a on fasting-induced gluconeogenesis in vivo. After an overnight fast (16 hours), Alb-Mir26a Tg mice had reduced expression of glucogenic genes (Pck1, G6pc) when compared with WT mice (Supplemental Figure 13C). Furthermore, treatment of mouse primary hepatocytes with dexamethasone (DEX), which mimics fasting signals, resulted in increased expression of G6pc and Pck1 in control cells; however, those increases were much less in miR-26a–overexpressing cells (Supplemental Figure 13D). Taken together, these data demonstrate that miR-26a suppresses gluconeogenesis, indicating that decreased gluconeogenesis may be the primary cause of the reduced glucose levels in Alb-Mir26a Tg mice fed a HFD.
miR-26a targets genes that regulate insulin signaling, lipid metabolism, and glucose metabolism.
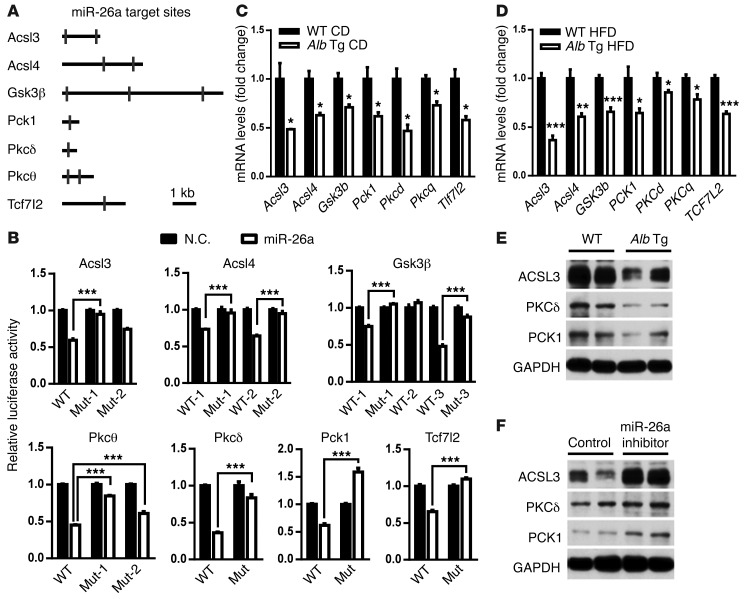
As described above, miR-26a controls insulin signaling, lipid metabolism, and glucose metabolism. We therefore investigated how the large repertoire of miR-26a target genes might contribute to the control of miR-26a in the liver. To search for potential targets, we performed gene ontology and biological association analyses using Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources (29) and looked for enrichment of specific target genes associated with insulin signaling and glucose and lipid metabolism. Several genes involved in these processes have predicted targets for miR-26a, including Pkcd, Pkcq, Gsk3b, Pten, Acsl3, Acsl4, Pck1, and Tcr7l2 (Figure 6A and Supplemental Figures 14 and 15). In addition to Pten, 7 genes that we believe to be novel miR-26a targets were experimentally verified. We generated reporter constructs in which the luciferase coding sequence was fused to the 3′ UTRs of these genes. miR-26a markedly repressed the luciferase activity of the Acsl3, Acsl4, Gsk3b, Pkcd, Pkcq, Pck1, and Tcf7l2 constructs (Figure 6B). Mutation of the miR-26a target sites abrogated miR-26a–associated repression in luciferase activity, suggesting a direct interaction of miR-26a with these sites.
Figure 6. miR-26a targets several genes critical for glucose metabolism, lipid metabolism, and insulin signaling.
(A) Schematic of mouse Acsl3, Acsl4, Gsk3b, Pck1, Pkcd, Pkcq, and Tcf7l2 3′ UTRs. Locations of the predicted miR-26a–binding sites are indicated. (B) Relative luciferase activity in HEK293 cells transfected with reporter constructs containing the 3′ UTR of target genes and cotransfected with either miR-26a mimics (miR-26a) or negative control (N.C.). Mut, mutant. (C) Expression of miR-26a target genes in the livers of Alb-Mir26a Tg and WT mice fed a CD. (D) mRNA levels of miR-26a target genes in livers of Alb-Mir26a Tg and WT littermate controls fed a HFD for 16 weeks. (E and F) Protein levels of selected miR-26a target genes in livers of Alb-Mir26a Tg and WT littermate controls fed a HFD for 16 weeks (E) and in Alb-Mir26a Tg mice fed a HFD for 16 weeks and treated with PBS (control) or LNA–miR-26a antisense (F). All experiments were performed in triplicate, and data are shown as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005, 2-tailed Student’s t test.
