Abstract
Glutaminase (GLS), which converts glutamine to glutamate, plays a key role in cancer cell metabolism, growth, and proliferation. GLS is being explored as a cancer therapeutic target, but whether GLS inhibitors affect cancer cell–autonomous growth or the host microenvironment or have off-target effects is unknown. Here, we report that loss of one copy of Gls blunted tumor progression in an immune-competent MYC-mediated mouse model of hepatocellular carcinoma. Compared with results in untreated animals with MYC-induced hepatocellular carcinoma, administration of the GLS-specific inhibitor bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide (BPTES) prolonged survival without any apparent toxicities. BPTES also inhibited growth of a MYC-dependent human B cell lymphoma cell line (P493) by blocking DNA replication, leading to cell death and fragmentation. In mice harboring P493 tumor xenografts, BPTES treatment inhibited tumor cell growth; however, P493 xenografts expressing a BPTES-resistant GLS mutant (GLS-K325A) or overexpressing GLS were not affected by BPTES treatment. Moreover, a customized Vivo-Morpholino that targets human GLS mRNA markedly inhibited P493 xenograft growth without affecting mouse Gls expression. Conversely, a Vivo-Morpholino directed at mouse Gls had no antitumor activity in vivo. Collectively, our studies demonstrate that GLS is required for tumorigenesis and support small molecule and genetic inhibition of GLS as potential approaches for targeting the tumor cell–autonomous dependence on GLS for cancer therapy.
Keywords: Hepatology, Metabolism, Oncology
Introduction
Cancer cells show profound metabolic changes, driven by oncogenes, such as RAS, PI3K, and MYC, or loss of tumor suppressors, such as p53, VHL, and LKB1 (1–5). Aerobic glycolysis or the Warburg effect is a metabolic hallmark of many cancer cells, which consume large amounts of glucose, but cancer cells also depend on many other nutrient sources for growth and proliferation (1, 5). While fatty acids, acetate, and other amino acids also serve as nutrients, a significant fraction of the biosynthetic needs of cancer cells in vitro are met by glutamine (6–8). Glutamine, which is the most abundant (0.5 mM) circulating amino acid in human plasma, is a significant nutrient source in vivo (6). It provides carbons to highly proliferative tumor cells to produce TCA cycle intermediates, glutathione, fatty acids, and nucleotides (6, 9, 10). Glutamine also contributes its amide nitrogen to produce hexosamines, nucleotides, and nonessential amino acids (6, 9, 10). Together with glucose, glutamine contributes to the building blocks for macromolecular synthesis and biomass accumulation in proliferating cancer cells (8, 11). Both the MYC oncogene (4, 12, 13) and p53 tumor-suppressor protein (14, 15) transcriptionally regulate glutamine metabolism so that its usage meets the biomass production and redox homeostasis requirements of cancer cells.
The central role of glutamine metabolism in cancer cell growth makes glutaminolytic enzymes attractive targets for anticancer therapy (9, 16–20). Glutaminase catalyzes the first step of glutamine metabolism by converting glutamine to glutamate and ammonia. Glutamate is further catabolized by glutamate dehydrogenase or transaminases to α-ketoglutarate, which is then oxidized via the TCA cycle (21). The major transaminases are glutamic-oxaloacetic transaminase (GOT) and glutamic pyruvate transaminase (GPT); both occur as cytoplasmic and mitochondrial isoforms. Two genes encode glutaminases in humans. The kidney type glutaminase, GLS, is encoded by GLS, and the liver type glutaminase, GLS2, is encoded by GLS2. Herein, the murine genes are termed Gls and Gls2. The role of GLS2, a p53 target, in cancer is controversial, with conflicting reports concerning its function as a tumor suppressor (14, 15, 22, 23). GLS, however, has emerged as a critical enzyme in a number of cancer types (9, 16, 17, 19). Its activity is increased by NF-κB (19), and its expression is induced by MYC, whose overexpression rendered transformed cells addicted to glutamine (9, 12, 13). GLS is expressed as a long mRNA splice variant, KGA, or a shorter form, GAC, which is increased in cancers relative to normal tissues (19, 24). siRNA-mediated knockdown of GLS slowed the growth of several different cancer types (13, 17, 19, 25–27), suggesting that pharmacological inhibition of GLS offers a potential therapeutic approach for treating cancer.
Glutamine analog inhibitors, such as azaserine and acivicin, can inhibit tumor growth, but they often have considerable off-target effects (6). The glutamine analog 6-diazo-5-oxo-l-norleucine (DON) inhibits a range of glutamine-dependent enzymes, such as glutamine fructose-6-phosphate amidotransferase and glutaminase, as well as other glutamine-dependent reactions (28, 29). Similarly, amino-oxyacetate (AOA), a transaminase inhibitor, has also been used to target a part of glutamine metabolism by inhibiting the production of α-ketoglutarate from glutamate, which is in turn derived from glutamine (12, 30). AOA, however, has been documented to inhibit a wide range of other pyridoxal-dependent enzymes in addition to GOT and GPT (31). Hence, AOA’s biological activity is also nonspecific.
The identification of an allosteric GLS-selective inhibitor, bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide (BPTES), raised the possibility of specifically inhibiting glutamine metabolism with minimal off-target effects (32). The crystal structure of BPTES-bound GLS reveals that BPTES docks in the GLS tetramer interfaces, locking GLS in an off mode and disabling phosphate-dependent activation of the enzyme (24, 33, 34). Other GLS inhibitors have been developed, including the BPTES-like drug candidate CB-839 and compound 968, which has a different mechanism of GLS inhibition (16, 35). We and others have demonstrated that pharmacological inhibition of GLS slowed proliferation in several cancer cell types in vitro and in xenograft models (9, 17, 19, 26, 36). However, previous studies have not directly addressed the mechanism of growth inhibition or whether off-target effects of BPTES, CB-839, or 968 could underlie their antitumor activity (9, 16, 19, 35).
GLS inhibition in vivo has been restricted to xenograft studies in immunocompromised mice, in which the potential negative effects on the immune system could not be measured. Activated T cells are known to use high levels of glutaminolysis for proliferation, and inhibition of glutaminase may hinder the natural immune response to the formation of new tumors (37). Whether or not GLS inhibition is effective in immunocompetent mice is not known, particularly since many metabolic pathways used by cancer cells are shared with normal activated T lymphocytes (38). Further, altered glutamine metabolism in the tumor stroma has been reported (39), raising the possibility that non–cell autonomous roles of GLS inhibition may underlie the effects of GLS inhibition in vivo.
In this report, we used an immunocompetent MYC-dependent genetically engineered model (LAP/MYC) of murine hepatocellular carcinoma (HCC) to determine the role of Gls in liver tumorigenesis. The animal age at the time of MYC activation in this model affects the biology, such that earlier MYC activation resulted in more aggressive tumors (40, 41). In utero MYC activation induces an aggressive hepatoblastoma-like disease upon birth, as compared with the HCCs induced when MYC is activated after birth (40). Activation of MYC 4 weeks after birth resulted in multinodular HCC and an overall mean survival time of 15 weeks (40). These tumors display increases in both glucose and glutamine metabolism (42, 43). Here, we report that animals derived from crosses of LAP/MYC mice with Gls (Gls+/–) heterozygotes (44) had delayed liver tumorigenesis and a trend toward prolonged survival. The genetic experiments were corroborated by our findings that inhibition of glutaminase with a small molecule inhibitor, BPTES, resulted in significantly (P < 0.0001) prolonged survival of LAP/MYC animals wild-type for Gls (Gls+/+) without obvious toxic side effects. We found that BPTES inhibited P493 lymphoma cell growth by causing DNA replication defects that triggered cell death and fragmentation. Ectopic overexpression of a GLS mutant, which resists inhibition by BPTES (24), could rescue P493-cultured cells or xenografts from BPTES-mediated growth inhibition, attesting to an on-target effect of BPTES. We further corroborated animal genetic and pharmacological studies using a human-specific Vivo-Morpholino that induces nonsense-mediated GLS mRNA decay and found that P493 tumor xenograft growth in mice could be markedly and specifically inhibited only by Vivo-Morpholino directed at human GLS, but not by that directed at mouse Gls.
Results
Glutaminase isoform switch in HCC.
We sought to determine the role of Gls in an inducible MYC-mediated murine model of HCC, which has been documented as undergoing enhanced glycolysis and glutaminolysis (42). Whether Gls is required for tumorigenesis and tumor progression has not been known. In this model, MYC expression is under the control of a tetracycline-off (Tet-off) system regulated by the Tet transactivating protein (tTA), which in turn is driven by the liver-activating protein (LAP) promoter in LAP/MYC mice (40, 41). Multifocal tumors in adult animals with varying aggressiveness can be induced according to the age at which MYC is activated by doxycycline withdrawal. The earlier the age at which MYC is activated, the more aggressively the disease develops (40). This model also offers a unique opportunity to compare frank tumor tissues with surrounding normal-appearing nontumor transgenic liver. In this study, we chose to investigate a more aggressive disease model by activating MYC at birth or 1 week after birth as compared with previous HCC metabolic studies, in which MYC was activated at 4 weeks (40, 42, 43).
