Abstract
Steroid-resistant nephrotic syndrome (SRNS) is a frequent cause of progressive renal function decline and affects millions of people. In a recent study, 30% of SRNS cases evaluated were the result of monogenic mutations in 1 of 27 different genes. Here, using homozygosity mapping and whole-exome sequencing, we identified recessive mutations in kidney ankyrin repeat-containing protein 1 (KANK1), KANK2, and KANK4 in individuals with nephrotic syndrome. In an independent functional genetic screen of Drosophila cardiac nephrocytes, which are equivalents of mammalian podocytes, we determined that the Drosophila KANK homolog (dKank) is essential for nephrocyte function. RNAi-mediated knockdown of dKank in nephrocytes disrupted slit diaphragm filtration structures and lacuna channel structures. In rats, KANK1, KANK2, and KANK4 all localized to podocytes in glomeruli, and KANK1 partially colocalized with synaptopodin. Knockdown of kank2 in zebrafish recapitulated a nephrotic syndrome phenotype, resulting in proteinuria and podocyte foot process effacement. In rat glomeruli and cultured human podocytes, KANK2 interacted with ARHGDIA, a known regulator of RHO GTPases in podocytes that is dysfunctional in some types of nephrotic syndrome. Knockdown of KANK2 in cultured podocytes increased active GTP-bound RHOA and decreased migration. Together, these data suggest that KANK family genes play evolutionarily conserved roles in podocyte function, likely through regulating RHO GTPase signaling.
Keywords: Cell Biology, Genetics, Nephrology
Introduction
Podocytes are polarized epithelial cells that are critical for the renal glomerular filtration barrier through their interdigitated foot processes. The filtration barrier is responsible for ultrafiltration of the blood and ensures that essential plasma proteins are retained. Therefore, dysfunction of podocytes is marked by the loss of proteins into the urine. Nephrotic syndrome (NS) is defined as the association of proteinuria, hypoalbuminemia, and edema and is the most common primary glomerular disease in children. The majority of children with NS respond to steroid therapy (steroid-sensitive NS [SSNS]). However, more than 20 percent of patients will fail to respond to steroid treatment (steroid-resistant NS [SRNS]) (1). SRNS inevitably progresses to end-stage kidney disease (ESKD) within a few years of onset, necessitating dialysis or renal transplantation for survival. It accounts for about 15% of all ESKD in children and has a 30% recurrence risk in renal transplants, leading again to ESKD (2). Monogenic mutations in 1 of 27 genes lead to SRNS. We have recently shown in a cohort of 1,700 families with SRNS that 30% of all cases are caused by mutations in one of these genes (3). Interestingly, most of the encoded gene products localize to podocytes, further confirming that podocytes are essential for maintaining the glomerular filtration barrier.
Drosophila pericardial nephrocytes are functionally analogous to human glomerular podocytes (4). Both cell types form highly specialized filtration structures called the “slit diaphragm” as well as the basement membrane. Both structures serve as the primary size- and charge-dependent filtration barriers. The function of pericardial nephrocytes can be readily monitored by assaying the filtration and uptake of secreted fluorescent proteins from the Drosophila hemolymph. This functional assay, in concert with nephrocyte-specific Gal4 drivers and RNAi transgenic libraries for individual Drosophila genes, makes it possible to screen and identify genes required for nephrocyte function (5). Previous studies have shown that many genes important for podocyte functions are also essential for nephrocyte function. The gene products include nephrin, NEPH1, and podocin at the slit membrane structures (4–6); ADCK4 in mitochondria function (7); and RHO GDI in the regulation of small GTPases (1). Many genes identified from Drosophila nephrocyte phenotypic screens have highly conserved human homologs that may play similar roles in podocytes (5).
To identify additional genes mutated in NS, here we performed homozygosity mapping (HM) and whole-exome sequencing (WES) in individuals with NS. We identified recessive mutations in kidney ankyrin repeat-containing protein 2 (KANK2) in two families with NS. In addition, we also discovered rare mutations in the related genes KANK1 and KANK4 in families with NS. KANK proteins are characterized by conserved ankyrin-repeat and coiled-coil domains and a motif (KN motif) at the N-terminus containing potential motifs for nuclear localization and export signals (8). In parallel, we identified what we believe to be a novel Drosophila gene, CG10249, as relevant for Drosophila nephrocyte function. This gene encodes a protein containing a KN motif, a coiled-coil motif, and 3 ankyrin repeats, similar to the domain composition of the 4 human KANK proteins. Therefore, we named it dKank. Here, we show that the KANK family genes are required for podocyte function in cultured podocytes, Drosophila nephrocytes, and zebrafish, suggesting that the KANK family genes play evolutionarily conserved roles in podocyte function and in the pathogenesis of NS.
Results
Mutations in KANK2 cause NS in humans.
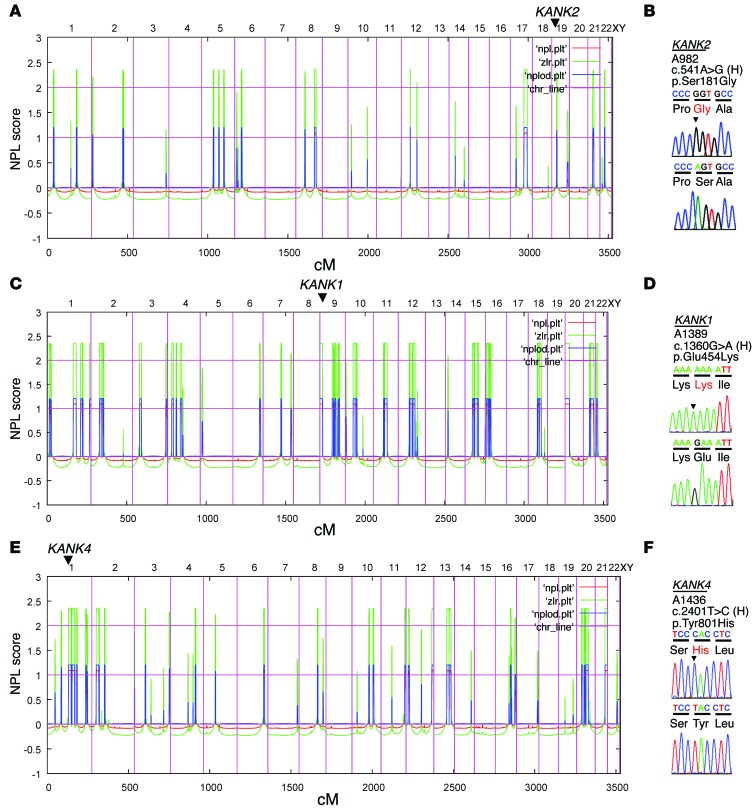
To identify additional genes mutated in NS, we performed HM followed by WES in families with NS. In a family (A982) of Arab origin, 2 siblings had early-onset SSNS with renal histology of minimal change disease (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI79504DS1). HM of one affected sibling (A982-21) yielded 14 regions of homozygosity by descent as candidate regions for a recessive mutation (Figure 1A). Following WES and filtering of variants from normal reference sequence (9), we detected a homozygous missense mutation (c.541A>G;p.S181G) in KANK2 (RefSeq accession NM_015393.6) on chromosome 19 (Figure 1B and Table 1). This variant was the only homozygous variant remaining from evaluation of the WES data (Supplemental Table 1) and segregated in a recessive way with the affected status in the family. In addition, this variant was absent from >6,500 controls (from 13,000 control chromosomes) in the Exome Variant Server ([EVS] http://evs.gs.washington.edu/EVS/). To discover additional mutations in KANK2, we performed exon resequencing of all 11 KANK2 exons using a high-throughput technique harnessing barcoded multiplex PCR and next-generation sequencing (10) in an additional 576 individuals with SSNS and 542 individuals with SRNS. In an individual with SSNS (A1751-21), we detected a homozygous missense mutation (c.2051C>T;p.S684F) in exon 9 of KANK2 (Table 1 and Supplemental Figure 1B). This variant is absent from the EVS and is predicted to be deleterious to protein function by publically available software programs (Table 1). For exclusion of known genetic causes of SRNS, 27 known genes previously linked to SRNS were screened in this individual, but no explanatory mutations were detected.