To investigate the effect of miR-26a on endogenous expression of these targets, we determined their expression in the livers of WT and Alb-Mir26a Tg mice fed a CD. QRT-PCR analysis revealed that expression was significantly lower in Alb-Mir26a Tg mice than in WT mice (Figure 6C). Similarly, expression of these miR-26a target genes was significantly reduced in Alb-Mir26a Tg mice fed a HFD for 16 weeks compared with WT littermate controls also fed the HFD (Figure 6D). Consistently, levels of ACSL3, PKCδ, and PCK1 proteins were decreased in Alb-Mir26a Tg mice (Figure 6E), but increased in locked nucleic acid–miR-26a (LNA–miR-26a) antisense–treated mice (Figure 6F). These data demonstrate that miR-26a directly targets several key genes crucial for insulin signaling and metabolism of lipid and glucose, including Acsl3, Acsl4, Gsk3b, Pkcd, Pkcq, Pck1, and Tcf7l2.
Inhibition of miR-26a in mice fed a CD.
On the basis of the above findings, we hypothesized that loss of miR-26a might lead to augmented lipid biosynthesis, increased glucose production, and decreased insulin sensitivity. Three distinct miR-26 loci (Mir26a1, Mir26a2, and Mir26ab) within the mouse genome render genetic loss of function difficult (27). Previous studies in mice and nonhuman primates have supported the feasibility of LNA antisense approaches to modulating miR pathways that have important physiological consequences in vivo (30, 31). We therefore intraperitoneally injected CD-fed WT mice with PBS (vehicle) or LNA–miR-26a antisense (miR-26a inhibitor). Forty-eight hours after a single injection of miR-26a inhibitor (6 mg/kg BW), mice were sacrificed and hepatic gene expression was determined. QRT-PCR analysis confirmed a 98% reduction in hepatic miR-26a levels in mice treated with LNA–miR-26a antisense when compared with PBS-treated mice (Supplemental Figure 16). Accordingly, miR-26a target genes (Acsl3, Acsl4, Gsk3b, Pck1, Pkcd, Pkcq, and Tcf7l2) were significantly induced in LNA–miR-26a antisense–treated mice, with Acsl4 being most sensitive to miR-26a inhibition (Supplemental Figure 16).
It has recently been shown that TCF7L2 plays a crucial role in hepatic metabolism, including lipid, glucose, and glycogen metabolism. Compared with PBS-treated mice, LNA–miR-26a antisense–treated mice had significantly increased expression of Tcf7l2 target genes, such as the well-known Wnt target gene axin 2, the liver-zonated gene Glul, and fasting response genes (Aldh3a2, Slc1a2, and St3gl5) (Supplemental Figure 16), suggesting that Tcf7l2 may be an important target gene mediating miR-26a function.
In line with enhanced expression of miR-26a target genes, expression of many key metabolic genes was also increased in LNA–miR-26a antisense–treated mice (Supplemental Figure 16). These genes are involved in gluconeogenesis (Fbp1, G6pc, and Pck1), lipid metabolism (Acc1, Acc2, Acly, Aldh3a2, Cpt1a, Dgat2, Ehhadh, Fasn, Ldlr, Lipc, and Srebf1), and glycogen metabolism (Gys2 and Pygl). Fasn, the rate-limiting enzyme for fatty acid synthesis, appeared most sensitive to miR-26a inhibition, as it was the most upregulated gene upon LNA–miR-26a antisense treatment.
Inhibition of miR-26a in DIO mouse model.