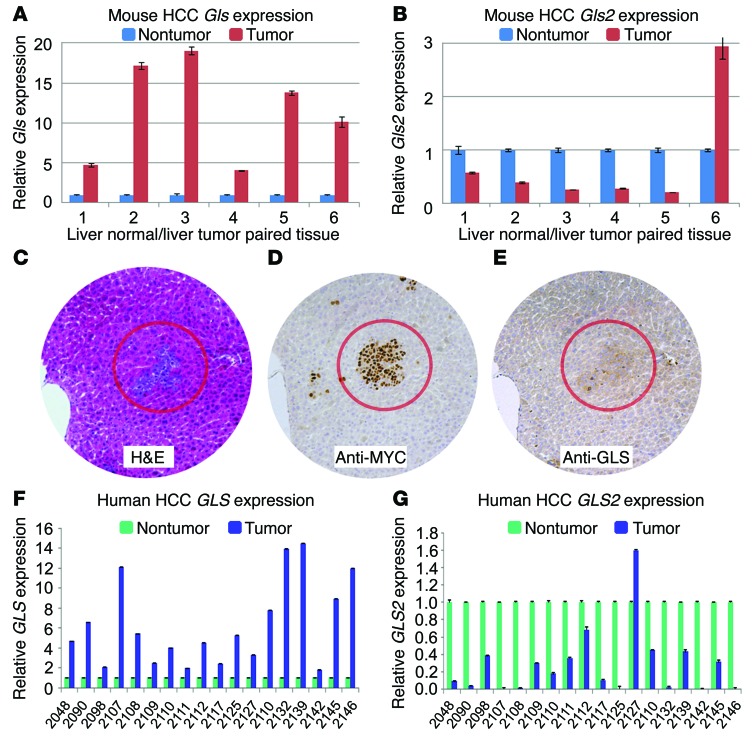
We determined the expression of glutaminase mRNAs in MYC-induced tumors and found that liver-type Gls2 mRNA expression was decreased in most tumors, while kidney type Gls mRNA was increased in the tumors when compared with surrounding nontumor liver (Figure 1, A and B). When MYC expression was induced 1 week after birth, mice exhibited small tumor nodules by 3 weeks after birth. We then studied MYC and GLS protein expression by immunohistochemistry (IHC). We found that MYC was highly expressed in the tumor nodules compared with the surrounding nontumor transgenic livers (Figure 1, C and D). Immunostaining revealed a slight increase in GLS protein levels in tumor nodules at 3 weeks of age (Figure 1E). These observations indicate that GLS is increased, while GLS2 is decreased, in this model of liver tumorigenesis.
Figure 1. Increased GLS and decreased GLS2 expression in mouse HCC model and human HCC samples.
(A) Gls mRNA levels determined by qPCR in LAP/MYC tumors compared with surrounding liver tissue (numbers at the bottom correspond to different mice). The test was done in triplicate; results represent mean ± SEM of technical triplicates. (B) Gls2 mRNA levels in LAP/MYC tumors compared with surrounding liver tissue. (C–E) Histology of consecutive sections of LAP/MYC tumors. H&E stain (C) shows early tumor formation in 3-week-old LAP/MYC mice with corresponding elevation of MYC (D) and GLS staining (E). Original magnification, ×10. (F) GLS mRNA levels determined by qPCR in human HCC tumors compared with surrounding nontumor tissue (numbers at the bottom represents different patients). (G) GLS2 mRNA levels in human HCC tumors compared with surrounding nontumor tissue. Test was done in triplicate; results represent mean ± SD. For F and G, numbers represent individual patients. Student’s t test was used.
To determine whether these observations could be generalized to human disease, we conducted a paired analysis of GLS and GLS2 levels in cancerous and noncancerous regions of liver in patients with HCC. We found that GLS2 mRNA was downregulated in most tumors (P < 0.05; Supplemental Figure 1B; supplemental material available online with this article; doi:10.1172/JCI75836DS1) and GLS mRNA was upregulated (P < 0.05; Supplemental Figure 1A) in primary human HCC (Figure 1, F and G). In another set of human HCC samples, we found with IHC that GLS protein levels were increased in 11 of 15 tumors (Supplemental Figure 1, E and F). GLS2 IHC, however, showed a less consistent pattern, with 1 nontumor sample having a GLS2 staining score of less than or equal to 2 and 7 tumors having scores less than or equal to 2 (Supplemental Figure 1, G and H). These observations suggest that GLS is increased in HCC and hence could play a role in tumorigenesis and tumor progression. It is notable that, while MYC mRNA levels were elevated in the tumors as compared with in normal-appearing liver (P < 0.05; Supplemental Figure 1C), they did not correlate directly with GLS mRNA levels (Supplemental Figure 1D), suggesting that factors other than MYC could affect HCC GLS mRNA levels. Nonetheless, the switch from GLS2 to GLS expression in human HCC samples was consistent across virtually all paired samples (Figure 1, F and G), underscoring the glutaminase isoform switch that occurred in mouse and human HCC tissues.
GLS is required for full MYC-induced murine liver cancer development.
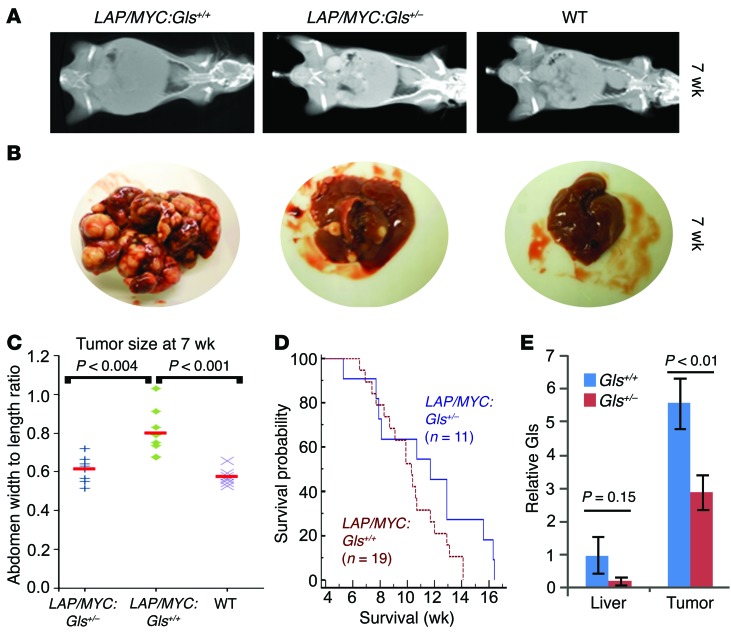
Because the glutaminase isoform switch (Gls2 to Gls) was conserved between mice and humans, we determined the role of Gls in tumorigenesis and tested to determine whether a 50% allelic reduction of Gls would affect MYC-mediated mouse HCC. We were unable to use Gls homozygous null mice because they died perinatally (44). Hence, we crossed the LAP/MYC mice with Gls+/– heterozygotes, which were previously documented to have decreased GLS protein and enzymatic activity (44). These Gls+/– heterozygotes developed normally, with subtle neurological findings (44, 45). We compared LAP/MYC heterozygous (LAP/MYC:Gls+/–) to homozygous (LAP/MYC:Gls+/+) animals. MYC expression was induced at 1 week postnatally by withdrawal of doxycycline. By 7 weeks of age, the LAP/MYC mice had visibly increased abdominal girth and tumor load, which was measured by CT imaging (Figure 2A and Supplemental Figure 2). The CT-estimated tumor load is consistent with representative images of livers retrieved from some of these animals at 7 weeks (Figure 2B). The estimated tumor load in heterozygous LAP/MYC:Gls+/– mice was significantly less than in the wild-type LAP/MYC:Gls+/+ mice (Figure 2C; n = 8; P < 0.001; see Supplemental Figure 2 for CT scans). Cohorts of LAP/MYC:Gls+/– (n = 11) and LAP/MYC:Gls+/+ (n = 19) mice were followed for survival. While cohorts of these sizes did not show a statistically longer survival of the LAP/MYC:Gls+/– animals (P = 0.12), there was a trend toward longer survival (mean survival time ~11.5 weeks vs. ~10.2 weeks) (Figure 2D). This suggests that a 50% allelic reduction of Gls can significantly delay early tumor progression, but may not ultimately prolong survival.
Figure 2. Allelic Gls reduction slows early tumor progression.
(A) Representative CT scans (for complete set, see Supplemental Figure 2) for LAP/MYC:Gls+/+, LAP/MYC:Gls+/–, and wild-type mice at 7 weeks of age. (B) Representative whole livers for LAP/MYC:Gls+/+, LAP/MYC:Gls+/–, and wild-type mice at 7 weeks of age. (C) Surrogate CT scan–based abdominal sizes in LAP/MYC:Gls+/+, LAP/MYC:Gls+/–, and wild-type mice at 7 weeks of age. (D) Kaplan-Meier plot of survival of LAP/MYC:Gls+/– mice (n = 11) compared with LAP/MYC:Gls+/+ mice (n = 19). P = 0.12. (E) Relative Gls mRNA expression in normal-appearing liver or liver tumors obtained from Gls homozygous (Gls+/+, blue) or heterozygous (Gls+/–, red) LAP/MYC mice (n = 3, each group). Values are shown as mean ± SD. Student’s t test was used.
To assess the transcriptional consequence of a 50% allelic reduction of Gls, we harvested tumors and paired normal-appearing liver tissue from Gls+/+ and Gls+/– LAP/MYC mice. Using quantitative PCR (qPCR), we found that Gls mRNA levels were significantly induced in homozygous Gls+/+ tumors as compared with normal-appearing surrounding livers (Figure 2E). The normal livers from heterozygous Gls+/– tumor-bearing mice had diminished Gls expression as compared with livers from homozygous mice (>70% reduction), indicating that allelic reduction was accompanied by decreased gene expression. The homozygous liver tumors displayed much higher levels (>5-fold) of Gls expression as compared with paired normal liver tissues (Figure 2E). Consistent with the 50% allelic reduction, the levels of Gls mRNA in heterozygous Gls+/– tumors were reduced by 2-fold relative to those in homozygous Gls+/+ tumors.
BPTES prolongs survival of a genetically engineered mouse model of liver cancer.