Figure 1. HM and WES identify genetic variants in the KANK1, KANK2, and KANK4 genes in families with NS.
(A, C, and E) Nonparametric lod scores (NPL) are plotted across the human genome. The x axis shows SNP marker positions on human chromosomes concatenated from p-ter (left) to q-ter (right). Genetic distance is given in cM. Maximum nonparametric lod score peaks indicate candidate regions of homozygosity by descent. (B, D, and F) Gene symbol (underlined), family number, variant, and predicted translational changes are given. Sequence traces are shown for the variant above normal control. Mutated nucleotides are indicated by arrowheads. (A) HM of an individual (A982-21) with SSNS. KANK2 is positioned (arrowhead) on chromosome 19 within one of the homozygous candidate regions. (B) Chromatogram of a KANK2 mutation identified in a family (A982) with SSNS (see also Table 1). (C) HM of an individual (A1389-21) with SSNS. KANK1 is positioned (arrowhead) on chromosome 9. (D) Chromatogram of a KANK1 variant identified in family A1389 (see also Table 1). (E) HM of an individual (A1436-24) with SRNS. KANK4 is positioned (arrowhead) with a homozygous region on chromosome 1. (F) Chromatogram of a KANK4 variant identified in family A1436 (see also Table 1).
Table 1. Rare variants identified in KANK1, KANK2, and KANK4 in individuals with NS.
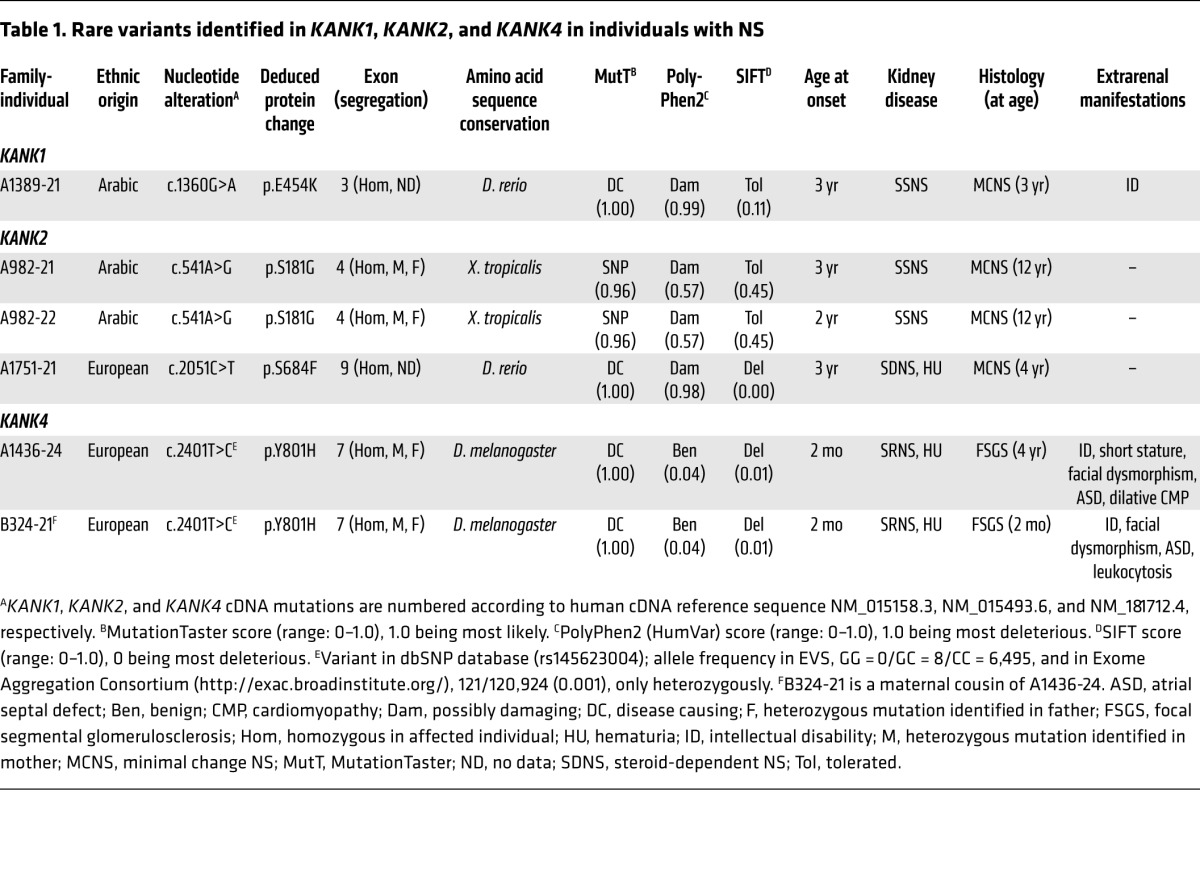
HM in an Arab family (A1389) yielded segments of homozygosity by descent with a cumulative genomic length of approximately 256 Mb (Figure 1C). Following variant filtering by HM and WES, 4 rare missense variants in KANK1, SOCS5, TBC1D2B, and GRIP1 remained (Supplemental Table 1). Mutations in the 27 known SRNS genes were excluded by evaluation of the WES data. The variant (c.1360G>A;p.E454K) in KANK1 (RefSeq accession NM_015158.3) was absent from >6,500 controls in the EVS (Figure 1D and Table 1). We examined an additional 576 individuals with SSNS and 542 individuals with SRNS to identify additional mutations in KANK1; however, we could not discover any additional mutation.
In a family from Slovakia, one child (A1436-24) and her maternal cousin’s grandchild (B324-21) had early-onset SRNS (Supplemental Figure 1C and Table 1). The renal histologies of A1436-24 and of B324-21 revealed focal segmental glomerulosclerosis (Supplemental Figure 1, D and E). HM in individuals A1436-24 and B324-21 yielded regions of homozygosity by descent with cumulative genomic lengths of approximately 228 Mb and 370 Mb, respectively (Figure 1E and Supplemental Table 1). Following variant filtering by HM and WES, two rare variants in KANK4 and THBS3 remained, and both variants segregated with the affected status in the family (Supplemental Table 1). Mutations in the 27 known SRNS genes were excluded by evaluation of the WES data. The variant (c.2401T>C;p.Y801H) in KANK4 (RefSeq accession NM_0181712.4) is reported as a SNP in the dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/); however, its minor allele frequency is less than 0.004, and it never occurred in the homozygous state (Figure 1F and Table 1). The KANK4 variant is predicted to be deleterious to protein function by publically available software programs and alters an amino acid residue conserved throughout evolution down to Drosophila melanogaster (Table 1).