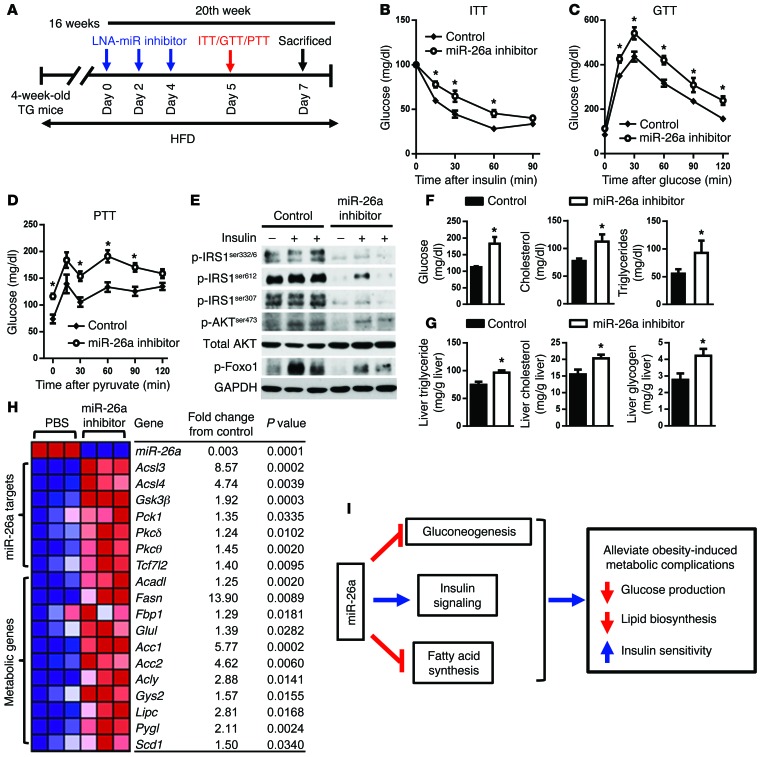
When fed a HFD, Alb-Mir26a Tg mice showed increased insulin sensitivity, decreased fatty acid synthesis, improved glucose tolerance, and decreased HGP (Figure 5). We tested to determine whether miR-26a inhibition in Alb-Mir26a Tg mice can abolish these effects. To this end, Alb-Mir26a Tg mice fed a HFD for 16 weeks were intraperitoneally injected with LNA–miR-26a antisense or PBS (Figure 7A). After 3 injections over 5 days, LNA–miR-26a antisense–treated mice had reduced insulin sensitivity (Figure 7B), decreased glucose tolerance (Figure 7C), and increased HGP (Figure 7D) when compared with PBS-treated mice. Consistent with this, LNA–miR-26a antisense–treated mice had decreased sensitivity to insulin compared with PBS-treated mice (Figure 7E). Levels of plasma glucose, total cholesterol, and triglycerides were significantly higher in LNA–miR-26a antisense–treated mice than in PBS-treated mice (Figure 7F). Hepatic triglyceride, cholesterol, and glycogen levels also were significantly increased in LNA–miR-26a antisense–treated mice (Figure 7G). These increases in glucose production and lipid synthesis in LNA–miR-26a antisense–treated mice were accompanied by augmented expression of miR-26a targets, glucogenic genes (Fbp1, Pck1, and Scd1), and lipogenic genes (Acadl, Acc1, Acc2, Acly, Fasn, Lipc, and Srebf1) (Figure 7H). In addition, miR-26a inhibition resulted in increased expression of Gys2 and Pygl, which are downstream of TCF7L2 and are involved in glycogen metabolism. These results strongly suggest that miR-26a inhibition in vivo contributes to insulin resistance and elevated HGP.
Figure 7. Silencing of miR-26a aggravates obesity-induced metabolic complications.
(A–H) Alb-Mir26a Tg mice were fed a HFD beginning at 4 weeks of age. Sixteen weeks later, mice received 3 intraperitoneal injections of LNA–miR-26a antisense or PBS over 5 days. (A) Protocol for LNA–miR-26a antisense delivery. (B–D) ITT (B), GTT (C), and PTT (D) analysis. (E) Insulin signaling in livers of mice infused with insulin (0.25 U/kg) through the portal vein. (F) Plasma glucose, cholesterol, and triglyceride levels. (G) Liver triglyceride, cholesterol, and glycogen levels. (H) Heat map of mRNA levels of hepatic genes. Red and blue depict higher and lower gene expression, respectively. Color intensity indicates magnitude of expression differences. (I) Proposed model for the role of miR-26a in known molecular pathways crucial for the development of T2D. Data are shown as mean ± SEM. *P < 0.05, 2-tailed Student’s t test.
Discussion
We have shown that miR-26a regulates liver metabolism and have implicated dysregulation of miR-26a in obesity-associated metabolic syndrome. miR-26a directly targets several key regulators involved in gluconeogenesis (TCF7L2 and PCK1), fatty acid synthesis (ACSL3 and ACSL4), and insulin signaling (GSk3β, PKCδ, PKCθ). These effects of miR-26a could be greatly consolidated and augmented by crosstalk between hepatic metabolism and insulin signaling, which suggests that a small change in miR-26a expression can sometimes have a large physiological effect (Figure 7I). In line with this idea, a minor decrease in miR-26a expression (about 2-fold) could contribute to the development of insulin resistance and T2D, whereas a modest increase in miR-26a expression in mice was sufficient to prevent obesity-associated complications. Our findings thus suggest that manipulating miR-26a expression could provide novel opportunities for treating obesity-associated metabolic syndrome.