Based on the observation that loss of 1 copy of Gls could delay tumorigenesis, we sought to determine whether pharmacological inhibition of GLS with BPTES could prolong survival in immunocompetent transgenic mice. We first determined the in vitro effect of BPTES (10 μM) on growth of a LAP/MYC-derived mouse HCC cell line (mHCC 3–4 cells), which displayed MYC-dependent expression of Gls mRNA and protein (Figure 3A and Supplemental Figure 3, A and B). We found that BPTES inhibited growth of mHCC 3–4 cells as did the clinical candidate drug CB-839 (ref. 16 and Figure 3, A and B) in a dose-dependent manner (Supplemental Figure 3C). While approximately 300 nM of BPTES (307 nM) did not affect growth, a similar concentration of CB-839 (333 nM) significantly diminished growth, indicating that CB-839 is a more potent inhibitor of GLS than BPTES (Supplemental Figure 3C and refs. 16, 32).
Figure 3. GLS inhibition prolongs survival in mouse HCC model.
(A) Effect of 10 μM BPTES on the in vitro growth of mHCC 3–4 cells derived from LAP/MYC tumors. n = 3. P < 0.001. (B) Effect of 10 μM GLS inhibitor CB-839 on the growth of mHCC 3–4 cells in vitro. n = 3. P < 0.001. (C) Kaplan-Meier plot of survival of BPTES-treated (n = 14) versus vehicle DMSO–treated LAP/MYC mice (n = 15). (D) Micrographs of representative fields of Ki-67–stained liver tumors from BPTES (n = 6) or vehicle-treated (n = 6) LAP/MYC mice. The far right graph depicts the quantification of area that is Ki-67 positive. Original magnification, ×5. (E) Relative glutamine and glutamate levels in tumors or normal livers from mice treated with BPTES or DMSO were measured (n = 5 each). Values are shown as mean ± SD. Student’s t test was used.
We then sought to test BPTES in vivo in an aggressive MYC-induced liver cancer model by activating MYC at birth, which normally results in the death of all animals by 11 weeks of age (40). We chose to treat with BPTES or vehicle at 3 weeks after birth because early tumor formation and increased GLS expression were evident by this time (Figure 1, C and E). Vehicle DMSO–treated LAP/MYC mice showed a median survival of approximately 7 weeks (n = 15) (Figure 3C). Remarkably, BPTES-treated LAP/MYC mice (n = 14) survived significantly longer than control mice, with a median survival of 11 weeks (P < 0.0001, Figure 3C). We performed IHC to determine whether BPTES might alter MYC protein levels rather than just inhibiting GLS and found no significant changes in MYC, GLS, or GLS2 levels with BPTES treatment (Supplemental Figure 3D).
To gain insight into the in vivo biological effects of BPTES, we studied its effects on glutamine and glutamate levels in tumors as well as its effects on proliferation, as determined by Ki-67 staining. For these studies, we collected tumors from 5-week-old cohorts of BPTES-treated and DMSO-treated LAP/MYC mice as well as control normal livers from identically treated 5-week-old littermate MYC mice (without the LAP activator). We readily noted that the livers from the BPTES-treated mice were smaller than those from the control-treated mice, and importantly, there were fewer and smaller tumor nodules (Supplemental Figure 4, A and B). We also found a significant decrease in Ki-67 staining, suggesting that blocking GLS in vivo slowed proliferation (Figure 3D). To determine the effect of BPTES treatment on glutaminase-mediated conversion of glutamine to glutamate in vivo, we extracted metabolites from liver tumors or the littermates’ normal livers. As expected, control DMSO–treated tumors had relatively higher levels of glutamate and lower levels of glutamine as compared with levels in BPTES-treated tumors, indicating that inhibition of GLS resulted in the accumulation of glutamine (P = 0.004, Figure 3E). Normal livers from littermate mice did not show a differential effect of BPTES versus DMSO on glutamine and glutamate levels. We surmise that normal liver tissues are largely dependent on normal hepatocyte GLS2, which is not inhibited by BPTES.
Collectively, our studies provide proof of concept that GLS inhibition offers a potential therapeutic approach to treating HCC, particularly since treatment with BPTES was well tolerated in immunocompetent mice. Histopathology of brain, heart, skeletal muscle, and kidney did not reveal any gross pathology in BPTES-treated versus DMSO-treated control animals (Supplemental Figure 5). Further, BPTES did not show any significant effects on body weight, blood chemistries, and hematology measurements (Supplemental Figure 6).
Biological effects and specificity of GLS inhibitors in vitro.
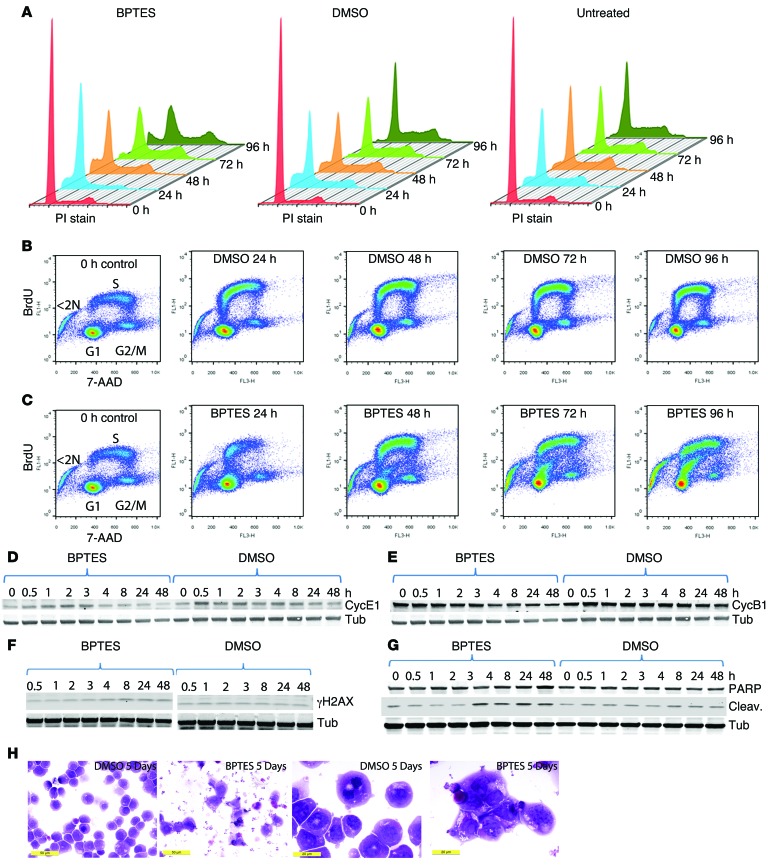
We sought to determine how BPTES inhibits cell proliferation based on our observations that it could diminish tumorigenesis and decrease Ki-67 levels (Figure 3D and ref. 7). For these studies, we used the human P493 B cell model of lymphoma, which we have previously shown to be BPTES sensitive (9). P493 cells were engineered with an EBV genome that includes the EBV EBNA2-estrogen receptor hormone-binding domain fusion gene and a Tet-repressible human MYC gene expression cassette (46). P493 cells were treated with Tet for 24 hours and then exposed to BPTES or DMSO and released from Tet to induce MYC. The cells were collected at various time points for cell-cycle analysis and BrdU incorporation studies. Tet-treated P493 cells arrested primarily in the G1 phase. Upon release from Tet and activation of MYC, DMSO-treated P493 cells progressively entered the cell cycle, as seen in Figure 4A. In contrast, BPTES-treated P493 cells slowly entered S phase and had increased cell death (<2 N DNA content) after several days. To determine the nature of cell death, we collected control or treated cells at various time points after MYC induction. The control or BPTES-treated P493 cells were then pulse labeled with BrdU and counterstained with 7-AAD for flow cytometry. As seen in Figure 4B, DMSO-treated cells progressively incorporated BrdU into DNA as cells entered S phase from G1 between 0 and 24 hours after MYC induction. Upon exiting S phase, pulse-labeled cells incorporated less BrdU as they entered G2, resulting in an overall arc of BrdU-labeled cell going from G1 into S phase and then to G2/M. In contrast with control cells, the BPTES-treated P493 cells had delayed and diminished incorporation of BrdU 24 hours after MYC induction. As time proceeded, the BPTES-treated P493 cells progressively incorporated less BrdU between 48 and 72 hours, resulting in a distinctly lower BrdU-labeled arc of cells that did not progress further into S phase (Figure 4C). In fact, there was a progressive increase in BrdU-labeled cells with less than 2 N DNA, particularly at 72 and 96 hours. These cells appeared to have aborted DNA synthesis and exited the cell cycle by dying.
Figure 4. Effects of GLS inhibition by BPTES on cell cycle, BrdU incorporation, cyclins, γH2AX, cleaved PARP levels, and morphology of P493 cells.
(A) Arrested P493 cells were washed to remove Tet to induce MYC expression and were untreated, DMSO treated, or BPTES treated. Cells at the indicated time points after wash were stained with propidium iodide to measure total DNA content by flow cytometry. (B) DMSO-treated control cells were pulse labeled with BrdU and then counterstained with 7-AAD for flow cytometry of samples obtained at the indicate times after MYC induction. Zero-hour control cells were untreated. (C) BPTES-treated cells were studied under conditions outlined in B. (D–G) Protein lysates from control or BPTES-treated unsynchronized cells at the indicated times after treatment were obtained and then immunoblotted for the following: (D) cyclin E1 (CycE1); (E) cyclin B1 (CycB1); (F) γH2AX; and (G) cleaved PARP. D and E share a tubulin blot from sequential staining of the same blot. (H) Photomicrographs of DMSO- and BPTES-treated P493 cells at low and high power. Scale bars: 50 μm (left panels); 20 μm (right panels).