Thus, our discovery of mutations in the KANK family genes by independent HM and WES provided genetic support for pathogenic roles of this group of proteins in NS. The KANK family proteins, KANK1–KANK4, are characterized by their unique structure, coiled-coil motifs in the N-terminal region, ankyrin repeats in the C-terminal region, and an additional motif, the KN motif, at the N-terminus (8).
KANK1, KANK2, and KANK4 localize to podocytes in glomeruli.
KANK family proteins are localized prominently to the cytoplasm of kidney cell lines (8). Because most of products of genes that cause NS if mutated are localized to podocytes, we examined localization of KANK1, KANK2, and KANK4 in adult rat kidney sections. In glomeruli, KANK1 was predominantly expressed in podocytes, as identified by the expression of nuclear WT1 (Figure 2A and Supplemental Figure 2A). KANK1 partially colocalized with synaptopodin, which regulates RHOA signaling (Figure 2A and ref. 11). For KANK2, a previous study showed that Kank2 mRNA is enriched in glomeruli rather than tubules in the mouse kidney and that KANK2 partially colocalizes with synaptopodin, but not with endothelial marker CD31, in human adult kidney (12). KANK2 localized to podocyte cell bodies as well as nuclei in rat glomeruli (Figure 2B and Supplemental Figure 2B). KANK proteins contain a KN motif, which includes potential nuclear localization and export signals (8), and were shown to shuttle between the nucleus and the cytoplasm via a process mediated by nuclear localization and export signals (13). KANK4 localized to podocyte cell bodies and primary processes (Figure 2C and Supplemental Figure 2C). KANK2 and KANK4 did not show any colocalization with synaptopodin (Figure 2, B and C). To investigate whether mutation of KANK proteins present in individuals with NS leads to a change in their subcellular localization, we performed immunofluorescence in cultured human podocytes upon overexpression (Supplemental Figure 3). However, none of the variant proteins exhibited any difference in subcellular localization compared with that of wild-type KANK proteins.
Figure 2. KANK1, KANK2, and KANK4 localize to podocytes in rat glomeruli.
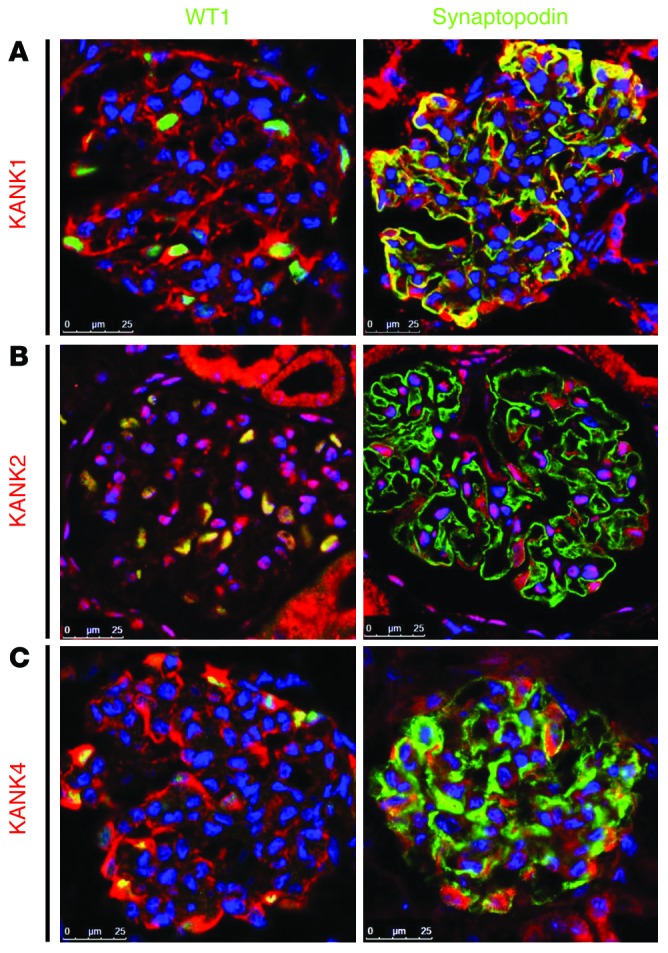
(A) Coimmunofluorescence of KANK1 with podocyte marker proteins in rat glomeruli. KANK1 is highly expressed in podocytes, as identified by the expression of nuclear WT1. KANK1 partially colocalizes with synaptopodin. (B) Coimmunofluorescence of KANK2 with podocyte marker proteins in rat glomeruli. KANK2 is expressed in cytoplasm and nuclei of podocytes. KANK2 does not colocalize with synaptopodin. (C) Coimmunofluorescence of KANK4 with podocyte marker proteins in rat glomeruli. KANK4 localizes to podocyte cell bodies and primary processes but does not colocalize with synaptopodin. Scale bar: 25 μm.
dKank is required for Drosophila cardiac nephrocyte function and cytoskeleton structure.
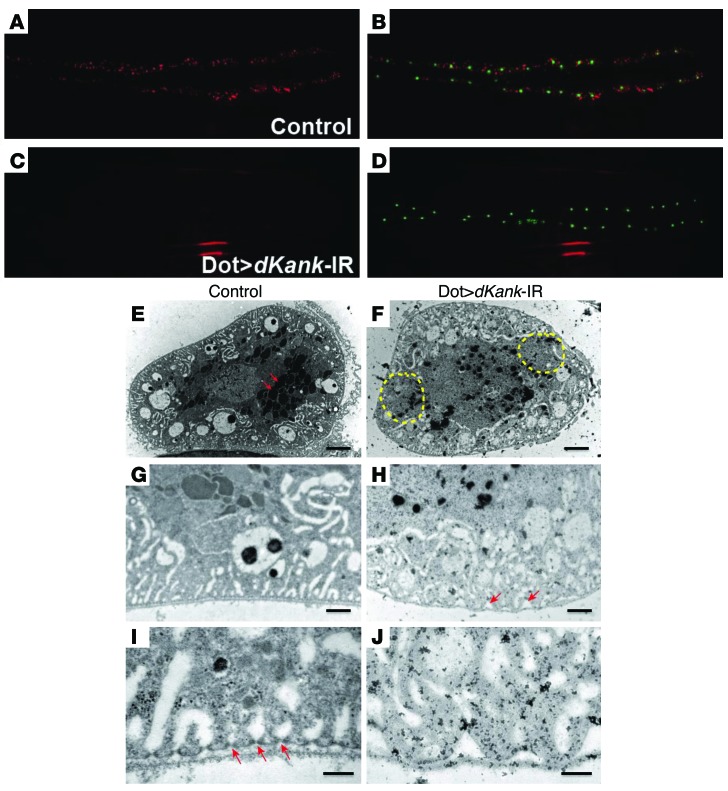
From a functional genetic screen for Drosophila cardiac nephrocytes, we identified a gene we believe to be novel, CG10249. This gene encodes the unique KANK family protein in Drosophila. Therefore, we named it dKank. The dKank protein contains a KN motif, a coiled-coil motif, and 3 ankyrin repeats, similar to the domain composition of the 4 human KANK proteins. Compared with wild-type nephrocytes (Figure 3, A and B), RNAi knockdown of dKank in nephrocytes substantially reduced the fluorescent protein uptake (as shown by red fluorescent protein fused with the atrial natriuretic factor [ANF-RFP] signal, Figure 3C) of cardiac nephrocytes, whereas the number and size of the cardiac nephrocytes remained the same (Figure 3D), as indicated by Hand-GFP labeling (green). Another independent RNAi line for dKank was tested to confirm that dKank is required for nephrocyte function. Significant reduction of fluorescent protein uptake was observed for both RNAi lines for dKank (Supplemental Figure 4). To examine in detail the exact roles of dKank in nephrocyte cellular function and structure, we performed transmission electron microscopy (TEM) analysis on third instar Drosophila larvae cardiac nephrocytes following dKank RNAi knockdown. Compared with wild-type nephrocytes (Figure 3, E, G, and I), dKank RNAi in cardiac nephrocytes led to uneven distribution of endocytic vacuoles and lacuna channels (Figure 3F). Wild-type cardiac nephrocytes showed evenly distributed and layered vesicles and vacuoles as well as a large number of lysosomes (Figure 3E, arrows). In contrast, cardiac nephrocytes with dKank RNAi showed diminished lacuna structures in many areas (Figure 3F, yellow dashed circles) as well as a reduced number of vacuoles and lysosomes (Figure 3F). High-magnification images showed that the surface of the cardiac nephrocyte upon knockdown was not as even and flat as that of wild-type nephrocytes (Figure 3, G and H), and marked gaps can be seen between the wave-shaped plasma membrane and the basement membrane of the nephrocytes with dKank RNAi (Figure 3H, arrows). A closer look at the slit diaphragm filtration structures showed that the evenly distributed slit diaphragm in wild-type nephrocytes (Figure 3I, arrows) was absent from nephrocytes with dKank RNAi (Figure 3J). In summary, compared with wild-type nephrocytes (Figure 3, G and I), lack of dKank led to uneven distribution of lacuna channels along the plasma membrane surface (Figure 3H) and abnormal lacuna shape (Figure 3J).