Hepatic miR-26a is consistently markedly reduced in obese mice and overweight humans. We further demonstrated that miR-26a regulates insulin signaling and glucose and lipid metabolism. Notably, miR-26a overexpression in mice abolishes HFD-induced hyperinsulinemia and hyperglycemia and diminishes hyperlipidemia. These results strongly suggest that reduced miR-26a expression is a primary cause, not merely a consequence, of obesity-associated metabolic abnormalities. This causative role of miR-26a was further confirmed by silencing of miR-26a in mice, which impaired insulin sensitivity, attenuated glucose homeostasis, and increased fatty acid synthesis. In addition, Alb-Mir26a Tg mice exhibited an ability similar to that of Hprt-Mir26a Tg mice to prevent obesity-induced metabolic abnormalities, indicating a dominant role of hepatic miR-26a. That is, hepatic overexpression of miR-26a is sufficient to prevent features of obesity-associated metabolic syndrome.
In line with these metabolic changes, overexpression of miR-26a substantially decreases the mRNA levels of many key genes that regulate lipid and glucose metabolism. Conversely, silencing of miR-26a causes remarkable increases in expression of these genes. We identified 7 miR-26a targets that regulate lipid metabolism (ACSL3 and ACSL4), glucose metabolism (PCK1 and TCF7L2), and insulin signaling (PKCδ, PKCθ, and GSK3β). ACSL3 and ACSL4 are essential enzymes for fatty acid metabolism (32). PCK1, the rate-limiting enzyme for gluconeogenesis, and TCF7L2, the WNT signaling effector, are crucial for glucose metabolism, especially for HGP (9, 10). Lipids or other inducers of insulin resistance stimulate the activation of PKCδ, PKCθ, and GSK3β, which phosphorylate IRS proteins on Ser residues, thereby attenuating insulin signaling (3, 33, 34). miR-26a directly represses the expression of PKCδ, PKCθ, and GSK3β, which at least partially contribute to the positive roles of miR-26a in insulin signaling.
In addition to these identified miR-26a targets, many metabolic genes that lack predicted miR-26a target sites show altered expression upon modulation of miR-26a expression. These genes are involved in lipid metabolism (Acc1, Acc2, Acly, Cpt1, Dgat2, Ehhadh, Fasn, Lipc, and Srebf1), glucose metabolism (Fbp1, G6pc, and Scd1), cholesterol metabolism (Cd36, Ldlr, and Srebf1), and glycogen metabolism (Gys2 and Pyg1). It is likely that these differentially expressed genes act downstream of miR-26a target genes. Indeed, ACSL3 has been shown to enhance expression of lipogenic genes, including Acc2, Cd36, and Scd1 (5). PKCδ increases expression of gluconeogenic and lipogenic enzymes, including Acc1, Fasn, Fbp1, Gck, Mixipl, Pck1, Scd1, and Srebf1 (35). Acting more broadly, TCF7L2 directly binds and regulates generic WNT target genes and many metabolic genes involved in gluconeogenesis, transport of fatty acids, and synthesis of ketone bodies, including PCK1, G6PC, ACADL, and ALDH3A2 (10). Although we cannot exclude the possibility that other known (such as PTEN) and unknown target genes of miR-26a contribute to liver metabolism, we speculate that coordinated regulation of the 7 miR-26a target genes identified here could profoundly alter gene-expression profiling related to liver metabolism, especially as regards lipid biosynthesis and glucose production, thereby modulating obesity-associated metabolic complications. The molecular mechanism underlying miR-26a’s function in liver metabolism awaits further investigation. For example, it would be interesting to clarify the roles of critical proteins that mediate miR-26a’s function in metabolic pathways.