We also treated unsynchronized P493 cells with DMSO and BPTES and collected cell lysates at various time points following treatment. The samples were immunoblotted for cell-cycle markers as well as cleaved poly-(ADP-ribose) polymerase (PARP), a marker of cell death. The immunoblots for cleaved PARP, γH2AX, and cyclins E1 and B1 corroborate the BrdU labeling results. As compared with control cells, BPTES-treated cells displayed decreased cyclins E1 and B1 levels (Figure 4, D and E), which are compatible with diminished S phase entry of unsynchronized cells. Consistent with the BrdU incorporation study, when compared with the controls, the BPTES-treated cells had a higher level of the DNA-damage marker γH2AX, which progressively increased from 4 to 24 hours following MYC induction (Figure 4F). Cleaved PARP, a marker of cell death, began to increase at 4 hours after MYC induction and remained elevated in BPTES-treated cells (Figure 4G). We also studied P493 cells by microscopy and found that BPTES-treated cells were dysmorphic and remarkably fragmented, with frequent bare nuclei as compared with the normal-appearing control DMSO-treated P493 cells (Figure 4H). These results together with the cell-cycle and BrdU incorporation studies indicate that BPTES caused DNA replication arrest, cell death, and fragmentation of P493 cells.
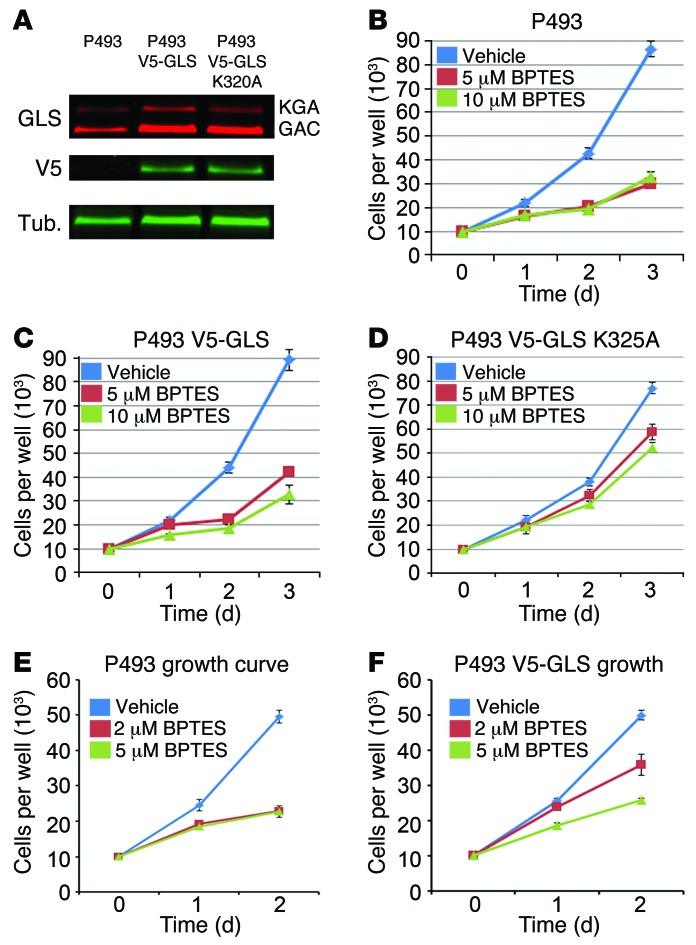
Because BPTES profoundly affected cell growth through inhibiting DNA replication, we used the P493 B cell lymphoma model to determine whether the effects of BPTES in vitro and in vivo resulted from off-target effects (9). Using lentiviruses expressing the V5-tagged GAC forms of GLS, we created stable P493 cell lines overexpressing either wild-type V5-GLS or the BPTES-resistant V5-GLS-K325A mutant (ref. 24 and Figure 5A). We observed that the P493 V5-GLS-K325A cell line resisted BPTES while the P493 parental and P493 V5-GLS cell growth rates were inhibited by 5 μM and 10 μM BPTES, respectively (Figure 5, B–D). These biological effects are consistent with our biochemical findings of the inhibitory effects of BPTES on various forms of engineered cellular GLS. When compared with GLS or V5-GLS, the glutaminase activity from the V5-GLS-K325A cells was most resistant to BPTES (Supplemental Figure 7, A–C). We observed similar biological effects of BPTES using PC-3 prostate cancer cells overexpressing V5-GLS or V5-GLS-K325A (Supplemental Figure 7, D–F), suggesting that the rescue by the mutant GLS-K325A is not a cell line–specific effect. We also tested the clinical candidate drug CB-839 and found that the GLS K325A-expressing PC3 cell line was more resistant to CB-839 treatment as compared with control PC-3 cells (Supplemental Figure 7G). Collectively, these results indicate that BPTES- and CB-839–mediated growth inhibition are on target.
Figure 5. On-target inhibition of GLS by BPTES in vitro.
(A) Western blot of V5-GLS and V5-GLS K325A expression in P493 cell lines. GLS, anti-GLS Ab; V5, anti-V5 tag Ab; Tub, anti-tubulin Ab. (B) Effects of 5 μM and 10 μM BPTES on the growth of the P493 parent cell line in vitro. P < 0.001. (C) Overexpression of V5-GLS does not rescue 5 μM or 10 μM BPTES inhibition of P493 growth in vitro. P < 0.001. (D) Overexpression of V5-GLS K325A strongly rescues 5 μM or 10 μM BPTES inhibition of P493 growth in vitro. P < 0.001. (E) 5 μM and 2 μM BPTES similarly inhibit P493 growth in vitro. 2 μM vs. 5 μM, P = 0.91. (F) V5-GLS overexpression partially rescues P493 growth at 2 μM BPTES but not at 5 μM BPTES in vitro, 2 μM vs. 5 μM, P = 0.02. Values are shown as mean ± SD. Student’s t test was used.
To further determine the target specificity of BPTES, we tested the ability of the overexpressed wild-type V5-GLS to decrease the sensitivity of P493 cells to intermediate doses of BPTES, because high target protein levels in cells have been shown to increase the IC50 of drugs (47). While both P493 and P493 V5-GLS had similarly diminished proliferation at 5 μM BPTES (Figure 5, E and F), both proliferated rapidly when exposed to DMSO vehicle control. At 2 μM BPTES, however, the P493 V5-GLS cell line proliferated markedly more than the parental P493 cell line, suggesting that GLS protein levels can affect BPTES sensitivity in vitro. Similarly, overexpression of V5-GLS shifted the dose-response curve in CB-839–treated PC-3 prostate cancer cells (Supplemental Figure 7G). These observations also attest to an on-target effect of BPTES and CB-839, because an increase in target protein levels can increase the IC50. Further, we found that neither BPTES nor CB-839 profoundly affected the activation of normal T lymphocytes as compared with metformin treatment (Supplemental Figure 8), supporting the lack of obvious side effects seen in BPTES-treated mice.
On-target inhibition of GLS by BPTES in vivo.
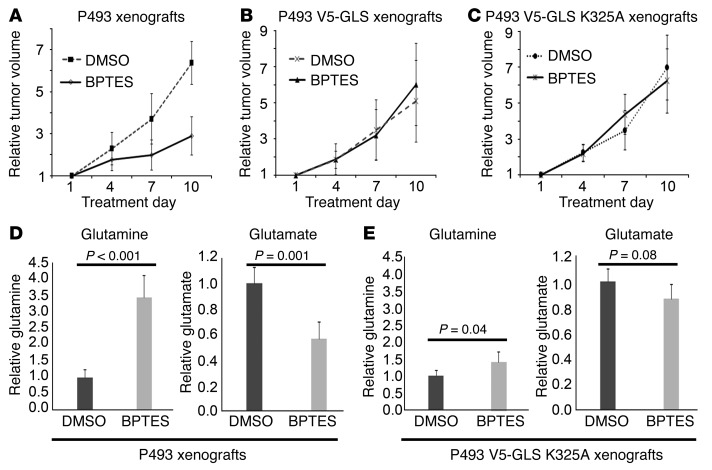
To determine the effects of GLS overexpression in vivo, xenografts of the P493, P493-V5-GLS, or P493-V5-GLS-K325A cell lines were grown in athymic nude mice. When the xenografts reached approximately 100 mm3, the mice were treated with BPTES every 3 days and tumor sizes were measured. BPTES reduced the growth of P493 xenografts by approximately 50% over a 10-day treatment period (Figure 6A), as we have previously reported (9). BPTES, however, did not inhibit the growth of P493 xenografts overexpressing wild-type V5-GLS or the BPTES-resistant mutant V5-GLS K325A (Figure 6, B and C). Metabolic analysis of the parental P493 xenografts showed that BPTES treatment elevated tumor glutamine levels (P < 0.001, Figure 6D) and decreased glutamate levels as expected with glutaminase inhibition (P = 0.001; Figure 6D). Further, the mutant V5-GLS K325A tumors treated with BPTES continued to have reduced glutamine and elevated glutamate levels, reflecting uninhibited GLS activity (Figure 6E). These observations collectively suggest that the lower levels of BPTES, which were likely achieved in vivo, could only inhibit the parental P493 xenografts, but were not sufficient to inhibit those overexpressing either wild-type or mutant GLS. These findings further underscore the on-target effect of BPTES, particularly in vivo.
Figure 6. On-target inhibition of GLS by BPTES in vivo.
(A) BPTES inhibited P493 xenograft growth in vivo. n = 5. Day 10, P = 0.001. (B) Measurement of tumor glutamine and glutamate ratios shows on-target effect of BPTES in P493 parental cell line xenografts in vivo. n = 5. Day 10, P = 0.57. (C) BPTES failed to inhibit the growth of P493 xenografts overexpressing V5-GAC. n = 5. P = 0.50. (D) BPTES treatment elevated tumor glutamine levels and decreased glutamate levels in wild-type P493 xenografts in vivo. n = 5. (E) V5-GLS K325A overexpression rescues BPTES increase in glutamine and decrease in glutamate in P493 xenografts in vivo. n = 5. Values are shown as mean ± SD. Student’s t test was used.
Antisense morpholino targeting of GLS diminishes in vivo cell-autonomous tumorigenesis.