Figure 3. dKank is required for Drosophila cardiac nephrocyte function and cytoskeleton structure.
(A and B) Control Drosophila larvae carrying myosin heavy chain promoter–driven ANF-RFP (red), which is secreted from muscles into hemolymph and then filtered and taken up by cardiac nephrocytes. Hand-GFP labels the nuclei of nephrocytes. (C) Nephrocytes with dKank RNAi knockdown using Dot-Gal4 (Dot>dKank-IR) show substantially reduced uptake of ANF-RFP, (D) whereas the number and size of nephrocytes remain the same as those of wild-type nephrocytes. (E) Control third instar Drosophila larvae nephrocytes show evenly distributed vesicles (arrows) and vacuoles in distinct layers of cytoplasm. (F) Nephrocytes with dKank RNAi exhibit uneven distribution of endocytic vacuoles and lacuna channels, with reduced numbers of vesicles and vacuoles in some areas (yellow circles) but increased vesicles and vacuoles in other areas as well as reduced numbers of lysosomes. (G) High-magnification image of control nephrocytes showing even and flat plasma membrane and adjacent basement membrane. (H) Nephrocytes with dKank RNAi show wave-shaped plasma membrane surface and gaps between the plasma membrane and the basement membrane (arrows). Distribution of lacuna channels close to the nephrocyte cell surface is disrupted. (I) High-magnification images of control nephrocytes showing evenly distributed electron-dense slit diaphragm (arrows) and lacuna channel structures. (J) Nephrocytes with dKank RNAi show wave-shaped plasma membrane surface, without slit diaphragm structures and abnormally shaped lacuna channels. Original magnification, ×20 (A–D). Scale bar: 4 μm (E and F); 1 μm (G and H); 0.2 μm (I and J).
Knockdown of kank2 in zebrafish results in proteinuria.
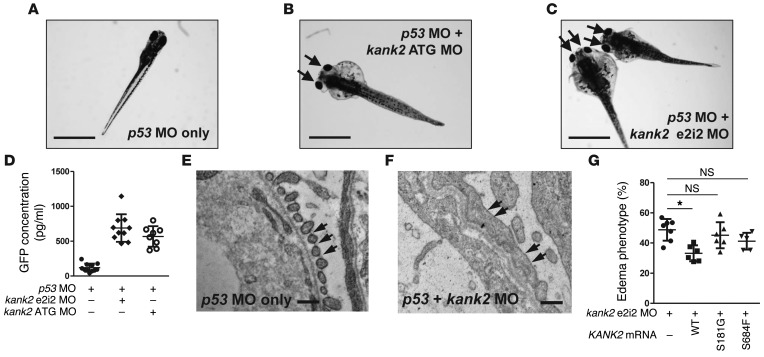
Because we have identified mutations in KANK2 in two families with NS, we performed knockdown of the KANK2 ortholog in zebrafish to further examine the role of KANK2 for glomerular function. Zebrafish kank2 has 48% amino acid sequence identity to human KANK2. A p53 morpholino oligonucleotide (MO) is known to reduce the cell death induced by off-target effects of MOs (14), and injection of p53 MO alone did not produce any phenotype in zebrafish (Figure 4A and Supplemental Table 2). Upon knockdown of kank2 by a MO targeting the translation initiation site of zebrafish kank2 (ATG MO), we observed periorbital edema and total body edema in 54.5% of larvae at 120 hours after fertilization, reminiscent of signs of NS in humans (Figure 4B). Similar results were obtained with a MO targeting the donor site of exon 2 (e2i2) of kank2 (periorbital edema and total body edema in 52.3% of larvae at 120 hours after fertilization) (Figure 4C and Supplemental Figure 5A). The proteinuric effect of both kank2 e2i2 and ATG MOs was confirmed using an established zebrafish proteinuria assay (15) by ELISA against a fusion protein of vitamin D–binding protein (VDBP) and GFP in I-fabp::VDBP-GFP transgenic zebrafish (Figure 4D).
Figure 4. Functional analysis of kank2 knockdown in zebrafish.
(A) Control zebrafish injected with p53 MO. p53 MO did not produce any phenotype until 168 hours after fertilization (n > 100). (B) Zebrafish coinjected with a MO targeting the translation initiation site of zebrafish kank2 (ATG MO) and p53 MO. At 120 hours after fertilization, kank2 morphants display the nephrosis phenotype of periorbital edema (arrows) and body edema in 54.5% of embryos (113 of 207). (C) Zebrafish coinjected with a MO targeting exon 2 splice donor site of kank2 (e2i2 MO) and p53 MO. e2i2 MO causes edematous phenotypes (arrows) in 52.3% of embryos (243 of 464). (D) Proteinuria assay in I-fabp::VDBP-GFP transgenic zebrafish. Note that knockdown of kank2 by either e2i2 or ATG MO causes significant proteinuria compared with that in control fish. (E and F) Electron microscopic structure of glomerular basement membrane and podocyte foot processes in 5-day-old (E) control and (F) kank2 morphant zebrafish. In the control, the foot processes are regularly spanned by slit diaphragms (arrows). In contrast, the foot processes of morphants are effaced and disorganized, with only occasional intercellular junctions (arrows in F). The glomerular basement membrane is disorganized. (G) Functional analysis of KANK2 mutations in zebrafish. Coinjection of kank2 e2i2 MO with a human wild-type KANK2 mRNA (156 of 491) partially rescued edematous phenotypes of kank2 morphants, whereas injection of KANK2 mRNA bearing the p.S181G (277 of 623) or p.S684F (187 of 447) mutation failed to rescue the phenotype. Error bars indicate the SD of more than 3 independent experiments. *P < 0.05, 2-tailed Student’s t test. Scale bar: 1 mm (A–C); 1 μm (E and F).