T2D is a complex metabolic disorder and accounts for about 90% of all diabetes deaths, which emphasizes the urgent need for new treatment strategies (36). Recently, miRs have gained considerable attention as therapeutic targets for human diseases, including diabetes (14, 37). For example, miR-122 has been identified as a key regulator of lipid metabolism, and an anti–miR-122 is in phase II clinical trials for the treatment of chronic hepatitis C virus (HCV) infection (38). We have found that miR-26a controls all 3 hallmarks of T2D, namely insulin resistance, excessive HGP, and elevated lipid synthesis. In addition to the multiple functions of miR-26a, it is important to note the following: (a) miR-26a is universally expressed at high levels in human tissues, except in the liver, which has the lowest miR-26a expression of any organ (39), indicating that modest overexpression of hepatic miR-26a could be safe; in support of this idea, physiological and pathological side effects of miR-26a overexpression were not observed in liver-specific miR-26a Tg mice of up to 2 years of age (data not shown); (b) global or hepatocyte-specific overexpression of miR-26a does not induce weight gain, circumventing the major side effect of increasing insulin sensitivity for diabetes therapies (40); and (c) miR-26a acts as a tumor suppressor and suppresses tumorigenesis and metastasis of many cancers, including melanoma and liver and prostate cancer (39). Thus, overexpressing miR-26a could decrease the risk of tumorigenesis. Given the potent role of miR-26a in liver metabolism as well as the above observations, miR-26a represents a promising target for the treatment of obesity-associated metabolic syndrome.
In summary, this study demonstrates the importance of hepatic miR-26a in glucose metabolism, lipid metabolism, and insulin signaling through regulation of critical metabolic genes. Downregulating miR-26a in obese mice contributes to obesity-induced metabolic abnormalities. Conversely, restoring miR-26a in mice prevents obesity-induced metabolic damage. These findings suggest miR-26a as a promising novel target for treatment of obesity-associated metabolic syndrome.
Methods
Animals.
Generation of Tg(CAG-Neo-STOPfl-Mir26a1) mice and Hprt-Mir26a Tg mice has been described previously (27). Hepatocyte-specific miR-26a Tg mice (Alb-Mir26a Tg) were generated by crossing Tg(CAG-Neo-STOPfl-Mir26a1) mice with Alb-Cre Tg mice. Alb-Mir26a Tg and littermate Tg(CAG-Neo-STOPfl-Mir26a1) mice were used for experiments.
Cell culture.
Huh7 cells were provided by Clifford J. Steer (University of Minnesota, Minneapolis, Minnesota, USA). Cells were cultured in DMEM with 10% FBS. Transfection of miR was performed using HiPerfect (QIAGEN) according to the manufacturer’s protocol.
Quantification of miR-26a in human liver samples.
Human liver samples were collected from patients undergoing abdominal surgery. Data and biopsy samples are part of the ABOS cohort, which is registered at ClinicalTrials.gov (NCT01129297).
Total RNA (10 ng) was reverse-transcribed using the High Capacity Reverse Transcription Kit (Applied Biosystems, Life Technologies), and miR-26a expression was quantified by QRT-PCR in an Applied 7000 QPCR System using TaqMan Universal PCR Master Mix (Applied Biosystems).
Metabolic measurements.
Serum insulin levels were determined using the Ultra Sensitive Mouse Insulin ELISA Kit (Crystal Chem Inc.). Blood glucose levels were determined using a portable glucose meter (Abbot Laboratories). For GTT and PTT, mice were fasted for 16 hours and then injected intraperitoneally with d-glucose (2 g/kg BW) or pyruvate (2 g/kg BW). For ITT, mice were fasted for 6 hours and then injected intraperitoneally with human insulin (Humulin R, Eli Lilly, 1 U/kg BW). Insulin resistance (HOMA-IR) was calculated as follows: fasting glucose (mg dl–1) × fasting insulin (μU ml–1)/405. Plasma concentrations of triglycerides, total cholesterol, and HDL cholesterol were measured at the City of Hope Helford Research Hospital. Hepatic concentrations of triglycerides, cholesterol, and glycogen were measured using commercial kits (Biovision) according to the manufacturer’s instructions. Hyperinsulinemic euglycemic clamp was performed at Baylor College of Medicine and analyzed as described in our previous publication (41).
In vivo insulin stimulation and analysis of insulin signaling.