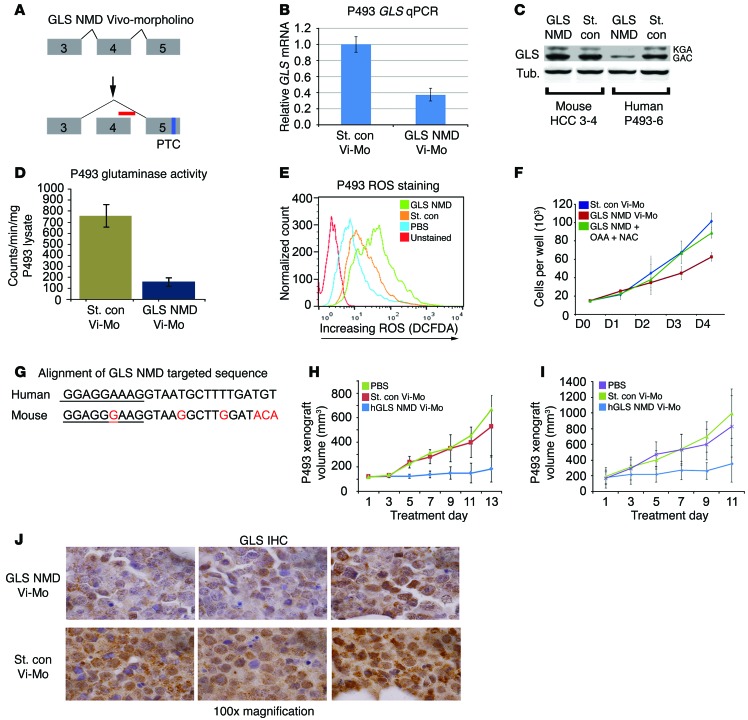
To further corroborate the requirement of GLS for in vivo P493 lymphomagenesis, we sought to inhibit GLS expression through Vivo-Morpholino–mediated targeting of gene expression. Vivo-Morpholinos, which are octaguanidine-modified antisense oligonucleotides capable of altering pre-mRNA processing in vivo (48), have been used in human cell line–derived xenografts (49). We designed a human GLS pre-mRNA targeting Vivo-Morpholino, which binds to the exon 4–intron 5 junction, blocks junction complex binding to the exon 4 splice donor, and causes exon 4 skipping. Skipping of exon 4 puts GLS out of frame, causing an exon 5 premature truncation codon, which triggers nonsense-mediated decay (NMD) of the frame-shifted GLS mRNA (Figure 7A). We termed this Vivo-Morpholino “NMD-VMorph” and the accompanying standard control (nontargeting) Vivo-Morpholino “Cont-VMorph.”
Figure 7. Anti-GLS Vivo-Morpholino inhibits xenograft growth cell autonomously.
(A) The GLS NMD Vivo-Morpholino (red bar) caused skipping of human GLS exon 4, resulting in a premature truncation codon (PTC) in exon 5. (B) 2 μM GLS NMD Vivo-Morpholino (Vivo-Mo) reduced GLS mRNA in P493 cells. Data are shown as mean ± SEM. n = 3. (C) 10 μM GLS NMD Vivo-Morpholino reduced GLS protein in human P493 cells but not mouse GLS in mHCC 3–4 cells. (D) 2 μM GLS NMD Vivo-Morpholino decreased glutaminase activity in P493 cells. P = 0.01. (E) GLS knockdown in P493 increased ROS as indicated by increased DCFDA staining. (F) 2 μM GLS NMD Vivo-Morpholino inhibited growth of P493 cells in vitro versus standard control morpholino. Growth is rescued with a combination of TCA cycle intermediate oxaloacetate (OAA) and the antioxidant NAC. Data are shown as mean ± SD. n = 3. GLS NMD Vivo-Morpholino vs. GLS NMD Vivo-Morpholino OAA + Nac, P < 0.01. (G) Mismatch between human and mouse sequence provides species specificity for human GLS NMD Vivo-Morpholino (exon, underlined; intron, not underlined). (H and I) The GLS NMD Vivo-Morpholino slowed growth of P493 xenografts when treatment was begun at a xenograft volume of (H) 100 mm3 (PBS and standard control, n = 5; GLS NMD, n = 6) or (H or I) 200 mm3 (PBS, n = 5; standard control, n = 4; GLS NMD Vivo-Morpholino, n = 6). SD of replicates. GLS NMD versus standard control final day for G and H. P ≤ 0.01. (J) GLS IHC of P493 xenografts treated with GLS NMD or standard control Vivo-Morpholino shows reduced GLS staining in GLS NMD Vivo-Morpholino–treated xenografts. Original magnification, ×100. Student’s t test was used.
Treating the P493 cells with NMD-VMorph caused an approximately 60% reduction in GLS mRNA level (Figure 7B) and a corresponding reduction of GLS protein level compared with treatment with Cont-VMorph (Figure 7C). Additionally, the NMD-VMorph–treated P493 cells showed a decrease in glutaminase activity compared with P493 cells treated with Cont-VMorph (Figure 7D). Consistent with the role of glutaminase in glutathione production, NMD-VMorph treatment also increased ROS in P493 cells, as evaluated by DCFDA flow cytometry (Figure 7E). Importantly, P493 cells treated with NMD-VMorph had significantly diminished proliferation as compared with P493 cells treated with Cont-VMorph (Figure 7F). To corroborate that the NMD-VMorph’s target was GLS, we performed rescue experiments with oxaloacetate, a TCA cycle intermediate derived from glutamine, and N-acetylcysteine (NAC), which titrates ROS and substitutes for diminished glutamate-dependent glutathione synthesis. Consistent with our previous observation of GLS knockdown in P493 cells (13), oxaloacetate and NAC were able to rescue growth (Figure 7F), supporting the hypothesis that NMD-VMorph diminished the contribution of glutamine to the TCA intermediates and to glutathione synthesis.
It is noteworthy that the NMD-VMorph does not target mouse Gls mRNA due to mismatches with the targeted human sequence (Figure 7G). We confirmed in mouse mHCC 3–4 cells that the human NMD-VMorph did not affect mouse GLS protein levels (Figure 7C). P493 xenografts (~100 mm3) were treated by subcutaneous injection every other day for 13 days with vehicle PBS control (n = 5), Cont-VMorph (n = 6), or GLS NMD-VMorph (n = 6). PBS- or Cont-VMorph–treated P493 xenografts showed similar growth, with tumor volume increasing approximately 6- to 7-fold over 13 days (Figure 7H). In contrast, NMD-VMorph–treated P493 xenografts showed a drastic reduction in tumor growth with signs of tumor regression (Figure 7H). Similar results were found with larger P493 xenografts (~200 mm3; Figure 7I). To confirm the in vivo effect of the NMD-VMorph, we immunostained for GLS levels in P493 xenografts (Figure 7J), which showed reduced GLS staining when compared with Cont-VMorph–treated xenografts. These experiments indicate that cell-autonomous reduction of P493 GLS levels with a human-specific Vivo-Morpholino in mice was sufficient for an antitumor effect.
We further studied the specificity of the NMD Vivo-Morpholino targeting human GLS with a mouse-specific Gls targeting Vivo-Morpholino (termed mGls-NMD). By binding to the exon 1–intron 1 boundary of the mouse Gls, the mGls NMD caused the retention of intron 1, resulting in NMD (Supplemental Figure 9A). Treatment of mHCC 3–4 cells with mGls-NMD Vivo-Morpholino decreased levels of Gls mRNA and protein (Supplemental Figure 9, B and C). Of the 25 bases in mGls NMD Vivo-Morpholino, 7 were mismatched with the homologous human sequence (Supplemental Figure 9D). Treatment of human P493 cells with 5 μM mGls-NMD did not decrease human GLS protein (Supplemental Figure 9E), and treatment by subcutaneous injections did not inhibit growth of human P493 xenografts (Supplemental Figure 9F). These observations further indicate that inhibition of the human tumor GLS in a cell-autonomous fashion is sufficient to diminish tumorigenesis.
Discussion
The in vivo and in vitro results of this work with MYC-inducible liver and lymphoma cancer models support the feasibility of using glutaminase inhibition as a therapeutic strategy. We addressed whether glutaminase, Gls, is required for a MYC-inducible transgenic model of liver cancer. We found that a 50% Gls allelic loss slowed liver tumorigenesis, indicating that Gls participated in tumorigenesis. This observation was further corroborated by pharmacological inhibition of GLS with the tool compound BPTES, which significantly (P < 0.0001) prolonged the survival of treated animals without obvious side effects.
The means by which BPTES inhibited growth was delineated through a time series experiment after MYC activation with BPTES-treated P493 cells. These studies document that BPTES caused a delay in G1-S cell-cycle transition and subsequent DNA replication arrest, cell death, and fragmentation. These findings are consistent with reports that Ras-transformed cells are also dependent on glutamine and glutaminase, such that glutamine deprivation caused DNA replication arrest (50–52). The effect of BPTES inhibition or glutamine withdrawal on DNA replication is consistent with the biochemical role of glutamine in de novo aspartate synthesis, required for pyrimidine biosynthesis. Using 13C-labeled glutamine for metabolic studies of P493 cells, we have found that glutamine provided the bulk of de novo aspartate synthesis that was mostly used for production of pyrimidine nucleotides (T.W. Fan et al., unpublished observations). These findings are compatible with the role of GLS in nucleotide synthesis, such that GLS inhibition would result in DNA replication arrest and subsequent cell death as we document in this report. Because the DNA replication damage response requires PARP activation and NAD+ for repair, our mechanistic studies with BPTES also provide the foundation for future studies on combination therapies. We surmise that the combination of BPTES or CB-839 with PARP inhibitors or inhibitors of NAMPT, which participates in NAD+ synthesis, would have synergistic antitumor effects.