We then performed TEM imaging of zebrafish pronephric glomeruli to permit a more direct evaluation of a nephrosis phenotype on the subcellular level. When compared with p53 MO–injected controls (Figure 4E and Supplemental Figure 5B), we observed characteristic alterations of nephrosis upon knockdown of kank2 (Figure 4F and Supplemental Figure 5, C and D). These included podocyte foot process effacement and disorganization, rarefaction of slit membranes, and disorganization of the glomerular basement membrane in zebrafish glomeruli (Figure 4F) as previously described in plce1 knockdown (16).
To test the pathogenicity of the identified mutations, we injected synthetic mRNA containing KANK2 missense mutations into zebrafish with kank2 e2i2 MO. Injection of wild-type human KANK2 mRNA partially rescued the edema phenotype (Figure 4G). In contrast, both mutant mRNAs failed to rescue the kank2 morphant phenotype (Figure 4G), indicating that these mutations cause loss of function.
KANK2 interacts with ARHGDIA.
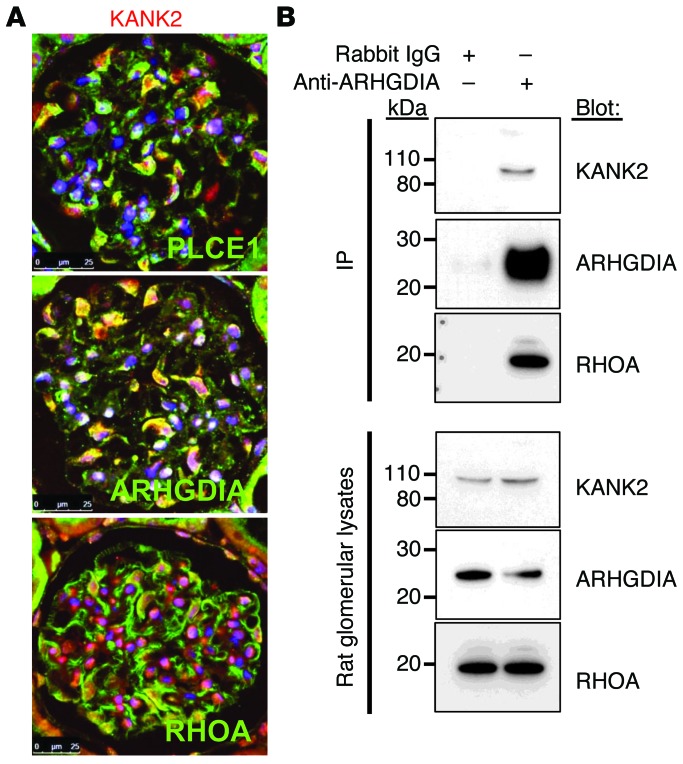
As knockdown of kank2 resulted in a nephrosis phenotype in zebrafish larvae, we further examined KANK2 expression in glomeruli of adult rat kidneys. In cytoplasm and nuclei of podocytes, KANK2 partially colocalized with ARHGDIA and PLCE1, two proteins of which genes are mutated in human NS (Figure 5A and refs. 1, 16). In addition, we found that KANK2 also partially colocalized with the RHO small GTPase RHOA in podocytes (Figure 5A). As KANK2 partially colocalized with ARHGDIA and RHOA and as we have previously shown that the protein complex precipitated by ARHGDIA in rat glomerular lysates includes RHOA, RAC1, and CDC42 (1), we examined whether this protein complex includes KANK2. We performed coimmunoprecipitation experiments using rat glomerular lysates. The anti-ARHGDIA antibody coimmunoprecipitated KANK2 as an interaction partner and also coimmunoprecipitated RHOA as previously shown (Figure 5B and ref. 1). We then performed coimmunoprecipitation in cultured human podocytes to examine whether the KANK2 variant (p.S181G), which we detected in the two siblings with NS of family A982, affects protein-protein interaction. The missense KANK2 variant showed stronger binding affinity to ARHGDIA compared with wild-type KANK2 when coexpressed and coimmunoprecipitated in cultured podocytes (Supplemental Figure 6A). We confirmed this finding by GST pull-down assay using lysates of podocytes transfected with WT KANK2 or p.S181G KANK2 (Supplemental Figure 6B). Interestingly, when KANK2 p.S181G was present, the binding affinity of ARHGDIA to RHOA, RAC1, and CDC42 seemed to be increased (Supplemental Figure 6, A and B). These data suggest that KANK2 may be related to RHO GTPase function, analogous to the regulation of RHOA by KANK1 via 14-3-3 (17).
Figure 5. KANK2 colocalizes with and interacts with ARHGDIA in rat glomeruli.
(A) Coimmunofluorescence of KANK2 with PLCE1, ARHGDIA, and RHOA. KANK2 partially colocalizes with PLCE1 and ARHGDIA, which are associated with NS. Anti-PLCE1 and anti-ARHGDIA antibodies were directly fluorescently labeled with Alexa Fluor 547. KANK2 also partially colocalizes with RHOA in rat glomeruli. Scale bar: 25 μm. (B) Coimmunoprecipitation of KANK2 in rat renal glomerular lysates. The protein complex precipitated by an anti-ARHGDIA antibody includes KANK2 as well as RHOA.
KANK2 knockdown in cultured podocytes causes increased levels of active RHOA and a decreased migratory phenotype.
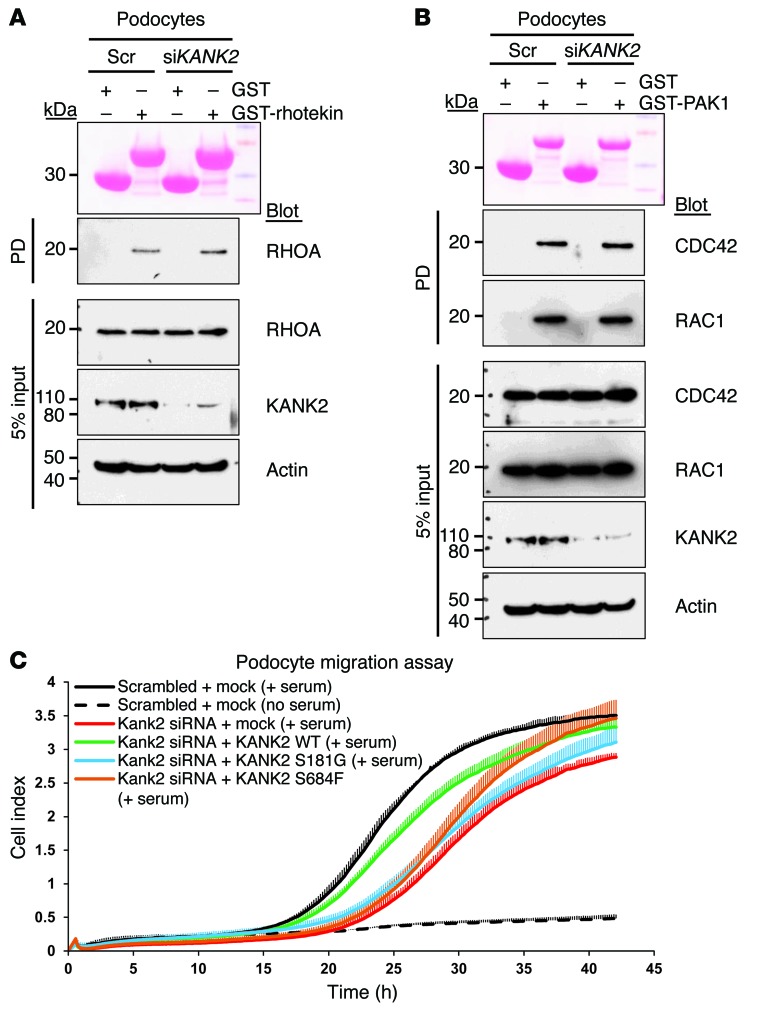
Previously, we have shown that mutations in ARHGDIA cause monogenic NS and that they change in the active states of RHO GTPases (1). Here, we showed that KANK2 interacts with the RHO GTPase regulator ARHGDIA. To test whether KANK2 is also involved in RHO GTPase signaling, we performed knockdown of KANK2 in cultured human podocytes. We used purified rhotekin to assay the active GTP-bound state of RHOA and used purified PAK1 to assay the active states of RAC1 and CDC42. Knockdown of KANK2 increased the GTP-bound form of RHOA (Figure 6A) but had no effect on RAC1 or CDC42 (Figure 6B). To test the pathogenicity of the identified mutations, we overexpressed wild-type and mutant KANK2 proteins in murine cultured podocytes. Regardless of whether wild-type or mutant protein was used, overexpression of KANK2 in itself did not change RHOA activity (Supplemental Figure 7A). However, similar to human cultured podocytes, knockdown of Kank2 resulted in increased active RHOA in murine podocytes (Supplemental Figure 7B). This increased RHOA activity was reversed by overexpression of wild-type KANK2 but not by overexpression of KANK2 p.S181G or p.S684F (Supplemental Figure 7B), suggesting that both mutations cause loss of function regarding the regulation of RHOA. Human KANK2 was resistant to Kank2 siRNAs used in this experiment (Supplemental Table 2). We then examined the effect of KANK2 on podocyte migration and found that knockdown of Kank2 in murine cultured podocytes caused a decreased migratory phenotype (Figure 6C), which was reversed by overexpression of human KANK2. In contrast, overexpression of KANK2 p.S181G failed to rescue the decreased migratory phenotype caused by Kank2 knockdown, whereas overexpression of KANK2 p.S684F resulted in partial rescue, indicating that this mutation is potentially hypomorphic in migration (Figure 6C).