In vivo insulin stimulation and analysis of insulin signaling were performed as described previously, with minor modifications (42). Antibodies used in Western blot analysis included anti–phospho-IRS1S332/336 (2580, Cell Signaling Technology [CST]), anti–phospho-IRS1S612 (3203, CST), anti–phospho-IRS1S307 (2381, CST), anti–phospho-AKTS473 (9271, CST), anti–phospho-AKTt308 (9275, CST), anti–phospho-FOXO1s256 (9461, CST), anti-AKT (9272, CST), anti-ACSL3 (sc-166374, Santa Cruz Biotechnology Inc.), anti-PKCδ (9616, CST), anti-PCK1 (12940, CST), anti-GAPDH (2118, CST) and anti-actin (A5316, Sigma-Aldrich). ImageJ (http://imagej.nih.gov/ij/) was used to quantify the phospho-protein/total protein ratio.
Proteomics analysis.
Total proteins from the livers of WT and Alb-Mir26a Tg mice fed a HFD for 16 weeks were extracted, followed by trichloroacetic acid precipitation and trypsin digestion. Samples were loaded on the nano-ACQUITY UPLC chromatographic system (Waters). LC-MSE data were processed with ProteinLynx GlobalServer v2.3 (Waters) and searched in the associated mouse protein database.
Mouse hepatocyte isolation and culture.
Primary hepatocytes were isolated and cultured as described previously (43). Glucose production in primary hepatocytes was determined by glucose colorimetric assay (Biovision).
Statistics.
Results of experiments are displayed as mean ± SEM. Two-tailed Student’s t test was used to determine differences between data groups unless otherwise indicated.
Study approval.
All human studies were conducted according to the principles of the Declaration of Helsinki, were approved by the Centre Hospitalier Régional Universitaire of Lille ethical committee, and complied with the French National Ethics Committee guidelines. All human subjects gave informed consent. All animal procedures followed NIH and City of Hope guidelines for the care and use of laboratory animals and were approved by the City of Hope IACUC.
Supplementary Material
Acknowledgments
We thank Barry Forman and Rama Natarajan for reviewing the manuscript; Kyle Sousa and other members of the Huang lab for helpful discussions; and Keely Walker for editing the manuscript. This work was supported by the National Cancer Institute (P30 CA033572 and NCI R01-139158), the American Cancer Society (RSG-11-132-01-CCE), and the National Natural Science Foundation of China (81441121 and 81123003). B. Staels is a member of the Institut Universitaire de France. This work was supported by grants from the EGID (ANR-10-LABX-46).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(6):2497–2509. doi:10.1172/JCI75438.
References
- 1.Copps KD, White MF. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia. 2012;55(10):2565–2582. doi: 10.1007/s00125-012-2644-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel S, Doble BW, MacAulay K, Sinclair EM, Drucker DJ, Woodgett JR. Tissue-specific role of glycogen synthase kinase 3beta in glucose homeostasis and insulin action. Mol Cell Biol. 2008;28(20):6314–6328. doi: 10.1128/MCB.00763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samuel VT, Shulman GI. Mechanisms for insulin resistance: common threads and missing links. Cell. 2012;148(5):852–871. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore DD. Nuclear receptors reverse McGarry’s vicious cycle to insulin resistance. Cell Metab. 2012;15(5):615–622. doi: 10.1016/j.cmet.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bu SY, Mashek MT, Mashek DG. Suppression of long chain acyl-CoA synthetase 3 decreases hepatic de novo fatty acid synthesis through decreased transcriptional activity. J Biol Chem. 2009;284(44):30474–30483. doi: 10.1074/jbc.M109.036665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Westerbacka J, et al. Genes involved in fatty acid partitioning and binding, lipolysis, monocyte/macrophage recruitment, and inflammation are overexpressed in the human fatty liver of insulin-resistant subjects. Diabetes. 2007;56(11):2759–2765. doi: 10.2337/db07-0156. [DOI] [PubMed] [Google Scholar]
- 7.Lin HV, Accili D. Hormonal regulation of hepatic glucose production in health and disease. Cell Metab. 2011;14(1):9–19. doi: 10.1016/j.cmet.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beale EG, Hammer RE, Antoine B, Forest C. Disregulated glyceroneogenesis: PCK1 as a candidate diabetes and obesity gene. Trends Endocrinol Metab. 2004;15(3):129–135. doi: 10.1016/j.tem.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Gomez-Valades AG, et al. Pck1 gene silencing in the liver improves glycemia control, insulin sensitivity, and dyslipidemia in db/db mice. Diabetes. 2008;57(8):2199–2210. doi: 10.2337/db07-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boj SF, et al. Diabetes risk gene and Wnt effector Tcf7l2/TCF4 controls hepatic response to perinatal and adult metabolic demand. Cell. 2012;151(7):1595–1607. doi: 10.1016/j.cell.2012.10.053. [DOI] [PubMed] [Google Scholar]