We addressed in this study whether the antitumorigenic effect of BPTES depended on its on-target GLS inhibitory activity or off-target effects. While previous studies suggest that targeting glutamine metabolism could have a positive therapeutic outcome, whether the possible inhibition of host GLS could diminish tumorigenesis was not addressed. For example, it is possible that host neovascular or immune cells could depend on glutaminase and systemic treatment targeting GLS could have antiangiogenic or immunomodulatory effects. We provide evidence that cell-autonomous inhibition of glutaminase is sufficient to inhibit tumorigenesis without invoking the potential effects of inhibitors on the tumor microenvironment. We also found that BPTES or CB-839 did not blunt T cell activation compared with metformin treatment. Further, rescue experiments using a BPTES inhibition–resistant GLS mutant provide evidence that BPTES is indeed a specific inhibitor and that its effects in vitro and in vivo are on target. The use of Vivo-Morpholinos specific for human glutaminase further corroborates that inhibition of tumor GLS is sufficient for a therapeutic effect. This was underscored by the finding that a mouse-specific GLS Vivo-Morpholino did not inhibit the growth of a human tumor xenograft. Our data also suggest that, in addition to small molecule inhibition of GLS, the use of morpholinos provided additional evidence for the requirement of GLS in P493 tumor xenograft growth.
Although BPTES is an effective tool compound for studying GLS, efforts have now yielded a new, more potent allosteric inhibitor of GLS, CB-839, that is being tested clinically (16). The challenge now lies in the ability to predict which tumors would respond to glutaminase inhibition and which combination therapy should be used in specific types of cancers (53). Previous work suggests that the presence of glutamine synthetase in breast cancers predicts independence from glutamine addiction and, hence, these tumors are not expected to respond to glutaminase inhibition (19). To date, triple-negative breast cancers have the features that suggest sensitivity to GLS inhibition (16). Further, a recent study, which requires validation, suggests that a subset of estrogen receptor–negative breast cancers with elevated MYC expression produced high levels of the oncometabolite 2-hydroxyglutarate (2HG) in a glutamine-dependent fashion (54). Since BPTES was demonstrated to inhibit the growth of an engineered gliomas cell line with a mutant isocitrate dehydrogenase (IDH), which produces 2HG in a glutamine-dependent fashion, 2HG could serve as a biomarker for this subset of breast cancers (55, 56). Several hematological malignancies, including multiple myeloma, are also expected to be responsive to GLS inhibition due to the prevalent role of MYC in their pathogenesis (7, 57, 58). In fact, myeloma may depend on GLS, since low circulating glutamine levels measured before therapy return to normal after standard treatment (59). With the emergence of new chemical GLS inhibitors heading for clinical use, our foundational study paves the way for exploring whether inhibition of GLS alone or in combination with other therapies will prove to be useful clinically.
Methods
Cell culture.
PC-3 and P493 cells were maintained in RPMI 1640 with 10% FBS and penicillin/streptomycin. mHCC 3–4 cells were maintained in DMEM high glucose, 10% FBS with sodium pyruvate, nonessential amino acids, and glutamine supplementation. Cells were counted using a hemocytometer.
Stable P493 cell line production.
pCDNA3.1 V5-GLS and pCDNA3.1 V5-GLS K325A (mutagenized with hGAC.K325A F-5′-GTGGACTAAGATTCAACGCACTATTTTTGAATGAAG-3′ and hGAC.K325A R-5′-CTTCATTCAAAAATAGTGCGTTGAATCTTAGTCCAC-3′) vectors were obtained as a gift from Andre Ambrosio (Laboratórios Nacionais de Biociências, Centro Nacional de Pesquisa em Energia e Materiais, Campinas, Brazil) and Sandra Gomes (Laboratórios Nacionais de Biociências) (24). The CMV promoter–GLS coding sequence and polyadenylation site were amplified using the primers F-5′-GATCCAGTTTGGTTAATTAATGCTTAGGGTTAGGCGTTTT-3′ and R-5′GCCGCTGCACCCTTAATTAAGGTTCTTTCCGCCTCAGAAG-3′. The CMV promoter–GLS coding sequence-polyadenylation site was cloned into the PacI site of the FUGW lentiviral (60) vector using the In-fusion HD Cloning Kit (Clontech). The vectors were confirmed by sequencing. FUGW V5-GLS and FUGW V5-GLS K325A vectors were used to make lentivirus using standard protocols in 293T cells using the pMD2.G and psPAX2 vectors. FUGW-V5 GLS or FUGW V5-GLS K325A virus was added to P493 or PC-3 cells with polybrene for 24 hours, as previously described (61). Cells were expanded, and then GFP-positive cells were isolated by fluorescence-activated cell sorting (FACS). Transgene expression was confirmed with Western blot for the V5 tag using an anti-V5 Ab (Invitrogen) and an anti-GLS Ab (GTX81012, GeneTex).
Transgenic mice: LAP/MYC model.
LAP-tTA/TRE-MYC (herein termed LAP/MYC; FVB/N background) transgenic mice that conditionally express human MYC cDNA in hepatocytes (40, 41) were used to generate mice with primary liver tumors. As TRE-MYC is integrated in the Y chromosome, TRE-MYC male mice were crossed with female mice carrying LAP-tTA, whereby the tTA express is driven in the liver by LAP. Mice possessing both LAP-tTA and TRE-MYC ectopically express MYC in the liver, while LAP/MYC mice treated with doxycycline or lacking 1 of the transgenes fail to express MYC. MYC expression was induced by removing doxycycline (100 μg/ml) from the drinking water of mice.
Transgenic mice: Gls mutant.
Previously described Gls mutant mice (44) were maintained on the 129SvEv × C57BL/6J background. Mice were genotyped using the 3-primer strategy as previously described (44).
LAP/MYC: Gls+/– murine survival study.
LAP/MYC mice (40, 41) were crossed with heterozygous Gls null mice (44). Male animals were genotyped to identify those carrying the desired combination. Mice were designated LAP/MYC:Gls+/+, LAP/MYC:Gls+/–, or wild type by genotyping and were tracked for survival (44). MYC was induced by doxycycline removal at 1 week after birth.
BPTES treatment of mouse HCC model.
MYC expression was induced in LAP/MYC mice by doxycycline removal on the day of birth. LAP/MYC mice were randomly assigned to the BPTES or vehicle treatment group. Intraperitoneal injections (12.5 mg/kg body weight every 3 days) of BPTES or vehicle control (10% DMSO in 200 μl of PBS every 3 days) were initiated 3 weeks after birth.
BPTES treatment of tumor xenografts.
P493 cells (2 × 107) were injected subcutaneously into the flank of athymic nude mice (Charles River Laboratories). When the tumor volumes reached approximately 100 mm3, intraperitoneal 0.2 ml injections of BPTES (200 μg) or vehicle control (10% DMSO in PBS) were initiated and carried out every 3 days for 10 days. The tumor volumes were measured using digital calipers every 3 days and calculated using the following formula: length (mm) × width (mm) × width (mm) × 0.52.
Vivo-Morpholino design.
The human GLS mRNA was analyzed for which exons would trigger NMD if skipped, with exon 4 being a candidate for triggering NMD. The exon 4–intron boundaries were submitted to the Gene Tools design service for morpholino design (http://www.gene-tools.com/). The GLS NMD Vivo-Morpholino sequence is 5′-ACATCAAAAGCATTACCTTTCCTCC-3′. The standard control nontargeting Vivo-Morpholino is the sequence designed and published by Gene Tools (CCTCTTACCTCAGTTACAATTTATA). The exon 1–intron 1 mouse boundary was submitted to Gene Tools design service for morpholino design. The mouse Gls NMD Vivo-Morpholino sequence is 5′-GCCTCACCCACGCAACTCACTTGTC-3′.
Vivo-Morpholino treatment of nude mice with xenografts.
Athymic nude mice (Charles River Laboratories) subcutaneously inoculated with P493 lymphoma cells as described above were allowed to form tumors with an average volume of approximately 100 or 200 mm3 before therapy was initiated. To test the therapeutic efficacy of Gls gene silencing, we randomly sorted mice bearing P493 tumors into 3 groups (n = 4–6 mice per group). Knockdown of human GLS gene was carried out with the GLS NMD Vivo-Morpholino obtained from Gene Tools. Equal volumes (60 μl) of PBS and standard control (nontargeting) Vivo-Morpholino (Gene Tools) were used as negative controls. Vivo-Morpholino or vehicle (PBS) was injected subcutaneously next to (~0.1 to 0.5 cm from the tumor), but not into, the xenografts every other day for 11 to 13 days. The tumor volumes were measured using digital calipers every other day and calculated using the following formula: length (mm) × width (mm) × width (mm) × 0.52. Complete regression was defined as absence of palpable tumor.
qPCR analysis of mouse tissue.
RNA was extracted from liver tumor or surrounding nontumor liver from LAP/MYC mice using the RNeasy Plus Mini-Kit (QIAGEN). RNA was reverse transcribed into cDNA with Superscript III Reverse Transcriptase Kit with random hexamers. Expression was analyzed using predesigned IDT PrimeTime primer/probe assays for mouse Gls, Gls2, and Actb using ABI TaqMan Gene expression master mix. Predesigned IDT PrimeTime primer/probe assay IDs were as follows: mouse Gls Mm.PT.45.6877164; mouse Gls2 Mm.PT.45.16450030.g; and mouse ActB Mm.PT.45.9990212.g.
Mouse tissue IHC.
Mouse IHC was performed essentially as described above. The slides were exposed to GLS2 Ab HPA038608 (Sigma-Aldrich; 1:100), GLS Ab ab156876 (Abcam; 1:100), anti-Ki67 Ab (ab16667), or the MYC Ab ab32072 (Abcam; 1:200).
CT imaging.