Figure 6. Effects of Kank2 knockdown on RHO GTPase activity and podocyte migration in cultured podocytes.
(A) Active GTP-bound RHOA precipitated from cultured human podocytes transfected with scrambled (Scr) or KANK2 siRNA using a GST-rhotekin (RBD) pull-down (PD) assay. Ponceau red staining shows the GST proteins used. Compared with control podocytes, podocytes transfected with KANK2 siRNA exhibit a relative increase in RHOA. The efficiency of knockdown by siRNA was confirmed by immunoblotting with an anti-KANK2 antibody. (B) Active GTP-bound forms of RAC1 and CDC42 precipitated from podocytes transfected with scrambled or KANK2 siRNA using a GST-PAK1 (CRIB) pull-down assay. Cells transfected with scrambled control siRNA versus KANK2 siRNA exhibited no significant difference in relative RAC1 and CDC42 activity. Pull-down experiments are representative of more than 3 experiments. (C) Effect of Kank2 knockdown on podocyte migration. Cultured murine podocytes transfected with Kank2 siRNA (red) exhibited less active migration compared with those transfected with scrambled siRNA (black). Decrease in podocyte migration by Kank2 knockdown was rescued by overexpression of human wild-type KANK2 in podocytes (green) but not by overexpression of the KANK2 p.S181G variant (blue). Overexpression of p.S684F resulted in partial rescue, indicating that this mutation is potentially a hypomorphic mutation. Error bars are shown in only one direction for clarity, and the result shown is representative of 3 experiments.
Furthermore, we examined whether KANK1 and KANK4 have effects on active states of RHO GTPases. Similar to KANK2, knockdown of KANK1 in cultured podocytes increased active RHOA and RAC1 but not CDC42 (Supplemental Figure 8). This is consistent with previous studies that have shown that knockdown of KANK1 in HeLa cells increases RHOA-GTP (17) and that KANK1 specifically inhibits the binding of IRSp53 with active RAC1 but not with active CDC42 (18). In contrast, knockdown of KANK4 in podocytes did not have any effect on the active states of RHOA, RAC1, and CDC42 (data not shown). In the podocyte migration assay, knockdown of KANK1 resulted in decreased migration, whereas knockdown of KANK4 did not change the migratory phenotype of podocytes compared with that of control podocytes (data not shown).
Discussion
Here, we described the function of KANK proteins in podocytes and we identified genetic mutations in the KANK family genes in families with NS by the combined approach of WES and HM. We showed that KANK1, KANK2, and KANK4 localize to podocytes in glomeruli and regulate RHO GTPase activity. Knockdown of KANK1 and KANK2 in podocytes resulted in increases of the GTP-bound form of RHOA and reduced their migratory phenotype. By functional screen in Drosophila nephrocytes and knockdown of kank2 in zebrafish, we demonstrated that the function of this gene is required for proper podocyte function. TEM analysis of both Drosophila and zebrafish showed that knockdown of kank genes led to disrupted slit diaphragm filtration structures, suggesting that in both model systems the kank genes are required for normal cytoskeleton structure, which in turn is required for the formation of slit diaphragm filtration structures. In addition, the identification of both rhoGDI and kank from the functional genetic screen in Drosophila cardiac nephrocytes indicates that these genes may play evolutionarily conserved roles in the function of nephrocytes and podocytes (1).
KANK was originally identified in a genomic region, showing a loss of heterozygosity in renal cell carcinoma. Reduced or loss of expression of KANK1 was observed in renal cell carcinoma tissues and cell lines, and overexpression of KANK1 resulted in disorganization of β-actin distribution in G-402 cells (19). KANK1 is known to regulate actin polymerization and cell migration, and this function of KANK1 is mediated through two signaling pathways: the regulation of RHOA activity through phosphoinositide 3-kinase/AKT (PI3K/AKT) signaling (17) and the regulation of RAC1 signaling by inhibition of IRSp53 (18). In this study, we showed that KANK2, as well as KANK1, regulates RHO GTPases and cell migration in podocytes and that KANK2 is in a complex with ARHGDIA.
We have identified by HM and WES mutations in KANK1 and KANK4 in a family with NS. It appears that mutation of KANK1 or KANK4 represents a very rare cause of NS, as screening of more than 1,100 individuals with NS using a high-throughput mutation analysis failed to identify any additional mutation in KANK1 or KANK4. This is consistent with the finding that some of the recently identified recessive genetic causes of NS are exceedingly rare (1, 20, 21). In addition, we identified two families with NS with KANK2 variants. Recently, Ramot et al. identified a homozygous missense mutation (p.A670V) in KANK2 in two families presenting with keratoderma and wooly hair and showed that localization of NCOA3 (SRC-3) was changed in individuals with p.A670V KANK2 mutation (22). Therefore, we have examined whether the identified mutations in KANK2 affect interaction with NCOA3. None of the KANK2 mutations identified in individuals with NS abrogated the interaction with NCOA3 (Supplemental Figure 9). Additional identification of genetic variants in the KANK family genes is necessary to determine the phenotypic spectrum of mutations.