- 11.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doria A, Patti ME, Kahn CR. The emerging genetic architecture of type 2 diabetes. Cell Metab. 2008;8(3):186–200. doi: 10.1016/j.cmet.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voight BF, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42(7):579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13(4):239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482(7385):347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quiat D, Olson EN. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest. 2013;123(1):11–18. doi: 10.1172/JCI62876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esau C, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Trajkovski M, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474(7353):649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 19.Jordan SD, et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol. 2011;13(4):434–446. doi: 10.1038/ncb2211. [DOI] [PubMed] [Google Scholar]
- 20.Kornfeld JW, et al. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature. 2013;494(7435):111–115. doi: 10.1038/nature11793. [DOI] [PubMed] [Google Scholar]
- 21.Ji J, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361(15):1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kota J, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137(6):1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dill H, Linder B, Fehr A, Fischer U. Intronic miR-26b controls neuronal differentiation by repressing its host transcript, ctdsp2. Genes Dev. 2012;26(1):25–30. doi: 10.1101/gad.177774.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu X, et al. miR-26a enhances miRNA biogenesis by targeting Lin28B and Zcchc11 to suppress tumor growth and metastasis. Oncogene. 2014;33(34):4296–4306. doi: 10.1038/onc.2013.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng Z, et al. miR-194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology. 2010;52(6):2148–2157. doi: 10.1002/hep.23915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38(3):323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Fu X, et al. MicroRNA-26a targets ten eleven translocation enzymes and is regulated during pancreatic cell differentiation. Proc Natl Acad Sci U S A. 2013;110(44):17892–17897. doi: 10.1073/pnas.1317397110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Postic C, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic β cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274(1):305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 29.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 30.Bernardo BC, et al. Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc Natl Acad Sci U S A. 2012;109(43):17615–17620. doi: 10.1073/pnas.1206432109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rottiers V, et al. Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR. Sci Transl Med. 2013;5(212):212ra162. doi: 10.1126/scitranslmed.3006840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellis JM, Frahm JL, Li LO, Coleman RA. Acyl-coenzyme A synthetases in metabolic control. Curr Opin Lipidol. 2010;21(3):212–217. doi: 10.1097/MOL.0b013e32833884bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson AM, Olefsky JM. The origins and drivers of insulin resistance. Cell. 2013;152(4):673–684. doi: 10.1016/j.cell.2013.01.041. [DOI] [PubMed] [Google Scholar]
- 34.Newgard CB. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012;15(5):606–614. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bezy O, et al. PKCΔ regulates hepatic insulin sensitivity and hepatosteatosis in mice and humans. J Clin Invest. 2011;121(6):2504–2517. doi: 10.1172/JCI46045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tahrani AA, Bailey CJ, Del Prato S, Barnett AH. Management of type 2 diabetes: new and future developments in treatment. Lancet. 2011;378(9786):182–197. doi: 10.1016/S0140-6736(11)60207-9. [DOI] [PubMed] [Google Scholar]
- 37.van Rooij E, Olson EN. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat Rev Drug Discov. 2012;11(11):860–872. doi: 10.1038/nrd3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janssen HL, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368(18):1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 39.Fu X, et al. miR-26a enhances miRNA biogenesis by targeting Lin28B and Zcchc11 to suppress tumor growth and metastasis. Oncogene. 2014;33(34):4296–4306. doi: 10.1038/onc.2013.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Badeanlou L, Furlan-Freguia C, Yang G, Ruf W, Samad F. Tissue factor-protease-activated receptor 2 signaling promotes diet-induced obesity and adipose inflammation. Nat Med. 2011;17(11):1490–1497. doi: 10.1038/nm.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong B, et al. Activation of nuclear receptor CAR ameliorates diabetes and fatty liver disease. Proc Natl Acad Sci U S A. 2009;106(44):18831–18836. doi: 10.1073/pnas.0909731106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Y, et al. Regulation of glucose homeostasis through a XBP-1-FoxO1 interaction. Nat Med. 2011;17(3):356–365. doi: 10.1038/nm.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen WD, et al. Neonatal activation of the nuclear receptor CAR results in epigenetic memory and permanent change of drug metabolism in mouse liver. Hepatology. 2012;56(4):1499–1509. doi: 10.1002/hep.25766. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.