CT images were acquired on an X-SPECT small animal SPECT/CT system (Gamma Medica-Ideas). Mice were induced and maintained with 3% and 1.5% isoflurane (v/v), respectively. Data were analyzed using ImageJ software (http://imagej.nih.gov/ij/). For estimation of abdominal girth as a measure of tumor load, the maximum abdominal width was divided by the length of the abdomen (defined as the top of the pelvis to the bottom of the sternum). Abdominal girth and abdomen length measurements were performed with genotypes blinded from the analyzer.
Hematology.
Blood samples from BALB/c mice treated for 2 weeks with BPTES (12.5 mg/kg body weight every 3 days) or vehicle (10% DMSO in 200 μl of PBS every 3 days) were analyzed using the Hemavet 950FS hematology system (Drew Scientific) using the mouse key.
Toxicity study.
After a 14-day quarantine period, BALB/c mice were assigned at random to 2 groups. The study included a treatment group (12.5 mg/kg body weight every 3 days) and a control group (10% DMSO in 200 μl of PBS every 3 days). Mice were treated for 10 days (4 injections on days 1, 4, 7, and 10). Observations were made twice daily for clinical signs of pharmacologic and toxicologic effects of the BPTES. Histopathologic evaluation of brain, heart, skeletal muscle, lung, and kidney was done by a pathologist without any animal group information.
Liver, tumor, or xenograft glutamine/glutamate measurements.
Xenografts, liver, or tumors from anesthetized mice were frozen using a liquid nitrogen cold clamp. Tumors were weighed and dissected without thawing, and metabolites were extracted with 80/20 cold methanol/water, with the volume of methanol/water 15 or more times the volume of the tumor (0.2–1 g); the extracts were cleared by centrifugation, evaporated with a nitrogen evaporator, and then lyophilized. Metabolites were resuspended in 1 ml or less PBS and analyzed on a YSI 7100 analyzer. Concentration values were derived from metabolite standard curves, corrected for the volume of PBS used for resuspension, and then divided by the tissue wet weight to obtain a surrogate value for tissue metabolite concentration.
RNA analysis of human samples.
Pairs of freshly snap-frozen human HCC and adjacent normal tissues (n = 18) were collected and histopathologically diagnosed at the First Affiliated Hospital of Sun Yat-sen University. The qPCR analysis was performed essentially as described above with the following primers: GLS F-TCTACAGGATTGCGAACGTCT; GLS R-CTTTGTCTAGCATGACACCATCT; GLS2 F-TCTCTTCCGAAAGTGTGTGAGC; GLS2 R-CCGTGAACTCCTCAAAATCAGG; C-MYC F-AACGATTCCTTCTAACAG; and C-MYC R-GGCTAAATCTTTCAGTCT.
Human tissue IHC.
The tumors and paired adjacent nontumorous tissues from 15 HCC patients were obtained from the National Cancer Centre Singapore Tissue Repository. Abs used for IHC were GLS2 Ab HPA038608 (Sigma-Aldrich) and GLS Ab ab156876 (Abcam). The tissue specimens were deparaffinized with xylene, dehydrated in alcohol, boiled in antigen retrieval buffer (Vector Labs, low pH), and treated with hydrogen peroxide for 5 minutes before incubation with primary Ab overnight at 4°C. Concentrations of Abs used were as follows: anti-GLS: 1:50; anti-GLS2: 1:200. The isotype control rabbit IgG (I-1000, Vector Labs) showed no positive staining. Semiquantitative scoring of IHC was performed by a pathologist using the 0, 1+, 2+, and 3+ system. Heterogeneity of randomly selected microscopic fields was scored by estimated percentage of the tissue section staining at various levels. Overall scoring was determined by weighted average in samples with staining heterogeneity.
Cytospin methods.
P493 cells were cytospun onto the slides through SS-215 Dual Sample Chambers with Fast Cytopads (Wescor) in a 7620 Cytopro Cytocentrifuge Centrifuge (Wescor) at 212 g for 7 minutes. The slides were stained with H&E.
Primary human T cell proliferative analysis.
Primary human T lymphocytes were cultured in RPMI 1640 (Gibco; Life Technologies) supplemented with 10% FBS (HyClone), 10 mM HEPES, 2 mM l-glutamine, 10 U/ml penicillin G, and 100 μg/ml streptomycin. T cells were activated with 4.5 μm microbeads containing immobilized anti-human CD3 and anti-human CD28 (Gibco; Life Technologies) at a ratio of 3 beads to 1 cell. After 72 hours of stimulation in the presence of either DMSO or BPTES (10 μM), T cells were counted by flow cytometry using CountBright Beads (BD Biosciences), Via-Probe (BD Biosciences), and mAbs to human CD4/CD8. Cell volume was assayed using a Multisizer III particle counter (Beckman-Coulter).
Indirect coating of tissue culture surfaces with Abs for T cell stimulation.
Twelve-well tissue culture plates were treated with 10 μg/ml goat anti-mouse IgG (Cappel) overnight at 4°C. The plates were then rinsed 3 times with PBS, and a blocking buffer containing 5% BSA in PBS was applied for 1 hour. After a series of washes, the plates were incubated with 5 μg/ml of Okt3 (BioLegend) for 2 hours. Following a final series of washes, T cells were seeded at 1 × 106 cells/well and supplemented with soluble 9.3 (Bio-X-Cell). Additionally, T cells were treated with either DMSO or BPTES (10 μM). After 72 hours, T cells were counted by flow cytometry using CountBright Beads (BD Biosciences) and Viaprobe (BD Biosciences). Cell volume was assayed using a Multisizer III particle counter (Beckman-Coulter).
ROS staining.
Intracellular ROS staining was performed by incubating cells with 5-(and-6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (carboxy-H2DCFDA; Molecular Probes) at 37°C for 20 minutes; cells were washed in PBS according to the manufacturer’s instructions. To quantify the ROS level, the fluorescence intensity in the cells was detected by FACScan flow cytometers (BD Bioscience).
Glutaminase activity.
Glutaminase activity measurements in P493 B cells were adapted from previously described protocols (62).
CB-839 preparation.
CB-839 was a gift from Calithera BioSciences (16). CB-839 doses were made by serial dilutions in DMSO, with all CB-839 experiments carried out with 0.1% DMSO in media.
Flow cytometry.
P493 cells were treated with Tet for 24 hours. They were then exposed to BPTES or DMSO and released from Tet to induce MYC. The cells were collected at various time points for cell-cycle and BrdU incorporation studies. For cell-cycle distribution, after treatment with BPTES or DMSO, P493 cells were harvested by centrifugation and washed with PBS. Cells were stained with propidium iodide after fixation with 70% ethanol at 4°C overnight. Labeled cells were washed with PBS and then analyzed on a BD FACSCalibur platform (Biosciences). CellQuest Pro software was used for data acquisition and evaluation. For the cell proliferation assay, the staining protocol from the BD Biosciences — Pharmingen BrdU Flow Kits instruction manual was followed. P493 cells were treated with either BPTES or DMSO and pulsed with BrdU for 1 hour; this was followed by staining with anti-BrdU FITC and 7-AAD. BrdU FITC-A versus DNA 7-AAD dot plots with gates for different cell-cycle phases were generated with BD CellQuest Pro software on a Mac. FlowJo software was used for the reanalysis of data. For each experiment, 100,000 events per sample were recorded.
Western blot analysis.
Cell pellets were harvested in M-PER reagent with phosphatase and protease inhibitors (Pierce) after washing with PBS. The protein concentration of lysates was determined by BCA assay (Pierce), and approximately 60 μg of protein per well was separated by SDS/PAGE and transferred by iBlot gel transfer stacks (Invitrogen). The membrane was probed with rabbit anti-cyclin B1 Ab (ab2949), anti-cyclin E1 Ab (ab3927), PARP Ab (sc-25780), and anti-γH2AX (ab11174); this was followed by peroxidase-conjugated anti-rabbit Ab (1:8000 dilution, Pierce). Blots were reprobed with mouse anti–α-tubulin to confirm equal protein loading. LI-COR Odyssey CLX infrared imaging system (LI-COR Biosciences) was used for detection. GLS Western blots were performed essentially as described above using Ab GTX81012 (GeneTex).
Statistics.
Values are shown as mean ± SD. Data were analyzed using Student’s 2-tailed t test, and significance was defined as P < 0.05. MedCalc was used in this project to perform survival analysis and generate Kaplan-Meier plots.
Study approval.
Human tissue collections were obtained and studied according to institutional guidelines using protocols approved by the Partners (Singapore General Hospital and National Cancer Centre, Singapore) Human Research Committee or the Institutional Research Ethics Committee of Sun Yat-sen University Cancer Center. Written, informed consent was obtained from study participants. Tumors and paired adjacent nontumorous tissues from 15 HCC patients were obtained from the National Cancer Centre Singapore Tissue Repository with prior approval from the SingHealth Centralized IRB (SingHealth CIRB no: 2008/440/B). Primary human T lymphocytes were obtained under approval by the University of Pennsylvania Institutional Review Board. All mouse experiments were carried out according to the NIH’s Guide for the Care and Use of Laboratory Animals (8th ed. The National Academies Press. 2011.), and protocols were approved by the Animal Care and Use Committees at the University of Pennsylvania and Johns Hopkins University.
Supplementary Material
Acknowledgments
We thank Zandra Walton and Mai Dang for assistance. Z.E. Stine is supported by National Cancer Institute (USA) grant 5F32CA174148. P. Gao is supported by National Natural Science Foundation of China grant 81372148. This work was supported by the Abramson Family Cancer Research Institute, Leukemia and Lymphoma Society grant 6106-14 (to C.V. Dang), and NCI grants R01CA057341 and P30CA16620 (to C.V. Dang).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(6):2293–2306. doi:10.1172/JCI75836.