RHO GTPases are key regulators of cytoskeletal dynamics in a wide range of cellular processes, including cell adhesion, migration, and cell division (23). RHO GTPases are tightly regulated, so at any given time, only a small fraction of all RHO GTPases present in the cell are in the active state (24). The cycling of RHO GTPases between active (GTP-bound) and inactive (GDP-bound) states is regulated by 3 sets of proteins: quinine nucleotide-exchange factors, GTPase-activating proteins, and guanine nucleotide-dissociation inhibitors (24). It has been shown that aberrant RHO GTPase signaling in podocytes results in proteinuria in humans and animal models (1, 25–30). Knockdown of KANK1 and KANK2 in cultured podocytes led to an increase in the GTP-bound form of RHOA. Previously, it was demonstrated that transgenic mice, which overexpress constitutively active RhoA in podocytes, exhibit foot process effacement and proteinuria (26). Our current data from Drosophila, zebrafish, and human patients demonstrate that KANK family proteins have essential roles in podocyte/nephrocyte function and regulate RHO GTPase activity. Because NS caused by KANK variants is due to dysregulation of RHO GTPase activity, it is worth considering that this genetic form of NS, due to mutations in the KANK family genes, may be treated with drugs that inhibit or promote RHO GTPase activity. Depletion of KANK4 in podocytes did not change the activity of RHO GTPases or cell migration, so the function of KANK4 in podocytes may be different from that of KANK1 or KANK2.
Methods
Study participants.
After obtaining informed consent, clinical data and blood samples were obtained from individuals with SRNS or SSNS (see Supplemental Methods). The diagnosis of NS was made by (pediatric) nephrologists, based on standardized clinical and renal histologic criteria (31). Renal biopsies were evaluated by renal pathologists. Clinical data were obtained using a questionnaire provided by Renal Genes, Boston Children’s Hospital (http://www.renalgenes.org).
RNAi-based Drosophila nephrocyte functional screen.
The functional readout, transgenic flies, and screen strategies were described previously (5). The first dKank RNAi line (dKank-IR1, BL-33432) was identified from the nephrocyte screen and then verified by another independent dKank RNAi line (dKank-IR2, v-15009). ImageJ (version 1.48) was used to quantify the intensity of the ANF-RFP signal in nephrocytes. Threshold was adjusted to each picture to remove the background first on both ANF-RFP (red) and Hand-GFP (green) channels. The ANF-RFP mean intensity in each nephrocyte was normalized by the Hand-GFP of the same cell. At least 20 nephrocytes were measured for each genotype.
TEM analysis of Drosophila cardiac nephrocytes.
The protocols and reagents were described previously (5, 32).
HM, whole-exome resequencing, and mutation calling.
HM, whole-exome resequencing, and mutation calling were performed as described previously (9).
Plasmids, siRNAs, cell culture, and transfection.
Human KANK1 (GenBank BC037495), KANK2 (BC098105), and KANK4 (GenBank BC060866) clones were purchased from Open Biosystems. Mutated clones were generated by a PCR-based site-directed mutagenesis method. The KANK-specific and control scrambled siRNAs were purchased from GE Dharmacon. The immortalized human podocytes were maintained in RPMI plus GlutaMAX-I (Gibco) supplemented with 10% fetal bovine serum, 50 IU/ml penicillin/50 μg/ml streptomycin, and insulin-transferrin-selenium-X (33). The immortalized murine podocytes were provided by Peter Mundel (Massachusetts General Hospital, Boston, Massachusetts, USA) and RPMI plus GlutaMAX-I supplemented with 10% fetal bovine serum, 50 IU/ml penicillin/50 μg/ml streptomycin, and IFN-γ (50 U/ml). Plasmids and siRNAs were transfected into podocytes grown at the permissive temperature of 33°C using Lipofectamine 2000 (Invitrogen).
Immunoblotting, immunoprecipitation, pull-down assay, and immunofluorescence staining.
Immunoblotting, immunoprecipitation, pull-down assay, and immunofluorescence staining were performed as described previously (1). GST-rhotekin RBD beads were purchased from Cytoskeleton Inc. GST-PAK1 CRIB domain and GST-ARHGDIA were purified from BL21(DE3) Escherichia coli strain. Anti-KANK2 (HPA015643), anti-KANK1 (HPA005539), and anti-KANK4 (HPA014030) (Sigma-Aldrich); anti-FLAG (2368S), anti-MYC (2276S), and anti-BiP (3177S) (Cell Signaling Technology); anti-RAC1 (610650) and anti-CDC42 (610928) (BD Transduction Laboratories); anti-ARHGDIA (sc-360), anti-RHOA (sc-418), and anti-V5 (sc-83849-R) (Santa Cruz Biotechnology); and anti-synaptopodin (American Research Products) were purchased from commercial sources. Anti-PLCE1 antibody was previously described by Hinkes et al. (16). Anti-ARHGDIA and anti-PLCE1 antibodies were labeled with the APEX Alexa Fluor Labeling Kit (Life Technologies). Alexa Fluor 488 Phalloidin (A12379) was purchased from Life Technologies. Coimmunoprecipitation was performed using rec-Protein A-Sepharose 4B Conjugate (Life Technologies) or EZview Red ANTI-FLAG M2 Affinity Gel (Sigma-Aldrich). Fluorescent images were obtained with a Leica SP5X laser scanning microscope.
Podocyte migration assay.
Real-time migration assay was performed using the xCELLigence system (ACEA Biosciences) in CIM-plate 16 according to the manufacturer’s instructions. Briefly, 24 hours after transfection, 4 × 104 cells were plated in serum-free media in the upper chamber. The lower chambers were plated with 10% FBS for chemoattraction or with serum-free media. The obtained data were analyzed using RTCA software. Results were presented as time versus cell index curve.
Zebrafish studies.
Zebrafish (Danio rerio) were maintained and reared as described previously (15). 0.2 mM MOs were injected into the AB* wild-type strain for phenotype analysis of morphants, TEM, and drug treatment. The proteinuria assay was performed using I-fabp::VDBP-GFP transgenic fish and the GFP ELISA Kit (Cell Biolabs) as described previously (15). For mRNA rescue in zebrafish, the wild-type and mutant KANK2-coding region was subcloned into the pCSmCherryDest expression vector (34) using Gateway technology (Invitrogen). Capped RNAs were synthesized from linearized vectors using the SP6 mMessage mMachine Kit (Ambion). RNA products were column purified with the RNeasy Mini Kit (Qiagen) prior to injection at 25- to 400-pg doses into 1-cell stage zebrafish embryos. For rescue of morphant phenotypes, MOs were injected into 2- to 4-cell embryos that had previously been injected with RNA.
Statistics.
Results are presented as mean ± SEM or SD for the number of experiments indicated in figure legends. Statistical analysis of continuous data was performed with 2-tailed Student’s t test or Tukey’s multiple comparison test, as appropriate. P < 0.05 was considered statistically significant.
Study approval.
Approval for human subjects research was obtained from the University of Michigan and the Boston Children’s Hospital Institutional Review Boards. All participants or their guardians provided written informed consent. Approval for zebrafish research was obtained from the University of Michigan and the Boston Children’s Hospital University Committees on Use and Care of Animals.
Supplementary Material
Acknowledgments
The authors thank the families who contributed to this study. We thank the Bloomington stock center and the Vienna Drosophila Resource Center for fly stocks. This research was supported by grants from the NIH to F. Hildebrandt (DK076683 and DK086542), to W. Zhou (DK091405), and to Z. Han (DK098410 and HL090801). F. Hildebrandt was supported by the NephCure Foundation. H.Y. Gee is supported by a American Society of Nephrology Nephcure Foundation for Kidney Research grant. W. Zhou is a Carl W. Gottschalk Scholar. F. Hildebrandt is an investigator of the Howard Hughes Medical Institute and an associate of the Manton Center.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(6):2375–2384. doi:10.1172/JCI79504.