References
- 1.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkers CR, Maddocks OD, Cheung EC, Mor I, Vousden KH. Metabolic regulation by p53 family members. Cell Metab. 2013;18(5):617–633. doi: 10.1016/j.cmet.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 4.Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2(10):881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123(9):3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altman BJ, Dang CV. Normal and cancer cell metabolism: lymphocytes and lymphoma. FEBS J. 2012;279(15):2598–2609. doi: 10.1111/j.1742-4658.2012.08651.x. [DOI] [PubMed] [Google Scholar]
- 8.DeBerardinis RJ, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104(49):19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le A, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15(1):110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W, et al. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci U S A. 2012;109(23):8983–8988. doi: 10.1073/pnas.1203244109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullen AR, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481(7381):385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wise DR, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105(48):18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458(7239):762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki S, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A. 2010;107(16):7461–7466. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci U S A. 2010;107(16):7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gross MI, et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther. 2014;13(4):890–901. doi: 10.1158/1535-7163.MCT-13-0870. [DOI] [PubMed] [Google Scholar]
- 17.Seltzer MJ, et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70(22):8981–8987. doi: 10.1158/0008-5472.CAN-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shukla K, et al. Design, synthesis, and pharmacological evaluation of bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide 3 (BPTES) analogs as glutaminase inhibitors. J Med Chem. 2012;55(23):10551–10563. doi: 10.1021/jm301191p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang JB, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18(3):207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vander Heiden MG. Exploiting tumor metabolism: challenges for clinical translation. J Clin Invest. 2013;123(9):3648–3651. doi: 10.1172/JCI72391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chandel NS. Navigating Metabolism. Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory Press; 2014. [Google Scholar]
- 22.Xiang L, et al. Knock-down of glutaminase 2 expression decreases glutathione, NADH, and sensitizes cervical cancer to ionizing radiation. Biochim Biophys Acta. 2013;1833(12):2996–3005. doi: 10.1016/j.bbamcr.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Giacobbe A, et al. p63 regulates glutaminase 2 expression. Cell Cycle. 2013;12(9):1395–1405. doi: 10.4161/cc.24478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferreira AP, et al. Active glutaminase C self-assembles into a supratetrameric oligomer that can be disrupted by an allosteric inhibitor. J Biol Chem. 2013;288(39):28009–28020. doi: 10.1074/jbc.M113.501346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qie S, Chu C, Li W, Wang C, Sang N. ErbB2 activation upregulates glutaminase 1 expression which promotes breast cancer cell proliferation. J Cell Biochem. 2014;115(3):498–509. doi: 10.1002/jcb.24684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Heuvel AP, Jing J, Wooster RF, Bachman KE. Analysis of glutamine dependency in non-small cell lung cancer: GLS1 splice variant GAC is essential for cancer cell growth. Cancer Biol Ther. 2012;13(12):1185–1194. doi: 10.4161/cbt.21348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szeliga M, Bogacinska-Karas M, Rozycka A, Hilgier W, Marquez J, Albrecht J. Silencing of GLS and overexpression of GLS2 genes cooperate in decreasing the proliferation and viability of glioblastoma cells. Tumour Biol. 2013;35(3):1855–1862. doi: 10.1007/s13277-013-1247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajagopalan KN, DeBerardinis RJ. Role of glutamine in cancer: therapeutic and imaging implications. J Nucl Med. 2011;52(7):1005–1008. doi: 10.2967/jnumed.110.084244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shapiro RA, Clark VM, Curthoys NP. Inactivation of rat renal phosphate-dependent glutaminase with 6-diazo-5-oxo-L-norleucine. Evidence for interaction at the glutamine binding site. J Biol Chem. 1979;254(8):2835–2838. [PubMed] [Google Scholar]
- 30.Thornburg JM, et al. Targeting aspartate aminotransferase in breast cancer. Breast Cancer Res. 2008;10(5):R84. doi: 10.1186/bcr2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DaVanzo JP, Kang L, Ruckart R, Daugherty M. Inhibition of pyridoxal phosphokinase by aminooxyacetic acid. Biochem Pharmacol. 1966;15(1):124–126. doi: 10.1016/0006-2952(66)90118-3. [DOI] [PubMed] [Google Scholar]
- 32.Robinson MM, et al. Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES). Biochem J. 2007;406(3):407–414. doi: 10.1042/BJ20070039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeLaBarre B, et al. Full-length human glutaminase in complex with an allosteric inhibitor. Biochemistry. 2011;50(50):10764–10770. doi: 10.1021/bi201613d. [DOI] [PubMed] [Google Scholar]
- 34.Thangavelu K, et al. Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism. Proc Natl Acad Sci U S A. 2012;109(20):7705–7710. doi: 10.1073/pnas.1116573109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stalnecker CA, et al. Mechanism by which a recently discovered allosteric inhibitor blocks glutamine metabolism in transformed cells. Proc Natl Acad Sci U S A. 2015;112(2):394–399. doi: 10.1073/pnas.1414056112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emadi A, Jun SA, Tsukamoto T, Fathi AT, Minden MD, Dang CV. Inhibition of glutaminase selectively suppresses the growth of primary acute myeloid leukemia cells with IDH mutations. Exp Hematol. 2014;42(4):247–251. doi: 10.1016/j.exphem.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 37.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35(6):871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macintyre AN, Rathmell JC. Activated lymphocytes as a metabolic model for carcinogenesis. Cancer Metab. 2013;1(1):5. doi: 10.1186/2049-3002-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ko YH, et al. Glutamine fuels a vicious cycle of autophagy in the tumor stroma and oxidative mitochondrial metabolism in epithelial cancer cells: implications for preventing chemotherapy resistance. Cancer Biol Ther. 2011;12(12):1085–1097. doi: 10.4161/cbt.12.12.18671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beer S, et al. Developmental context determines latency of MYC-induced tumorigenesis. PLoS Biol. 2004;2(11):e332. doi: 10.1371/journal.pbio.0020332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shachaf CM, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431(7012):1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 42.Yuneva MO, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012;15(2):157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu S, et al. 13C-pyruvate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell Metab. 2011;14(1):131–142. doi: 10.1016/j.cmet.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masson J, et al. Mice lacking brain/kidney phosphate-activated glutaminase have impaired glutamatergic synaptic transmission, altered breathing, disorganized goal-directed behavior and die shortly after birth. J Neurosci. 2006;26(17):4660–4671. doi: 10.1523/JNEUROSCI.4241-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaisler-Salomon I, et al. Glutaminase-deficient mice display hippocampal hypoactivity, insensitivity to pro-psychotic drugs and potentiated latent inhibition: relevance to schizophrenia. Neuropsychopharmacology. 2009;34(10):2305–2322. doi: 10.1038/npp.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuhmacher M, et al. Control of cell growth by c-Myc in the absence of cell division. Curr Biol. 1999;9(21):1255–1258. doi: 10.1016/S0960-9822(99)80507-7. [DOI] [PubMed] [Google Scholar]
- 47.Billiard J, et al. Quinoline 3-sulfonamides inhibit lactate dehydrogenase A and reverse aerobic glycolysis in cancer cells. Cancer Metab. 2013;1(1):19. doi: 10.1186/2049-3002-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morcos PA, Li Y, Jiang S. Vivo-Morpholinos: a non-peptide transporter delivers Morpholinos into a wide array of mouse tissues. Biotechniques. 2008;45(6):613–614. doi: 10.2144/000113005. [DOI] [PubMed] [Google Scholar]
- 49.Zammarchi F, et al. Antitumorigenic potential of STAT3 alternative splicing modulation. Proc Natl Acad Sci U S A. 2011;108(43):17779–17784. doi: 10.1073/pnas.1108482108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaglio D, et al. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol Syst Biol. 2011;7:523. doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaglio D, Soldati C, Vanoni M, Alberghina L, Chiaradonna F. Glutamine deprivation induces abortive s-phase rescued by deoxyribonucleotides in k-ras transformed fibroblasts. PLoS One. 2009;4(3):e4715. doi: 10.1371/journal.pone.0004715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107(19):8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fendt SM, et al. Metformin decreases glucose oxidation and increases the dependency of prostate cancer cells on reductive glutamine metabolism. Cancer Res. 2013;73(14):4429–4438. doi: 10.1158/0008-5472.CAN-13-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terunuma A, et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest. 2014;124(1):398–412. doi: 10.1172/JCI71180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DiNardo CD, et al. Serum 2-hydroxyglutarate levels predict isocitrate dehydrogenase mutations and clinical outcome in acute myeloid leukemia. Blood. 2013;121(24):4917–4924. doi: 10.1182/blood-2013-03-493197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang JH, et al. Prognostic significance of 2-hydroxyglutarate levels in acute myeloid leukemia in China. Proc Natl Acad Sci U S A. 2013;110(42):17017–17022. doi: 10.1073/pnas.1315558110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yustein JT, Dang CV. Biology and treatment of Burkitt’s lymphoma. Curr Opin Hematol. 2007;14(4):375–381. doi: 10.1097/MOH.0b013e3281bccdee. [DOI] [PubMed] [Google Scholar]
- 58.Slack GW, Gascoyne RD. MYC and aggressive B-cell lymphomas. Adv Anat Pathol. 2011;18(3):219–228. doi: 10.1097/PAP.0b013e3182169948. [DOI] [PubMed] [Google Scholar]
- 59.Puchades-Carrasco L, et al. Multiple myeloma patients have a specific serum metabolomic profile that changes after achieving complete remission. Clin Cancer Res. 2013;19(17):4770–4779. doi: 10.1158/1078-0432.CCR-12-2917. [DOI] [PubMed] [Google Scholar]
- 60.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295(5556):868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 61.Chang TC, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci U S A. 2009;106(9):3384–3389. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Engler JA, Gottesman JM, Harkins JC, Urazaev AK, Lieberman EM, Grossfeld RM. Properties of glutaminase of crayfish CNS: implications for axon-glia signaling. Neuroscience. 2002;114(3):699–705. doi: 10.1016/S0306-4522(02)00357-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.