References
- 1.Gee HY, et al. ARHGDIA mutations cause nephrotic syndrome via defective RHO GTPase signaling. J Clin Invest. 2013;123(8):3243–3253. doi: 10.1172/JCI69134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith JM, Stablein DM, Munoz R, Hebert D, McDonald RA. Contributions of the Transplant Registry: The 2006 Annual Report of the North American Pediatric Renal Trials and Collaborative Studies (NAPRTCS). Pediatr Transplant. 2007;11(4):366–373. doi: 10.1111/j.1399-3046.2007.00704.x. [DOI] [PubMed] [Google Scholar]
- 3.Sadowski CE, et al. A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol. doi: 10.1681/ASN.2014050489. [published online ahead of print October 27, 2014]. doi: 10.1681/ASN.2014050489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weavers H, et al. The insect nephrocyte is a podocyte-like cell with a filtration slit diaphragm. Nature. 2009;457(7227):322–326. doi: 10.1038/nature07526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang F, Zhao Y, Han Z. An in vivo functional analysis system for renal gene discovery in Drosophila pericardial nephrocytes. J Am Soc Nephrol. 2013;24(2):191–197. doi: 10.1681/ASN.2012080769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhuang S, Shao H, Guo F, Trimble R, Pearce E, Abmayr SM. Sns and Kirre, the Drosophila orthologs of Nephrin and Neph1, direct adhesion, fusion and formation of a slit diaphragm-like structure in insect nephrocytes. Development. 2009;136(14):2335–2344. doi: 10.1242/dev.031609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashraf S, et al. ADCK4 mutations promote steroid-resistant nephrotic syndrome through CoQ10 biosynthesis disruption. J Clin Invest. 2013;123(12):5179–5189. doi: 10.1172/JCI69000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Y, Kakinuma N, Wang Y, Kiyama R. Kank proteins: a new family of ankyrin-repeat domain-containing proteins. Biochim Biophys Acta. 2008;1780(2):128–133. doi: 10.1016/j.bbagen.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 9.Gee HY, et al. Whole-exome resequencing distinguishes cystic kidney diseases from phenocopies in renal ciliopathies. Kidney Int. 2014;85(4):880–887. doi: 10.1038/ki.2013.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halbritter J, et al. High-throughput mutation analysis in patients with a nephronophthisis-associated ciliopathy applying multiplexed barcoded array-based PCR amplification and next-generation sequencing. J Med Genet. 2012;49(12):756–767. doi: 10.1136/jmedgenet-2012-100973. [DOI] [PubMed] [Google Scholar]
- 11.Asanuma K, et al. Synaptopodin regulates the actin-bundling activity of alpha-actinin in an isoform-specific manner. J Clin Invest. 2005;115(5):1188–1198. doi: 10.1172/JCI23371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X, et al. Expression of novel podocyte-associated proteins sult1b1 and ankrd25. Nephron Exp Nephrol. 2011;117(2):e39–e46. doi: 10.1159/000320049. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Kakinuma N, Zhu Y, Kiyama R. Nucleo-cytoplasmic shuttling of human Kank protein accompanies intracellular translocation of β-catenin. J Cell Sci. 2006;119(pt 19):4002–4010. doi: 10.1242/jcs.03169. [DOI] [PubMed] [Google Scholar]
- 14.Robu ME, et al. p53 activation by knockdown technologies. PLoS Genet. 2007;3(5):e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou W, Hildebrandt F. Inducible podocyte injury and proteinuria in transgenic zebrafish. J Am Soc Nephrol. 2012;23(6):1039–1047. doi: 10.1681/ASN.2011080776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinkes B, et al. Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat Genet. 2006;38(12):1397–1405. doi: 10.1038/ng1918. [DOI] [PubMed] [Google Scholar]
- 17.Kakinuma N, Roy BC, Zhu Y, Wang Y, Kiyama R. Kank regulates RhoA-dependent formation of actin stress fibers and cell migration via 14-3-3 in PI3K-Akt signaling. J Cell Biol. 2008;181(3):537–549. doi: 10.1083/jcb.200707022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy BC, Kakinuma N, Kiyama R. Kank attenuates actin remodeling by preventing interaction between IRSp53 and Rac1. J Cell Biol. 2009;184(2):253–267. doi: 10.1083/jcb.200805147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarkar S, Roy BC, Hatano N, Aoyagi T, Gohji K, Kiyama R. A novel ankyrin repeat-containing gene (Kank) located at 9p24 is a growth suppressor of renal cell carcinoma. J Biol Chem. 2002;277(39):36585–36591. doi: 10.1074/jbc.M204244200. [DOI] [PubMed] [Google Scholar]
- 20.Gee HY, et al. Mutations in EMP2 cause childhood-onset nephrotic syndrome. Am J Hum Genet. 2014;94(6):884–890. doi: 10.1016/j.ajhg.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gbadegesin RA, et al. Mutations in the gene that encodes the F-actin binding protein anillin cause FSGS. J Am Soc Nephrol. 2014;25(9):1991–2002. doi: 10.1681/ASN.2013090976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramot Y, et al. Mutation in KANK2, encoding a sequestering protein for steroid receptor coactivators, causes keratoderma and woolly hair. J Med Genet. 2014;51(6):388–394. doi: 10.1136/jmedgenet-2014-102346. [DOI] [PubMed] [Google Scholar]
- 23.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9(9):690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Mata R, Boulter E, Burridge K. The ‘invisible hand’: regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol. 2011;12(8):493–504. doi: 10.1038/nrm3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu L, Jiang R, Aoudjit L, Jones N, Takano T. Activation of RhoA in podocytes induces focal segmental glomerulosclerosis. J Am Soc Nephrol. 2011;22(9):1621–1630. doi: 10.1681/ASN.2010111146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, et al. Mechanisms of the proteinuria induced by Rho GTPases. Kidney Int. 2012;81(11):1075–1085. doi: 10.1038/ki.2011.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott RP, et al. Podocyte-specific loss of cdc42 leads to congenital nephropathy. J Am Soc Nephrol. 2012;23(7):1149–1154. doi: 10.1681/ASN.2011121206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blattner SM, et al. Divergent functions of the Rho GTPases Rac1 and Cdc42 in podocyte injury. Kidney Int. 2013;84(5):920–930. doi: 10.1038/ki.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu H, et al. Rac1 activation in podocytes induces rapid foot process effacement and proteinuria. Mol Cell Biol. 2013;33(23):4755–4764. doi: 10.1128/MCB.00730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akilesh S, et al. Arhgap24 inactivates Rac1 in mouse podocytes, and a mutant form is associated with familial focal segmental glomerulosclerosis. J Clin Invest. 2011;121(10):4127–4137. doi: 10.1172/JCI46458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.[No authors listed]. Primary nephrotic syndrome in children: Clinical significance of histopathologic variants of minimal change of diffuse mesangial hypercellularity: A Report of the International Study of Kidney Disease in Children. Kidney Int. 1981;20(6):765–771. doi: 10.1038/ki.1981.209. [DOI] [PubMed] [Google Scholar]
- 32.Zhang F, Zhao Y, Chao Y, Muir K, Han Z. Cubilin and amnionless mediate protein reabsorption in Drosophila nephrocytes. J Am Soc Nephrol. 2013;24(2):209–216. doi: 10.1681/ASN.2012080795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saleem MA, et al. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J Am Soc Nephrol. 2002;13(3):630–638. doi: 10.1681/ASN.V133630. [DOI] [PubMed] [Google Scholar]
- 34.Kwan K, et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236(11):3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.